Abstract
Black caraway is of great importance for its terpene compounds. Many genes are involved in the biosynthesis of secondary metabolites in medicinal plants. For this study, black caraway seeds were collected from five different regions, i.e. [Isfahan; Kerman (Khabr); Semnan; Kerman (Sirch); and Hormozgan]. The black caraway seed oil was extracted and analyzed by means of the gas chromatography method. There was a negatively significant correlation (p ≤ 0.05) observed between cuminaldehyde and gammaterpinene compounds. 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) and isopentenyl pyrophosphate isomerase (IPI) play an important role in the biosynthesis of secondary metabolites. Appropriate primers were designed for these genes based on the conserved regions in other plants. Amplified fragments were then sequenced. Blastn results indicated the similarity of the high RNA sequences between new sequences and other HMGR and IPI gene sequences in GenBank, and it also identified the HMGR and IPI gene sequences of B. persicum. A fragment of the HMGR gene with KJ143741 number was recorded in the gene bank. Quantitative PCR showed that the relative expression of two genes in different growth stages of B. persicum was significantly different between the germination stage and the multi-leaf stage, and also between the germination stage and the flowering stage (p < 0.05); however, there was no significant difference observed between the flowering stage and the multi-leaf stage. The results indicated that the expression of HMGR increased from the germination stage to the adult plant, and then it got stable until the flowering stage; in the same vein, the expression of IPI increased continuously from the germination stage to the flowering stage. The expression of HMGR and IPI genes occurred differently at the germination stage of five ecotypes. The Hormozgan ecotype showed the least expression rate.
Keywords: Black caraway, Bunium persicum, Secondary metabolites, HMGR, IPI
Introduction
Being considered as a non-crop and non-domestic plant, black caraway or mountainous Black Zira (Bunium persicum) is one of the economical and important medicinal plants in Iran and across the world (Sofi et al. 2009; Jalilzadeh-Amin et al. 2011). This plant belongs to the Apiaceae (Umbelliferae) family and naturally grows in temperate and arid regions of Iran and some other countries (Jalilzadeh-Amin et al. 2011). Terpene compounds are the major active ingredients contained in B. persicum. These compounds have a large amount of γ-terpinene (Oroojalian et al. 2010) that is a monoterpene. In another study, the antioxidant properties of B. persicum and thyme were examined and they were attributed to the compounds in the essential oils of these plants that contained terpene, especially γ-terpinene (Shahsavari et al. 2008). The results of the data analysis using geNorm and Bestkeeper-1 software tools showed that the elongation factor 1-alfa (EF1A) gene is expressed more stable than the two other genes and that the standard error (SE) was higher in β-actin (ACT) and Ubiquitin (UBC) genes; it was also demonstrated that they are expressed less than EF1A. Considering the studies done on the stability, and uniformity and stability of the EF1A gene expression in different ecotypes of B. persicum, it is recommended that the data of the target gene expression be normalized (Darvishi Zeidabadi et al. 2015).
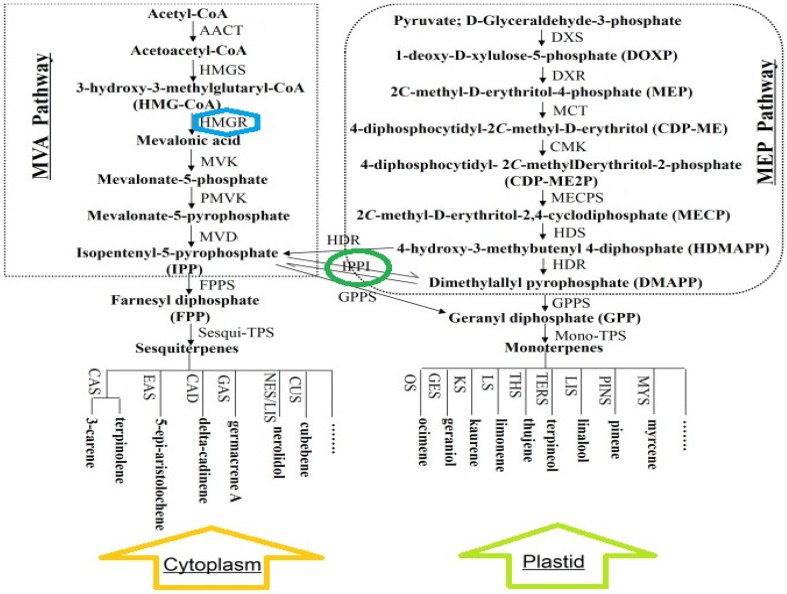
Isoprenoids are generally made through one of the two pathways, i.e., either the mevalonate pathway is used by the cells of eukaryotes and some prokaryotes, or the alternate path of 2-C-methyl-d-erythritol 4-phosphate (MEP) that is used by most prokaryotes (Campos et al. 2001; Goldstein and Brown 1990; Hecht et al. 2001; McAteer et al. 2001). The enzyme of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR, EC 1.1.1.34) catalyzes the NADPH-dependent reduction of HMG-CoA to mevalonate, being considered as the first committed step in the isoprenoid pathway to produce the largest group of contemporary natural products (Wu et al. 2012) (Fig. 1).
Fig. 1.
Biosynthetic pathway of monoterpenes and sesqui terpenes
Terpenoids are derived from the repeated condensation of isoprenoids among which mevalonic acid acts as a precursor. Within plant cells, the concentration of mevalonic acid (MVA) is strictly controlled by the HMGR activity. HMGR is one of the most heavily regulated enzymes ever identified (Goldstein and Brown 1990).
Enzyme HMGR converts 3-hydroxy-3-methylglutaryl coenzyme A to mevalonic acid and is a significant key regulator and controller of isoprene metabolism in mammals and fungi (Chappell et al. 1995).
Studies indicate that HMGR is an important control point for the mevalonate pathway in plants (Stermer et al. 1994). Since HMGR is a key enzyme in the pathway that leads to compounds with diverse and important functions in plants, it is not surprising that the HMGR’s activity in plants is controlled by a variety of developmental and environmental signals. The higher levels of the HMGR’s activity are usually associated with the rapidly growing parts of the plant, including apical buds and roots, with a much reduced activity observed in mature tissues (Brooker and Russell 1975; Bach et al. 1980). As an instance, the relative activity of microsomal HMGR observed in the mature leaves of pea seedlings was shown to be only 7% of the activity observed in the apical buds (Brooker and Russell 1975).
The HMGR’s activity is controlled not only at the mRNA level, but also post-translationally. Some studies have suggested that the HMGR’s activity in plants is regulated by reversible phosphorylation, as it is done in animals (Russell et al. 1985; Sipat 1982). IPI catalyzes the interconversion of isopentenyl diphosphate (IDP) and dimethylallyl diphosphate (DMAPP). It has been reported that the S. cerevisiae IPI gene (IDI) is an essential and single-copy gene, and that the disruption of the IDI gene results in a lethal phenotype (Mayer et al. 1992). Based on the relevant knowledge, the conversion of IPP to DMAPP by IPI in plants is thought to be necessary for isoprenoid biosynthesis through the cytosolic MVA pathway (Heintz et al. 1972; Nes and Venkatramesh 1999; Lange and Ghassemian 2003). IPP and DMAPP are synthesized in plants via the cytosolic mevalonate (MVA) pathway and the plastidic methylerythritol phosphate (MEP) pathway, respectively (Okada et al. 2008). IPP is consequently condensed to DMAPP to yield the short-chain isoprenoid precursors geranyl diphosphate, farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP) that are further metabolized to monoterpenes (C10), sesquiterpenes (C15), and diterpenes (C20). IPP isomerase (IPI; EC 5.3.3.2) catalyzes the interconversion of IPP to DMAPP, being regarded an essential starter moiety for the condensation reactions (Xuan et al. 1994).
The transcriptional co-regulation of the genes involved in secondary metabolite pathways is an important mechanism to regulate such pathways in general. To understand the correlation between the expression pattern of various genes and the content of terpenoid metabolites involved in artemisinin biosynthesis, the qPCR method was used (Yang et al. 2015).
In this study, two genes (HMGR and IPI) of Black Zira and their relative expressions in various stages of the plant growth were identified, and they were also analyzed in the 5-ecotype germination stage. The GC–MS analysis was used to assess the ingredients of the seeds of five Black Zira ecotypes.
Materials and methods
Plant material
Five B. persicum ecotypes [Isfahan, Keman (khabr), Semnan, Kerman (sirch) and Hormozgan] were used in the experiments. The seeds were surface-sterilized and germinated synchronously based on the protocol developed by Sharifi and Pouresmael (2006). The seeds were sterilized with 0.2% (v/v) sodium hypochlorite for 20 min, followed by 70% (v/v) ethanol for 1 min. Having been washed, the seeds were treated with hormones (gibberellic 100 µm acid and thidiazuron 6 µm) for 24 h, and they were then washed with the sterile water. The seeds were transferred into a sterile Petri dish and incubated in the dark at 4 °C for an additional 3 weeks. The seeds subsequently started to be germinated. The seeds of all the ecotypes were grown in a greenhouse. Following 15 days, a sample of seedlings was kept at − 80 °C for a further analysis. Other samples of the multi-leaf stage and the flowering stage were taken and kept for further tests at − 80 °C.
GC–MS analysis
The ground dry seeds (10 g) of each of the five ecotypes (Isfahan, Keman-khabr, Semnan, Kerman-sirch and Hormozgan) were extracted using 120 ml distilled water for approximately 3 h in the Clevenger instrument. The essential oil of each of the ecotype seeds was analyzed using capillary GC on a Thermo-UFM gas chromatograph (Thermo, Inc.) equipped with an HP-5 ms column with 5% phenyl methyl silo hexane capillary (0.4 µm film thicknesses, 10 m × 0.1 mm i.d.) and an Chrom-Card A/T Network Mass Selective Detector. The oven temperature was programmed for 1 min at 60 °C and 60–280 °C, and 80 °C per minute, and a final time of 4 min. Helium was used as the carrier gas. The sample size was 1 µl in the split-less mode. The terpenoids were identified by comparing the mass spectra and the retention indices with the wiley-5 library.
Polymerase chain reaction and gene detection
HMGR and IPI sequences were searched through the GenBank. The sequences of HMGR and IPI in the other family, especially Apiaceae, were analyzed by MegAlign 5.00 software (© 1993–2001 DNASTAR Inc.) and were simulated using the Clustal W method. The conserved areas for HMGR and IPI genes were identified, and appropriate primers were designed for those areas. The required features of the primers were examined by OligoAnalyser software. The primers were then designed in the conserved regions of the genes (Table 1).
Table 1.
Primers used in qPCR, RACE PCR, and gene cloning
| Primer no. | Primer name | Use | Primer sequence |
|---|---|---|---|
| 1 | IPI1(forward) | qPCR | 5′-GAT GTG AAA GTA AAT CCC AAC CCT G-3′ |
| 2 | IPI1(reverse) | qPCR | 5′-GAA TCG TTT CCA TAT CAG CAA CTT CC-3′ |
| 3 | HMGR1(forward) | qPCR | 5′-GAT GCD ATG GGA ATG AAC ATG GT-3′ |
| 4 | HMGR1(reverse) | qPCR | 5′-GCA CAG TGG TTT TCA AYA CCT TCT TCA C-3′ |
| 10 | EF1A(forward) | qPCR | 5′-CTG GTG GTT TTG AAG CTG GT-3′ |
| 11 | EF1A(reverse) | qPCR | 5′-TGT TGT CAC CCT CGA ATC CA-3′ |
For RNA isolation, 50–100 mg of the plant tissue was ground and homogenized with the lysis buffer of the Ribospin plant Cat. no: 307–150 Kit, according to the manufacturer’s instructions (GeneAll, Korea). The RNA was treated with DNase I (Fermentas) to remove the remaining genomic DNA. The RNA quality was examined using agarose gel 1/5%, and its quantity was measured using Nanodrop (Epoch™ Multi-Volume Spectrophotometer). RNA (1 µg) was reverse transcribed using RevertAid™H Minus-MuLV reverse transcriptase (Fermentas) and 0.1 µg oligo (dT) 18 primer in accordance with the manufacturer’s instructions. Complementary DNA (cDNA) was performed by the RevertAid™ First strand cDNA synthesis kit from fermentase.
Tissues from different growth stages were collected and used for the amplification and analysis of the expression levels of IPI and HMGR genes. For each sample, the reaction was set up with the 10 µl Ampliqon PCR Master Mix, 1 µl forward (10 pmol) primer, 1 µl reverse (10 pmol) primer, and 7 µl dH2O and 1 µl (5 ng) cDNA as the template. The PCR program was used within the temperature range (gradient) for checking the optimal temperature of the primers. The cycling program for the PCR included 94 °C 4 min, 1 cycle, and 94 °C 30 s, 57 °C 45 s, 72 °C 1 min, 35 cycles; 72 °C 10 min, for HMGR and IPI. The quality of the PCR product was examined using agarose gel 1%. The target gene fragments were amplified by specific primers, and after observing the electrophoresis pattern and ensuring the reproduction quality, they were submitted for purifying and sequencing the PCR product. After getting sequenced, the fragments were aligned and assembled. The sequencing results were analyzed by SeqMan and BLAST software.
Gene expression
Tissues of different ecotypes from the germination stage were collected to analyze the correlation between the expression levels of the genes encoding the biosynthetic material and the secondary metabolites content. RNA was extracted using the Ribospin plant Cat. no: 307–150 Kit, according to the manufacturer’s instructions (GeneAll, Korea). The single-stranded cDNA was synthesized by RevertAid™ H Minus-Mulv reverse transcriptase (Fermentas), using 0.1 µg Oligo (dT)18-primer. ABio-Rad iCycler Real-time PCR system (MiniOpticon) was used to perform the amplification task. For each sample, the reaction was set up with 1 µl EvaGreen® qPCR Real Time Master Mix with ROX (Solis BioDyne), 1 µl forward primer, 1 µl reverse primer, and 7 µl dH2O and 1 µl cDNA as the template. The thermal cycling program included 95 °C 15 min, 1 cycle; 95 °C 15 s, 57 °C 60 s, 72 °C 30 s, 40 cycle; 95 °C 15 s, 60 °C 1 min, 95 °C 15 s, 1 cycle. The primers for each gene are listed in Table 1.
The relative fold changes in the gene expression were measured based on the comparative method (Livak and Schmittgen 2001; Sehringer et al. 2005; Cikos et al. 2007) with three replications. In this method, the levels of the target gene amplification in an experimental sample are compared with the levels of the target gene amplification in another sample or standard, both of which being first normalized to the amplification levels of a normalizing gene. For this study, the elongation factor 1-alfa (EF1A) gene was used as the normalizing factor. Before examining HMGR and IPI, the relative expression efficiency of each specific gene and the housekeeping gene was measured, and then the relative expression levels were analyzed using the REST 2009 software V. 2.0.13 (Qiagen, Hilden, Germany) (Pfaffl et al. 2002).
Results and discussion
GC–MS analysis
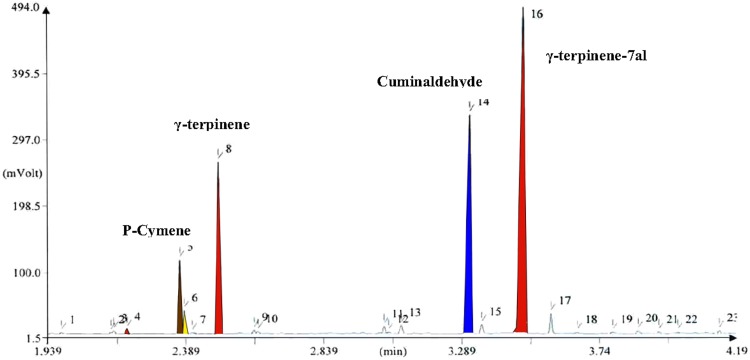
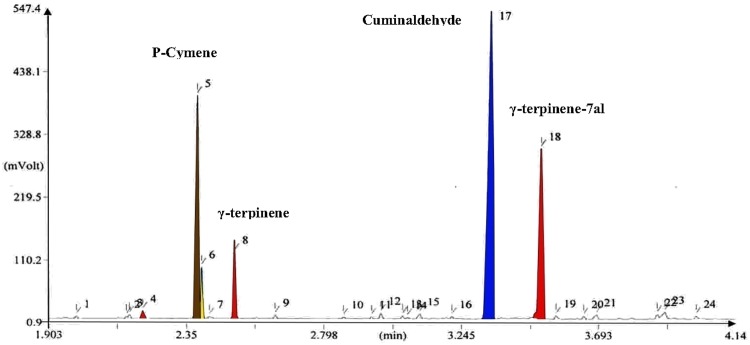
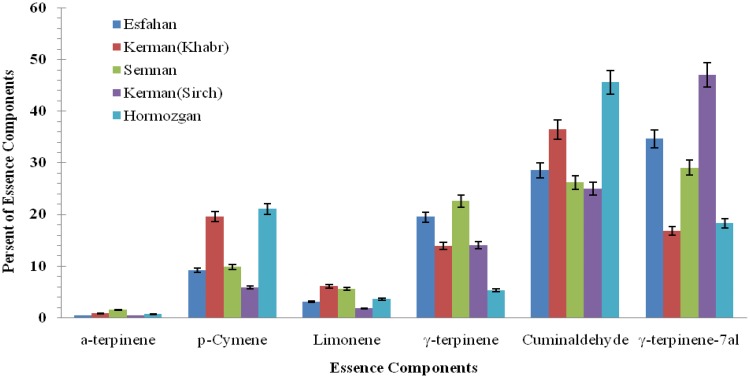
The analysis results of the essential oil from five ecotype seeds, making use of GC–MS, showed component differences in various ecotypes. Four components showed to have a higher content rate from among all other components (i.e., P-cymene, γ-terpinene, cuminaldehyde, and γ-terpinene-7al). Cuminaldehyde and γ-terpinene-7al had higher quantities in comparison with the three other components (Figs. 2, 3, 4).
Fig. 2.
GC–MS chromatogram of the essence analysis for Kerman (sirch) ecotype of B. persicum
Fig. 3.
GC–MS chromatogram of the essence analysis for Hormozgan ecotype of B. persicum
Fig. 4.
Different essence components of five ecotype seeds of B. persicum
A DNA fragment encoding the HMG-CoA reductase was obtained by PCR from HMGR1(forward) (5′-GAT GCD ATG GGA ATG AAC ATG GT-3′) and HMGR1(reverse) (5′-GCA CAG TGG TTT TCA AYA CCT TCT TCA C-3′) as primers; also for IPI, IPI1(forward) was (5′-GAT GTG AAA GTA AAT CCC AAC CCT G-3′) and IPI1(reverse) was (5′-GAA TCG TTT CCA TAT CAG CAA CTT CC-3ʹ).
The sequencing results showed that 217 bases had been sequenced from HMGR and 214 bp from IPI genes (Fig. 5).
Fig. 5.
Partial sequences from A-HMGR and B- IPI
The nucleotide sequences of both genes were analyzed using BLASTN software. The HMGR nucleotide sequence showed 85% similarity with Artemisia annua HMGR, 82% similarity with Tanacetum parthenium HMGR, and some similarities with a few other nucleotide fragments of different plants in GenBank. The IPI nucleotide sequence showed over 93% similarity with Dacus carruta IPI (JX100860), 83% similarity with Medicago truncatula IPI (XM-003624130.1), and some similarities with a few other nucleotide fragments of different plants in GenBank. The partial gene fragment of HMGR was registered for the first time for B. persicum under number KJ143741 in GenBank (NCBI).
Gene expression analysis
The sensitive quantitative Real Time PCR was performed using the total RNA isolated from various tissues of B. persicum. EF1A was used as the housekeeping gene. This gene should be highly, stably, and constitutively expressed in all conditions and tissues to be analyzed (Deprez et al. 2002; Thellin et al. 1999; Schmittgen and Zakrajsek 2000; Brunner et al. 2004); besides, EF1A is expressed in a higher and more uniformed form, compared with other genes in B. persicum (Darvishi Zeidabadi et al. 2015).
However, the gene is expressed highly in the leaves and abundantly in the flowers and roots. The efficiency of the three gene primers was calculated using the REST-2009 software. The rates were 0.98, 0.91, and 0.95 for EF1A, HMGR, and IPI, respectively. The expression of HMGR and IPI could be detected in all tissues from the germination stage to the flowering stage.
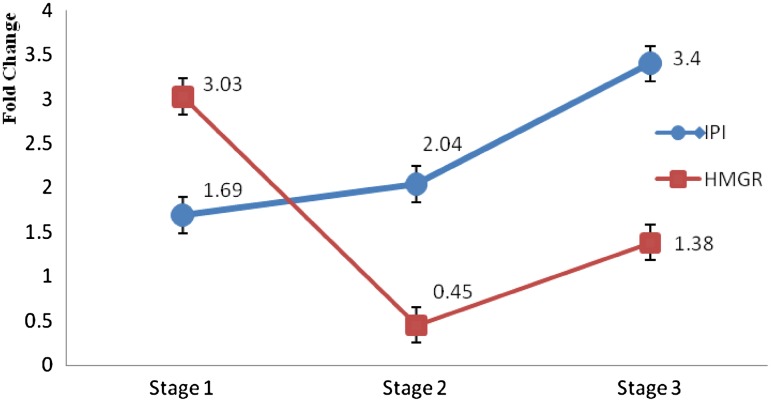
The results of the relative expression analysis of HMGR and IPI genes showed a significant difference between different stages. The HMGR gene showed a significant expression difference between the germination stage and the multi-leaf stage (3.03-fold), and also between the germination stage and the flowering stage (1.38-fold) (p < 0.05); however, there was no significant difference between the expression in the flowering stage and the multi-leaf stage (Fig. 6).
Fig. 6.
Relative expression (fold change) of HMGR and IPI genes in the three stages of the B. persicum growth in Kerman (sirch) ecotype: stage 1—germination, stage 2—multi-leaf, and stage 3—flowering
On the other hand, the results showed that the expression of HMGR increases from the germination stage to the mature-plant stage, and it then becomes stable until the flowering stage. The IPI gene showed no significantly different expression between the multi-leaf stage and the germination stage, but there was a significant difference between the flowering stage and the multi-leaf stage (2.04-fold), and also between the flowering stage and the germination stage (3.4-fold). The higher levels of the HMGR activity are usually associated with the rapid growth of the plant parts, including apical buds and roots, with a much reduced activity observed in mature tissues (Brooker and Russell 1975; Bach et al. 1980). HMGR also exhibited a lower expression rate in old leaves in comparison with other tissues in A. annua L. (Olofsson et al. 2011). The overexpression of both HMGR and ads genes in A. annua L. plants results not only in an increase in the artemisinin content, but it also enhances the synthesis of other isoprenoids, including the essential oils (Alam et al. 2014). The stability of the HMGR expression at the multi-leaf stage to the flowering stage might have happened since these stages are very important and plants need a highly strong defense system. The plastidic pathway was predicted to play an important role in the shift from the vegetative growth stage to the reproductive growth stage (Vail 2008).
HMGR expression is highly upregulated in the plants that shift into the reproductive growth stage (Vail 2008). However, HMGR is known to be regulated post-transcriptionally (Hey et al. 2006). During the flowering stage, the plant is in a vital and vulnerable phase; therefore, the need for the defensive compounds is by far higher (Vail 2008). Isoprenoids produced in the plastid are important for floral pigmentation (carotenoids) and fragrances (monoterpenes) (Vail 2008). Furthermore, Towler and Weathers (2007) demonstrated that the inhibition of the MEP pathway reduced the artemisinin production significantly.
However, the results showed that the mRNA levels of key isoprenoid biosynthetic genes in the plastid remained fixed in flower-budding plants compared to vegetative plants, and that they were also downregulated during the full flowering stage. Photosynthesis in the shoot is likely to assume a weaker role in the plant shifts to the reproductive phase. Cytosolic IPP is also likely to provide a crosstalk source of isoprene (IPP) biosynthesis in the plastid, since the cytosolic pool of IPP is probably very large due to the high increases in HMGR transcripts.
MEP pathway genes are known to be regulated post-transcriptionally (Sauret-Gueto et al. 2006); therefore, it is possible that although the levels of transcripts are unchanged as the plant shifts to the reproductive stage and the flowering stage, the post-translational regulation may occur.
Page et al. (2004) realized that Nicotiana benthamiana leaves where the IPI expression was downregulated by the tobacco rattle virus-mediated gene silencing exhibited a mottled whitish pale green phenotype and an 80% reduction in the level of chlorophyll compared with control leaves. They concluded that although not absolutely required, IPI plays a significant role in plastidic isoprenoid biosynthesis in higher plants (plants of relatively complex or advanced characteristics, especially vascular plants, including flowering plants).
The atipi1atipi2 double mutant conditionally showed a 20% decrease in chlorophyll and carotenoids compared with control plants under the continuous light. Thus, a pale green phenotype seems to be a common feature of plants defective in the IPI activity, although the extent of variegation varies significantly among plant species. However, the appearance of a pale green phenotype was greatly absent from the Arabidopsis ipi double mutant under LD (long day) conditions, indicating that the IPI activity is required for the production of photosynthetic pigments using the MEP pathway under conditions necessitating an increased isoprenoid production rate (Okada et al. 2008). In contrast to the atipi1atipi2 double mutant, neither the atipi1 single mutant nor the atipi2 single mutant demonstrates any visible phenotype. This observation was interesting given that AtIPI1 was localized in the cytoplasm, whereas AtIPI2 was targeted at mitochondria (Okada et al. 2008). The observations also indicated that cytoplasmic AtIPI1 was sufficient for the synthesis of isoprenoids in mitochondria, and that IPP, DMAPP and other prenyl diphosphates such as FPP were capable of moving between the mitochondria and the cytosol (Okada et al. 2008).
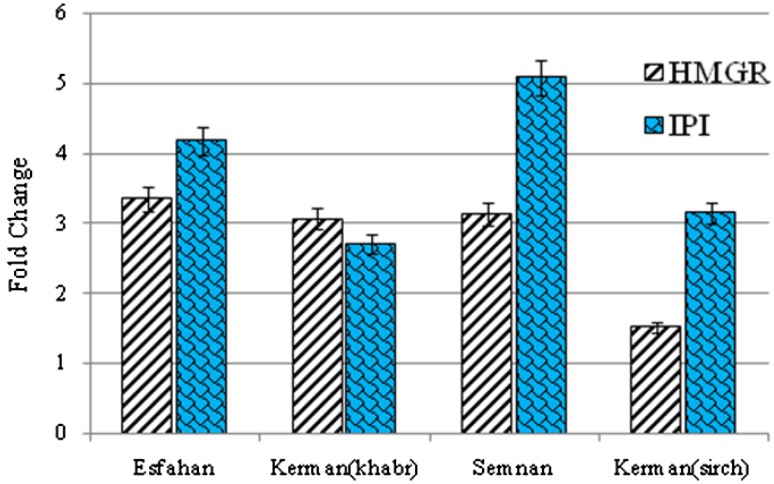
The analysis of the expression of HMGR and IPI for five ecotypes of B. persicum in the germination stage demonstrated a significant difference among ecotypes. These results show a variation among ecotypes concerning the expression factor. The Hormozgan ecotype was observed to have less expression than other ecotypes, so it was measured as the expression for other ecotypes (Fig. 7). The results showed that the Semnan ecotype had the highest expression of IPI.
Fig. 7.
Relative expression (fold change) of HMGR and IPI genes in four different ecotypes to Hormozgan ecotype of B. persicum
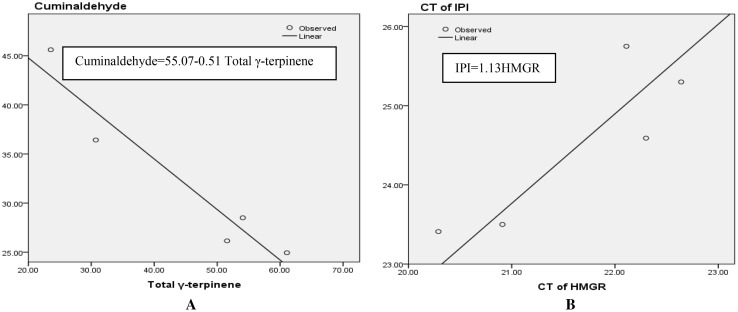
There was a negatively significant correlation (p ≤ 0.05) between cuminaldehyde and γ-terpinene-7al that shows a variation among the five ecotypes (Fig. 8a). There was a positive correlation between the HMGR expression and the IPI expression (Fig. 8b). The expressions of two genes in different ecotypes were correlated; hence, it can be concluded that the IPI expression has also been highly similar to the high expression of HMGR. The overexpression of HMGR, considered as a rate-limiting enzyme, upgrades the flux rate of the MVA pathway and leads to an increase in the terpenoid biosynthesis and IPI (isopentenyl diphosphate isomerase) (Qaderi et al. 2014). Cytosolic IPI increases the pool of the precursor DMAPP in plastid. The accumulation of DMAPP in plastid (the first precursor in the production of diterpenoids) leads to an increase in the amount of diterpenoids (Roberts 2007).
Fig. 8.
The curve of regression between: a total γ-terpinene and cuminaldehyde, and b CT of HMGR and CT of IPI
The correlation of HMGR and IPI CT with cuminaldehyde and γ-terpinene-7al was not significant. This indicates that the production of secondary metabolites such as γ-terpinene and cuminaldehyde has undergone the posttranscriptional regulation (Table 2). Although the levels of HMGR transcripts have remarkably increased, a consequent increase in the enzyme activity and the carbon flux may not necessarily follow the increased transcript levels (Re et al. 1995).
Table 2.
Pearson correlation between important essence components and specific genes CT
| Cuminaldehyde | γ-Terpinene | IPIct | HMGRct | |
|---|---|---|---|---|
| Cuminaldehyde | 1 | |||
| γ-Terpinene | − 0.833* | 1 | ||
| IPIct | 0.305 | − 0.361 | 1 | |
| HMGRct | − 0.259 | 0.079 | 0.611 | 1 |
*Correlation is significant at the 0.05 level
Compliance with ethical standards
Conflict of interest
We declare that the authors of this paper have no conflicting interests.
References
- Alam P, Kamaluddin Khan MA, Mohammad A, Khan R, Malik ZA. Enhanced artemisinin accumulation and metabolic profiling of transgenic Artemisia annua L. plants over-expressing by rate-limiting enzymes from isoprenoid pathway. J Plant Interact. 2014;9(1):655–665. doi: 10.1080/17429145.2014.893030. [DOI] [Google Scholar]
- Bach TJ, Lichtenthaler HK, Retey J. Properties of membrane-bound 3-hydroxy-3-methylglutaryl-coenzyme A reductase (EC. 1.1.1.34) from radish seedlings and some aspects of its regulation. In: Mazliak P, Benveniste P, Costes C, Douce R, editors. Biogenesis and function of plant lipids. Amsterdam: Elsevier; 1980. pp. 355–362. [Google Scholar]
- Brooker JD, Russell DW. Properties of microsomal 3-hydroxy-3-methylglutaryl coenzyme A reductase from Pkum sativum seedlings. Arch Biochem Biophys. 1975;167:723–729. doi: 10.1016/0003-9861(75)90517-2. [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004;4:14–24. doi: 10.1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos N, Rodríguez-Concepción M, Seemann M, Rohmer M, Boronat A. Identification of gcpE as a novel gene of the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 2001;488:170–173. doi: 10.1016/S0014-5793(00)02420-0. [DOI] [PubMed] [Google Scholar]
- Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C. Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme a reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol. 1995;109:1337–1343. doi: 10.1104/pp.109.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikos S, Bukovska A, Koppel J. Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Mol Biol. 2007;8:113–123. doi: 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvishi Zeidabadi D, Javaran MJ, Dehghani H, Monfared SR, Baghizadeh A. Selection of suitable housekeeping genes for gene expression study in caraway (Bunium persicum) J Biodivers Environ Sci. 2015;7(3):50–58. [Google Scholar]
- Deprez RHL, Fijnvandraat AC, Ruijter JM, Moorman AFM. Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depends on cDNA synthesis conditions. Anal Biochem. 2002;307:63–69. doi: 10.1016/S0003-2697(02)00021-0. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F (2001) Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. In: Proceedings of the National Academy of Sciences of the USA, vol 98, pp 14837–14842 [DOI] [PMC free article] [PubMed]
- Heintz R, Benveniste P, Robinson WH, Coates RM. Plant sterol metabolism. Demonstration and identification of a biosynthetic intermediate between farnesyl PP and squalene in a higher plant. Biochem Biophys Res Commun. 1972;49:1547–1553. doi: 10.1016/0006-291X(72)90517-7. [DOI] [PubMed] [Google Scholar]
- Hey SJ, Powers SJ, Beale MH, Hawkins ND, Ward JL, Halford NG. Enhanced seed phytosterol accumulation through expression of a modified HMG-CoA reductase. Plant Biotechnol J. 2006;4:219–229. doi: 10.1111/j.1467-7652.2005.00174.x. [DOI] [PubMed] [Google Scholar]
- Jalilzadeh-Amin G, Maham M, Dalir-Naghadeh B, Kheiri F. Effects of Bunium persicum (Boiss.) essential oil on the contractile responses of smooth muscle (an in vitro study) Vet Res Forum. 2011;2:87–96. [Google Scholar]
- Lange BM, Ghassemian M. Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol. 2003;51:925–948. doi: 10.1023/A:1023005504702. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(∆∆CT) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Hahn FM, Stillman DJ, Poulter CD. Disruption and mapping of IDI1, the gene for isopentenyl diphosphate isomerase in Saccharomyces cerevisiae. Yeast. 1992;8:743–748. doi: 10.1002/yea.320080907. [DOI] [PubMed] [Google Scholar]
- McAteer S, Coulson A, McLennan N, Masters M. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J Bacteriol. 2001;183:7403–7407. doi: 10.1128/JB.183.24.7403-7407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes WD, Venkatramesh M. Enzymology of phytosterol transformations. Crit Rev Biochem Mol Biol. 1999;34:81–93. doi: 10.1080/10409239991209219. [DOI] [PubMed] [Google Scholar]
- Okada K, Kasahara H, Yamaguchi S, Kawaide H, Kamiya Y, Nojiri H, Yamane H. Genetic evidence for the role of isopentenyl diphosphate isomerases in the mevalonate pathway and plant development in Arabidopsis. Plant Cell Physiol. 2008;49(4):604–616. doi: 10.1093/pcp/pcn032. [DOI] [PubMed] [Google Scholar]
- Olofsson L, Engström A, Lundgren A, Brodelius PE. Relative expression of genes of terpene metabolism in different tissues of Artemisia annua L. BMC Plant Biol. 2011;11:45. doi: 10.1186/1471-2229-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroojalian F, Kasra-Kermanshahi R, Azizi M, Bassami MR. Synergistic antibacterial activity of the essential oils from three medicinal plants against some important food-borne pathogens by microdilution method. Iran J Med Aromat Plants. 2010;26(2):133–146. [Google Scholar]
- Page JE, Hause G, Raschke M, Gao W, Schmidt J, Zenk MH, Kutchan TM. Functional analysis of the final steps of the 1-deoxy-d-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiol. 2004;134:1401–1413. doi: 10.1104/pp.103.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qaderi A, Omidi M, Zebarjadi A, Hajiaghaee R. Isolation and characterization of gene encoding 3-hydroxy-3-methylglutaryl-CoA reductase from Iranian Hazel (Corylus avellana L.) Int J Biosci. 2014;4(5):216–225. [Google Scholar]
- Re EB, Jones D, Learned RM. Co-expression of native and introduced genes reveals cryptic regulation of HMG CoA reductase expression in Arabidopsis. Plant J. 1995;7:771–784. doi: 10.1046/j.1365-313X.1995.07050771.x. [DOI] [PubMed] [Google Scholar]
- Roberts SC. Production and engineering of terpenoids in plant cell culture. Nat Chem Biol. 2007;3:387–395. doi: 10.1038/nchembio.2007.8. [DOI] [PubMed] [Google Scholar]
- Russell DW, Knight JS, Wilson TM. Pea seedling HMG-CoA reductases: regulation of activity in vitro by phosphorylation and Ca2+, and posttranslational control in vivo by phytochrome and isoprenoid hormones. In: Randall DD, Blevins DG, Larson RL, editors. Current topics in plant biochemistry and physiology, vol4. Columbia: University of Missouri; 1985. p. 191206. [Google Scholar]
- Sauret-Gueto S, Botella-Pavia P, Flores-Perez U, Martinez-Garcia JF, San Roman C, Leon P, Boronat A, Rodriguez-Concepcion M. Plastid cues posttranscriptionally regulate the accumulation of key enzymes of the methylerythritol phosphate pathway in Arabidopsis. Plant Physiol. 2006;141:75–84. doi: 10.1104/pp.106.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/S0165-022X(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Sehringer B, Zahradnik HP, Deppert WR, Simon M, Noethling C, Schaefer WR. Evaluation of different strategies for real-time RT-PCR expression analysis of corticotropin-releasing hormone and related proteins in human gestational tissues. Anal Bioanal Chem. 2005;383:768–775. doi: 10.1007/s00216-005-0067-9. [DOI] [PubMed] [Google Scholar]
- Shahsavari N, Barzegar M, Sahari MA, Naghdibad H. Antioxidant activity and chemical characterization of essential oil of Bunium persicum. Plant Foods Hum Nutr. 2008;63:183–188. doi: 10.1007/s11130-008-0091-y. [DOI] [PubMed] [Google Scholar]
- Sharifi M, Pouresmael M. Breaking seed dormancy in Bunium persicum by stratification and chemical substances. Asian J Plant Sci. 2006;5(4):695–699. doi: 10.3923/ajps.2006.695.699. [DOI] [Google Scholar]
- Sipat AB. Hydroxymethylglutaryl CoA reductase (NADPH) in the latex of Hevea brasiliensis. Phytochemistry. 1982;21:2613–2618. doi: 10.1016/0031-9422(82)83087-2. [DOI] [Google Scholar]
- Sofi PA, Zeerak NA, Singh P. Kala zeera (Bunium persicum Bioss.): a Kashmirian high value crop. Turk J Biol. 2009;33:249–258. [Google Scholar]
- Stermer BA, Bianchini GM, Korth K. Regulation of HMG-CoA reductase activity in plants. J Lipid Res. 1994;35:1133–1140. [PubMed] [Google Scholar]
- Thellin O, Zorzi W, Lakaye BD, Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–295. doi: 10.1016/S0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- Towler MJ, Weathers PJ. Evidence of artemisinin production from IPP stemming from both the mevalonate and the nonmevalonate pathways. Plant Cell Rep. 2007;26:2129–2136. doi: 10.1007/s00299-007-0420-x. [DOI] [PubMed] [Google Scholar]
- Vail DR (2008) Artemisnin biosynthesis: developmental and sugar regulation of mRNA levels. A thesis of Masters of Science in Biology and Biotechnology, Worcester Polytechnic Institute, p 100
- Wu Q, Sun C, Chen S. Identification and expression analysis of a 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from American ginseng. Plant Omics J. 2012;5(4):414–420. [Google Scholar]
- Xuan JW, Kowalski J, Chambers AF, Denhardt DT. A human promyelocyte mRNA transiently induced by TPA is homologous to yeast IPP isomerase. Genomics. 1994;20:129–131. doi: 10.1006/geno.1994.1139. [DOI] [PubMed] [Google Scholar]
- Yang K, Rashidi Monafared S, Wang H, Lundgren A, Brodelius PE. The activity of the artemisinic aldehyde ∆11 (13) reductase promoter is important for artemisinin yield in different chemotypes of Artemisia annua L. Plant Mol Biol. 2015 doi: 10.1007/s11103-015-0284-3. [DOI] [PubMed] [Google Scholar]