Abstract
Background
We aim to investigate the utility of serial gene mutation tracking for locally advanced CRC in those who underwent curative resection following neoadjuvant chemotherapy.
Methods
We prospectively collected 10 locally advanced CRC cases for which curative resection was performed following preoperative neoadjuvant chemotherapy. Tissues from the primary tumour, distant metastatic tumours, and blood plasma were obtained during serial treatment. Comprehensive mutation analysis of 47 cancer-associated genes was performed using a pre-designed gene panel and next-generation sequencing.
Results
All cases showed a partial response to neoadjuvant chemotherapy, and pathological R0 resection was accomplished. In primary tumours, non-synonymous mutations were detected at between 1 and 14 sites before chemotherapy and at between 1 and 2 sites after. Founder mutations were precisely detected in blood plasma and metastatic tumours during longitudinal treatment.
Conclusions
Serial mutational analysis indicated that subclonal selection occurs during chemotherapy and that plasma can substitute for tumourous tissue in mutational analysis for drug selection and treatment decisions.
Subject terms: Surgical oncology, Colorectal cancer
Introduction
Surgical resection is the only curative treatment for CRC, but locally advanced cancer and distant metastasis prevent CRC patients from undergoing curative resection. Considering the high response rate of cytotoxic and molecular-targeted agents for metastatic CRC,1,2 neoadjuvant chemotherapy (NAC) designed to down-stage initially non-resectable or marginally resectable locally advanced CRC could allow curative resection and bring a significant survival benefit to patients.3 However, there is no standard treatment protocol for locally advanced CRC because systematic studies have not yet been conducted.4,5
It is estimated that there are 138 driver genes of CRC, however, one case usually contains only 2–8 driver gene alterations, with the remainder being “passenger” gene defects which have no effect on the neoplastic process.6,7 Activating mutations in the RAS gene family are well-established biomarkers for a lack of response to anti-epidermal growth factor receptor antibody therapy.8–10 Several new biomarkers such as BRAF V600E mutation, microsatellite instability, HER2/neu overexpression, and ERBB2 amplification are under investigation for targeted therapies.11 We previously revealed the clonal evolution and intra-tumour heterogeneity of CRC with comprehensive exome sequencing and multiregional analysis.12 Understanding intra-tumour heterogeneity is clinically important for NAC because it could cause therapeutic failure by fostering evolutionary adaptation.13 Although good responses are obtained in patients who receive cytotoxic and molecular targeted therapy for metastatic CRC, most tumours will ultimately progress.10 This progression may be attributed to continued clonal evolution and diversification, including resistance to therapy resulting from selective pressure.14 Furthermore, the mutational profile from the primary tumour tissue at one time point may not represent the true genomic alteration in the patient, thus it is necessary to identify mutations during serial treatment to maximise the effect of NAC.
To explore and conquer these issues, we obtained tumour and plasma samples from 10 patients with locally advanced CRC. They were acquired consecutively within a prospective tissue collection protocol both at diagnosis and at surgery following oxaliplatin-containing NAC. We tracked serial genomic evolution in locally advanced CRC during disease evolution and following cytotoxic and targeted therapy.
Materials and methods
Patients and sample collection
Ten CRC patients with locally advanced primary tumours were recruited into this study. Their primary tumours invaded adjacent organs or tissues and were diagnosed as T4a or T4b using the TNM classification system of the Union for International Cancer Control (UICC). All patients underwent primary tumour resection following oxaliplatin-based NAC at Kyushu University Beppu Hospital. Samples were obtained after written informed consent. The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethical Review Board of Kyushu University. Detailed clinical characteristics and the sampling protocol are provided in Table S1 and Fig. S1. Tissues were obtained from 3 to 5 locations in the primary tumour by endoscopic biopsy prior to NAC, and DNA was extracted from these samples combined. After NAC, cancer tissues were obtained from surgically resected specimens. In two cases with distant metastatic tumours, tissues were obtained from resected specimens. In two cases that agreed the research with informed consent, 10 mL blood samples were obtained in EDTA tubes and centrifuged to collect the plasma at the time of surgery.
DNA extraction
Genomic DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue, fresh frozen tumour, adjacent normal intestinal mucosa, and plasma using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany), the AllPrep DNA/RNA Mini Kit (Qiagen), or the QIAamp Circulating Nucleic Acid Kit (Qiagen) as described elsewhere.15
Identification of genomic mutations by next-generation sequencing
Target-enriched libraries for 47 cancer-associated genes were prepared using the ClearSeq Cancer Panel (Agilent Technologies) according to the manufacturer’s instructions (Table S2). Genomic DNA was digested using restriction enzymes, the HaloPlex probe library (Agilent Technologies) was hybridised in the presence of the indexing primer cassette, which resulted in genomic DNA fragment circularisation. The HaloPlex probes were biotinylated, so target DNA-probe hybrids were captured using streptavidin-coated magnetic beads. Next, the target DNA-probe hybrids were ligated closed and PCR amplified to produce a target-enriched library for targeted deep sequencing. The target-enriched libraries were subjected to high-throughput sequencing using a Hiseq 2500 (Illumina, San Diego, CA, USA), and sequencing reads were aligned to the NCBI Human Reference Genome Build 37 hg19 with BWA version 0.7.10 using default parameters (http://bio-bwa.sourceforge.net/). Read depth of the sequencing was provided in Table S3, and mean coverage was between 1284 and 28021. The sequence data were processed using an in-house pipeline (http://genomon.hgc.jp/exome/). The sequencing reads were aligned to the NCBI Human Reference Genome Build 37 hg19 with BWA version 0.5.10 using default parameters (http://bio-bwa.sourceforge.net/). Mutation calling was conducted using the following parameters: (i) mapping quality score ≥ 25, (ii) base quality score ≥ 15, (iii) mismatched bases ≤ 5, (iv) both tumour and normal depths ≥ 100, (v) variant allele frequencies in tumour samples ≥ 0.001 (tissue) or 0.00 (plasma), (vi) Fisher’s exact test P-values < 0.05.12,15,16 After mutation calling, the Integrative Genomics Viewer (IGV) (http://software.broadinstitute.org/software/igv/) was used to visualise the mutations identified with next-generation sequencing (NGS).17
Results
Clinical response of locally advanced CRC to NAC
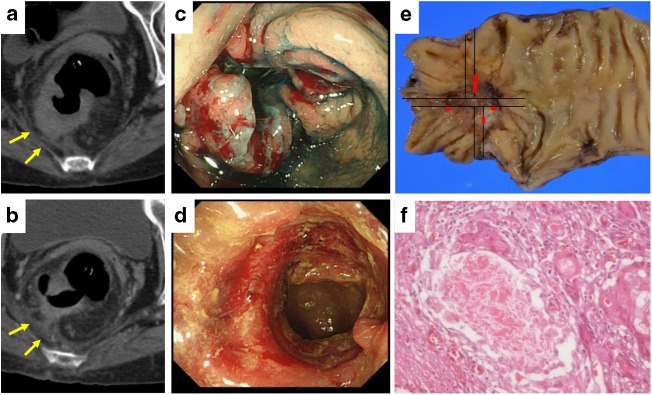
All 10 patients showed a partial response in computed tomography and endoscopic colonoscopy according to the revised Response Evaluation Criteria in Solid Tumours (RECIST) guidelines (Fig. 1a–d) 18 and underwent complete primary tumour resection without either macroscopic or microscopic residual tumour. The pathological response of the primary tumour was evaluated using the percent of the area showing fibrosis and necrosis (Fig. 1e, f). Five cases (50%) showed >66% tumour regression (grade 2), 3 cases (30%) showed 34–66% regression (grade 1b), and 2 cases (20%) showed <33% regression (grade 1a) according to pathological findings.
Fig. 1.
Treatment response of primary tumour to preoperative neoadjuvant chemotherapy. a, b Computed tomography of primary tumour. The tumour had invaded adjacent retroperitoneum at diagnosis (a), and significant shrinkage and fibrosis was seen after chemotherapy (b). c, d Endoscopic findings of the primary tumour. The tumour occupied the whole lumen of the rectum (c), and significant response was seen after chemotherapy (d). e Resected primary tumour specimen. The viable cancer cells are indicated with red lines. f Pathological findings of the primary tumour after chemotherapy. More than 2/3 was replaced by fibrosis
Gene mutational tracking during preoperative chemotherapy
Mutations were identified in between 1 and 14 genes in primary tumour obtained by endoscopic biopsy before treatment (Table 1). A mutation in TP53 was identified in 9 of the 10 cases (90%), and a mutation in KRAS or NRAS was identified in 5 cases (50%). After NAC, the number of mutations decreased, and only 1 or 2 gene mutations, which were RAS and TP53 mutations exclusively found in the primary tumour biopsy before NAC, were detected in the primary tumours (Table 1). In 1 case, mutations in KRAS and PTEN were detected before chemotherapy, but no mutations were detected after chemotherapy. In 2 metastatic tumours in the liver and spleen, the same mutations were found as in the resected primary tumours (Table 1).
Table 1.
Genes identified nonsynonymous mutations by serial mutational analysis of locally advanced colorectal cancer during longitudinal treatment
| Case no. | Primary tumour biopsy (prechemotherapy) gene name (VAF) | Primary tumour resection (postchemotherapy) gene name (VAF) | Plasma (postchemotherapy) gene name (VAF) | Metastatic tumour gene name (VAF) |
|---|---|---|---|---|
| 1 | TP53 S109F (0.249), FGFR3 P250R (0.028), NOTCH1 E1567X (0.056), ERBB2 R849Q (0.062) | TP53 S109F (0.550) | no data | no data |
| 2 | TP53 G113S (0.174), ERBB2 G776V (0.117), ERBB4 R938H (0.060), FGFR3 P250R (0.019), EGFR V845M (0.117), RUNX1 A384S (0.138), FANCA E950X (0.083) | TP53 G113S (0.013) | no data | TP53 G113S (0.621) |
| 3 | KRAS(0.200),Q61L PTENE288fs | no mutation | no data | no data |
| 4 | KRAS G12D (0.337), TP53 H61L (0.113) | KRAS G12D (0.319), TP53 H61L (0.526) | KRAS G12D (0.003), TP53 H61L (0.002) | no data |
| 5 | KRASG13D(0.421), TP53,105_108del FGFR3A265E(0.079), MAP2K4D276Y(0.078) | KRAS G13D (0.780), TP53 105_108del | no data | KRAS G13D (0.463), TP53 105_108del |
| 6 | TP53 R43H (0.202) | TP53 R43H (0.412) | TP53 R43H (0.022) | no data |
| 7 | TP53 P59P (0.052), FGFR3 T264M (0.024) | TP53 P59P (0.052) | no data | no data |
| 8 | KRAS G12V (0.387), TP53 R116Q (0.047), RET P766L (0.040), SMAD4 R445X (0.033), STK11 E256K (0.027), ALK R1275X (0.022), RUNX1 R180Q (0.026), PIK3CA N345K (0.165), FGFR3 A391V (0.028), MET E168D (0.113), CDKN2A R71C (0.053), FANCG D437H (0.028), ABL1 D276E (0.048), NOTCH1 R1598R (0.056) | TP53 R116Q (0.096), MET E168D (0.352) | no data | no data |
| 9 | NRAS Q61L (0.219), TP53 R116Q (0.030), MAP2K4 I113T (0.256) | NRAS Q61L (0.200), TP53 R116Q (0.021) | no data | no data |
| 10 | TP53 C110F (0.280), PIK3R1 L340L (0.038) | TP53 C110F (0.395) | no data | no data |
VAF variant allele frequency
Tracking mutations in circulating tumour DNA during NAC
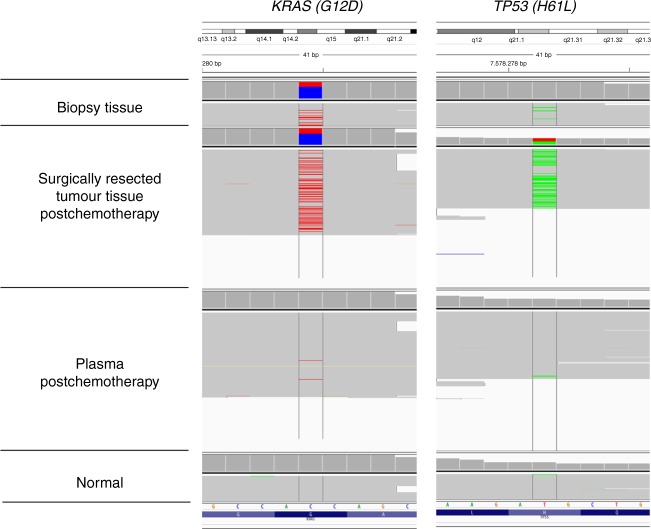
Serial mutational analysis may be necessary in patients with conversion surgery following NAC because various genomic modifications could result from chemotherapy. We explored mutant circulating tumour DNA (ctDNA) in blood samples after NAC in two patients. In one case, mutations in KRAS and TP53 were detected from a biopsy specimen before chemotherapy. After 4 courses of CAPOX therapy, the same mutations were detected in ctDNA (Fig. 2). Those mutations were also verified from surgically resected tissues after NAC. In another case, a driver mutation in TP53 coincident with the primary tumour tissue was also detected in ctDNA. These results indicate that plasma can be used for repeated and noninvasive serial monitoring of mutational changes.
Fig. 2.
Visualization of gene mutations in primary tumour and plasma with Integrative Genomics Viewer. A representative mutation in the primary tumour at diagnosis, in the surgically resected tumour, in plasma, and in normal samples. Nonsynonymous mutations in KRAS (chr12_25398284) and TP53 (chr17_7578271) are visualized
Discussion
This is the first clinical series showing serial gene mutation tracking in patients with locally advanced CRC. The tumours responded well to NAC, and all patients eventually underwent pathologically complete resection. Our results indicate that down-staging by NAC could be a standard strategy for treating locally advanced CRC. It is important to maximise the anti-tumour effect with the optimal regimen and duration of chemotherapy, otherwise it is possible that the patients will miss their chance for surgical resection because of an adverse event or tumour progression during preoperative chemotherapy. Serial tracking of gene mutations can be a useful biomarker for preoperative chemotherapy because it can monitor treatment response, tumour burden, and resistant clones.19,20
In this study, the number of mutations decreased after NAC. Most founder mutations such as KRAS and TP53 remained throughout chemotherapy, but other passenger mutations were not found after exposure to cytotoxic and molecular-targeted agents. These results suggest that the certain subclones are diminished by a massive response to NAC. In one case, mutations found in the primary tumour at diagnosis were not detected after NAC. In this case, massive fibrosis and necrosis were found in the pathological analysis, suggesting the strong selection of subclones or small content of cancer cells from neoadjuvant treatment may cause changes in the mutational phenotype. Our data indicate that serial tracking of gene mutations could detect dynamic genomic evolution during chemotherapy maximise the anti-tumour effects of molecular profile-based precision medicine.
The source of ctDNA appears to be mainly cell death, thus ctDNA from tumours in blood has been reported as a biomarker for early detection, tumour burden, and response to treatment.21–27 We found founder mutations consistent with the primary tumour in plasma samples after chemotherapy, indicating that ctDNA could be used instead of tissue samples as a ‘liquid biopsy’ to detect tumour-specific mutations for better patient selection and treatment individualisation throughout multiple lines of therapy.28 Using ctDNA has advantages for the serial monitoring of genetic and epigenetic alterations because samples can be obtained noninvasively and repeatedly. The other issue to be highlighted regarding ctDNA is that it could be beneficial to evaluate molecular profiles beyond intra- and inter-tumour (primary and metastatic, etc.) molecular heterogeneity.12,29 We analysed a post-NAC plasma, but the serial mutational tracking of pre- and post-NAC plasma would be warranted for further studies.
The main limitation of this study is the small patient cohort size. Although this is a preliminary study, to our knowledge this is the first published data of serial mutational tracking during NAC and surgery for locally advanced CRC. Our serial mutational analysis illustrates that dynamic mutational changes occur during subclonal NAC for locally advanced CRC. These findings show the utility of mutational tracking for drug selection and treatment decisions.
Electronic supplementary material
Acknowledgements
We thank K. Oda, M. Kasagi, T. Kawano, K. Imamura, and Y. Ishikawa for their technical assistance.
Author contributions
Study conception and design: K.S., S.S., K.M., Y.M. Acquisition of data: K.S., S.S., S.K., T.I., H.E., T.M., Y.S. Analysis and interpretation of data: K.S., S.S., A.N., H.H., Y.S. Drafting of manuscript: K.S., S.S., K.M. Critical revision: K.S., S.S., K.M., M.M., Y.T.
Competing interests
The authors declare no competing interests.
Funding
This work was supported in part by the following grants and foundations: Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Science Research (grant numbers 15571128, 25430111, 25461953, 25861199, 25861200, 26861085 and 15H05707), Grant-in-Aid for Scientific Research on Innovative Areas (15H0912), Priority Issue on Post-K computer (hp170227, hp170227 and hp160219), and The Shinnihon Foundation of Advanced Medical Treatment Research.
Ethics approval and consent to participate
Samples were obtained after written informed consent from the patients. The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethical Review Board of Kyushu University.
Electronic supplementary material
Supplementary information is available for this paper at 10.1038/s41416-018-0208-5.
References
- 1.Douillard JY, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J. Clin. Oncol. 2010;28:4697–4705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg BA, Hartley ML, Salem ME. Precision medicine in metastatic colorectal cancer: relevant carcinogenic pathways and targets-PART 1: biologic therapies targeting the epidermal growth factor receptor and vascular endothelial growth factor. Oncology. 2017;31:539–548. [PubMed] [Google Scholar]
- 3.Adam R, et al. Patients with initially unresectable colorectal liver metastases: is there a possibility of cure? J. Clin. Oncol. 2009;27:1829–1835. doi: 10.1200/JCO.2008.19.9273. [DOI] [PubMed] [Google Scholar]
- 4.Jakobsen A, et al. Neoadjuvant chemotherapy in locally advanced colon cancer. A phase II trial. Acta Oncol. 2015;54:1747–1753. doi: 10.3109/0284186X.2015.1037007. [DOI] [PubMed] [Google Scholar]
- 5.Foxtrot Collaborative G. Feasibility of preoperative chemotherapy for locally advanced, operable colon cancer: the pilot phase of a randomised controlled trial. Lancet Oncol. 2012;13:1152–1160. doi: 10.1016/S1470-2045(12)70348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149:1177–1190.e3. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misale S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz LA, Jr., et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douillard JY, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 11.Sartore-Bianchi A, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738–746. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 12.Uchi R, et al. Integrated multiregional analysis proposing a new model of colorectal cancer evolution. PLoS Genet. 2016;12:e1005778. doi: 10.1371/journal.pgen.1005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietrantonio F, et al. Heterogeneity of acquired resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer. Clin. Cancer Res. 2017;23:2414–2422. doi: 10.1158/1078-0432.CCR-16-1863. [DOI] [PubMed] [Google Scholar]
- 14.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 15.Ueda M, et al. Somatic mutations in plasma cell-free DNA are diagnostic markers for esophageal squamous cell carcinoma recurrence. Oncotarget. 2016;7:62280–62291. doi: 10.18632/oncotarget.11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiraishi Y, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41:e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Murugaesu N, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015;5:821–831. doi: 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barault, L. et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 2017. 10.1136/gutjnl-2016-313372. [DOI] [PMC free article] [PubMed]
- 21.Diehl F, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl Acad. Sci. USA. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tie J, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015;26:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cree IA, et al. The evidence base for circulating tumour DNA blood-based biomarkers for the early detection of cancer: a systematic mapping review. BMC Cancer. 2017;17:697. doi: 10.1186/s12885-017-3693-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spindler KG, et al. Cell-free DNA in metastatic colorectal cancer: a systematic review and meta-analysis. Oncologist. 2017;22:1049–1055. doi: 10.1634/theoncologist.2016-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamauchi M, et al. Serial profiling of circulating tumor DNA for optimization of anti-VEGF chemotherapy in metastatic colorectal cancer patients. Int. J. Cancer. 2017;142:1418–1426. doi: 10.1002/ijc.31154. [DOI] [PubMed] [Google Scholar]
- 26.Bhangu JS, Taghizadeh H, Braunschmid T, Bachleitner-Hofmann T, Mannhalter C. Circulating cell-free DNA in plasma of colorectal cancer patients - a potential biomarker for tumor burden. Surg. Oncol. 2017;26:395–401. doi: 10.1016/j.suronc.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Jia S, Zhang R, Li Z, Li J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget. 2017;8:55632–55645. doi: 10.18632/oncotarget.17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz LA. The promise of liquid biopsy in colorectal cancer. Clin. Adv. Hematol. Oncol. 2014;12:688–689. [PubMed] [Google Scholar]
- 29.Siravegna G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015;21:827. doi: 10.1038/nm0715-827b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.