Abstract
The aim of this study was to establish a system for the efficient expression and purification of new subtype of antioxidant peptide from Pinctada fucata meat (NPFMAP), which is designed by molecular modification technology based on the sequence of purified and identified antioxidant peptide from Pinctada fucata meat (PFMAP, Gly-Ala-Gly-Leu-Pro-Gly-Lys-Arg-Glu-Arg), and to better understand the relationship between structure and antioxidant activity. Meanwhile, gene codon usage was optimized and the glutathione S-transferase (GST) tag of pGEX-6P-1 was added to facilitate expression and purification NPFMAP in Escherichia coli. The results of antioxidant activity assay in vitro showed a higher antioxidant activity in NPFMAP than that in enzymatic hydrolysis digested or chemically synthesized PFMAP. In particular, the DPPH scavenging radical activity increased by about 4.7 times after molecular modification. Structural bioanalysis indicated that new subtype antioxidant peptide had spatial conformation and good hydrophilic after modification, which was confirmed by antioxidant activity assays. Thus, the proposed method could be used to obtain NPFMAP with high antioxidant activity.
Keywords: Pinctada fucata peptide, Molecular modification, Expression, Purification, Antioxidant activity
Introduction
Free radicals such as the superoxide anion (O·−2) and hydroxyl radical (·OH) can cause protein damage and DNA mutations, which can initiate several diseases, including coronary heart diseases and diabetes (Ahn et al. 2014). Thus, antioxidants that can scavenge free radicals play an important role in biological systems. At present, food-derived bioactive antioxidant peptides are being actively investigated. Examples include antioxidant peptides obtained by the digestion of proteins from animal and plant products, such as egg yolk (Zambrowicz et al. 2014), corn gluten meal (Zhuang et al. 2013), egg white (Chen et al. 2011), porcine plasma (Liu et al. 2010), royal jelly (Guo et al. 2009), and canola (Cumby et al. 2008). In addition, proteins from marine organisms have also been shown to be a good source of antioxidant peptides; these include Pacific hake hydrolysates (Cheung et al. 2012), skipjack tuna meat (Nalinanon et al. 2011), silver carp (Zhong et al. 2011), and hoki frame (Kim et al. 2007). However, researchers concentrated in the isolation and purification of antioxidant peptides from the tissues or organs protein using enzymatic hydrolysis techniques, but rarely in preparing antioxidant peptides based on these functional peptides sequence.
Pinctada fucata belongs to the family of pearl oysters from the phylum Mollusca. It is distributed mainly in Guangxi and Guangdong provinces, Hainan island, the Taiwan Strait, and coastal areas of Japan. During pearl extraction, Pinctada fucata meat is discarded or used as feedstock (Wu et al. 2012). Over the past 10 years, enzymatic hydrolysis has become a widely applied method for preparing antioxidant peptides (Bougatef et al. 2012). Antioxidant peptides from Pinctada fucata meat (PFMAP) were prepared by enzymatic hydrolysis using alkaline protease, hydrolysates were separated by molecular weight, and the purified peptides’ primary structures were identified by liquid chromatography-mass spectrometry (Zhang et al. 2010; Wu et al. 2013a, b). Currently, antioxidant peptides from fish and shellfish protein hydrolysates are used in cosmetics, health food, and pharmaceutical industries (Wu et al. 2013a). Moreover, antioxidant activity is related to peptide chain length, amino acid composition and sequence, the degree of amino acid side chain glycosylation, the size of side chain structures, and molecular weight. This opens up the possibility of applying genetic engineering (Lee et al. 2014) for the production of PFMAP, and the evaluation of the corresponding free radical scavenging activities in vitro. The key advantage of such an approach is the ability to merge the same or different antioxidant peptides together into new antioxidant agents, whose spatial conformation may boost antioxidant activity. Besides high expression and easy purification, this strategy may also substantially enhance antioxidant activity.
Here, we focused on the preparation, purification, and characterization of the small-molecule antioxidant peptide Gly-Ala-Gly-Leu-Pro-Gly-Lys-Arg-Glu-Arg from Pinctada fucata meat (PFMAP). We generated a series of modified antioxidant peptide sequences, and used molecular biology software to optimize the corresponding mRNAs. This biotechnological approach allowed us to express and characterize novel subtypes of new antioxidant peptides from Pinctada fucata (NPFMAP). Furthermore, NPFMAP antioxidant activities were determined by three assay: (1) DPPH radical scavenging assay; (2) superoxide radical scavenging assay; (3) hydroxyl radical scavenging assay. In summary, this study tried to improve antioxidant activity, by modifying the structure of NPFMAP and optimizing production in recombinant engineered bacteria. Compared with traditional enzymatic processes, this method could benefit from several advantages in terms of purity, speed of preparation, cost, and degree of automation for the production of single peptides.
Materials and methods
Pinctada fucata was purchased from Hainan Aquaculture Base, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences. Rosetta Escherichia coli (DE3) and the pEGX-6P-1 expression vectors were from our laboratory. NPFMAP sequence and primers were synthesized by Sangon (Shanghai, China). The pfu DNA Polymerse, MiniBEST DNA Fragment Purification Kit Ver. 4.0 and MiniBEST Plasmid Purification Kit Ver. 4.0 were from TaKaRa (Kusatsu, Japan). Chelating Sepharose Fast Flow column and glutathione S-transferase (GST)-Sefinose™ Resin were from Sangon. DNA markers of 500 bp and 2000 bp, T4 DNA ligase, and restriction endonucleases (BamH I and Xho I) were from TaKaRa. Ampicillin, chloramphenicol, isopropyl-β-d-1-thiogalactopyranoside (IPTG), (2, 2-diphenyl-1-picrylhydrazyl) DPPH, tryptone, and yeast extract were from Solarbio (Beijing, China).
Sequence analysis and DNA constructs
For further functional expression and preparation, NPFMAP programme physicochemical properties were obtained according to the literature Yu et al. (2014) that are used for the analysis about predicted hydrophilicity scales at the websites http://web.expasy.org/protparam/ and http://web.expasy.org/protscale/. Meanwhile, the antioxidant peptides NPFMAP sequence was deposited in the GenBank and could be accessed through accession number MG680495. Next, the DNA sequence encoding the 61 amino acids of NPFMAP was synthesized and codon-optimized for E. coli expression, using Primer Premier 5.0 (Tong et al. 2015).Meanwhile, the NPFMAP sequence was optimized using high-usage E. coli codons to ensure a higher yield. DNA constructs were generated by overlap extension polymerase chain reaction (OE-PCR) (Xiao and Pei 2011). OE-PCR-specific steps included, first, P1–10 primer synthesis and, second, two rounds of PCR. The 50-µL reaction contained 0.4 µL of each primer P1–10, 1 µL of 25 mM dNTPs mix, 2 µL of each primer P1/P2, and 0.5 µL of pfu DNA polymerase. PCR conditions were as follows: 95 °C for 3 min; 18 cycles at 95 °C for 22 s, 59 °C for 20 s, 72 °C for 25 s; and a final extension at 72 °C for 5 min. After completing the first round of PCR, the second round was performed in a 50-µL reaction containing 2 µL of each primer P1/P10, 2 µL of first-round PCR product, 1 µL of 25 mM dNTPs mix, and 0.5 µL of pfu DNA polymerase. PCR conditions were as follows: 95 °C for 3 min; 20 cycles at 95 °C for 24 s, 56 °C for 20 s, 72 °C for 25 s; and a final extension at 72 °C for 5 min. Primer 1–10 sequences used in this study were listed in Table 1.
Table 1.
Primer 1–10 sequences used in this study
| Primer name | Primer sequence | Tm (°C) |
|---|---|---|
| P-1 | 5′-GACACGGATCCATGAATGGT-3′ | 54.8 |
| P-2 | 5′-GTTTACCAGGCAGACCTGCACCACCATTCATGGATCCGTG-3′ | 68.8 |
| P-3 | 5′-CAGGTCTGCCTGGTAAACGTGAGCGTAATGGTGGTGCTGG-3′ | 69.7 |
| P-4 | 5′-TGCGTTCACGTTTACCTGGCAGGCCAGCACCACCATTACG-3′ | 71.1 |
| P-5 | 5′-CAGGTAAACGTGAACGCAACGGTGGCGCAGGTCTGCCGGG-3′ | 73.3 |
| P-6 | 5′-CAGCACCACCGTTACGTTCGCGCTTGCCCGGCAGACCTGC-3′ | 74.5 |
| P-7 | 5′-CGTAACGGTGGTGCTGGTCTGCCGGGTAAACGCGAGCGCA-3′ | 73.9 |
| P-8 | 5′-TTGCCAGGCAGACCCGCACCACCGTTGCGCTCGCGTTTAC-3′ | 74.5 |
| P-9 | 5′-GGGTCTGCCTGGCAAACGTGAACGTAATGGTTAACTCGAG-3′ | 67.6 |
| P-10 | 5′-GTGTCCTCGAGTTAACCATTACGTTC-3′ | 57.3 |
Recombinant plasmid construction
pEGX-6P-1 vectors containing the gene of interest, flanked by BamH I and Xho I restriction sites, were digested with the corresponding enzymes and ligated with NPFMAP. Following ligation, the resulting pGEX-6P-1-NPFMAP (pGEX-NPFMAP) vectors were transformed into chemically competent DE3 cells and plated on ampicillin- and chloramphenicol-containing agar plates. Single colonies were screened for successful ligation via PCR (forward primer: 5′-GGGCTGGCAAGCCACGTTTGGTG-3′; reverse primer: 5′-CCGGGAGCTGCATGTGTCAGAGG-3′), restriction digestion, and DNA sequencing (Sangon). Positive clones were transformed into competent DE3 cells for subsequent protein expression.
NPFMAP expression in vitro
Cells carrying the recombinant pGEX-NPFMAP vector (DE3-pGEX-NPFMAP) (1%, v/v) were inoculated in sterile Luria–Bertani (LB) medium containing ampicillin (50 µg/mL) and chloramphenicol (34 µg/mL). Samples were pre-cultured at 37 °C in a rotary shaker (220 rpm) until OD600 reached 0.8–1.0. Then, IPTG was added at a final concentration of 0.5 mM, cells were cultured for 4 h at 37 °C, and for an additional 12 h at 20 °C to induce production of the target protein. A negative control without IPTG was included. At the end of the induction period, the thallus was harvested by centrifugation at 5000 rpm for 10 min at 4 °C and sonicated to dissolve the target protein in the buffer. After centrifugation at 12,000 rpm for 20 min at 4 °C, lysates and supernatant were collected and separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie blue staining and western blot analysis to determine expression levels.
To analyze solubility of the target proteins, DE3-pGEX-NPFMAP strains were transferred (1%, v/v) into nine flasks containing 200 mL LB medium supplemented with ampicillin (50 µg/mL) and chloramphenicol (34 µg/mL), and grown at 37 °C in a rotary shaker (220 rpm) until exponential phase. Then, all flasks were induced with IPTG at a final concentration of 0.5 mM and individually cultured at varying temperatures (20, 22, 24, 26, 28, 30, 32, 35, and 37 °C) for further expression. Next, the thallus (20 mL) from each flask was harvested by centrifugation at 5000 rpm for 10 min at 4 °C and sonicated. Finally, soluble and insoluble fractions were collected and analyzed by 12% SDS-PAGE to verify the solubility of the GST-NPFMAP fusion construct.
NPFMAP purification
For NPFMAP purification, pre-cultured samples were inoculated (1%, v/v) in fresh LB broth containing ampicillin (50 µg/mL) and chloramphenicol (34 µg/mL) at 37 °C for 3 h. Cells (1 L) of DE3-pGEX-NPFMAP were induced with 0.5 mM IPTG at 37 °C for 4 h in a rotary shaker (220 rpm). After centrifugation at 5000 rpm for 10 min at 4 °C, the cells were resuspended in 300 mL of buffer A (1 × phosphate-buffered saline (PBS), 0.2% v/v glycerin, 20% v/v Triton X-100, 2 mM dithiothreitol (DTT) pH 7.4) and sonicated at 400 W for 20 min, with 2 s on and 6 s pause, until the solution became clear. Following centrifugation at 12,000 rpm for 20 min at 4 °C, the supernatant was collected. The GST-Sefinose™ Resin affinity chromatography column was balanced with ten volumes of binding buffer B (1 × PBS, 20% v/v glycerin, 2 mM DTT pH 7.4). The supernatant was loaded on the 10-mL column at a flow rate of 2 mL/min. Next, the column was washed with ten volumes of binding buffer B to remove non-specifically bound hybrid proteins. The target protein was eluted with 20 mM glutathione in binding buffer B, using four volumes for each step. Fractions containing the target protein were identified via SDS-PAGE and collected. Then, the protein was dialyzed into binding buffer B for 12 h in the presence of PreScission protease.
After completing the first GST-NPFMAP purification step, a second purification of NPFMAP was required. This differed from the first step in that we collected the effluent and analyzed the fractions by 12% Tricine-SDS-PAGE. Protein concentration at each step was determined by the BCA protein assay.
Antioxidant activities assay
DPPH radical scavenging activity
NPFMAP antioxidant activity was determined by the DPPH radical scavenging assay according to the method reported by Rajapakse et al. (2005). The experimental group (Ai) protein sample (0.2 mL) was added to a sterile 2-mL centrifuge tube, mixed with 0.2 mL deionized water and 0.4 mL DPPH solution (0.2 mM in anhydrous ethanol), and vortexed for 10 s. The tube was then incubated at room temperature for 20 min in the dark, and antioxidant activity was detected at 517 nm. The blank (Aj) and control (A0) groups were assessed with the same method under the same conditions. The blank group contained 0.4 mL anhydrous ethanol in place of DPPH solution, and 0.2 mL deionized water in place of protein sample. The control group contained 0.2 mL deionized water in place of protein sample. Butylated hydroxytoluene (BHT) and vitamin C were used as positive controls. DPPH radical scavenging activity was calculated as follows: DPPH radical scavenging activity (%) = [1 − (Ai − Aj)/A0] × 100.
Superoxide radical scavenging activity
Superoxide radical scavenging activity was measured by means of improved pyrogallol autoxidation (Xia et al. 2012) with some modifications. Briefly, the sample was mixed with a 25-fold volume of Tris–HCl buffer (pH 8.3) in a clean centrifuge tube. The mixture was incubated in a 25 °C water bath for 10 min, an equal volume of pyrogallic acid was added, and absorbance was then measured at 325 nm every 30 s for 5 min. The blank contained an equal volume of deionized water instead of sample. BHT and vitamin C were used as positive controls. Superoxide radical scavenging activity was calculated as follows: superoxide radical scavenging activity (%) = [1 − Vi/V0] × 100. Vi and V0 represent the slope of the absorbance line for sample and blank, respectively.
Hydroxyl radical scavenging activity
Hydroxyl radical scavenging activity was measured according to the method by Zhuang et al. (2013) with some modifications. Briefly, the experimental group (Ai) NPFMAP sample (1.5 mL) was mixed with 0.5 mL of 3 mM ferrous sulfate and 0.5 mL of 3 mM salicylic acid, 0.5 mL of 3 mM H2O2 solution was added, the mixture was incubated at 37 °C for 30 min. and absorbance was measured at 510 nm. The blank (Aj) and control (A0) groups were measured with the same method under the same conditions. The blank group contained 0.5 mL deionized water in place of salicylic acid. The control group contained 1.5 mL deionized water in place of protein sample. BHT and vitamin C were used as positive controls. Hydroxyl radical scavenging activity was calculated as follows: hydroxyl radical scavenging activity (%) = [1 − (Ai − Aj)/A0] × 100.
Statistical analysis
All experiments were measured in three independent batches. Statistical analysis was performed using Excel (Microsoft Ltd., USA). Numerical values were shown as averages ± SD.
Results
Sequence analysis, DNA and recombinant plasmid constructs
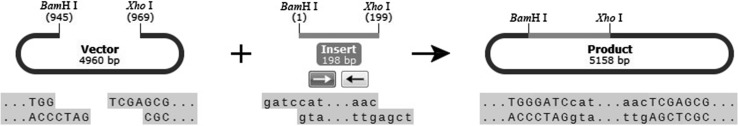
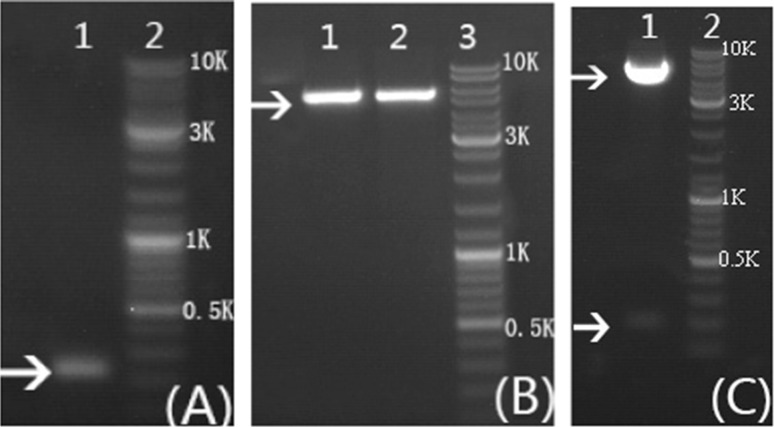
The sequence analysis results showed that the molecular formula, theoretical molecular weight, instability index (II) and isoelectric point of NPFMAP are C251H431N103O79, 6155.83 Da, 43.33 and 11.84, respectively. Meanwhile, the total number of negatively charged residues (Asp + Glu) and positively charged residues (Arg + Lys) are 5 and 15, respectively. These results indicated that the higher molecular weight of NPFMAP, the greater the opportunity for NPFMAP to adopt secondary structures such as coiled spatial conformations for a stable structure and antioxidant activity. To improve solubility of the target protein in E. coli, we fused the N-terminus of NPFMAP to a GST tag, which had been shown to increase solubility, facilitate purification, and cause no interference with the physiological activity of the target protein (Hwang et al. 2014). The NPFMAP fragment was inserted into the pGEX-6P-1 vector at the BamH I and Xho I restriction sites, with the GST tag at the N-terminus of NPFMAP (Fig. 1). A 200-bp band was observed by 1% agarose gel electrophoresis (Fig. 2a), indicating that the 204-bp NPFMAP gene had been successfully generated by OE-PCR. Furthermore, double enzymatic digestion of the pGEX-6P-1 (Fig. 2b) and recombinant pGEX-NPFMAP (Fig. 2c) plasmids revealed that NPFMAP had been successfully ligated into the expression vectors.
Fig. 1.
Scheme representing the generation of pGEX-6P-1-NPFMAP recombinant plasmids. The NPFMAP sequence was inserted into the vector at the BamH I and Xho I restriction sites. The pGEX-6P-1-NPFMAP plasmid contained a GST tag at the insert’s N-terminus
Fig. 2.
Agarose gel electrophoresis analysis of NPFMAP double-stranded DNA, pGEX-6P-1 vector, and pGEX-6P-1-NPFMAP recombinant plasmid. a Lane 1: NPFMAP PCR product after two rounds of overlap extension PCR; lane 2: DNA ladder of 10,000 bp. b Lanes 1–2: single digestion of the pGEX-6P-1 vector by BamH I (1) and Xho I (2); lane 3: DNA ladder of 10,000 bp. c Lane 1: double digestion of the pGEX-6P-1-NPFMAP recombinant plasmid by BamH I and Xho I; lane 2: DNA ladder of 10,000 bp. Arrows indicate the PCR (a) and digestion (b, c) products
Expression of fusion protein GST-NPFMAP
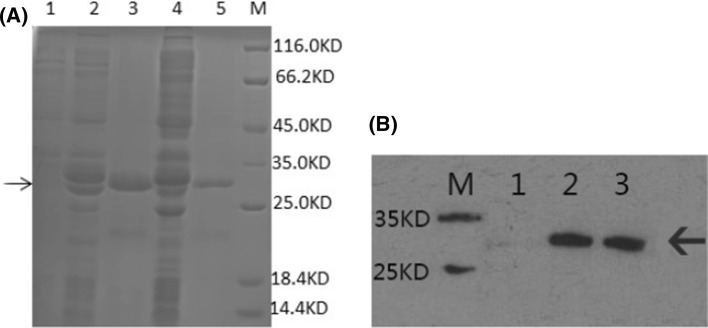
Results (Fig. 3a) indicated that the protein band under the 32,000 Da vicinity, corresponding to the target protein theoretical molecular weight (6155.83 Da) together with GST-tag theoretical molecular weight (26,000 Da), was observed only after IPTG induction. The result confirmed successful expression of the GST-NPFMAP fusion protein, and was corroborated by western blot analysis (Fig. 3b).
Fig. 3.
Expression of the GST-NPFMAP recombinant fusion protein. a 12% SDS-PAGE analysis showing expression of GST-NPFMAP at 20 °C and 37 °C. Lane 1: non-induced lysates; lane 2: lysates induced for 16 h at 20 °C; lane 3: supernatant induced for 16 h at 20 °C; lane 4: lysates induced for 4 h at 37 °C; lane 5: supernatant induced for 4 h at 37 °C; lane M: protein molecular weight marker. b Western blot analysis of GST-NPFMAP induced at 37 °C and probed using an anti-GST tag antibody. Lane M: protein molecular weight marker; lane 1: non-induced supernatant and lysates; lane 2: induced lysates; lane 3: induced supernatant. Arrows denote the position of the fusion proteins
Solubility of the fusion protein GST-NPFMAP
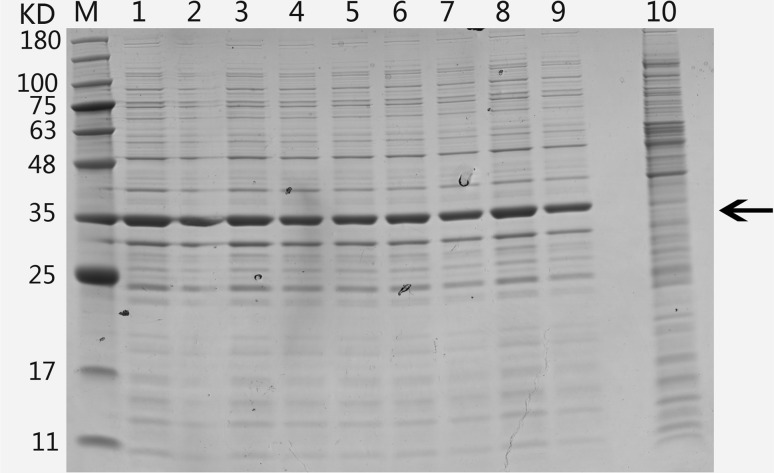
To evaluate the solubility of the GST-NPFMAP fusion protein, expression was induced at different temperatures using liquid cultures. Generally, a higher growth temperature leads to reduced solubility and increased aggregation of target proteins in E. coli expression systems. On the contrary, a lower growth temperature extends the time required to produce foreign proteins during fermentation. SDS-PAGE analysis indicated that GST-NPFMAP was soluble at all induction temperatures (Fig. 4), confirming that NPFMAP had a good solubility in this expression system. Considering induction time and cost, 37 °C was used for the following purification experiments.
Fig. 4.
SDS-PAGE analysis showing expression of GST-NPFMAP recombinant fusion proteins, following induction at 20, 22, 24, 26, 28, 30, 32, 35, and 37 °C Lane M: protein molecular weight marker; Lanes 1–9: lysates induced at 20, 22, 24, 26, 28, 30, 32, 35, and 37 °C, respectively; Lane 10: non-induced lysates at 37 °C. The arrow denotes the position of the fusion protein
Purification of NPFMAP
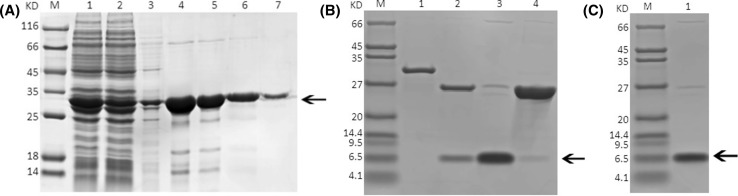
Given the high affinity of the GST tag, contaminants were removed by a pre-loaded GST agarose gel column. SDS-PAGE analysis indicated that the eluates containing the target protein also contained a few low-molecular weight contaminants (Fig. 5a); these were successfully removed following further purification. However, this additional step also decreased target protein yield. After desalting and concentration, purified GST-NPFMAP was obtained. The GST tag could then be removed by adding the PreScission protease to the concentration step, and NPFMAP was collected. At this point the purified target protein contained only a few high-molecular weight contaminants (Fig. 5b). Finally, the target protein molecular weight was confirmed by 12% Tricine-SDS-PAGE (Fig. 5c).
Fig. 5.
SDS-PAGE analysis comparing expression of GST-NPFMAP and NPFMAP at various stages of purification, a SDS-PAGE analysis of eluates from three purification steps lane M: protein molecular weight marker; lane 1: lysates of GST-NPFMAP induced at 37 °C; lane 2: effluent obtained while the sample was in the column; Lane 3: eluate resulting from washing out the column with binding buffer; lanes 4–7: eluate containing GST-NPFMAP. b Lane M: protein molecular weight marker; lane 1: GST-NPFMAP not digested with PreScission protease; lane 2: GST-NPFMAP digested with PreScission protease; lane 3: effluent obtained while the sample was in the column; lane 4: eluate obtained after washing out the column with elution buffer; c lane M: protein molecular weight marker; lane 1: NPFMAP following a second affinity chromatography purification step. Arrows denote the position of the fusion (a) and target (b, c) proteins
Antioxidant activity assays
DPPH radical scavenging capacity
The antioxidant activities of NPFMAP and GST-NPFMAP were investigated by measuring DPPH radical scavenging activity (Table 2). The results indicated that both NPFMAP and GST-NPFMAP had bioactive properties in vitro, with DPPH IC50 values of 0.028 mg/mL and 0.264 mg/mL, respectively. At the same time, DPPH IC50 values of natural-PFMAP, chemosynthesis-PFMAP, BHT, glutathione and vitamin C were also measured under the same experimental conditions, and amounted to 0.132 mg/mL, 0.385 mg/mL, 0.025 mg/mL, 0.033 mg/mL and 0.003 mg/mL, respectively. NPFMAP obtained by this method exhibited higher DPPH radical scavenging activity than PFMAP prepared by chemical synthesis or natural enzymatic extraction (p < 0.05), as well as preformed higher DPPH radical scavenging than glutathione that was commonly used in cosmetics yield as antioxidants (p < 0.05). Although the DPPH IC50 value of NPFMAP was lower than that of vitamin C, it was nevertheless nearly equal to that of BHT (p > 0.05).
Table 2.
Antioxidant activity of different antioxidant concluding NPFMAP, GST-NPFMAP, natural PFMAP, chemosynthesis-PFMAP, glutathione, BHT and Vitamin C
| Antioxidant | DPPH radical scavenging activity (IC50, mg/mL) | Superoxide radical scavenging activity (IC50, mg/mL) | Hydroxyl radical scavenging activity (IC50, mg/mL) |
|---|---|---|---|
| NPFMAP | 0.028 ± 0.002b | 0.105 ± 0.008b | 0.065 ± 0.005b |
| GST-NPFMAP | 0.264 ± 0.005e | 0.902 ± 0.051e | 0.688 ± 0.062e |
| Natural PFMAP | 0.132 ± 0.005d | 1.046 ± 0.03e | 0.268 ± 0.045c |
| Chemosynthesis-PFMAP | 0.385 ± 0.008f | 1.175 ± 0.063f | 0.975 ± 0.082f |
| Glutathione | 0.033 ± 0.001c | 0.386 ± 0.005d | 0.326 ± 0.004d |
| BHT | 0.025 ± 0.002b | 0.30 ± 0.003c | 0.27 ± 0.005c |
| Vitamin C | 0.003 ± 0.001a | 0.026 ± 0.001a | 0.020 ± 0.002a |
Values represent mean ± SD (n = 3), different letters (a, b, c, d, e, f) indicate significant differences (p < 0.05). IC50: the antioxidant concentration that scavenges free radical by 50%
Superoxide radical scavenging capacity
Superoxide radical scavenging activities of NPFMAP and GST-NPFMAP were reflected in Table 2. The results showed that NPFMAP, natural PFMAP and chemosynthesis PFMAP had antioxidant activity in vitro, and the IC50 values for superoxide radical scavenging activity were 0.105 mg/mL, 1.406 mg/mL and 1.175 mg/mL, respectively. These activity results indicate that NPFMAP exhibited higher superoxide radical scavenging activity than that prepared via chemosynthesis or enzymatic extraction method (p < 0.05). In particular, the superoxide radical scavenging activity was ten times higher than that of natural PFMAP, which were prepared using enzymatic extraction techniques before modification. At the same time, BHT, glutathione and vitamin C IC50 values were also measured under the same experimental conditions. NPFMAP showed a higher activity of superoxide radical scavenging compared to BHT (p < 0.05), as well as to glutathione (p < 0.05). However, vitamin C (0.026 mg/mL) exhibited higher superoxide radical scavenging activity compared with NPFMAP (p < 0.05). These superoxide radical scavenging activity results indicated that NPFMAP could be used in cosmetics and pharmaceutical industries compared to glutathione.
Hydroxyl radical scavenging capacity
Hydroxyl radical scavenging abilities of NPFMAP, GST-NPFMAP, natural-PFMAP, chemosynthesis-PFMAP, BHT, glutathione and vitamin C were measured under the same experimental conditions. The corresponding IC50 values were 0.065 mg/mL, 0.688 mg/mL, 0.268 mg/mL, 0.975 mg/mL, and 0.270 mg/mL, 0.326 mg/mL and 0.020 mg/mL, respectively (Table 2). Results indicate that both target NPFMAP and GST-NPFMAP had hydroxyl radical scavenging ability in vitro. In particular, NPFMAP showed a higher activity of hydroxyl radical scavenging compared to glutathione (p < 0.05), and the same is true of BHT, natural PFMAP and chemosynthesis PFMAP. Similarly, NPFMAP exhibited lower hydroxyl radical scavenging activity compared to vitamin C (p < 0.05).
Discussion
Several antioxidant peptides from marine organisms have been previously separated, purified, and characterized by enzymatic methods in vitro. However, these peptides presented low yield and low antioxidant activity (Wu et al. 2013b). For example, the chemical synthesis peptide were obtained using solid phase technologies, based on the sequence which isolated from Pinctada fucata by enzyme hydrolysis in previous study, to clear the relationship between primary structure of the peptide and its antioxidant activity. However, the L-conformation type amino acids were commonly chosen to synthesize peptide in the previous work (Guo et al. 2009; Wu et al. 2017). So this could make chemosynthesis peptide forming a linear structure according to the identified peptide sequence, and this primary structure could be different compared with natural peptide by enzyme hydrolysis method. Therefore, radical scavenging activity should be different between natural peptide and chemosynthesis peptide, and the natural peptide prepared using enzyme hydrolysis method might have some random coil structure. Next, it is well understood that peptides have better activity by enzyme hydrolysis method than chemical synthesis peptide functional activity, as well as demonstrate that the the spatial conformation like random coil formed in preparing peptides using enzyme hydrolysis method. Meanwhile, natural peptides PFMAP were also chemical synthesized with L-conformation type and R-conformation type amino acids, but if every amino acid site in peptide chain was selected one conformation (L-conformation or R-conformation type), then 210 peptide chains should be synthesized respectively to obtain the correctly conformation which was correspond with the enzyme hydrolysis peptide chain conformation. Moreover, the cost, safety and complexity level in chemical process of the peptide chain containing L-conformation type and R-conformation type amino acids was higher. Up till now, antioxidant peptides such as glutathione and L-carnosine which containing three or two amino acids, were commonly used additives in cosmetics industry replacing chemical BHT and BHA, but rarely reported peptide chain containing ten or more amino acids. So, exploring a new preparation method to obtain higher antioxidant activity and purity peptides using molecular modification is bound to open a breakthrough for higher value applications and industrialization applications. Compared with traditional separation and purification methods, such as enzymatic hydrolysis and chemical synthesis, genetic engineering allows for structural modification and large-scale production of antioxidant peptides. E. coli expression systems, in particular, enable rapid expression (within 12 h), high yields, low costs, and simple scale-up procedures (Peciak et al. 2014). Additionally, heterologous proteins are well expressed in E. coli, as exemplified by human interferon alpha, which was purified to homogeneity in a single step (Bis et al. 2014; Kralicek 2014). In the present work, the GST tag augmented protein synthesis in E. coli, whereas molecular modifications of the amino acid peptide chain of PFMAP could be used to boost antioxidant activity. Hence, a bacterial expression system appears suitable for the production of NPFMAP, which can then be used in research or clinical applications.
Here, DNA sequence of code-optimized NPFMAP was introduced into the E. coli host system and heterologous protein expression was driven by the IPTG inducible lac promoter. Recombinant protein production was optimal 4 h after induction with IPTG at a final concentration of 0.5 mM. The NPFMAP DNA sequence was generated by OE-PCR (Fig. 2a), and successful recombinant plasmid construction was confirmed by double enzymatic digestion and DNA sequencing (Fig. 2c). Our result was in agreement with that from a previous study that also used this vector (Yang et al. 2015). Unlike traditional separation and purification methods, which consume substantial manpower and material resources while optimizing time and temperature for enzymatic hydrolysis, the present strategy provides functional oligopeptides production with high purity (Fig. 5c). Accordingly, once the engineered strain is successfully constructed, large-scale NPFMAP production by fermentation can begin within a short time.
The 26-kDa GST tag is commonly used to label target proteins in E. coli. Given that NPFMAP had a molecular weight of about 6 kDa, the expected molecular weight of the GST-NPFMAP fusion protein was around 32 kDa. As shown in Fig. 3a, the fusion protein was recovered from the supernatant after induction with IPTG, and bands of the correct molecular weight were detected; these were absent from non-induced lysates. It is generally assumed that the solubility of heterologous proteins in E. coli increases at lower induction temperatures (Peciak et al. 2014). However, SDS-PAGE revealed that the solubility of recombinant GST-NPFMAP (Fig. 4) did not change much with increasing temperature. To maximize fusion protein production time and solubility, the appropriate temperature should be chosen (Vargas-Cortez et al. 2016). Western blot results (Fig. 3b) confirmed expression of the recombinant fusion protein and demonstrated that it reacted against antibodies. Significantly, only antibodies recognized the recombinant protein on western blots, whereas no reactivity with control was observed. The GST-NPFMAP fusion protein localized to the soluble fraction after sonication, which enabled its purification by affinity chromatography. SDS-PAGE analysis (Fig. 5a) confirmed the presence of only a few low-molecular weight contaminants. To prevent the GST tag from interfering with the target protein antioxidant activity, we cut off the tag using 3C protease, and concentrated the purified target protein (Fig. 5b). The final NPFMAP yield was somewhat lower because of the loss of the GST tag (Camper and Viola 2009).
Currently, several GST-tagged functional peptides can be expressed in E. coli, and protein production may be further increased by optimizing medium composition, IPTG dosage, temperature, and pH. Additionally, yield could be further improved by growing the culture to high cell density by fermentation (Rosano and Ceccarelli 2014). In the present study, double-step purification by affinity chromatography yielded about 90% pure protein (Fig. 5c). A comparison of the antioxidant activities of NPFMAP, GST-NPFMAP, natural-PFMAP, chemosynthesis-PFMAP, BHT, glutathione and vitamin C (Table 1) showed that both target and fusion proteins had bioactivity properties. Whereas co-expression of NPFMAP with the highly soluble GST tag facilitated expression and may ease purification during pilot production, the antioxidant activity of GST-NPFMAP was nevertheless lower than that of NPFMAP. Indeed, the DPPH IC50 value of NPFMAP was 0.028 mg/mL, which was ten times higher than that of GST-NPFMAP (0.264 mg/mL), and this result means that the type and size of the GST tag may affect peptide NPFMAP antioxidant activity in vitro experiment. Furthermore, DPPH radical scavenging activity of NPFMAP performed higher activity than other marine antioxidant peptides, such as Theragra (Jia et al. 2010), Salmon (Ahn et al. 2014), Bluefin (Chi et al. 2015) were 2.5 mg/ml, 1.63 mg/ml and 10 mg/ml, respectively (p < 0.05). Meanwhile, it was important that results showed NPFMAP had a better superoxide scavenging activity than other marine antioxidant peptides, for example, the IC50 of superoxide radical scavenging activity of antioxidant peptides from zein (Li et al. 2010), corn (Jin et al. 2016) were 12.82 mg/ml and 3.6 mg/ml (p < 0.05). Moreover, NPFMAP had a better hydroxyl scavenging activity than that reported in recent years, the hydroxyl radical scavenging activity of antioxidant peptides from Oreochromis niloticus (Ngo et al. 2010), Bluefin leatherjacket (Chi et al. 2015) were 2 mg/ml, 0.263 mg/ml, 1 mg/ml, and 1.04 mg/ml, respectively (p < 0.05). These radical scavenging activity results indicated that NPFMAP had a higher value applications and industrialization applications in cosmetics as a new small molecule functional oligopeptide. Moreover, preparation NPFMAP, which using molecular modification provide insight into the relationship between structure and activity, as well as establish the new way to open a breakthrough for the next step industrialization application like glutathione which was commonly used in cosmetics industrial application. Additionally, NPFMAP exhibited higher radical scavenging activity compared to natural PFMAP, and the same is true for natural PFMAP and chemosynthesis PFMAP. These results suggested that antioxidant peptides had random coil which exhibiting antioxidant activity in peptide chain. This random coil which performing second structure or spatial conformation might play an important role in its antioxidant activity compared with its peptide chain length, amino acid composition, amino acid sequence, degree of side chain amino acids glycosylation, size of the side chain structure and molecular weight. Furthermore, these activity properties are also confirmed antioxidant peptides which obtained by enzymatic hydrolysis and chemical synthesis method (Wu et al. 2013a), and the corresponding DPPH IC50 values were 0.132 mg/mL and 0.385 mg/mL, respectively. Compared with the enzymatic hydrolysis and chemical synthesis method, our approach is clearly advantageous in terms of bioactivity and preparation time. Finally, the proposed protocol could serve as a basis for large-scale expression and purification of other recombinant proteins, their characterization, and downstream application.
In this study, we generated new antioxidant peptides from Pinctada fucata meat (NPFMAP), using recombinant E. coli, in which five PFMAP amino acid peptide chains were genetically modified. In contrast to traditional separation and purification methods, the NPFMAP purification process was simple, less time-consuming (12 h), and resulted in higher yield and elevated antioxidant activity. In addition, for the next step practical application in cosmetic or food formulation, the shelf stability of NPFMAP, as well as the stability in vitro and in vivo tests will be further researched in future studies. In summary, the proposed method supplied the test data and theoretical basis on preparing NPFMAP for the next step research or its potential applications in cosmetics and health food industries.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31371800), the Special Scientific Research Funds for Central Non-profit Institutes, Chinese Academy of Fishery Sciences (No. 2016ZD10), Special Project of Marine fishery technology and industrial development in Guangdong province (No. Z2015008).
Contributor Information
Yanyan Wu, Phone: +86-20-34063583, Email: wuyygd@163.com.
Yongkai Ma, Email: yongkai_ma@126.com.
Laihao Li, Email: laihaoli@163.com.
Xianqing Yang, Email: yxqgd@163.com.
References
- Ahn C-B, Kim J-G, Je J-Y. Purification and antioxidant properties of octapeptide from salmon byproduct protein hydrolysate by gastrointestinal digestion. Food Chem. 2014;147:78–83. doi: 10.1016/j.foodchem.2013.09.136. [DOI] [PubMed] [Google Scholar]
- Bis RL, Stauffer TM, Singh SM, Lavoie TB, Mallela KMG. High yield soluble bacterial expression and streamlined purification of recombinant human interferon α-2a. Protein Expr Purif. 2014;99:138–146. doi: 10.1016/j.pep.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougatef A, Balti R, Haddar A, Jellouli K, Souissi N, Nasri M. Protein hydrolysates from Bluefin Tuna (Thunnus thynnus) heads as influenced by the extent of enzymatic hydrolysis. Biotechnol Bioprocess Eng. 2012;17(4):841–852. doi: 10.1007/s12257-012-0053-y. [DOI] [Google Scholar]
- Camper DV, Viola RE. Fully automated protein purification. Anal Biochem. 2009;393(2):176–181. doi: 10.1016/j.ab.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chi Y-J, Zhao M-Y, Lv L. Purification and identification of antioxidant peptides from egg white protein hydrolysate. Amino Acids. 2011;43(1):457–466. doi: 10.1007/s00726-011-1102-0. [DOI] [PubMed] [Google Scholar]
- Cheung IWY, Cheung LKY, Tan NY, Li-Chan ECY. The role of molecular size in antioxidant activity of peptide fractions from Pacific hake (Merluccius productus) hydrolysates. Food Chem. 2012;134(3):1297–1306. doi: 10.1016/j.foodchem.2012.02.215. [DOI] [PubMed] [Google Scholar]
- Chi C-F, Wang B, Wang Y-M, Zhang B, Deng S-G. Isolation and characterization of three antioxidant peptides from protein hydrolysate of bluefin leatherjacket (Navodon septentrionalis) heads. J Funct Foods. 2015;12:1–10. doi: 10.1016/j.jff.2014.10.027. [DOI] [Google Scholar]
- Cumby N, Zhong Y, Naczk M, Shahidi F. Antioxidant activity and water-holding capacity of canola protein hydrolysates. Food Chem. 2008;109(1):144–148. doi: 10.1016/j.foodchem.2007.12.039. [DOI] [PubMed] [Google Scholar]
- Guo H, Kouzuma Y, Yonekura M. Structures and properties of antioxidative peptides derived from royal jelly protein. Food Chem. 2009;113(1):238–245. doi: 10.1016/j.foodchem.2008.06.081. [DOI] [Google Scholar]
- Hwang WH, Lee WK, Ryoo SW, Yoo KY, Tae GS. Expression, purification and improved antigenicity of the Mycobacterium tuberculosis PstS1 antigen for serodiagnosis. Protein Expr Purif. 2014;95:77–83. doi: 10.1016/j.pep.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhou Y, Lu J, Chen A, Li Y, Zheng G. Enzymatic hydrolysis of Alaska pollack (Theragra chalcogramma) skin and antioxidant activity of the resulting hydrolysate. J Sci Food Agric. 2010;90:635–640. doi: 10.1002/jsfa.3861. [DOI] [PubMed] [Google Scholar]
- Jin D-X, Liu X-L, Zheng X-Q, Wang X-J, He J-F. Preparation of antioxidative corn protein hydrolysates, purification and evaluation of three novel corn antioxidant peptides. Food Chem. 2016;204:427–436. doi: 10.1016/j.foodchem.2016.02.119. [DOI] [PubMed] [Google Scholar]
- Kim S-Y, Je J-Y, Kim S-K. Purification and characterization of antioxidant peptide from hoki (Johnius belengerii) frame protein by gastrointestinal digestion. J Nutr Biochem. 2007;18(1):31–38. doi: 10.1016/j.jnutbio.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Kralicek A. A cell-free expression screen to identify fusion tags for improved protein expression. Methods Mol Biol. 2014;1118:35–54. doi: 10.1007/978-1-62703-782-2_3. [DOI] [PubMed] [Google Scholar]
- Lee PY, Yong VC, Rosli R, Gam LH, Chong PP. Cloning, expression and purification of squalene synthase from Candida tropicalis in Pichia pastoris. Protein Expr Purif. 2014;94:15–21. doi: 10.1016/j.pep.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Li HM, Hu X, Guo P, Fu P, Xu LI, Zhang XZ. Antioxidant properties and possible mode of action of corn protein peptides and zein peptides. J Food Biochem. 2010;34:44–60. doi: 10.1111/j.1745-4514.2009.00292.x. [DOI] [Google Scholar]
- Liu Q, Kong B, Xiong YL, Xia X. Antioxidant activity and functional properties of porcine plasma protein hydrolysate as influenced by the degree of hydrolysis. Food Chem. 2010;118(2):403–410. doi: 10.1016/j.foodchem.2009.05.013. [DOI] [Google Scholar]
- Nalinanon S, Benjakul S, Kishimura H, Shahidi F. Functionalities and antioxidant properties of protein hydrolysates from the muscle of ornate threadfin bream treated with pepsin from skipjack tuna. Food Chem. 2011;124(4):1354–1362. doi: 10.1016/j.foodchem.2010.07.089. [DOI] [Google Scholar]
- Ngo D-H, Qian Z-J, Ryu B, Park JW, Kim S-K. In vitro antioxidant activity of a peptide isolated from Nile tilapia (Oreochromis niloticus) scale gelatin in free radical-mediated oxidative systems. J Funct Foods. 2010;2(2):107–117. doi: 10.1016/j.jff.2010.02.001. [DOI] [Google Scholar]
- Peciak K, Tommasi R, J-W Choi, Brocchini S, Laurine E. Expression of soluble and active interferon consensus in SUMO fusion expression system in E. coli. Protein Expr Purif. 2014;99:18–26. doi: 10.1016/j.pep.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Rajapakse N, Mendis E, Jung W-K, Je J-Y, Kim S-K. Purification of a radical scavenging peptide from fermented mussel sauce and its antioxidant properties. Food Res Int. 2005;38(2):175–182. doi: 10.1016/j.foodres.2004.10.002. [DOI] [Google Scholar]
- Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Yang H, Xin Y, Zhang L, Wang W. Novel integration strategy coupling codon and fermentation optimization for efficiently enhancing sarcosine oxidase (SOX) production in recombinant Escherichia coli. World J Microbiol Biotechnol. 2015;31(5):707–716. doi: 10.1007/s11274-014-1795-9. [DOI] [PubMed] [Google Scholar]
- Vargas-Cortez T, Morones-Ramirez JR, Balderas-Renteria I, Zarate X. Expression and purification of recombinant proteins in Escherichia coli tagged with a small metal-binding protein from Nitrosomonas europaea. Protein Expr Purif. 2016;118:49–54. doi: 10.1016/j.pep.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Wu YY, Li LH, Duan ZH, Yang XQ, Shang J, Chen SJ. Application of response surface methodology to optimise preparation high antioxidant activity product from Pinctada fucata muscle. Adv Mater Res. 2012;396–398:1341–1348. [Google Scholar]
- Wu Y, Tian Q, Li L, Khan MN, Yang X, Zhang Z, Hu X, Chen S. Inhibitory effect of antioxidant peptides derived from Pinctada fucata protein on ultraviolet-induced photoaging in mice. J Funct Foods. 2013;5(2):527–538. doi: 10.1016/j.jff.2013.01.016. [DOI] [Google Scholar]
- Wu YY, Wang J, Li LH, Yang XQ, Hu X. Desalination of antioxidant peptides from Pinctada fucata using C18 chromatographic column. Adv Mater Res. 2013;781–784:1508–1512. doi: 10.4028/www.scientific.net/AMR.781-784.1508. [DOI] [Google Scholar]
- Wu YY, Wang J, Li LH, Yang XQ, Wang JX, Hu X. Purification and identification of an antioxidant peptide from Pinctada fucata muscle. CyTA J Food. 2017;16(1):11–19. doi: 10.1080/19476337.2017.1332099. [DOI] [Google Scholar]
- Xia Y, Bamdad F, Gänzle M, Chen L. Fractionation and characterization of antioxidant peptides derived from barley glutelin by enzymatic hydrolysis. Food Chem. 2012;134(3):1509–1518. doi: 10.1016/j.foodchem.2012.03.063. [DOI] [PubMed] [Google Scholar]
- Xiao YH, Pei Y. Symmetric overlap extension PCR method for site-directed mutagenesis. Methods Mol Biol. 2011;421(687):277–282. doi: 10.1007/978-1-60761-944-4_20. [DOI] [PubMed] [Google Scholar]
- Yang F, Pan Y, Chen Y, Tan S, Jin M, Wu Z, Huang J. Expression and purification of Canis interferon α in Escherichia coli using different tags. Protein Expr Purif. 2015;115:76–82. doi: 10.1016/j.pep.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Yu Y, Song J, Guo X, Wang S, Yang X, Chen L, Wei J. Characterization and structural analysis of human selenium-dependent glutathione peroxidase 4 mutant expressed in Escherichia coli. Free Radic Biol Med. 2014;71:332–338. doi: 10.1016/j.freeradbiomed.2014.03.032. [DOI] [PubMed] [Google Scholar]
- Zambrowicz A, Pokora M, Setner B, Dąbrowska A, Szołtysik M, Babij K, Szewczuk Z, Trziszka T, Lubec G, Chrzanowska J. Multifunctional peptides derived from an egg yolk protein hydrolysate: isolation and characterization. Amino Acids. 2014;47(2):369–380. doi: 10.1007/s00726-014-1869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang H, Wang L, Guo X, Wang X, Yao H. Isolation and identification of antioxidative peptides from rice endosperm protein enzymatic hydrolysate by consecutive chromatography and MALDI-TOF/TOF MS/MS. Food Chem. 2010;119(1):226–234. doi: 10.1016/j.foodchem.2009.06.015. [DOI] [Google Scholar]
- Zhong S, Ma C, Lin YC, Luo Y. Antioxidant properties of peptide fractions from silver carp (Hypophthalmichthys molitrix) processing by-product protein hydrolysates evaluated by electron spin resonance spectrometry. Food Chem. 2011;126(4):1636–1642. doi: 10.1016/j.foodchem.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Zhuang H, Tang N, Yuan Y. Purification and identification of antioxidant peptides from corn gluten meal. J Funct Foods. 2013;5(4):1810–1821. doi: 10.1016/j.jff.2013.08.013. [DOI] [Google Scholar]