Abstract
High magnesium (Mg2+) in some extreme serpentine soils or semi-arid regions is an important factor affecting crop growth and development. Specific loci that form the genetic framework underlying high Mg2+ homeostasis, however, are not well understood. By using GWA mapping on 388 accessions of Arabidopsis thaliana selected from a worldwide collection and genotyped at approximately 250,00 SNPs, we successfully identified 109 and 74 putative genetic regions associated in nutrient traits under normal (1,000 µM) and high Mg2+ (10,000 µM), respectively. Above 90% SNPs associated with nutrients including Mg2+ and only two SNPs shared between normal and high Mg2+. A single strong peak of SNPs associated with Ca concentration corresponding to candidate gene At1g60420 ARABIDOPSIS NUCLEOREDOXIN (AtNRX1) under high Mg2+ was further determined. Compared with wildtype, mutants of Atnrx1-1 and Atnrx1-2 supplied with high Mg2+ had higher Ca concentrations in the plant, and higher cytosolic Ca2+ concentrations during root elongation, as well as higher fresh weight and lateral-root number. This suggests that AtNRX1 was a critical gene negatively regulating Ca uptake under high Mg2+ conditions. The discovery could help to breed/select crops that can adapt to high-Mg2+ soils such as serpentine soils (high ratio of Mg2+: Ca2+) or Mars soil with high levels of magnesium sulfate.
Introduction
Magnesium (Mg) is the 8th most abundant mineral element on earth and the fourth abundant mineral element in plants following nitrogen (N), potassium (K) and calcium (Ca)1. Magnesium in soils originates from source rock material containing various types of silicates and carbonates. In extreme serpentine (high magnesium: calcium) soils2 and in semi-arid regions, where water stress conditions3 can lead to Mg accumulation. In addition, Mars regolith (unconsolidated surface material) is a potential medium for plant growth in bioregenerative life support systems. However, analyses by the Mars Exploration Rover Landers at Meridiani Planum and Gusev Crater have also suggested there are particularly high levels of magnesium sulfate minerals in outcrops and soils4–6.
Although plants rely on a sufficient supply of Mg2+ for normal growth and development, excessive Mg2+ accumulation often causes toxicity to plant cells. Plants grown in high-Mg2+ soils like serpentine or Mars soils may alleviate Mg toxicity by limiting internal Mg accumulation, Mg excretion from leaves and/or increasing plant tolerance to high Mg2+ concentrations via altering leaf size, sclerophylls, stature and root systems7,8. Previous reciprocal transplant experiments between serpentine populations (or species) and their spatially adjacent populations (or sister species) growing on serpentine soils demonstrated unequivocally that adaptation to serpentine soils had a genetic basis9–12. Several quantitative trait loci that control contrasting adaptive traits in serpentine-tolerant Microseris douglasii and serpentine-intolerant M. bigelovii have also been identified13. A high degree of polymorphism has been reported among plant species in terms of growth tolerance to serpentine soils14–16.
This present study employed genome-wide association studies (GWAS), an approach that has been applied successfully to identify many different traits ranging from genetically simple traits to more complex features17, to identify candidate genes likely to be involved in adaptation to high Mg2+, based on 388 accessions of Arabidopsis thaliana selected from a worldwide collection. Arabidopsis thaliana has a broad geographical distribution and consequently is subject to varying nutritional environments, which makes it a useful model species for studying nutrient homeostasis18,19 in a wide variety of environments20. Importantly, more than 1300 distinct accessions have been genotyped21 and to date more than 1000 inbred lines have been ‘fully sequenced’. Meanwhile, automated phenotyping platforms have been developed22–24, allowing for precise and high-throughput measurements of plant growth. These developments enhance our ability to map causal genetic polymorphisms through GWAS. Moreover, high Mg2+ decreased the growth of Arabidopsis including biomass production, root hair development and the absorption of other nutrients25–28. To further identify the likely candidate genes that are involved in adaptation to high-Mg2+ soils, we used a chemically-defined nutrient solution that contained 10,000 μM MgSO4 of maximum authentic high Mg2+ soils to perform a viability screen on a large collection of wild-type accessions of Arabidopsis thaliana. This study aimed to (1) investigate natural variations in growth response to high Mg2+, and (2) identify novel genes tolerant to Mg2+ excess.
Materials and Methods
Plant material and growth conditions
Seeds of 388 accessions with accessions CS76636, CS76427, CS78885 and CS22660 and two T-DNA insertion lines SALK_100357C and SALK_113401C were derived from the Arabidopsis Biological Resources Center stock center (http://nasc.nott.ac.uk). The accessions were selected without any a priori consideration of their natural habitat, including geographical distribution or precipitation levels, and were collected from the wild and available in the stock center or soon-to-be-released collections. Most of these accessions were originated from Europe including Sweden, Germany, France, Czech Republic and United Kingdom, and from North America and Middle Asia, representing the sampling strategy of the 1,307 worldwide accessions21. This was consistent with the distribution of the 1,307 Arabidopsis lines collected in RegPanel.
Arabidopsis cultivation
The 388 accessions of Arabidopsis thaliana were grown under 2 Mg2+ treatments, which were achieved by altering the concentration of MgSO4 in the basal medium. To minimise maternal effects prior to phenotyping, natural accessions were grown for one generation during 2015 under controlled greenhouse conditions at Zhejiang University (N30°18′25, E120°04′54). For surface sterilization, seeds of various accessions were placed for 1 h in open 200-μL PCR tubes in a sealed box producing chlorine gas generated from 13 mL of 10% sodium hypochlorite and 350 μL of 37% hydrochloric acid. Sterile seeds were then put on the surface of 30 mL agar media, containing 1.2% (w/v) agar and 0.6% (w/v) sucrose (A-1296; Sigma-Aldrich; http://www.sigmaaldrich.com) in 10 × 10-cm2 plates with grid schematic engraved below the plate. Plates were positioned in racks and oriented in a vertical position, and were kept at 4 °C for 48 h in the dark for seed stratification. Thereafter, the racks containing the plates were transferred to a growth chamber under a 10-h light/14-h dark photoperiod at constant temperature of 22 °C, 60% relative humidity and light intensity of 120 µmol photons m−2 s−1. Twelve seedlings were grown in each plate and each treatment received at least four independent replicates. The racks were removed to the image acquisition room once per day and then immediately returned to the growth chamber. Throughout the experiments, the plate position within the box and box position in the growth chamber were re-randomized daily. For assays on agar plates, studies were performed on 8-d-old plants; that is, at an early stage of their stability and homogeneity growth phase. Moreover, many previous publications adopted 6- to 8-d-old Arabidopsis seedlings under stresses for determining Arabidopsis morphology, physiology, and development as well as responses to nutrient stresses29–31.
Mg2+ Treatments
The basal medium (Normal Mg2+, N), which was used as control, contained (µM) 1500 KNO3, 500 NaH2PO4, 1000 CaCl2, 250 (NH4)2SO4, 1000 MgSO4, 25 Fe-EDTA, 10 H3BO3, 0.5 MnSO4, 0.5 ZnSO4, 0.1 CuSO4 and 0.1 (NH4)6Mo7O24. The normal and high Mg2+ treatments contained 1,000, and 10,000 µM MgSO4, respectively, which was in accordance with Mg2+ concentrations in soil solutions27. 10,000 μM Mg2+was selected in the present study for the quantitative analysis of the natural variation of Arabidopsis in response to high Mg2+ stress because this concentration had an adverse effect on growth without toxic symptoms of most natural accessions27,32. Besides, the concentrations of Mg2+ in the control were 1000 µM, that has been adopted for Arabidopsis growth by many plant biologists33–35. Though the concentration of SO42− increased 10 times comparing with that in the normal medium, SO42− had hardly affected morphogenesis of Arabidopsis27,36. The pH of the growing media was adjusted to pH 5.8 with MES (N-morpholino) ethane-sulphonic acid)-KOH buffer before autoclaving.
Phenotype analysis
Each line had four biological replicates for each treatment. Seeds that had not germinated at day 6 were discarded from further analysis, resulting in approximately 3–4 seedlings analyzed per genotype per treatment. The 21 traits under each treatment were classified into 2 categories: 7 morphological traits and 14 nutrient traits. Seedlings were photographed with a high-resolution digital camera (Sony RX100, Japan) daily for determination of root and shoot growth. Root or shoot biomass, and the number of days from seeding to emergence (>50% seedlings having the first radicle or cotyledon), were recorded. Meanwhile, photographs after 8 days of treatment were analyzed and quantified for phenotype using the public domain image analysis program Image J version 1.43 (http://rsb.info.nih.gov/ij/)27. The length of primary roots, rosette diameter and epicotyl were determined across the median seedling using Image J. Lateral-root number was determined by counting the number of true roots (>1 mm long lateral root primordia) per primary root. For each condition, an independent sample accession consisted of four plants at the same growth stage, and the time of sampling was the end of the light period of day 8.
Analyses of mineral homeostasis
After 8-d growth at various Mg2+ concentrations, plants were harvested. All fully-expanded seedlings without lesions were collected, and weighed. The results were the average value across all available replicates. Seedlings were washed thoroughly with ultrapure water and dried in an oven at 75 °C for 24 h. The dried root and shoot samples were then wet-digested in the concentrated HNO3/H2O2 at 90, 120 and 140 °C for 2 h, respectively, until there was no brown fume, and then further digested at 180 °C until the digest became clear. Concentrations of potassium (K), calcium (Ca), Magnesium (Mg), sulfur (S), iron (Fe), manganese (Mn) and sodium (Na) in the digests were analyzed by ICP-MS (Inductively coupled plasma mass spectrometer, Agilent 7500a, USA), the contents per plant were calculated. Mineral analysis was done in the whole-plant.
Fluo 4-AM loading and confocal imaging of cytosolic Ca2+ concentration in root
The Ca2+-sensitive fluorescent dye 1-[2-amino-5-(2,7-difluoro-6-hydroxy-3-oxo-9-xanthenyl)phenoxy] -2-(2-amino-5-methylphenoxy)ethane-N,N,N′,N′-tetraacetic acid, pentaacetoxymethyl ester (Fluo-4/AM) ester was used to estimate cytosolic Ca2+ concentration in the roots. The Fluo-4/AM ester was loaded into the root at 37 °C in the dark at a final concentration of 5 μM Fluo 4-AM dye and Pluronic F-127 (0.05%) in the Hank’s Balanced Salt Solution (HBSS) solution. After 1-h incubation, the roots were washed with the standard HBSS medium three times and left at 25 °C for 30 min. The samples were mounted and photographed with a Zeiss LSM 780 (Zeiss Co., Germany). Fluorescence was detected using 488 nm excitation and 505–545 nm band pass filter. Roots including root hairs were placed on the stage of a Zeiss Axiovert inverted microscope attached to a LSM780 laser scanning confocal microscope, and imaged using a Zeiss 20×, N.A., wet objective. Each frame represents a 7-s scan of the laser.
Genome-Wide Association (GWA) mapping, SNP selection, and statistics
The GWA mapping was performed on the values of epicotyl length, rosette width, primary root length, and lateral root number of the median seedling of each accession. For traits of and nutrients, means values of 4 biological replicates were used. We linked 21 traits under normal or high Mg2+ (Supporting Information Table S1) to published genomic data on accessions from a 250 K SNP chip. The GWA analysis was performed in the GWAPP web (http://gwas.gmi.oeaw.ac.at/) interface using the mixed model algorithm method37. A kinship matrix was generated using the identity in state of SNPs between each pair of accessions38. The P-value above bias due to population stratification was evaluated with Q–Q plots. Candidate loci were selected based on the minor allele frequency, LOD score and trait-associated SNPs and their position in the genome to pinpoint candidate genes within 5 kb up- and down-stream of the identified SNP (Supporting Information Table S2). The putative candidate genes corresponding to each SNP are listed in Supporting Information Table S3. For each of the candidate genes, the annotations were retrieved from TAIR10 (Arabidopsis.org).
Extraction of total RNA and quantitative PCR
Samples were collected from 8-day-old Arabidopsis after grown at normal and high Mg2+ treatments. For each condition, four samples were taken on independent seedling materials from eight different plants at the same growth stage and the time of sampling was the end of the light period of day 8. Total DNA was extracted by Plant DNA Isolation Reagent (Takara) from about 80 mg of fresh seedling tissues of four independent biological replicates. Two homozygous T-DNA insertion lines (SALK_100357C, Atnrx1-1) and (SALK_113401C, Atnrx1-2) was conducted by PCR with gene-specific primers SALK_100357C LP: 5′-AACCCAACCATCTTTGGACTC-3′; SALK_100357C RP: 5′-TCAAGACTTCAAGACCAAGCC-3′ and T-DNA border primer LBb1.3:5′-ATTTTGCCGATTTCGGAAC-3′; SALK_113401C, LP: 5′-ATCCACTTTTGTCGTTGAACG-3′; SALK_113401C RP: 5′-CGATCGCAACTTCTTCTGATC-3′ and T-DNA border primer LBb1.3:5′-ATTTTGCCGATTTCGGAAC-3′; Total RNA was extracted by RNAisoPlus (Takara, Otsu, Shiga, Japan) from about 70 mg of fresh seedling tissues of four independent biological replicates. All RNA samples were checked for DNA contamination before cDNA synthesis. cDNA was synthesized, and possible residual genomic DNA contamination was verified as described in our previous study39. The mRNA levels of AT1G60420 in two lines of SALK_100357C and SALK_113401C were detected by the SYBR Green RT-PCR kit (Takara) with following pairs of gene-specific primers: AT1G60420, fw: 5′- TTCTCGGCCCTGATGGAAAAACC-3′, rv: 5′- GCTTCATTCCGCTCCTTTATCTGCT-3′ A pair of housekeeping gene of UBQ10 (fw: 5′-GGTTCGTACCTTTGTCCAAGCA-3′, rv: 5′-CCTTCGTTAAACCAAGCTCAGTATC-3′) was used for a control PCR. Melting curve analysis and gel electrophoresis of the PCR products were used to confirm the absence of non-specific amplification products. Relative expression levels were calculated by subtracting the threshold cycle (Ct) values for UBQ10 from those of the target gene (to give ∆Ct) and then calculating 2−∆Ct, where Ct refers to the cycle number at which the fluorescence rises above the set threshold qPCR. UBQ10 was chosen as the housekeeping reference40.
Phylogenetic and Statistical Analyses
DNA sequences were obtained from the 1001 Genomes project (www.1001genomes.org). All statistical analyses were conducted with DPS software (Stirling Technologies Inc., China). Means were compared by the t test or the Fisher’s least significant difference test at P = 0.05.
Results
Distribution and correlation of different 21 traits under normal and high Mg2+
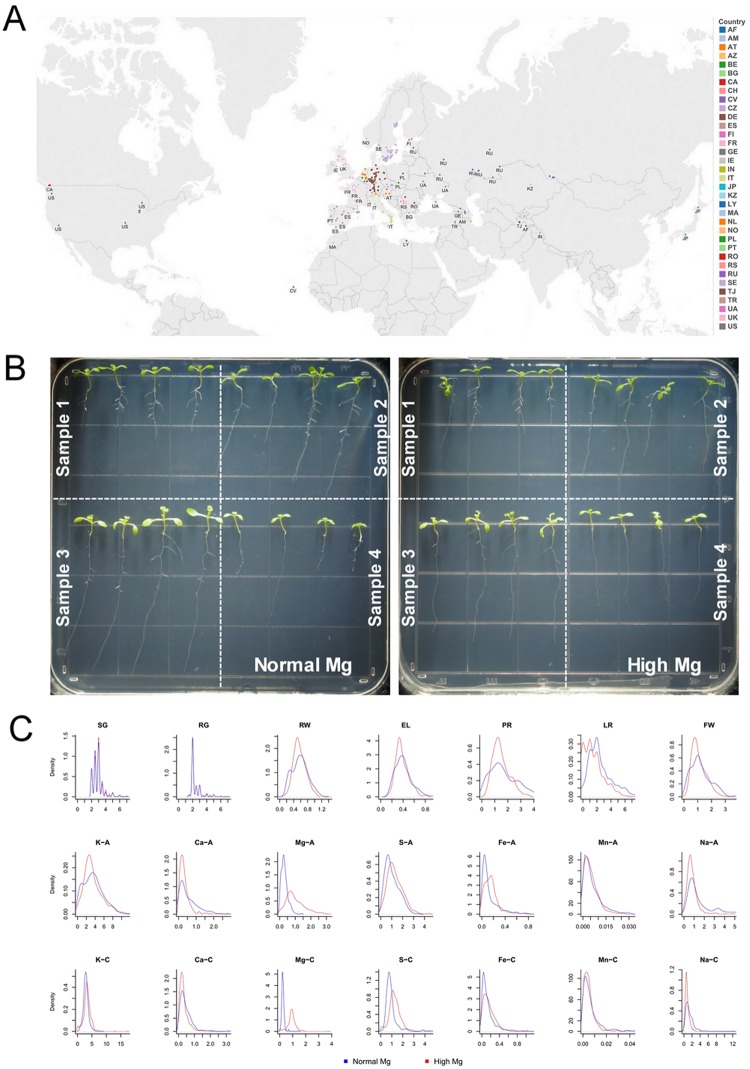
Distribution of 388 Arabidopsis accessions collected from the wild and available in the stock center or soon-to-be-released collections is shown in Fig. 1A. In general, high Mg2+ supply (10,000 μM) had an adverse effect on the growth of most natural accessions. This high Mg2+ concentration induced a large degree of phenotypic variation and thus was selected for the quantitative analysis of the natural variation of Arabidopsis in response to high Mg2+ stress (Fig. 1B). A subset of 388 accessions (250 K SNPs) was characterized phenotypically for multiple traits, including root emergence (RG), shoot emergence (SG), primary root length (PR), lateral root number (LR), epicotyl length (EL), rosette width (WD), fresh weight (FW) of the plants and fourteen nutrient traits, including absorption per plant and concentration of K, Ca, Mg, S, Fe, Mn and Na under normal and high Mg2+ supplies (see Table 1 and ‘Phenotype analysis’ in Materials and Methods).
Figure 1.
Worldwide diversity of 388 A. thaliana natural accessions and cultivation used in this study according to their reported country origins and frequency distributions of non-normalized data of all traits. (A) The data were exported for visualization and geographical mapping in Tableau Desktop 8.1 (http://www.tableausoftware.com/). (B) Cultivation for 388 Arabidopsis inbred lines at Zhejiang University, Hangzhou, China. Each line was treated with low (1 µM), normal (1,000 µM), and high Mg2+ (10,000 µM). Each line had four biological replicates, and the median value was used in the analysis. (C) Frequency distributions of non-normalized data of all traits in the 388 population. Blue and Red line indicates normal and high Mg2+, respectively.
Table 1.
Summary for 21 Arabidopsis traits.
| Traits | Full name of the trait | Unit | Trait description |
|---|---|---|---|
| RG | Days to root germination | day | The number of days from seeding until emergence, with more than half of the seedlings having a first radicle. |
| PR | Primary root length | cm | After 8 days of growth under the two treatment conditions, the plants were flattened directly on agar and imaged using a camera. The primary root lengths and lateral root number were determined across the median seedling using ImageJ. |
| LR | Lateral root number | ||
| SG | Days to seed germination | day | The number of days from seeding until emergence, with more than half of the seedlings having the first cotyledon. |
| EL | Epicotyl length | cm | After 8 days of growth under the two treatment conditions, the plants were flattened directly on agar and imaged using a camera. The epicotyl length and rosette width were determined across the median seedling using ImageJ. |
| RW | Rosette width | cm | |
| FW | Fresh weight of plants | mg | All fully-expanded seedlings without lesions were collected from each of 4 plants for each accession and weighed to obtain the fresh weight. The results are the average value across all available replicates. |
| K-A | The amount of potassium absorbed per plant | mg K/plant | Elemental analysis was performed with an ICP-MS (Agilent 7500a, USA). All samples were normalized to calculated weights as previously described. The ‘Nutrient absorption(A)’ was expressed as total amount of nutrient per plant; ‘Nutrient concentration (C)’ was calculated by total amount of nutrient per plant /plant fresh weight (FW). The results are the average value across all available replicates. |
| K-C | Potassium concentration per fresh weight | mg K/g (FW) | |
| Ca-A | The amount of calcium absorbed per plant | mg Ca/plant | |
| Ca-C | Calcium concentration per fresh weight | mg Ca/g (FW) | |
| Mg-A | The amount of magnesium absorbed per plant | mg Mg/plant | |
| Mg-C | Magnesium concentration per fresh weight | mg Mg/g (FW) | |
| S-A | The amount of sulfur absorbed per plant | mg S/plant | |
| S-C | Sulfur concentration per fresh weight | mg S/g (FW) | |
| Fe-A | The amount of iron absorbed per plant | mg Fe/plant | |
| Fe-C | Iron concentration per fresh weight | mg Fe/g (FW) | |
| Mn-A | The amount of manganese absorbed per plant | mg Mn/plant | |
| Mn-C | Manganese concentration per fresh weight | mg Mn/g (FW) | |
| Na-A | The amount of sodium absorbed per plant | mg Na/plant | |
| Na-C | Sodium concentration per fresh weight | mg Na/g (FW) |
Before performing the quantitative genetic analyses, we assessed the distributions of all traits based on all raw data (Supporting Information Table S1). The phenotypic analysis of the 388 accessions showed that the frequency distribution of all traits was close to normality (Fig. 1C; Table 1). Further detailed analysis showed that distribution of Mg-A-H (Mg2+ absorption under high Mg2+) and Mg-C-H (Mg2+ concentration under high Mg2+) showed a great right-skewed probability density with the right-skewed being greater for Mg-C-H than that for Mg-A-H, suggesting that high Mg2+ supply increased Mg concentration of most Arabidopsis accessions at least in the early phase of development. However, only slight skewness and kurtosis was observed for S absorption and concentration (Fig. 1C). Distribution of RW, EL, PR, FW, Ca-A (Ca absorption), Ca-C (Ca concentration), Na-A (Na absorption) and Na-C (Na concentration) presented a standard normal distribution but with a sharp peak upon high Mg2+ as compared with normal Mg2+ (Fig. 1C), indicating that the high Mg2+ treatment resulted in a more focused response in those traits. Notably, the right side of the curve of absorption and concentration of Ca, Fe and Mn under both normal and high Mg2+ fitted a normal distribution, but the left-hand side deviated significantly from the normal distribution, displaying a decapitation tail (Fig. 1C).
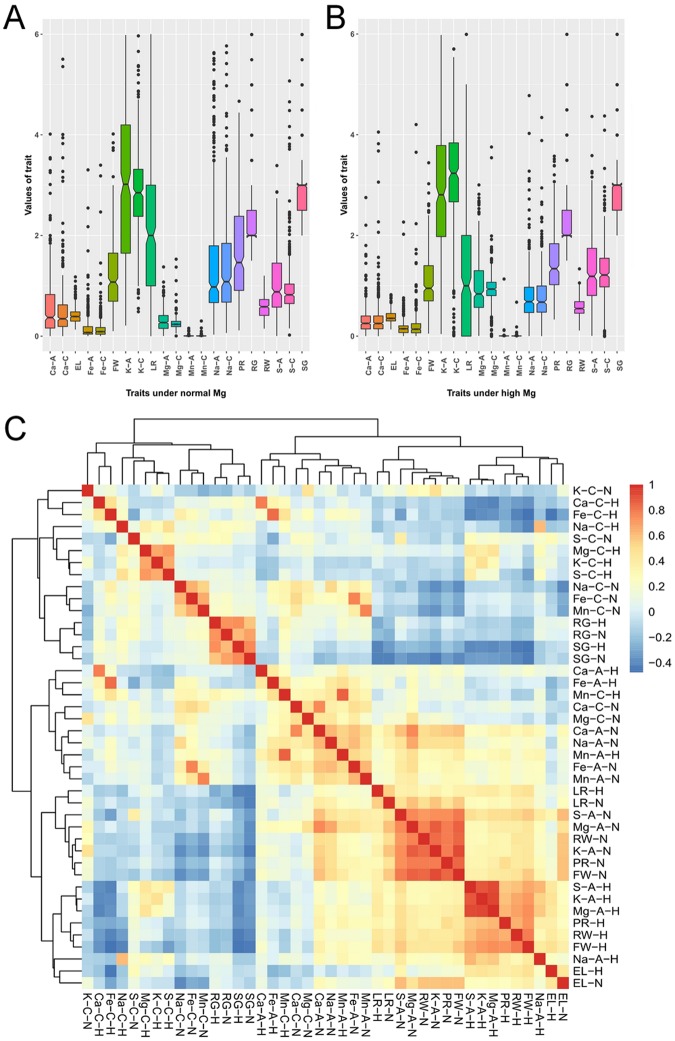
Data of the traits are close to normal distribution reflecting the homogeneity of the investigated population. However, what was the response of each individual domain to high Mg2+? According to the date (Supporting Information Table S1), the effects of high Mg2+ on all phenotypes can be divided into two types. First, high Mg2+ had a similar promoting effect on LR, Mg-A, Mg-C, S-A and S-C of most accessions. Second, high Mg2+ enhanced traits of PR, EL, RW, FW and absorption and concentrations of K, Ca, Fe and Mn in the accessions with low value while reduced them in the accessions with high value as compared with normal Mg2+ supply. Furthermore, the initial comparison of 21 plant traits between normal and high Mg2+ was performed using statistical analysis by a paired two-tailed Student t test, with a P-value < 0.01 (Table 2). Accordingly, the above first type traits were consistently changed by high Mg2+ as compared with normal Mg2+. Apart from this, the above second traits were inconsistently altered by high Mg2+ (Table 2, Fig. 2A,B).
Table 2.
Minimum (Min), maximum (Max), mean and standard deviation (SD) values of individual traits for 21 phenotypes under high (10,000 µM) and normal Mg2+ (1,000 µM).The correlations (r2) between values of individual traits of the plants grown at high and normal Mg2+ are also shown.
| Normal Mg2+ | Min | Max | Mean | SD | p-value (normality test)a | ||
|---|---|---|---|---|---|---|---|
| RG | 1.5000 | 7.0000 | 2.4000 | 0.8200 | 1.07E-28 | ||
| PR | 0.1155 | 4.6749 | 1.6712 | 1.0430 | 4.61E-10 | ||
| LR | 0 | 9.0000 | 2.4845 | 1.7004 | 2.00E-16 | ||
| SG | 2.0000 | 7.0000 | 2.8814 | 0.7949 | 6.07E-22 | ||
| EL | 0.1305 | 1.1846 | 0.4088 | 0.1590 | 1.73E-12 | ||
| RW | 0.1493 | 1.2021 | 0.5823 | 0.2241 | 7.13E-05 | ||
| FW | 0.1000 | 4.025 | 1.2242 | 0.7383 | 3.26E-11 | ||
| K-A | 0.2105 | 14.185 | 3.6072 | 2.3203 | 8.73E-11 | ||
| K-C | 0.4935 | 8.5100 | 2.9159 | 0.9256 | 2.17E-11 | ||
| Ca-A | 0.0010 | 4.0225 | 0.5932 | 0.6245 | 2.49E-22 | ||
| Ca-C | 0.0009 | 7.4220 | 0.5646 | 0.8206 | 1.01E-31 | ||
| Mg-A | 0.0033 | 1.3830 | 0.3042 | 0.2049 | 6.52E-13 | ||
| Mg-C | 0.0118 | 1.5367 | 0.2547 | 0.1369 | 1.50E-27 | ||
| S-A | 0.0030 | 3.3967 | 1.0466 | 0.6320 | 1.62E-10 | ||
| S-C | 0.0300 | 6.7483 | 0.9736 | 0.6864 | 1.85E-28 | ||
| Fe-A | 0.0008 | 3.3113 | 0.1664 | 0.2977 | 1.45E-32 | ||
| Fe-C | 0.0007 | 8.7300 | 0.1826 | 0.5313 | 4.24E-37 | ||
| Mn-A | 0.0000 | 0.1647 | 0.0075 | 0.0134 | 2.73E-33 | ||
| Mn-C | 0.0000 | 0.3088 | 0.0084 | 0.0201 | 7.84E-36 | ||
| Na-A | 0.0300 | 5.6400 | 1.4760 | 1.2287 | 2.86E-21 | ||
| Na-C | 0.0643 | 21.200 | 1.4744 | 1.6282 | 3.50E-31 | ||
| High Mg 2+ | Min | Max | Mean | SD | p -value (normality test) a | r 2 (H vs N) | p -value (t-test) b |
| RG | 1.5000 | 7.0000 | 2.4526 | 0.8379 | 2.54E-27 | 0.53 | 0.3607 |
| PR | 0.3301 | 3.5816 | 1.4777 | 0.6374 | 5.06E-10 | 0.3115 | 0.0019 |
| LR | 0.0000 | 7.0000 | 1.4845 | 1.3780 | 2.51E-17 | 0.4146 | 1.86E-18 |
| SG | 2.0000 | 7.0000 | 2.9021 | 0.7731 | 2.05E-20 | 0.5337 | 0.7143 |
| EL | 0.1327 | 0.8300 | 0.3825 | 0.1190 | 6.46E-10 | 0.2886 | 0.0092 |
| RW | 0.1111 | 1.3434 | 0.5714 | 0.1754 | 0.000113 | 0.2034 | 0.4492 |
| FW | 0.0500 | 3.4500 | 1.0945 | 0.5354 | 4.64E-12 | 0.3430 | 0.0052 |
| K-A | 0.0413 | 12.0925 | 3.4778 | 2.0909 | 2.89E-12 | 0.1070 | 0.4147 |
| K-C | 0.0148 | 17.2500 | 3.3030 | 1.4688 | 1.05E-20 | 0.0102 | 1.3130 |
| Ca-A | 0.0003 | 22.1050 | 0.3740 | 1.1442 | 1.54E-38 | 0.0057 | 0.0010 |
| Ca-C | 0.0002 | 36.8417 | 0.4576 | 1.9330 | 5.79E-39 | 0.0023 | 0.3163 |
| Mg-A | 0.0048 | 3.0103 | 0.9847 | 0.5822 | 2.78E-11 | 0.1679 | 2.13E-73 |
| Mg-C | 0.0021 | 3.7629 | 0.9260 | 0.3694 | 2.96E-19 | 0.0204 | 3.08E-129 |
| S-A | 0.0063 | 4.3700 | 1.3066 | 0.6900 | 6.40E-08 | 0.1911 | 6.01E-08 |
| S-C | 0.0015 | 4.3800 | 1.2760 | 0.5470 | 4.75E-12 | 0.0307 | 2.34E-11 |
| Fe-A | 0.0010 | 7.0500 | 0.2013 | 0.4963 | 2.72E-37 | 0.1023 | 0.2353 |
| Fe-C | 0.0007 | 4.2090 | 0.2164 | 0.3608 | 4.00E-33 | 0.0222 | 0.3012 |
| Mn-A | 0.0000 | 1.1415 | 0.0085 | 0.0585 | 1.17E-39 | 0.0224 | 0.7351 |
| Mn-C | 0.0001 | 0.6815 | 0.0077 | 0.0363 | 4.85E-39 | 0.0101 | 0.7021 |
| Na-A | 0.0025 | 4.7840 | 0.7881 | 0.5499 | 1.24E-21 | 0.0520 | 6.06E-22 |
| Na-C | 0.0011 | 12.2300 | 0.8342 | 0.7908 | 8.93E-31 | 0.0119 | 9.15E-12 |
aSharipo test was used for normality test; bHigh and normal date-sample student’s t-test at p value < 0.01.
Figure 2.
Natural variation in traits of Arabidopsis accessions grown at normal (1,000 µM) (A) and high Mg2+ (10,000 µM). (B) The average value per accession was calculated, and values of individual traits were plotted using the ‘ggplot2’ package in R. Accessions representing outliers for a trait are indicated by closed circles. (C) Heat map represents cluster of the pearson correlation coefficient between each pair of 42 phenotypes, the pearson’s correlation coefficients were normalized (z-score) and represented with blue-red color scheme.
Pearson’s correlation coefficients (r2) were used to examine the natural variation within the population of quantified traits between normal and high Mg2+ conditions (Table 2). The RG and SG showed positive correlations between normal and high Mg2+ treatments (r2 = 0.53) (Table 2). A weak correlation was observed for nutrient traits, especially Ca, Fe and Mg concentrations between normal and high Mg2+ treatments (r2 < 0.2), suggesting different physiological functions of nutrients under different Mg2+ supplies.
Cluster of Pearson’s correlation coefficient between phenotypes (Heat map) showed a negative correlation between Ca-C-H, Fe-C-H and Na-C-H and Mg-C-H (Fig. 2C), suggesting that Mg2+ would compete for uptake with Ca2+, Fe2+ and Na2+ in most Arabidopsis accessions when supplied with high Mg2+. Increased Mg2+ absorption would decrease the uptake of these ions and hence plant growth. However, Mg-A, Mg-C, S-A, S-C, K-A and K-C were positively correlated only under high Mg2+ (Fig. 2C), indicating a common mechanism or genetic pathway between Mg2+, S and K under high Mg2+ (Fig. 2C). In contrast, there were highly significant positive correlations between PR, RW, LR, FW as well as S-A, Mg-A and K-A of the phenotypes under normal but not high Mg2+ supply (Fig. 2C).
GWAS analysis for nutrient metabolism in response to high Mg2+
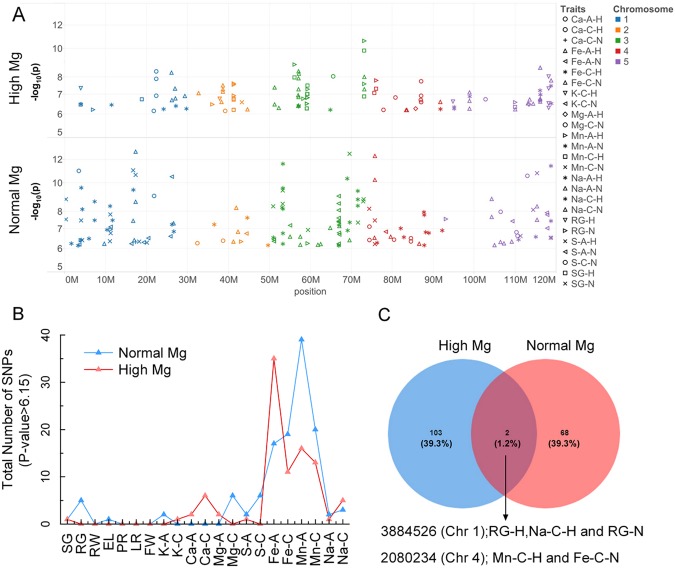
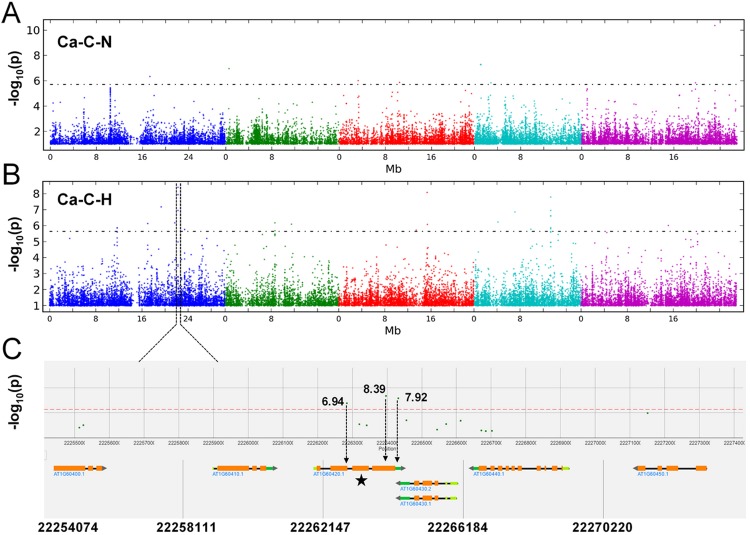
The phenotypic data collected from normal and high-Mg2+ conditions (Supporting Information Table S1) were used as an input for genome-wide association study (GWAS). All SNPs were converted to homozygote or heterozygote according to the genotype, and tri-allelic or tetra-allelic SNPs were removed. Candidate loci were selected based on the minor allele frequency, LOD score and number of significantly associated SNPs within one locus. Using −log10(p) > 6.15 as the nominal genome-wide significance threshold, the GWASs had 109 hits in total, 32, 10, 25, 18 and 24 located on chromosomes (Chr) 1, 2, 3, 4, and 5, respectively, under normal Mg2+, and 79 hits in total, 14, 13, 18, 10 and 19 located on chromosomes 1, 2, 3, 4, and 5, respectively, under high Mg2+ (Fig. 3A,B). There were 2 shared hits between normal and high Mg2+ supplies (Fig. 3C).
Figure 3.
GWASs for 21 traits under normal (1,000 µM) and high Mg2+ (10,000 µM) conditions. The x-axis is the chromosomal coordinates for Arabidopsis, the y-axis represents p-value. The GWASs hits observed at high Mg2+ supply were presented on the top; the hits observed at normal Mg2+ were represented at the bottom panel (A). GWAS hits and chromosome were represented by different symbols and coloured, respectively. GWAS hits were shared between normal and high Mg2+ were ordered by their longitudinal (x-axis) position. A SNP GWAS hit was defined as the one had −log10(p) > 6.15. (B) Total number of traits that were scared for SNPs with log10(p) > 6.15. The x-axis represents scared traits for Arabidopsis, the y-axis is the p-value. Blue and Red lines represent normal and high Mg2+, respectively. (C) Overlap between total number SNP hits for 21 traits at normal and high Mg2+. Venn diagrams display overlap between SNP number at normal Mg2+ (blue) and high Mg2+ (red).
Under normal Mg2+ supply, 41 and 22 loci were associated with Mn absorption (Mn-A-N) and concentration (Mn-C-N), and accounted for 7.47% and 2.67% of the total phenotypic variance, respectively. 17 and 21 loci were associated with Fe absorption (Fe-A-N) and concentration (Fe-C-N), and accounted for 3.60% and 5.04% of total phenotypic variance individually (Fig. 3B, Supporting Information Table S3). In comparison, under high Mg2+ supply, 16 and 13 loci were associated with Mn absorption (Mn-A-H) and concentration (Mn-C-H), and accounted for 2.93% and 12.04% of total phenotypic variance individually. Thirty three and 11 loci were associated with Fe absorption (Fe-A-H) and Fe concentration (Fe-C-H), which accounted for only 5.07% and 2.60% of total phenotypic variance individually (Fig. 3B, Supporting Information Table S2). The GWAS analysis confirmed that only 2 common loci were detected differed between normal Mg2+ and high Mg2+ (Fig. 3C).
Single nucleotide polymorphisms (SNP) 1_3884526 (Chromosome 1, 3884526 bp) was found to associate with SG-H, RG-H, RG-N and Na-C-N. It had a Minor allele frequency (MAF) of 0.0633, and is within the OST3A, a gene related to oligosaccharyl transferase complex/magnesium transporter family (Supporting Information Table S2). SNP 4_2080234 was associated with Mn-C-H and Fe-C-N. It had a MAF of 0.050 and is within AT4G042960 and AT4G04293, both transposable element genes.
No significant associations were observed for the Mg2+ (Mg-A) and Ca absorption (Ca-A) under normal Mg2+, but 2 loci were identified to associate with these traits under high Mg2+. Furthermore, no significant associations were found for Mg2+ (Mg-C) and S concentration (S-C) under high Mg2+, whereas 7 and 6 loci were identified to associate with these traits under normal Mg2+ (Fig. 3B). No significant associations for K absorption (K-A) were identified under normal and high Mg2+.
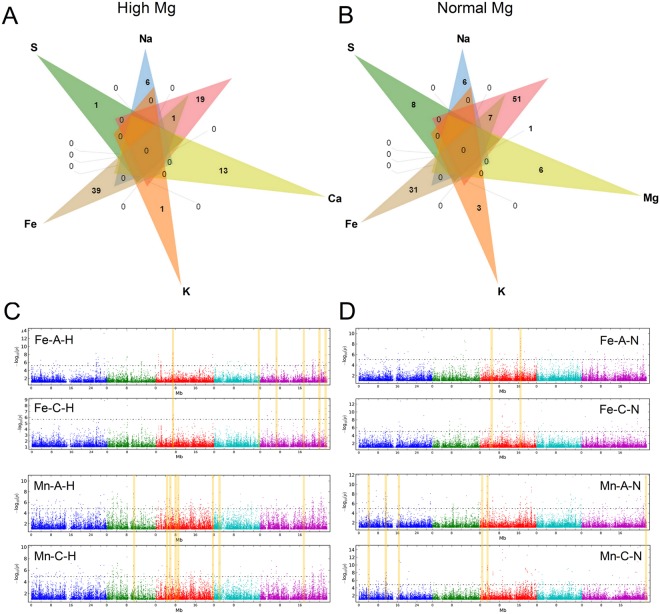
We speculated that the absorption and concentration of nutrients should share many candidate loci associated with the same element under the same Mg2+ level. However, this was not the case in our data set, as no common candidate loci were observed to associate with absorption and concentrations of K, Ca, Mg, S and Na under either normal or high Mg2+ (Supporting Information Table S2). Though common loci were found between the absorption and concentration of Fe or Mn in plants grown under both normal and high Mg2+, these only accounted for a small part of the variance in attitudes. While six common loci were detected between the traits of Fe-A-H and Fe-C-H, only 2 common SNPs were for Fe-A-N and Fe-C-N. Likewise, 10 common loci were associated with Mn-A-H and Mn-C-H and 6 common loci associated with Mn-A-N and Mn-C-N (Fig. 4A,B). Furthermore, manhattan plot showed the detailed distribution of the GWAS hits for Fe and Mn (Fig. 4C,D).
Figure 4.
Venn diagrams representing differentially nutrient (including absorption and concentration) GWAS hits was defined as the one had −log10(p) > 6.15 in high Mg2+ (A) and normal Mg2+ (B) of Arabidopsis. Each list in the venn diagram denotes by a transparent shape and overlaps shape indicates elements shared between lists or more often the corresponding counts. Four lists of input data of Fe-A, Fe-C, Mn-A and Mn-C were highlighted for both high Mg2+ and normal Mg2+ in green, blue, pink and yellow, respectively. Genome-wide association mapping results for the absorption and concentration of Fe and Mn in Arabidopsis under high Mg2+ (C) and normal Mg2+. (D) The ‘Manhattan setting’ plate for customized chromosome color, fixed pixel size, or dynamic pixel size proportional to p-values in Manhattan plot. The 5 chromosomes are represented by different shades of gray with the x-axis as the physical position. The highlighted positions represent the physical positions of the regions sharing common SNP between absorption and concentration of Fe and Mn.
Characterization of Atntrl mutants in response to high Mg2+ treatment
GWAS is conducted to identify causative/predictive factors for a given trait, or to determine aspects of the genetic architecture of the trait (i.e. the number of loci that contribute to the phenotype)41. A single strong peak of SNPs 22262848 between position of 22262147 and 22266184 bp on chromosome, which is associated with Ca concentration, was identified under high but not normal Mg2+ (Fig. 5A,B). This SNP accounted for 0.326% of the total variance of an estimated 2.20% of the total variance in Ca concentration under high Mg2+. It contained three neighboring significant SNP hits (−log10(p) > 6.9) corresponding to only a candidate gene At1g60420 (AtNRX1) (Fig. 5C), and was calcium locus CD-domain containing protein.
Figure 5.
Manhattan plot illustrating the GWAS mapping of the Ca concentration in 388 Arabidopsis accessions grown under supply of normal (1,000 µM) (A) and high Mg2+ (10,000 µM). (B) Chromosomes are shown in different colors. (C) Detailed plot of the region shown in the red box in (B). The positions on the chromosome are on the x axis and the score on the y axis. The dots in the scatterplot represent SNPs. A horizontal dashed line shows the 5% threshold.
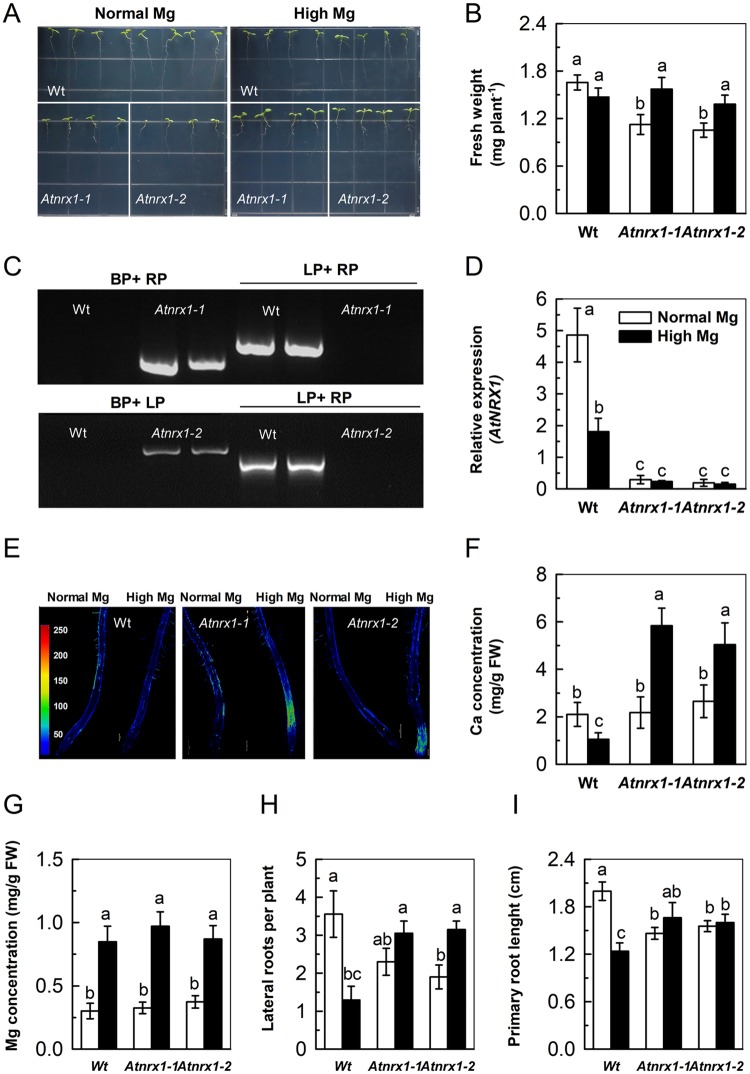
To validate the GWAS results in the present study, the above putative significant candidate gene was determined through a collection of well-characterized mutants (Atnrx1-1 and Atnrx1-2) and physiological and morphological comparisons in mutants and wild-types. Before this, we further verified that the mutant stocks were homozygous T-DNA insertion lines, by PCR with gene-specific primers LP, RP and LB. As expected, the results confirmed that both Atnrx1-1 and Atnrx1-2 were only mutation in At1g60420 (Fig. 6A). Moreover, the expression of AT1G60420 were greatly reduced in two mutants under both normal and high Mg2+ conditions as compared to wildtype (Fig. 6B). Compared with normal Mg2+, high Mg2+ increased the fresh weight of Atnrx1-1 and Atnrx1-2 but did not affect that of wildtype. It is worth mentioning that compared with wildtype, fresh weight of two mutants was not changed under high Mg2+ but was reduced under normal Mg2+ (Fig. 6C). It is suggested that high Mg2+ reduced wildtype growth but had little effect on the growth of Atnrx1 mutant.
Figure 6.
Validation and physiological phenotypes of Atnrx1 mutant. Image of mutant Atnrx1-1 and Atnrx1-2 and wild plants in media with normal or high Mg2+ (A); results of T-DNA insertion mutant Atnrx1-1 and Atnrx1-2 by PCR with gene-specific primers LP, RP and LB (B); relative expression levels (C); fresh weight (D); confocal images of cytosolic Ca2+ concentration distribution (Fluo-4/AM imaging) during the initiation and tip growth of root hairs (E); concentrations of Ca (F); concentrations of Mg2+ (G); number of lateral root per plant (H) and the length of primary root (I) of wild-type and Atnrx1 mutant (8-day-old seedlings) grown at supply of normal (1,000 µM) and high Mg2+ (10,000 µM). Levels of cytosolic Ca2+ concentration have been color-coded according to the inset scale (Quantitative values under the scale in nM). Bar = 200 μm applies to all photographs in D. The images are representative of at least three independent experiments. Data are means ± SD (n = 5). Means followed by a common letter within a root segment are not significantly different at P < 0.05. FW, fresh weight.
As shown in Fig. 6D, a tip-focused [Ca2+]c gradient was formed during the initiation and elongation of roots of both wildtype and mutants, and such a tip-focused gradient of [Ca2+]c was weak under normal Mg2+. In contrast, under high Mg2+, Atnrx1-1 and Atnrx1-2 but not wildtype formed a strong tip-focused [Ca2+]c gradient in the initiation and elongation of root of the mutant (Fig. 6D). Furthermore, high Mg2+ greatly enhanced concentration of Ca in Atnrx1-1 and Atnrx1-2 but decreased it in wildtype as compared with normal Mg2+ (Fig. 6E). Meanwhile, when compared with wildtype, Ca concentration of high Mg-fed Atnrx1-1 and Atnrx1-2 mutants was greater while that of normal Mg-fed Atnrx1-1 and Atnrx1-2 mutants was similar (Fig. 6E). As expected, high Mg2+ enhanced Mg concentrations in both mutants and wildtype (Fig. 6F).
Compared with the wildtype, both Atnrx1-1 and Atnrx1-2 had shorter primary root and less number of lateral roots under normal Mg2+ but had greater length of primary roots and a similar number of lateral roots under high Mg2+ (Fig. 6G,H). Under high Mg2+, there is no significant difference in K, Fe, Mn and S concentrations in plants between the two mutants and wildtype, but the mutant had a higher Na content (Data not shown). These results indicate that AtNRX1 is a special and critical negative gene regulating Ca uptake and probably the plant growth under high Mg2+ stress.
Discussion
To adapt to the constantly high availability of Mg2+ in the environment, plants have evolved mechanisms of reduced ion uptake and/or sequestering excessive ions in the vacuole or developing higher Mg2+ requirement for the maximum growth, which finally improves the tolerance to the stress of certain elements. By linking the mapping with reverse genetic experiments, we identified the causal gene and established that polymorphisms at AtNRX1 were the major genetic determinant for the variation in Ca concentration in this global A. thaliana population (Fig. 6). Atnrx1 mutants have most of the phenotypes that would associate with the tolerance to serpentine soils, including survival in solutions with a low Ca2+: Mg2+ ratio; requirement for a high concentration of Mg2+ for maximum growth; greater biomass and more lateral roots, reduced leaf tissue concentration of Mg; and poor growth performance on ‘normal’ levels of Mg2+.
Shoot and root morphology variation in response to high Mg2+
In order to understand molecular mechanisms of plant adaptation to high Mg2+ stress, we grew 388 wild Arabidopsis accessions at normal and high Mg2+ concentrations, and observed a substantial quantitative variation in plant growth between these accessions (Fig. 1; Table 1). We also showed a diversity in shoot and root morphology across two levels of Mg2+ supply and among these natural accessions. High Mg2+ supply resulted in an overall reduction in primary root length, lateral root number, epicotyl length and fresh weight but its effect on the rosette width varied among the accessions (Fig. 2; Table 2). The phenotypic data indicate that Arabidopsis thaliana has evolved some capacities to adapt to high Mg2+ stress and that the number of lateral roots of seedlings is a suitable index of high-Mg2+ responses.
By using the phenotypic data of 388 Arabidopsis accessions (Supporting Information Table S1) and GWAS, this study showed that a great extent of variation in the length of primary and lateral roots, rosette growth and hypocotyl length among the accessions tested was strongly Mg-dependent. As we known, the growth of primary and lateral roots, hypocotyl and rosette is determined by a number of extrinsic and intrinsic factors in addition to Mg2+ supply. The significant associations with the leaf or root growth-related phenotypes according to −log10(p) > 6.15 reflects the high complexity of the underlying genetic architecture or that these traits were not influenced by any of the SNPs alone. This notion is supported by the presence of obvious sub-threshold peaks associated with shoot and root morphology under both normal and high Mg2+ supply. Meanwhile, the low heritability of investigated traits in the present study suggested a weak genetic component determining the observed phenotypic variability. Hopefully, it is known that many traits of Arabidopsis are polygenic with small effect size, and that increasing the sample size would improve the power to recover meaningful associations42. Therefore, increasing sampling size was likely to underestimate the range natural variation in photomorphogenic responses or growth-related pathways and processes that strongly interact with the high Mg2+ stress.
Arabidopsis showed remarkable natural genotypic variation for ionomics under high Mg2+
The present study showed that most of the SNPs were derived from the nutrient concentration and uptake, SNPs differed between normal and high Mg2+, the effective number of SNPs (−log10(p) > 6.15) was also different (Fig. 3; Supporting Information Table S2). For example, positive SNPs associated with Ca-A and Mg-A were located only under high Mg2+ (Fig. 3B). Likewise, no significant associations were observed for either Mg-C and S-C under high Mg2+ but seven and six significant SNPs were associated with them under normal Mg2+ (Fig. 3), indicating that different genetic systems were responsible for Mg2+ and S uptake under normal and high Mg2+. Meanwhile, the SNP significance tests showed that different traits were generally associated with different genes or gene regions, but many of the traits also shared common genes or gene regions. The significant score >10 association was for the SNP on Mn under high Mg2+ in a narrow region of chr3, where multiple SNPs were associated with different traits in Chr1, 3, 4 and 5 under normal Mg2+ (Fig. 3).
Though not all cases, the chromosomal regions harbouring the significantly associated SNPs are coincided with the location of the gene(s) co-expressed with known-function genes that control these traits. Of the 105 loci under normal Mg2+, the majority of the loci were associated with Mn-A and Mn-C then with Fe-A and Fe-C. Among these, SNP 1_3953597, which is within the SS3, a gene encoding a starch synthase, is co-expressed with SPLICING ENDONUCLEASE 1 (SEN1)43. Likewise, SNP 3_19424421 of p-value 12.55, associated with Mn-C under the normal Mg2+ condition, had a MAF of 0.058 and was within gene AT3G52380 CHLOROPLAST RNA-BINDING PROTEIN 33 (CP33) that is co-expressed with SEN1. SEN1 gene was reported encoding an integral membrane protein homologous to Glycine max nodulin-21, and also to CCC1, a vacuolar Fe/Mn transporter of Saccharomyces cerevisiae of Arabidopsis41. Besides, SNP 1_210424, associated with Fe-C, had a MAF of 0.055 and was within gene FRO2 which encodes the low-Fe-inducible ferric chelate reductase. It is suggested to be the major Fe(III) chelate reductase in Arabidopsis44. The most SNP hits under high Mg2+ were identified associations with Fe-A and Fe-C and then Mn-A and Mn-N. SNP 1_26930918 had a MAF of 0.074 and was located within AT1G71480 and AT2G33410 genes which are involved in nuclear transport factor 2 family gene, and are co-expressed with Fe transporter gene FERRIC REDUCTION OXIDASE 6 (FRO6) and 7 (FRO7)45 and 1 (FRO1)46. Furthermore, SNP 2890675, associated with Fe-C under the high Mg2+ on Chromosome 5, had a MAF of 0.074 and was located within AT5G09310 gene and co-expressed with Fe transporter gene FRO146. Likewise, SNP 3_6977829 of P-value 8.40, associated with Fe-A, within gene AT3G20015 (ASPG2), and was co-expressed with H( + )-ATPASE 2 (AHA2), a main actor in Fe uptake and signaling47.
In addition, SNP 4_772594 associated with Mg + C under the normal Mg2+, had a MAF of 0.079 and was located within AT4G01800 ALBINO OR GLASSY YELLOW 1 (AGY1) gene which was co-expressed with MAGNESIUM CHELATASE I2 (CHLI2) while CHLI2 regulates the function of Mg2+ chelatase48. It should be mentioned that SNP 1_21762851 of P-value 9.05, associated with S-C under the normal Mg2+, had a MAF of 0.090 and contained only one gene, AT1G51980, which encodes an insulinase and is co-expressed with SULFATE TRANSPORTER 1;2 (SULTR1;2) that is involved in sulfur metabolism49. As discussed above, changing the Mg2+ availability in the soil would alter absorption and utilization of other elements, and transporters, channels, or/and the genes that encode and regulate them in Arabidopsis. However, a few of the genes in proximity of the most significant associations were among the major known regulators of ion uptake or transport, highlighting the potential of GWAS to identify previously unknown regulators50.
Changes in nutrient availability influence plant vegetative growth and phenotypic traits of A. thaliana, but the effect may vary among populations or genotypes51–53. Decreases in nutrient availability result in phenotypic responses (such as smaller leaf areas, shorter primary root and reduced lateral root formation) that vary across A. thaliana genotypes.
A novel candidate gene AtNRX1 negatively regulates Ca concentration at high Mg2+ supply
Calcium is an essential plant macronutrient with key structural and signaling roles. Calcium ions (Ca2+) act as an osmoticum within vacuoles, a stabilizing element in the membranes, a strengthening agent in cell walls, and a secondary messenger for a multitude of signals54–56. By using GWA mapping on the 388 A. thaliana accessions, we successfully identified a single strong peak of SNP 1_22263976/22264331/22262848 (Chromosome 1, 22263976 bp) associated with Ca concentration under high Mg2+ (Fig. 5). Mutants of Atnrx1-1 and Atnrx1-2 supplied with high Mg2+ had higher cytosolic Ca2+ concentrations ([Ca2+]c) during root elongation, meanwhile produced higher biomass and more lateral roots as compared with wildtype (Fig. 6). This suggested that the increase in Ca concentration in mutants of Atnrx1-1 and Atnrx1-2 by high Mg2+ cannot be attributed to increased biomass or photosynthesis alone, but also to the improved Ca nutrition of the plants through morphological, physiological and molecular responses to high Mg2+.
It has been known that AtNRX1 is involved in DC1 domain-containing protein, which is co-expressed with calcineurin B-like Ca sensor 1–protein kinase gene CBL-INTERACTING PROTEIN KINASE 23 (CIPK23)57,58. Study of Tang et al.59 showed that tonoplast CBL–CIPK calcium signaling network regulated ion homeostasis and vacuolar sequestration of Mg2+, thereby protecting plants from Mg2+ toxicity59,60. Besides, loss-of-function cax1 (gene encoding vacuolar H+/Ca2+) mutations could produce phenotypes characteristic of plants adapted to serpentine8.
This locus has a MAF of 0.0712 and is within the AT1G60420 (AtNRX1), a gene involved in catalytic activity of NADP-NADPH and disulfide-reductase activity61. In special cases, toxic ROS molecules are largely accumulated under excess stresses and diverse enzymes played as ROS scavengers. Arabidopsis contains 3 NADPH-dependent thioredoxin reductases (NTRs) for scavenging ROS and Nucleoredoxins (NRX) are potential nuclear TRX found in most eukaryotic organisms61. Recently, excellent research presents evidence that activity of the plant stresses-inducible TRX superfamily member, NRX1 is necessary for the integrity of antioxidant systems62. Meanwhile, it is the fact that many publications indicated ROS produced by NADPH oxidase activates Ca2+ channels at the plasma membrane, leading to an increase in Ca2+ concentration and a tip-focused Ca2+ concentration gradient and subsequent signaling inherent to plant growth32,63–66. Withal, our previous study also showed that high Mg2+ could inactivate NADPH and the production of ROS27 probably through regulating ABA–DELLA signaling28,67.
To sum up, it is likely that under high Mg, the wild-type plant is associated with cellular accumulation of ROS and redox imbalance, which triggers class of antioxidant enzymes activation and subsequently results in a decrease of intracellular ROS. ROS restrains ion channels and Ca2+ concentration; While in the nrx1 mutants, ROS could not be effectively removed without protection of NRX1 due to NRX1 mutation, which leads to ROS accumulation. Then ROS activate Ca2+-channels at the plasma membrane and promote cytosolic Ca2+ concentration. When both wild-type and nrx1 mutants under normal Mg conditions, the cell is normally controlled with H2O2 and redox balance, thus the protective effect of NRX1 gene is not prominent, or maybe even little dedication.
Notably, Mg2+ concentrations in Atnrx1-1 and Atnrx1-2 mutants was increased compared to that in the wildtype plants under high Mg2+ supply. This implied that nrx1 mutants required for a high concentration of Mg2+for the maximum growth associating with tolerance to high Mg2+ soils. The AtNRX1 gene is co-expressed with MAGNESIUM TRANSPORTER 4 (MRS2-3), a transport gene related to Mg2+ nutrition68. These indicate that Mg2+ transporter genes have some interactions with AtNRX1. However, there is no significant difference in K, Fe, Mn and S concentration in plants between the two mutants and wildtype, but the mutant had a higher Na content (Data not shown). These was consistent with other study showed that the cbl2 cbl3 double mutant was hypersensitive to a handful cations but not to Na+ 59. Further work will be directed to identify other key components in this novel of NRX-signaling pathway in regulation of Ca uptake and how the positive effects of AtNRX1 can be maximized through plant adaptive strategies to improve the tolerance to high Mg2+.
To sum up, our study suggests that AtNRX1 was a critical gene negatively regulating Ca uptake and probably improving the plant tolerance to high Mg2+ soils. The discovery of the functions of AtNRX1 gene helps to breed/select crops that can adapt to high-Mg2+ soils such as serpentine soils (high Mg2+: Ca) and those in semi-arid regions or even serpentine or Mars soils with high levels of magnesium sulfate. Also, new genes associated with natural variation in A. thaliana can be used for future bioengineering.
Electronic supplementary material
Acknowledgements
This work was financially supported the Natural Science Foundation of China (NSFC, No. 31601277), the Ninth Special Post-Doctoral Fund of China (No. 2016T90156) and the China Postdoctoral Science Foundation to YFN (2014M551754 and 2015M581216). We are thankful to Hangzhou Guhe Information and Technology Co., Ltd for the help with bioinformatics analysis and the 985-Institute of Agrobiology and Environmental Sciences of Zhejiang University for providing the experimental equipment.
Author Contributions
Y.F.N. developed the concept and designed the study; Y.F.N., P.C. and Y.Z. performed the experiments; Y.F.N. and G.L.J. analyzed the data; Z.W.W. and S.K.H. interpreted the results of the experiments; C.X.T. and L.B.G. edited and revised the manuscript; All authors approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yaofang Niu, Ping Chen and Yu Zhang contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31950-0.
References
- 1.Maguire ME, Cowan JA. Magnesium chemistry and biochemistry. Biometals. 2002;15:203–210. doi: 10.1023/A:1016058229972. [DOI] [PubMed] [Google Scholar]
- 2.Brady KU, Kruckeberg AR, Bradshaw HD., Jr. Evolutionary ecology of plant adaptation to serpentine soils. Annu. Rev. Ecol. Evol. Syst. 2005;36:243–266. doi: 10.1146/annurev.ecolsys.35.021103.105730. [DOI] [Google Scholar]
- 3.Hawkesford, M. et al. Functions of macronutrients. In ‘Marscher’s mineral nutrition of higher plants’. 3rd edn (Ed. P Marschner) pp. 135–190 (2012).
- 4.Christensen PR, et al. Mineralogy at meridiani planum from the Mini-TES experiment on the opportunity rover. Science. 2004;306:1733–1739. doi: 10.1126/science.1104909. [DOI] [PubMed] [Google Scholar]
- 5.Haskin LA, et al. Water alteration of rocks and soils on Mars at the Spirit rover site in Gusev crater. Nature. 2005;436:66–69. doi: 10.1038/nature03640. [DOI] [PubMed] [Google Scholar]
- 6. Ming, D. W. et al. Geochemical and mineralogical indicators for aqueous processes in the Columbia Hills of Gusev crater, Mars. J. Geophys. Res.: Planets, 111(E2) (2006).
- 7.Tyndall, R. W. & Hull, J. C. Vegetation, flora, and plant physiological ecology of serpentine barrens of eastern North America. In: Anderson, R. C., Fralish, J. S. & Baskin, J. M. eds Savannas, Barrens, and Rock Outcrop Plant Communities of North America. Cambridge, UK: Cambridge University Press 67–82 (1999).
- 8.Bradshaw HD., Jr. Mutations in CAX1 produce phenotypes characteristic of plants tolerant to serpentine soils. New Phytol. 2005;167:81–88. doi: 10.1111/j.1469-8137.2005.01408.x. [DOI] [PubMed] [Google Scholar]
- 9.Kruckeberg, A. R. An experimental inquiry into the nature of endemism on serpentine soils PhD thesis, University of California, Berkeley, CA, USA (1950).
- 10.Kruckeberg AR. The ecology of serpentine soils: a symposium. III. Plant species in relation to serpentine soils. Ecology. 1954;35:267–274. [Google Scholar]
- 11.Rajakaruna N, Siddiqi MY, Whitton J, Bohm BA, Glass AD. Differential responses to Na+/K+ and Ca2+/Mg2+ in two edaphic races of the Lasthenia californica (Asteraceae) complex, a case for parallel evolution of physiological traits. New Phytol. 2003;157:93–103. doi: 10.1046/j.1469-8137.2003.00648.x. [DOI] [PubMed] [Google Scholar]
- 12.Nyberg Berglund AB, Dahlgren S, Westerbergh A. Evidence for parallel evolution and site‐specific selection of serpentine tolerance in Cerastium alpinum during the colonization of Scandinavia. New Phytol. 2003;161:199–209. doi: 10.1046/j.1469-8137.2003.00934.x. [DOI] [Google Scholar]
- 13.Gailing O, Macnair MR, Bachmann K. QTL mapping for a trade-off between leaf and bud production in a recombinant inbred population of Microseris douglasii and M. bigelovii (Asteraceae, Lactuceae): a potential preadaptation for the colonization of serpentine soils. Plant Biol. 2004;6:440–446. doi: 10.1055/s-2004-817958. [DOI] [PubMed] [Google Scholar]
- 14.Mengoni A, et al. Chloroplast genetic diversity and biogeography in the serpentine endemic Ni-hyperaccumulator Alyssum bertolonii. New Phytol. 2003;157:349–356. doi: 10.1046/j.1469-8137.2003.00664.x. [DOI] [PubMed] [Google Scholar]
- 15.Patterson TB, Givnish TJ. Geographic cohesion, chromosomal evolution, parallel adaptive radiations, and consequent floral adaptations in Calochortus (Calochortaceae): evidence from a cpDNA phylogeny. New Phytol. 2004;161:253–264. doi: 10.1046/j.1469-8137.2003.00951.x. [DOI] [Google Scholar]
- 16.Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- 17.Huard-Chauveau C, et al. An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet. 2013;9:e1003766. doi: 10.1371/journal.pgen.1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevalier F, Pata M, Nacry P, Doumas P, Rossignol M. Effects of phosphate availability on the root system architecture, large-scale analysis of the natural variation between Arabidopsis accessions. Plant, Cell Environ. 2003;26:1839–1850. doi: 10.1046/j.1365-3040.2003.01100.x. [DOI] [Google Scholar]
- 19.Chevalier F, Rossignol M. Proteomic analysis of Arabidopsis thaliana ecotypes with contrasted root architecture, in response to phosphate efficiency. Plant Physiol. 2011;168:1885–1890. doi: 10.1016/j.jplph.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann MH. Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae) J. Biogeogr. 2002;29:125–134. doi: 10.1046/j.1365-2699.2002.00647.x. [DOI] [Google Scholar]
- 21.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 2012;44:212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tisné S, et al. Phenoscope: an automated large‐scale phenotyping platform offering high spatial homogeneity. Plant J. 2013;74:534–544. doi: 10.1111/tpj.12131. [DOI] [PubMed] [Google Scholar]
- 23.Wuyts N, Dhondt S, Inzé D. Measurement of plant growth in view of an integrative analysis of regulatory networks. Curr. Opin. Plant Biol. 2015;25:90–97. doi: 10.1016/j.pbi.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Flood PJ, et al. Phenomics for photosynthesis, growth and reflectance in Arabidopsis thaliana reveals circadian and long-term fluctuations in heritability. ethod. 2016;12:14. doi: 10.1186/s13007-016-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visscher AM, et al. Growth performance and root transcriptome remodeling of Arabidopsis in response to Mars-Like levels of Magnesium sulfate. PLoS ONE. 2010;5:e12348. doi: 10.1371/journal.pone.0012348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamanaka T, et al. MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol. 2010;152:1284–1296. doi: 10.1104/pp.109.147371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu YF, et al. Phosphorus and magnesium interactively modulate the elongation and directional growth of primary roots in Arabidopsis thaliana (L.) Heynh. J. Exp. Bot. 2015;66:3841–3854. doi: 10.1093/jxb/erv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo WL. The remodeling of seedling development in response to long-term magnesium toxicity and regulation by ABA–DELLA signaling in Arabidopsis. Plant Cell Physiol. 2014;55:1713–1726. doi: 10.1093/pcp/pcu102. [DOI] [PubMed] [Google Scholar]
- 29.Weber AP, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czarnecki O, et al. An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell. 2011;23:4476–4491. doi: 10.1105/tpc.111.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greco M, Chiappetta A, Bruno L, Bitonti MB. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012;63:695–709. doi: 10.1093/jxb/err313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu YF, et al. Magnesium availability regulates the development of root hairs in Arabidopsis thaliana (L.) Heynh. Plant, Cell Environ. 2014;37:2795–2813. doi: 10.1111/pce.12362. [DOI] [PubMed] [Google Scholar]
- 33.Lanquar V, et al. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell. 2009;21:3610–3622. doi: 10.1105/tpc.109.068593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, et al. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol. 2013;162:720–731. doi: 10.1104/pp.113.214940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa M, et al. Expression-based and co-localization detection of arabinogalactan protein 6 and arabinogalactan protein 11 interactors in Arabidopsis pollen and pollen tubes. BMC Plant Biol. 2013;13:1. doi: 10.1186/1471-2229-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruber BD, Giehl RF, Friedel S, von Wirén N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013;163:161–179. doi: 10.1104/pp.113.218453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seren Ü, et al. GWAPP, a web application for genome-wide association mapping in Arabidopsis. Plant Cell. 2012;24:4793–4805. doi: 10.1105/tpc.112.108068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbredlines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu YF, et al. Auxin modulates the enhanced development of root hairs in Arabidopsis thaliana (L.) Heynh. under elevated CO2. Plant, Cell Environ. 2011;34:1304–1317. doi: 10.1111/j.1365-3040.2011.02330.x. [DOI] [PubMed] [Google Scholar]
- 40.Gan YB, et al. GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell. 2006;18:1383–1395. doi: 10.1105/tpc.106.041533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hakoyama T, et al. The integral membrane protein SEN1 is required for symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol. 2012;53:225–236. doi: 10.1093/pcp/pcr167. [DOI] [PubMed] [Google Scholar]
- 42.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS, a review. Plant Methods. 2013;9:29. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao CR, et al. Comparative transcriptomic characterization of aluminum, sodium chloride, cadmium and copper rhizotoxicities in Arabidopsis thaliana. BMC Plant Biol. 2009;9:32. doi: 10.1186/1471-2229-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 2003;133:1102–1110. doi: 10.1104/pp.103.025122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong J, Connolly EL. Iron uptake mechanisms in plants, functions of the FRO family of ferric reductases. Plant Sci. 2009;176:709–714. doi: 10.1016/j.plantsci.2009.02.011. [DOI] [Google Scholar]
- 46.Waters BM, Blevins DG, Eide DJ. Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol. 2002;129:85–94. doi: 10.1104/pp.010829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mamidi S, et al. Genome-wide association analysis identifies candidate genes associated with iron deficiency chlorosis in soybean. Plant Genome. 2011;4:154–164. doi: 10.3835/plantgenome2011.04.0011. [DOI] [Google Scholar]
- 48.Du SY, et al. Roles of the different components of magnesium chelatase in abscisic acid signal transduction. Plant Mol. Biol. 2012;80:519–537. doi: 10.1007/s11103-012-9965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibagaki N, et al. Selenate–resistant mutants of Arabidopsis thaliana identify SULTR1; 2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 2002;29:475–486. doi: 10.1046/j.0960-7412.2001.01232.x. [DOI] [PubMed] [Google Scholar]
- 50.Meijón M, Satbhai SB, Tsuchimatsu T, Busch W. Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat. Genet. 2014;46:77–81. doi: 10.1038/ng.2824. [DOI] [PubMed] [Google Scholar]
- 51.Myerscough PJ, Marshall JK. Population dynamics of Arabidopsis thaliana (L.) Heynh. Strain “Estland” at different densities and nutrient levels. New Phytol. 1973;72:595–617. doi: 10.1111/j.1469-8137.1973.tb04410.x. [DOI] [Google Scholar]
- 52.Pigliucci M, Whitton J, Schlichting CD. Reaction norms of Arabidopsis. I. Plasticity of characters and correlations across water, nutrient and light gradients. J. Evol. Biol. 1995;8:421–438. doi: 10.1046/j.1420-9101.1995.8040421.x. [DOI] [Google Scholar]
- 53.Dorn LA, Pyle EH, Schmitt J. Plasticity to light cues and resources in Arabidopsis thaliana: testing for adaptive value and costs. Evolution. 2000;54:1982–1994. doi: 10.1111/j.0014-3820.2000.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 54.Dayod M, Tyerman SD, Leigh RA, Gilliham M. Calcium storage in plants and the implications for calcium biofortification. Protoplasma. 2010;247:215–231. doi: 10.1007/s00709-010-0182-0. [DOI] [PubMed] [Google Scholar]
- 55.Gilliham M, et al. Calcium delivery and storage in plant leaves: exploring the link with water flow. J. Exp. Bot. 2011;62:2233–2250. doi: 10.1093/jxb/err111. [DOI] [PubMed] [Google Scholar]
- 56.Qin Y, et al. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 2009;5:e1000621. doi: 10.1371/journal.pgen.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheong YH, et al. Two calcineurin B‐like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007;52:223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- 58.Luan S. The CBL–CIPK network in plant calcium signaling. Trends in Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 59.Tang RJ, et al. Tonoplast CBL–CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. P NATL ACAD SCI USA. 2015;112:3134–3139. doi: 10.1073/pnas.1420944112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang RJ, et al. Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Research. 2012;22:1650–1665. doi: 10.1038/cr.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchal C, et al. NTR/NRX define a new thioredoxin system in the nucleus of Arabidopsis thaliana cells. Mol Plant. 2014;7:30–44. doi: 10.1093/mp/sst162. [DOI] [PubMed] [Google Scholar]
- 62.Kneeshaw S, et al. Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. P NATL ACAD SCI USA. 2017;114:8414–8419. doi: 10.1073/pnas.1703344114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foreman J, et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–426. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- 64.Miller G, et al. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009;2:ra45–ra45. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- 65.Samaj J, Baluska F, Menzel D. New signalling molecules regulating root hair tip growth. Trends Plant Sci. 2004;9:217–220. doi: 10.1016/j.tplants.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 66.Wrzaczek M, Brosché M, Kangasjärvi J. ROS signaling loops—production, perception, regulation. Curr. Opin. Plant Biol. 2013;16:575–582. doi: 10.1016/j.pbi.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 67.Guo WL, et al. Magnesium stress signaling in plant: Just a Beginning Plant Signal. Behav. 2015;25:e992287-1–e992287-5. doi: 10.4161/15592324.2014.992287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gebert M, et al. A root-expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell. 2009;21:4018–4030. doi: 10.1105/tpc.109.070557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.