Abstract
In this report, we investigate small proteins involved in bacterial alternative respiratory systems that improve the enzymatic efficiency through better anchorage and multimerization of membrane components. Using the small protein TorE of the respiratory TMAO reductase system as a model, we discovered that TorE is part of a subfamily of small proteins that are present in proteobacteria in which they play a similar role for bacterial respiratory systems. We reveal by microscopy that, in Shewanella oneidensis MR1, alternative respiratory systems are evenly distributed in the membrane contrary to what has been described for Escherichia coli. Thus, the better efficiency of the respiratory systems observed in the presence of the small proteins is not due to a specific localization in the membrane, but rather to the formation of membranous complexes formed by TorE homologs with their c-type cytochrome partner protein. By an in vivo approach combining Clear Native electrophoresis and fluorescent translational fusions, we determined the 4:4 stoichiometry of the complexes. In addition, mild solubilization of the cytochrome indicates that the presence of the small protein reinforces its anchoring to the membrane. Therefore, assembly of the complex induced by this small protein improves the efficiency of the respiratory system.
Introduction
A few years ago, Storz et al.1 claimed that “small proteins can no longer be ignored”. Indeed, the importance of small proteins in both prokaryotes and eukaryotes rises according to their number and their involvement in various cellular functions. Small proteins are defined as proteins of around 50 amino acids that are the product of small open reading frames in the genome and are not derived by proteolytic processes1. Often it is difficult to decipher the precise role and the accurate mechanism of action of the small proteins. Due to their size, it is accepted that small proteins do not possess an enzymatic activity. However, functions of these small proteins are versatile. For instance, small proteins are involved in the sporulation process like SpoVM of B. subtilis2, affect signal transduction like MgrB and Sda3,4, act like chaperones as described for MntS5 or FbpB6 and regulate inner membrane transporter like MgtS7 as well as the activity of membranous enzymatic complexes like CydX8–12. They are located in the various compartments of the cell, soluble in the cytosol like MntS in Escherichia coli5 and Sda in Bacillus subtilis3, membrane-associated like SpoVM in B. subtilis2 and embedded in the membrane as MgtR13 and CydX11 in Salmonella enterica and E. coli. CydX optimizes the activity of the cytochrome bd oxidase CydAB complex by interacting with it. In E. coli, deletion of the gene cydX leads to a decrease of the respiratory activity of the complex while it is abolished in Brucella abortus cydX deletion mutant8,9. Furthermore, it is proposed that CydX has a direct role in the positioning and stability of the hemes contained in the CydAB complex but no influence on the complex stability10,11. CydX is a hydrophobic, small structured alpha-helical protein integrally located in the membrane, and surprisingly its topology in the membrane differs from one organism to another8,10,11.
Recently, a small protein called TorE has been reported in the aquatic bacterium S. oneidensis14. The 56 amino acid protein TorE has been reported to be part of the Tor respiratory system that reduces trimethylamine oxide (TMAO) into the volatile and odorous trimethylamine (TMA)15. TMAO, widely spread in marine environments, has an important role in fish to counteract urea and pressure damages and it is considered as a kosmotropic chaperone16–18. TMAO is also used as a respiratory substrate by various bacteria and especially Shewanella sp.15,19,20. TMAO reduction is mainly due to the Tor system, which production is strictly controlled by the presence of TMAO in the bacterium environment14,15. It is made up by the periplasmic TMAO reductase TorA and the pentahemic membrane-anchored c-type cytochrome TorC15,21. The latter is organized in two domains; the tetrahemic domain anchored to the membrane belongs to the NapC/NirT cytochrome family22, it interacts with the TMAO reductase TorA, receives the electrons from the quinone pool and transfers them to the second domain which in turn feeds TorA catalytic site23,24. In addition to the genes encoding TorA, TorC and TorE, the tor operon contains a fourth gene, torD that codes for the specific chaperone of TorA25,26. It has been shown previously that TorD is required for TorA stability and that it belongs to the family of chaperones specifically dedicated to molybdoenzymes of the DMSO reductase family to which TorA belongs27,28. In S. oneidensis, the absence of TorD leads to the absence of TorA activity and degradation of the protein14. In contrast, in the absence of the small protein TorE the activity of the Tor system is significantly decreased, although the maturation, concentration and location of TorA and TorC are not modified14. Furthermore, a fitness experiment revealed that TorE presence gives a strong selective advantage to the bacteria harboring TorE14.
Here we report that TorE interacts with the c-type cytochrome TorC and allows for the formation of a 220 kDa-membrane complex, with a potential 4:4 stoichiometry that gives TorC a better anchor into the membrane stabilizing the Tor system, thereby increasing its catalytic efficiency. Thus, TorE plays indirectly a key role in the electron transfer of the Tor system. Moreover, TorE and its homolog NapE from the periplasmic nitrate reductase respiratory system are part of a family of small proteins that are required to stabilize membrane anchored respiratory systems of certain proteobacteria.
Results
Synteny study of TorE family proteins
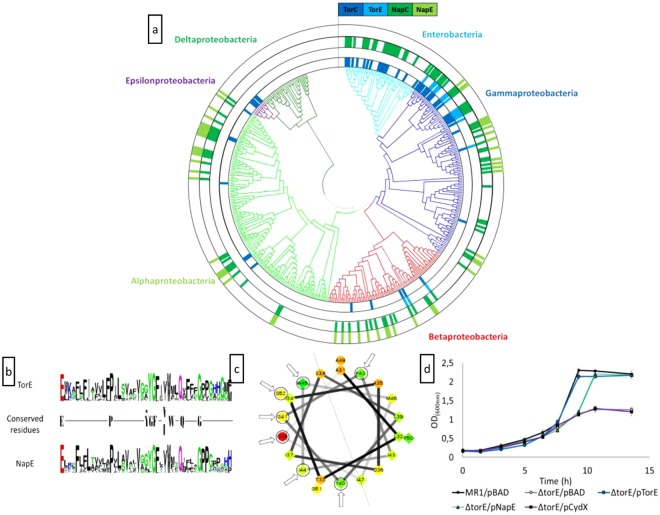
The small protein TorE contains 56 amino acids. It is a hydrophobic protein with a calculated molecular mass of 6119.29 Da and a pI of 9.52 (Expasy tools29–31). As many integral membrane small proteins, TorE is predicted to contain a single hydrophobic α helix spanning through the membrane (TMHMM tool)32,33. A bioinformatic analysis using TorE from S. oneidensis as a probe indicated that TorE homologs are found in synteny with a multihemic c-type cytochrome of the NapC/NirT family (Fig. 1a) and a molybdoenzyme. TorE homologs can be divided into two subfamilies: first, the TorE orthologs in synteny with pentahemic c-type cytochromes homolog to TorC and second, the TorE homologs, NapE, in synteny with tetrahemic c-type cytochrome NapC (Fig. 1a). NapE and NapC are part of the alternative nitrate respiratory system Nap, which terminal reductase NapA allows the reduction of nitrate in the periplasmic space of bacteria. The absence of NapE leads to a decrease of the Nap specific activity34. It is noteworthy that TorE orthologs are found in various genders of β and γ classes of Proteobacteria, whereas in α and ε-Proteobacteria the systems are present in a few genders and do not possess any TorE homolog. Compared to the Tor system, the Nap system presents a higher distribution in all division of proteobacteria except the epsilon and delta proteobacteria, with the latter not synthetizing either of these two systems. Except for enterobacteria, the Nap system contains in most cases NapE (NapENapC/NapC ratio of 68%), while TorE is less distributed in the phylogenetic tree (TorETorC/TorC ration of about 43%). Surprisingly, while in Enterobacteria, the Tor and Nap systems are well distributed, with none of them containing the small TorE/NapE proteins. One hypothesis could be that these proteins were lost during the evolution under conditions that do not require a highly efficient respiratory system. The comparison of the consensus sequences of both subfamilies shows that the proteins share homologies only in their hydrophobic stretch and that nine amino acids are highly conserved (Fig. 1b). Moreover, using a simulation program, it seems that in both proteins these residues are located on the same side of the α-helix with the sole exception of the highly conserved residue Phe42 (Fig. 1c). This indicates that the sequence variability is high for these proteins as long as the location of the conserved residues is maintained in the structure.
Figure 1.
(a) Occurrence of torC, torE, napC and napE genes among the proteobacteria. The presence of each gene is represented by block coloured in dark blue, light blue, dark green and light green, respectively. The phylogenetic tree has been constructed using the GyrB protein sequence from a bacterium of each genus of proteobacteria. Branches are coloured according to the class of proteobacteria. Within the gammaproteobacteria class, the enterobacteria order was specifically indicated. (b) Sequence logo of the TorE and NapE proteins. The conserved residues in the two sequences are shown. The black star indicated that the tyrosine residue is highly conserved in the NapE proteins except in bacteria that also possess the TorE protein. (c) Helical projection of the hydrophobic stretch of the TorE protein of S. oneidensis. These residues (from 31 to 52) were selected because of their location between two prolines. The conserved residues are indicated by arrows. The centreline of the helix is represented. (d) Complementation experiments. Growth curves of the wild type strain (MR1/pBAD) and ΔtorE mutant harbouring either pBAD, pTorE, pNapE or pCydX plasmid. Cells were grown anaerobically in LB medium in the presence of TMAO, arabinose and chloramphenicol. Each point represents the average ± standard error of three independent experiments.
NapE improves the growth of a torE deleted strain
To corroborate these observations, we tested whether it is possible to replace TorE by NapE in the Tor system. Since S. oneidensis has no NapE and NapC proteins, we used napE gene from S. massilia15. Thus, a torE mutant was generated that was complemented with either torE or napE. The recombinant and mutated strains were incubated with TMAO as sole substrate for respiration and their growth was followed and compared to that of the wild type strain. The torE mutant shows a growth defect that was complemented with the torE gene as previously shown14. Interestingly, the torE mutation was also complemented by napE, although a growth delay was observed. Nevertheless, this complementation indicates that NapE can replace TorE in the functionality of the Tor system. To verify that this result is specific to TorE/NapE family, we tried to complement the torE mutation by cydX coding for a small transmembranous protein involved in the cytochrome bd oxidase CydABX of S. oneidensis11. In the latter case, the growth defect of the recombinant strain is still observable, showing that CydX in contrast to NapE cannot replace TorE in the Tor system. This is also in agreement with the fact that no CydX protein was found among TorE or NapE homologs, since they do not share any conserved residues although CydX is a small membranous protein involved in a respiratory system.
Membrane localization of TorE
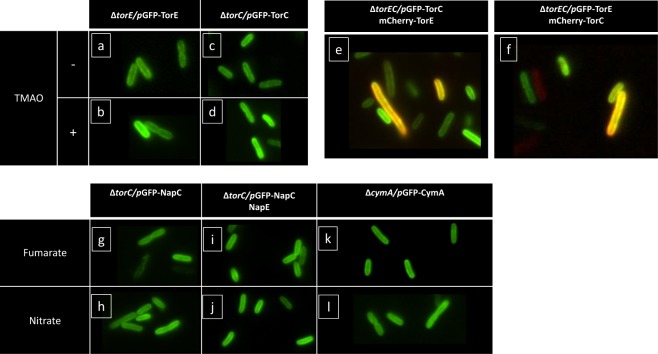
At the moment, many researches are focused on how proteins are organized in the bacterial membrane. In the membrane of prokaryotes, microdomains are structured to increase the efficiency of the response to environmental cues. They are made up of lipid rafts harboring various proteins like flotillin and proteins involved in signaling and transport35. In Pseudomonas aeruginosa, denitrification occurs through a membrane located supercomplex containing the four multisubunit enzymes allowing the production of molecular nitrogen from nitrate reduction and also many other enzymes and proteins36. In E. coli, the nitrate reductase NarGHI has been described as present in the membrane mostly at the cell poles under anoxia, allowing a higher nitrate reduction rate under certain conditions37. Since the presence of TorE increases the efficiency of the respiratory system, we wondered whether TorE could play a role in the targeting to a particular localization of the Tor system in the cell membrane. In a first approach, a functional chimeric protein GFP-TorE was engineered and produced in a strain deleted of torE gene (Figure S1). The subcellular localization of the GFP-labeled TorE was characterized by fluorescence microscopy imaging on exponentially growing S. oneidensis cells under fumarate-respiring conditions (Fig. 2a). The fluorescence signal was only detected at the edge of the bacteria and it was evenly distributed. By fractionation of the cells, we confirmed that the fluorescence was entirely in the membrane fraction and not in the periplasm. To test whether the presence of the other components of the Tor system may influence the localization of TorE, cells were grown in the presence of TMAO, the latter allowing the production of the TorA and TorC proteins from the chromosomal gene expression. The fluorescence signal was similarly distributed and no specific cluster of TorE was obvious (Fig. 2b). To corroborate these results, the same approach was performed with functional TorC labelled with GFP and produced in a strain deleted of torC (Figure S1). The fluorescence signal was well distributed along the membrane of cells grown in the presence or absence of TMAO, indicating that the cytochrome has no particular localization in the membrane whatever the presence of TorE and of TorA which production is strictly induced by TMAO14,15 (Fig. 2c,d). Finally, when TorE and TorC linked to the GFP and the mCherry tags were produced concomitantly from a plasmid without torA induction, their distribution along the membrane was still unchanged indicating that the absence of the terminal reductase TorA has no effect on the distribution of either TorC or TorE. Altogether these results indicate that the Tor respiratory system has no special spatiolocalization in the membrane as seen with the NarGHI complex37. Moreover, the presence of TorE does not induce a clustering of the Tor system in the cell membrane. To extend these conclusions, a similar approach was performed with NapE and NapC of S. massilia produced in S. oneidensis. The experiments were done with GFP labelled-NapC produced in the presence or absence of NapE. Fluorescence microscopy imaging indicated that NapC was uniformly distributed along the membrane and that the concomitant production of NapE has no effect on it. Finally, the distribution of CymA, another tetrahemic c-type cytochrome of the NapC/NirT family, was tested. CymA has no known small protein associated and is involved in several respiratory chains in S. oneidensis acting as the quinol oxidase during the reduction of fumarate, DMSO, nitrate and other electron acceptors, except TMAO38–41. The presence and localization of CymA was tested in cells grown with various substrates showing that it was evenly distributed whatever the nature of the electron acceptor (Fig. 2h).
Figure 2.
Fluorescence microscopy of S. oneidensis MR1 cells producing GFP and mCherry fused respiratory proteins. The genetic context of the strains, the plasmids harboured (pBAD33 derivatives) and the fluorescent chimeras are indicated in each panel. Cells were grown in a rich medium supplemented with fumarate (panels: a, c, e, f, g, i, k) or with electron acceptors inducing specific respiratory systems of S. oneidensis: TMAO (panels b and d); nitrate (panels h, j, l) and that are indicated on the edge of the panels. TMAO (−) stands for no TMAO added in the medium preventing chromosomal gene expression and thus the production of the Tor components.
TorE reinforces TorC anchoring to the membrane
We have shown in a previous work that TorE increases the efficiency of the Tor system and gives a selective advantage to the strain when competing with a torE mutant14. This suggests that the electron transfer is more efficient in the presence of TorE which can be due to a direct effect of TorE on either TorC prosthetic groups i.e. one of the five hemes, on the quinol binding site of TorC or on the stabilization of TorC and the whole respiratory system. To determine whether TorE acts also on the spectral properties of TorC, we expressed either torC or torEC in E. coli grown anaerobically with fumarate as sole source of electron acceptor. Under these conditions, the unique c-type cytochrome produced would be TorC of S. oneidensis42. Since the redox spectrum of TorC was similar for membranes where TorE was present or absent, this suggested that TorE has no effect on TorC spectrum. The stabilization of the Tor system by TorE was also investigated. We looked for conditions under which TorC anchoring into the membrane was reinforced by the presence of TorE. To do so, the membranes were treated either with a non-ionic detergent (n-Dodecyl-β-D-maltoside) or with a salt (NaBr) and the supernatants obtained after the solubilization were loaded on SDS-gel to decipher the amount of TorC released in the presence or absence of TorE (Fig. 3). We observed that when the membranes were incubated with DDM for a short time (20 min), the amount of TorC recovered in the solubilized fractions is higher when TorE is absent than in the wild type strain. Moreover, when membranes were treated with NaBr, allowing the release of proteins weakly anchored and the solubilized fraction submitted to SDS-PAGE, a band corresponding to TorC is observed only in the sample in which TorE was absent. This result suggests that the release of TorC by salt extraction or after a rapid solubilization is facilitated in the absence of TorE and thus, that TorE improves the anchoring of the cytochrome into the membrane.
Figure 3.
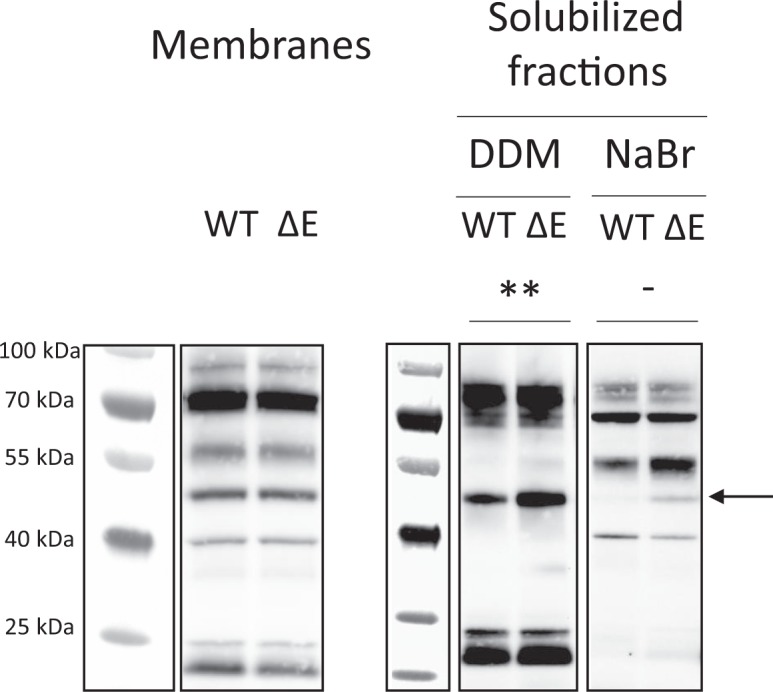
Solubilization of the TorC cytochrome under mild conditions. Peroxidase activity of c type hemes of cytochromes from membranous and membrane extracted proteins of S. oneidensis wild type cells (WT) or ΔtorE deletion mutant (ΔE) either after 2% DDM solubilization or 1.5 M NaBr. Ten microgram proteins were loaded in each lane of the 3–14% SDS gel. After electrophoresis, c-type cytochromes were revealed by their peroxidase activity. TorC is indicated by an arrow. **T test analysis performed by GraphPad Prism Software shows that data from three replicates are significantly different (p = 0.0058). No band can be detected in the WT lanes for three replicated experiments, preventing statistical analyses Separated portions of the same blots are represented with their respective molecular weight markers (see Figure S2).
TorE and TorC constitute a membranous complex
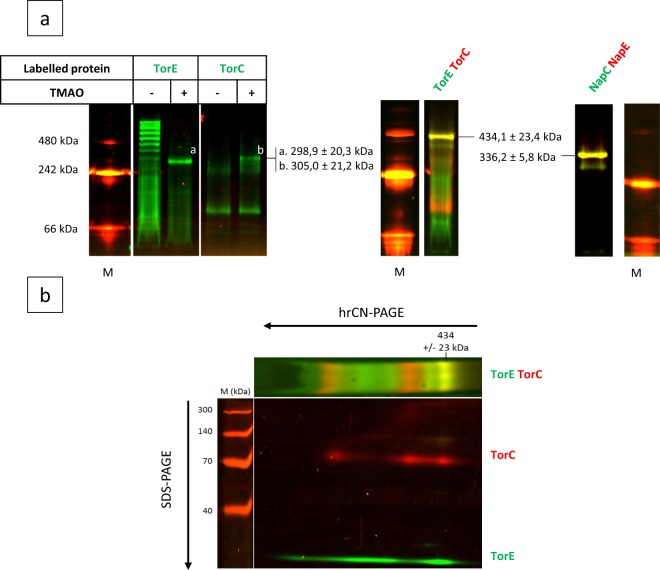
Taking into account the solubilization results and the fact that TorE spans the membrane, our hypothesis is that TorE could stabilize TorC and better anchor it to the membrane by interacting with it. We decided to visualize the interaction of the two proteins directly on membranous extracts. In a strain deleted of torE gene, we produced GFP-labelled TorE in the presence or not of TMAO and the solubilized membrane fraction was loaded on a 3–14% high resolution Clear Native (hrCN) gel. After electrophoresis, the GFP-labelled TorE was revealed by appropriate laser scanning. When the membrane extract was from the cells grown without TMAO, the fluorescence obtained indicated that TorE presents numerous states of oligomerization, while when the extract was from cells grown in the presence of TMAO, only one fluorescent band with a molecular weight of about 300 kDa was observed (Fig. 4). Since the addition of TMAO during the cell growth induces the expression of the remaining tor genes, i.e. torCAD, and since only TorC is anchored to the membrane, our hypothesis was that GFP-TorE interacts with TorC to create a complex. To verify this, the reverse experiment was performed. In this experiment, the torC deleted strain harboring a plasmid allowing the production of GFP-TorC was grown in the presence or absence of TMAO corresponding to the presence or absence of TorE in the extract, respectively. When GFP-TorC was produced without the other Tor proteins, two bands were observed: a major band at 70 kDa corresponding to the chimeric GFP-TorC and a faint band that could correspond to a trimer of GFP-TorC (Fig. 4a). When TorE is produced in the extract a third band appears on the track corresponding to a complex with a molecular weight of ≈305 kDa. To corroborate these results, we produced concomitantly both proteins labelled with GFP and mCherry for TorE and TorC, respectively. In addition to the bands corresponding to the mCherry-TorC and the GFP-TorE, a band with a combined fluorescence was revealed that corresponded to the complex of the two chimeras with a mass of about 434 kDa (Fig. 4a). To gain insight into the protein profile of the complex, a two-dimensional hrCN/SDS-PAGE was run which showed the presence of the two chimeras at the complex level. Moreover, the other bands of the hrCN gel that corresponded to each fluorescent protein migrated on the SDS-gel at the molecular weight estimated for the protein and the fluorochrome, i.e. 75 and 33 kDa for mCherry-TorC and GFP-TorE, respectively (Fig. 4b). Finally, the bands corresponding to the complex and to each chimera were cut from the hrCN-gel and were analyzed by mass spectroscopy. As expected, since a crude membrane extract was loaded on a gel, many proteins were present but recombinant TorC and TorE were significantly identified in the two replicates with high sequence coverages (Table 1). This analysis indicates that TorE and TorC co-migrate in the gel and that they interact to form a complex. According to the estimated sizes of the complex obtained in the various experiments (434 kDa, 305 kDa) and according to the size of labelled TorE and TorC, we propose that the complex is made up of 4 TorE and 4 TorC (Table 2). A similar experiment was performed with the proteins of the Nap system. NapC and NapE were labelled with GFP and mCherry, respectively, and produced together in S. oneidensis. The membrane extract of the recombinant strain was submitted to 3–14% hrCN-PAGE. Analysis of the fluorescence of the gel indicated that a complex is formed by the two proteins as visualized by the yellow color of the band. Based on the molecular weight of each labelled protein (47.6 and 32.5 kDa for GFP-NapC and mCherry-NapE, respectively) the complex has a mass of 336 kDa corresponding to a stoichiometry of 4NapE:4NapC. This result corroborates those obtained with TorE and TorC and indicates that both TorE and NapE are involved in the anchoring of their cognate cytochrome into the membrane.
Figure 4.
In vivo identification of the TorE-TorC complex. (a) hrCN-gels, first dimension. The strains were growth in anaerobiosis. The fluorescent labelled proteins were solubilized from the membrane extracts of the various recombinant strains using DDM (2%, v/v). The solubilized fractions were loaded on hrCN gel (3–14%) and in-gel detection of fluorescent labelled proteins and complexes was performed by scanning. Recombinant strains ΔtorE/pGFP-TorE (TorE) and ΔtorC/pGFP-TorC (TorC) were grown in rich medium supplemented with TMAO to induce the other components of the Tor system (+). The strain ΔtorEC carrying either pGFP-TorE-mCherry-TorC (TorE-TorC) or pmCherry-NapE;GFP-NapC (NapE-NapC) were grown in rich medium. Separated gels are represented with their respective molecular weight markers. White lines separate portions of the same initial gel (see Figure S3). (b) SDS-Gel, second dimension. A band of hrCN gel corresponding TorE-TorC was excised and after treatment with SDS and β-mercaptoethanol was submitted to SDS-12%-PAGE. The direction of the migration is indicated by arrows. (a,b) Fluorescence was revealed by scanning. Complexes TorE-TorC and NapE-NapC are indicated by arrows. M is for molecular weight markers. The pictures are representative of three independent experiments.
Table 1.
Protein content identified by mass spectrometry in the 430 kDa band sample.
| Ordera | Accessionb | Descriptionc | Coverage %d | PSMe | Unique Peptidesf | Scoreg |
|---|---|---|---|---|---|---|
| 1 | Q8E8H2 | proton-coupled multidrug efflux pump permease component VmeB | 29 | 45 | 21 | 137 |
| 3 | — | mCherry_TagGS_TorC | 39 | 39 | 19 | 114 |
| 13 | — | GFP_tagGS_TorE | 25 | 12 | 4 | 31 |
aOrder in the identified proteins.
bUniprot accession number.
cUniprot Description.
dProtein sequence coverage by the matching peptides.
ePeptide spectral matches given by the algorithm corresponding to the total number of identified peptide sequences.
fNumber of different peptides matching to protein sequence and unique to this protein.
gProtein score given by Sequest algorithm.
Table 2.
Theoretical molecular weights (kDa) of the complex with various stoichiometries of the subunits.
| GFP-TorE + TorC or GFP-TorC + TorE | GFP-TorE + mCherry-TorC | |
|---|---|---|
| 1: 1 | 77,69 | 104,67 |
| ΔMW | 221,23 | 329,41 |
| 2: 2 | 155,38 | 209,33 |
| ΔMW | 143,54 | 224,75 |
| 3: 3 | 233,07 | 314,00 |
| ΔMW | 65,85 | 120,08 |
| 4: 4 | 310,77 | 418,67 |
| ΔMW | −11,84 | 15,41 |
“ΔWM” stands for the difference between the experimental and the theoretical size of the complex (305 kDa, 434 kDa).
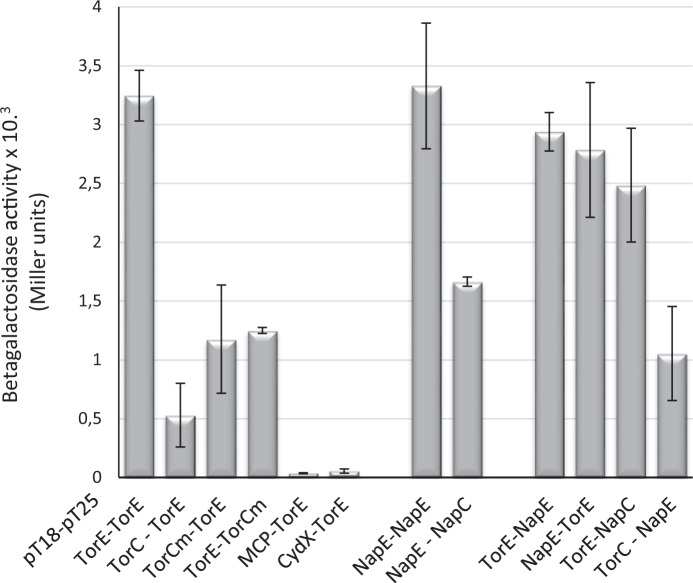
Through a two-hybrid experiment, the dimerization of TorE was confirmed as well as the interaction between TorE and the membrane anchor of TorC (Fig. 5). Moreover, it showed that TorE interacts with NapE to form heterodimers and with NapC. NapE dimerizes and interacts with NapC and with TorC. The specificity of the interactions was ascertained using CydX, as a small protein control, and MCP2117, as a membranous protein unrelated to respiratory systems. In both cases, interaction was seen with neither TorC nor TorE (Fig. 5). Altogether these data confirm that TorE and TorC can be organized as a complex as seen on hrCN gels, and explain why NapE can complement a torE mutation and not CydX.
Figure 5.
In vivo interaction of the proteins using bacterial two-hybrid system. Coding sequences of the studied genes were cloned in bacterial two-hybrid plasmids pT18 and pT25. Recombinants plasmids were introduced in E. coli BTH101 in different combinations as indicated on the graph. β-galactosidase activity was assayed on cells grown overnight in the presence of IPTG and was expressed in arbitrary units62.
Discussion
The cellular role of small proteins is still not fully understood. Concerning small proteins related to membranous complexes, they seem to act in the stabilization of the complex although direct experiments are sometimes lacking. However, in various cases like in the Photosystem II of Cyanobacteria, small proteins included in the complex were reported as involved essentially in the stabilization, the assembly, the dimerization or the photoprotection of the complex43. In the case of the K+ transporter system KdpABC of E. coli, it was described that the presence of the small protein KdpF is required for the complex activity in vitro, can be replaced by an increase of the lipid concentration in the sample and is not necessary for the activity in vivo, strongly suggesting that KdpF stabilizes the KdpABC complex in the membrane44. CydX is another example of a small protein involved in a membranous complex, CydAB, which is a respiratory system present in many bacteria. According to the bacterium, the role of CydX differs according to the authors from acting on the stability of the complex to a direct role in the quinol binding site of the complex or the coordination of a heme1,11,12.
Here we deciphered the role of TorE, the small protein involved in the Tor system that allows S. oneidensis growth on TMAO. It was established previously that TorE increases the efficiency of the electron transfer during TMAO reduction14. The present study shows that TorE stabilizes the pentahemic c-type cytochrome TorC in vivo by organizing a complex that contains 4 TorE and 4 TorC (Fig. 4). TorE forms multimers as seen on hrCN gel in the absence of TorC while in the presence of the cytochrome high levels of oligomerization are lost. This indicates that in vivo TorE interacts with TorC rather than forming homo-oligomeric clusters in the membrane. In contrast, TorC can only weakly oligomerize according to the results obtained in both hrCN gels. This indicates that TorE is necessary to support TorC-TorC interaction. Since the size of the complex in the membrane corresponds to four monomers of each subunit, there are at least two possibilities for the complex formation. One in which the tetramerization of TorE forms the core of the complex followed by the interaction of each TorE with one monomer of TorC; a second in which the four TorE-TorC dimers are made before being organized in the whole complex. In both cases, the final organization leads to a stronger anchorage of TorC to the membrane (Fig. 6).
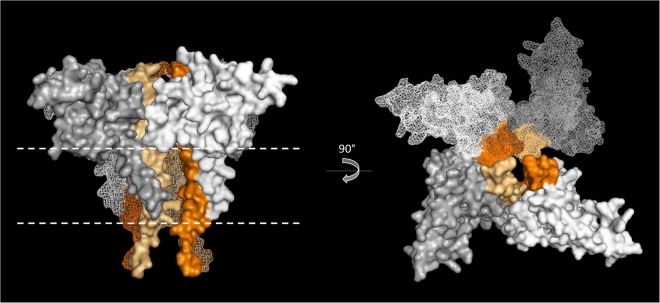
Figure 6.
Structure modelling of the TorC – TorE complex. TorC and TorE structures have been modelled with SWISS MODEL63–67 based on NrfH an PilN crystallographic structures68,71. The presented modeled complex is composed of 4 TorE -TorC heterodimers predicted by Memdock70 The TorC proteins are colored in gradient of grey and the TorE in a gradient of orange. Position of the membrane, determined by the PPM server69, was represented by dashed white lines.
The presence of the same highly conserved residues in the hydrophobic stretch of TorE and NapE and their localization on one side of the alpha helix suggest that this region is important probably for an interaction with a partner. This is in agreement with the experiment of complementation of a torE mutant with napE. The comparison of sequences of the hydrophobic N-terminal part of the various cytochromes TorC indicates that we can distinguish those that interact with TorE from the others by the presence of conserved amino acid residues. These residues are localized in one side of the alpha helix of TorC and in a loop located in its soluble part (Figure S4a and S4b). Using a modelling tool to construct a putative model of the TorE:TorC complex, it appears that the conserved motifs of TorE and TorC could be involved in the binding of the two proteins (Figures S4b and S4c). Using these results, a model of the 4TorE:4TorC complex is proposed (Fig. 6). However, this latter is just a static picture of the complex, one can imagine that the binding of the proteins leads to structural modifications that are not taking into account in Fig. 6. Interestingly, NapC sequence alignment indicates that the same conserved motif is found in all NapC proteins (Figure S4a). Although NapE is able to replace TorE, the growth curves are slightly different. This point suggests that the two proteins TorE and NapE have co-evolved with their partner leading to the sequence variations between them.
Since TorC is organized in two domains, one interacting with TorA and the second feeding the reductase TorA with electrons probably by a flip-flop movement, one can imagine that this stronger anchorage increases the efficiency of the electron transfer and that the TorE-TorC complex can transfer electrons to four TorA enzymes at the same time. Moreover, TorC transfers one electron at a time to TorA, while the reduction of TMAO by the molybdenum cofactor requires two electrons45. Another possibility could be that the 4TorE:4TorC complex introduces electrons to two TorA enzymes allowing the simultaneous transfer of two electrons to TorA. We suggest that one or the other of these organizations of the different components of the Tor system improve the electron transfer to TorA. Our conclusion is that the complex is made up of four monomers for each subunit. The hypothesis of the presence of another protein in the TorE:TorC complex was discarded in light of the results obtained. Indeed, as shown in Table 2, none of the various simulated stoichiometries is compatible with an additional component. This supports the hypothesis of a complex containing only a tetramer of two components TorE and TorC.
Recently, it was shown that, in E. coli, NarGHI complex that allows anaerobic growth on nitrate reduction was localized at the pole of the bacterium37. Since the clustering of enzymes involved in a same metabolism is described to increase its efficiency, we wondered if TorE could influence the location of the Tor respiratory system in a peculiar region of the inner membrane. Fluorescence microscopy experiments indicate clearly that the TorEC complex is evenly distributed all along the membrane and that the presence of TorE has no effect on the localization of TorC. Moreover, the presence of the terminal reductase TorA and of the substrate (TMAO) do not modify the distribution of the Tor system, meaning that its activity does not act on the location of the respiratory system contrary to what was observed with the NarGHI respiratory system of E. coli. Finally, since the NapEC complex and the cytochrome CymA present a similar homogenous distribution in the membrane and their terminal reductase is constitutively produced in anoxia, it seems that at least in S. oneidensis, the respiratory systems are not clustered in a special region of the membrane in anaerobiosis and that a quite simple complex is sufficient to support an efficient electron transfer.
In conclusion, this study unveils the role of TorE, a small protein present in the Tor respiratory system of various bacteria. TorE by stabilizing the anchoring of the system in the membrane increases the efficiency of TMAO respiration of S. oneidensis. By extending the study to NapE, we showed that TorE and NapE belong to the same family of small proteins involved in major respiratory systems. They are required to organize the protein complex that increases the anchorage of these systems to the membrane. A rapid adaptation to its environment and an improved energetic reactivity is of crucial importance for the bacterium especially when living in a competitive medium. We thus propose that in respiratory pathways, small proteins of this family are starter that organize the quaternary structuration of the system allowing a positive regulation of the catalytic activity at the complex level.
Methods
Synteny analysis
The TorE protein sequence from S. oneidensis was used as a query for a tBLASTn46 analysis in each of the groups of proteobacteria. This analysis allows to find annotated or unannotated torE and napE genes. The two homologs have been identified according to the presence of torC and napC genes in the torE genomic environment. The tor and nap operons that do not contain torE and napE, respectively, were identified using S. oneidensis TorC sequence as a query for another tBLASTn analysis. Data from both analyzes were manually compared. Degenerated sequences of torC and napC genes were also taken into account. The DNA sequences have then been translated using Emboss Transeq online tool47.
For construction of the phylogenic tree of proteobacteria, GyrB protein sequence from S. oneidensis was the query for a tBLASTn analysis of each of the groups of proteobacteria. Only sequences from complete genomes were used. The sequence of the best match was manually taken and translated using the Emboss Transeq online tool46. Only one random sequence per genus was kept for construction of the tree and the name of the genus only is indicated. The minimum evolution tree was constructed with the MEGA program48 using default parameters. The group of proteobacteria of each species was assigned according to the taxonomy option on the NCBI website49,50.
The Sequence Logo has been constructed using the Weblogo online tool51 with default parameters using the conserved E residues as first residue.
Helical projection
The helical projection of residues was carried out using the “Helical Wheel projection” online tool (default parameters) using the sequence of S. oneidensis TorE protein from residue 31 to 52. The conserved residues were selected according to the Sequence Logo of the different TorE and NapE proteins.
Bacterial strains and growth conditions
The E. coli strains C60052 and 1047/pRK201353 used for conjugation were routinely grown in Lysogeny Broth (LB) medium at 37 °C.
All S. oneidensis strains used in this study are derivatives of strain MR1-R54 (referred as WT and used instead of MR-1 to allow counter-selection by rifampicin in the conjugation experiments) and were routinely grown at 28 °C in LB medium under agitation. The strains used in this article are listed in Table 3. Deletion of the torC gene was done as detailed previously55.
Table 3.
Strains and plasmids used in this study.
| Strains | Characteristics | Source |
|---|---|---|
| C600 | F- tonA21 thi-1 thr-1 leuB6 lacY1 glnV44 rfbC1 fhuA1 λ− | Ref.52 |
| Bth101 | F′ cya-99 araD139 galE15 galK16 rpsL1 (StrR) hsdR2 mcrA1 mcrB1 relA1 | Ref.60 |
| MR1-R | S. oneidensis, wild type strain, Rif R | Ref.54 |
| ΔtorE | MR1-R strain deleted of torE gene. | Ref.14 |
| ΔtorC | MR1-R strain deleted of torC gene. | This study |
| ΔtorEC | MR1-R strain deleted of torEC genes. | This study |
| ΔcymA | MR1-R strain deleted of cymA gene. | This study |
| Plasmids | Characteristics | Source |
| pBAD33 | Vector containing pBAD promoter with a p15A origin of replication | Ref.72 |
| pTorE | Coding sequence of the S. oneidensis TorE inserted in the pBAD33 vector | Ref.14 |
| pNapE | Coding sequence of the S. massilia NapE inserted in the pBAD33 vector | This study |
| pCydX | Coding sequence of the S. oneidensis CydX inserted in the pBAD33 vector | This study |
| pGFP-TorE | Coding sequence of the sfGFP fused to the S. oneidensis TorE with GSGGSG spanner and inserted in the pBAD33 vector | This study |
| pGFP-TorC | Coding sequence of the sfGFP fused to the S. oneidensis TorC and inserted in the pBAD33 vector | This study |
| pGFP-TorE; mCherry-TorC | Coding sequence of the sfGFP fused to the S. oneidensis TorE with GSGGSG spanner and and coding sequence of the mCherry fused to the S. oneidensis TorC with GSGGSG spanner, inserted in the pBAD33 vector | This study |
| pGFP-TorC; mCherry-TorE | Coding sequence of the sfGFP fused to the S. oneidensis TorC with GSGGSG spanner and and coding sequence of the mCherry fused to the S. oneidensis TorE with GSGGSG spanner, inserted in the pBAD33 vector | This study |
| pGFP-NapC | Coding sequence of the sfGFP fused to the S. massilia NapC with GSGGSG spanner and inserted in the pBAD33 vector | This study |
| pGFP-NapC; NapE | Coding sequence of the sfGFP fused to the S. massilia NapC with GSGGSG spanner and coding sequence of S. massilia NapE inserted in the pBAD33 vector | This study |
| pGFP-NapC; mCherry-NapE | Coding sequence of the sfGFP fused to the S. massilia NapC with GSGGSG spanner and coding sequence of mCherry fused to S. massilia NapE with GSGGSG spanner inserted in the pBAD33 vector | This study |
| pGFP-CymA | Coding sequence of the sfGFP fused to the S. oneidensis TorE and inserted in the pBAD33 vector | This study |
| pEC86 | CcmABCDEFGH genes from E. coli cloned in the pACYC184 vector | Ref.73 |
| pEB355 | pUT18C derivative, coding for the T18 domain of the adenylate cyclase of B. pertussis | Ref.60 |
| pT18 TorE | Sequence coding for S. oneidensis TorE cloned in frame at the 3 extremity of the sequence coding for the T18 domain into pEB355 | This study |
| pT18 TorC | Sequence coding for S. oneidensis TorC cloned in frame at the 3 extremity of the sequence coding for the T18 domain into pEB355 | This study |
| pT18 TorCm | Sequence coding for the 1- amino acids of S. oneidensis TorC cloned in frame at the 3 extremity of the sequence coding for the T18 domain into pEB355 | This study |
| pT18 MCP | Sequence coding for the SO_2117 MCP from S. oneidensis cloned in frame at the 3 extremity of the sequence coding for the T18 domain into pEB355 | This work |
| pT18 CydX | Sequence coding for S. oneidensis CydX cloned in frame at the 3 extremity of the sequence coding for the T18 domain into pEB355 | This work |
| pT18 napE | Sequence coding for S. massilia NapE cloned in frame at the 3 extremity of the sequence coding for the T18 domain into pEB355 | This work |
| pEB354 | pKT25 derivative, coding for the T25 domain of the adenylate cyclase of B. pertussis | Ref.60 |
| pT25 TorE | Sequence coding for S. oneidensis TorE cloned in frame at the 3 extremity of the sequence coding for the T25 domain into pEB354 | This work |
| pT25 torCm | Sequence coding for the 1- amino acids of S. oneidensis TorC cloned in frame at the 3 extremity of the sequence coding for the T25 domain into pEB354 | This work |
| pT25 NapE | Sequence coding for S. massilia NapE cloned in frame at the 3 extremity of the sequence coding for the T25 domain into pEB354 | This work |
| pT25 NapC | Sequence coding for S. massilia NapC cloned in frame at the 3 extremity of the sequence coding for the T25 domain into pEB354 | This work |
When required, media were solidified by adding 17 g.L−1 agar. When needed, chloramphenicol and rifampicin were used at 25 μg.mL–1 and 10 μg.mL–1, respectively. Plasmid induction was carried out with routinely 0.02% arabinose. When indicated, TMAO (40 mM), fumarate (20 mM) and KNO3 (2 mM) were added in the medium as alternative electron acceptors. Except in bacterial two-hybrid experiments, all growths were performed in a medium buffered with MOPS (25 mM) to avoid alkalization by TMA production. Arabinose was added in the growth media for induction of WT/pBAD, ΔtorE/pBAD, ΔtorE/pTorE at 0.02% and for induction of plasmids ΔtorE/pNapE and ΔtorE/pCydX at 0.2%.
Plasmid construction
All PCR amplifications were performed with Q5 DNA polymerase (New England Biolabs), from S. oneidensis MR1-R chromosomal DNA for torE, torC, cymA, cydX and mcp2117 and S. massilia15 chromosomal DNA for napC, napE. For fluorescence experiments, the primers contain a common spanner sequence containing ggaagtggaggaagtgga at its 5′-ends. Genes coding for mCherry and sfGFP were amplified from plasmids with complementary spanner sequence at their 3′ ends. Chimeric sequences were constructed by a second PCR by mixing an equimolar amount of the corresponding PCR products and convergent primers. The gene was cloned between the SalI and the SphI restriction sites of the pBAD33 vector, under the control of the inducible Para promoter. The resulting plasmids were introduced into strain C60052 and controlled by DNA-sequencing before conjugation in S. oneidensis strains. When two genes were carried by the plasmid, a gene amplification was performed and the gene of interest was cloned between XmaI and XbaI restriction sites. The plasmids used in this article are listed in Table 3.
Fluorescence microscopy
For fluorescence microscopy, cells were grown anaerobically to mid-exponential phase at 28 °C. Living cells are then shifted in buffer and 2 μl was mounted on microscope slide and coverslip. Images were acquired with a Nikon TiE PFS inverted epifluorescence microscope (100 × oil objective NA 1.45 Phase Contrast) and a Hamamatsu Flash4 sCMOS camera. Images were collected with NIS elements software.
Solubilization experiments
Cells were grown anaerobically at 28 °C in a medium supplemented with MOPS and TMAO in 1L-bootles until exponential phase. Membranes were prepared as previously described14 and washed with 20 mM phosphate buffer pH 7.4, 2% glycerol (Buffer A) in the presence of protease inhibitors and re-ultracentrifuged at 110,000 g for 1 h. Pellets were re-suspended in Buffer A supplemented either with 1.5 M NaBr salt incubated for 30 min or with mild nonionic detergent n-dodecyl-β-D-maltoside (DDM) in the same buffer A with a DDM/protein ratio of 2 (g/g) incubated for 20 min and ultracentrifuged. All steps were done at 4 °C. NaBr supernatant was dialyzed using Slide-A-Lyser Dialysis Cassette 3500 MWCO (Thermo Scientific) against Buffer A supplemented with 0.02% DDM. All chemicals were purchased from SIGMA-Aldrich.
Protein concentrations were determined with Lowry modified assay using bovine serum albumin as standard56.
For c-type cytochromes detection, unheated membrane and solubilized fractions (10 µg) were submitted to 3–14% SDS-PAGE. Apparent molecular masses were estimated using protein ladder (Lonza ProSieve QuadColor). Proteins were transferred onto nitrocellulose membranes with Pierce Power Blotter (Thermo Scientific) using Mixed Range Molecular Weight program. After a one –hour saturation with PBS 3% BSA, the peroxidase activity of hemes was revealed with the SuperSignal West Pico Kit (Thermo Scientific) on Image Quant Las4000 apparatus (GE Healthcare).
High resolution Clear Native Electrophoresis (hrCNE) and two dimensional (2D) hrCNE/SDS-PAGE
Conserved frozen DDM-solubilized supernatants were supplemented with the same loading buffer used for Blue Native electrophoresis (BNE)57 except for the Coomassie Blue G250 dye which was omitted avoiding the quenching of fluorescent dye protein. 20% Glycerol was added to each sample and 0.001% Ponceau S serves as a marker for the front of the gel. To preserve oligomeric states and avoid protein aggregation during electrophoresis, anionic detergent sodium deoxycholate (DOC) 0.05% was added to the cathode buffer (50 mM Tricine, 7.5 mM imidazole/pH 7, 0.02% DDM). Anode buffer was 25 mM imidazole/HCl pH758. hrCN were carried out using a 3–14% linear polyacrylamide gradient and gels were run with constant current (10 mA) for 3 hours (Mini-Protean II; Bio-Rad). After electrophoresis, protein bands were visualized by fluorescence then by c type heme detection and finally with PageBlue (Fermentas) and cut out from the gel for mass spectrometry analysis.
For two-dimensional hrCNE/SDS-PAGE, a hrCN lane was cut out of the gel, washed three times for 10 min with migration buffer (Tris 25 mM, Glycine 192 mM) supplemented with 2% SDS and 0.1% 2-mercaptoethanol then washed in the same buffer without any SDS and 2-mercaptoethanol and applied on the top of 12% SDS-PAGE. After electrophoresis, gels were scanned using the Typhoon FLA9500 laser scanner (GE Healthcare) with excitation at 473 and 535 nm, and filter LPB and LPG (GE Healthcare) for GFP and mCherry, respectively.
Mass spectrometry
Trypsin digestion and peptide mass fingerprints by MALDI-ToF (Matrix Assisted Laser Desorption Ionisation-Time-of-Flight) were performed as previously described59. LC-MSMS analyses (Liquid chromatography coupled to tandem mass spectrometry) (Table 1) were performed on a Q-Exactive plus mass spectrometer (ThermoFisher Scientific, Villebon sur Yvette, France) as previously described59.
Bacterial Two-Hybrid experiment
Plasmids used for two-hybrid experiments are listed in Table 3. Bacterial two-hybrid experiments were performed as described60,61 with the following modifications. The PCR fragments were cloned into pEB354 and pEB355 vectors, using the EcoRI and XhoI restriction sites, in frame at the 3′-extremity of the sequences coding for the T18 and T25 domains of the adenylate cyclase from B. pertussis. For interaction experiments, combination of plasmids coding for the T18 and T25 fusions were transformed into BTH101 strains, and the plates (LB agar, kanamycin (50 μg/ml), and ampicillin (100 μg/ml)) were incubated at 30 °C for 2 days. Ten isolated colonies from each plates were used to inoculate 3 ml of LB containing kanamycin (50 μg/ml), ampicillin (100 μg/ml), and isopropyl β-D-thiogalactopyranoside (0.5 mM). After overnight growth at 30 °C, β-galactosidase activity was measured as described before62
Protein structure modelling, protein docking prediction and model construction
The S. oneidensis TorC and TorE structures were predicted using the SWISS model tool, the structures used being the NrfH cytochrome from Desulfovibrio vulgaris Hildenborough and the transmembrane part of PilN from Myxococcus xanthus, respectively63–68. The TorC and TorE structures were predicted from residue 23 to residue 184 and from residue 1 to residue 50, respectively. The membrane position was determined in TorC and TorE structure models using the PPM server using default parameters69. Membrane docking prediction was determined using the Memdock server, also using default parameters70. The model has been constructed using the PyMOL Molecular Graphics system.
Electronic supplementary material
Acknowledgements
We would like to thank Wolfgang Nitschke for fruitful discussions and support on this work. We also thank the BIP03 team and especially Olivier Genest and Sandra Angelini for interesting discussions. We are grateful to Regine Lebrun from The Proteomic Platform of the Mediterranean Institute of Microbiology (IMM) that is part of MaP (Marseille Protéomique, IBiSA) for mass determination and the “Cascales-lab” for providing GFP and mCherry containing vectors. We acknowledge Kevin Copp for reviewing of the paper. This work was supported by the Centre National de la Recherche Scientifique (CNRS) and Aix-Marseille Université (AMU). O.N. L. was supported by a MESR fellowship, AMU and CNRS. This study was supported by the “BIP Project Funding”. We thank the Aix-Marseille Université for supporting S.L. with a visiting professor position.
Author Contributions
C.I.N., O.N.L. conception and design of this study; O.N.L., P.I. data acquisition; O.N.L., A.A.C. strain constructions; O.N.L., L.E. microscopy experiments; all authors analysis and interpretation of the data; C.I.N. writing of the manuscript; V.M., S.L., M.T.G.O., O.N.L., P.I. critical revision of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31851-2.
References
- 1.Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annu. Rev. Biochem. 2014;83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J. Bacteriol. 2003;185:1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/S0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 4.Miyashiro T, Goulian M. Stimulus-dependent differential regulation in the Escherichia coli PhoQ PhoP system. Proc. Natl. Acad. Sci. USA. 2007;104:16305–16310. doi: 10.1073/pnas.0700025104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters LS, Sandoval M, Storz G. The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J. Bacteriol. 2011;193:5887–5897. doi: 10.1128/JB.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smaldone GT, Antelmann H, Gaballa A, Helmann JD. The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases. J. Bacteriol. 2012;194:2586–2593. doi: 10.1128/JB.05567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, et al. Increasing intracellular magnesium levels with the 31-amino acid MgtS protein. Proc. Natl. Acad. Sci. USA. 2017;114:5689–5694. doi: 10.1073/pnas.1703415114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y-H, et al. The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front. Cell. Infect. Microbiol. 2012;2:47. doi: 10.3389/fcimb.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.VanOrsdel CE, et al. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J. Bacteriol. 2013;195:3640–3650. doi: 10.1128/JB.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoeser J, Hong S, Gehmann G, Gennis RB, Friedrich T. Subunit CydX of Escherichia coli cytochrome bd ubiquinol oxidase is essential for assembly and stability of the di-heme active site. FEBS Lett. 2014;588:1537–1541. doi: 10.1016/j.febslet.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Luo Q, Yin J, Gao T, Gao H. Evidence for the requirement of CydX in function but not assembly of the cytochrome bd oxidase in Shewanella oneidensis. Biochim. Biophys. Acta. 2015;1850:318–328. doi: 10.1016/j.bbagen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Allen RJ, et al. Conservation analysis of the CydX protein yields insights into small protein identification and evolution. BMC Genomics. 2014;15:946. doi: 10.1186/1471-2164-15-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alix E, Blanc-Potard A-B. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 2008;27:546–557. doi: 10.1038/sj.emboj.7601983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemaire ON, et al. Efficient respiration on TMAO requires TorD and TorE auxiliary proteins in Shewanella oneidensis. Res. Microbiol. 2016;167:630–637. doi: 10.1016/j.resmic.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Dos Santos JP, Iobbi-Nivol C, Couillault C, Giordano G, Méjean V. Molecular analysis of the trimethylamine N-oxide (TMAO) reductase respiratory system from a Shewanella species. J. Mol. Biol. 1998;284:421–433. doi: 10.1006/jmbi.1998.2155. [DOI] [PubMed] [Google Scholar]
- 16.Yancey PH, Rhea MD, Kemp KM, Bailey DM. Trimethylamine oxide, betaine and other osmolytes in deep-sea animals: depth trends and effects on enzymes under hydrostatic pressure. Cell. Mol. Biol. Noisy–Gd. Fr. 2004;50:371–376. [PubMed] [Google Scholar]
- 17.Yancey PH, Gerringer ME, Drazen JC, Rowden AA, Jamieson A. Marine fish may be biochemically constrained from inhabiting the deepest ocean depths. Proc. Natl. Acad. Sci. USA. 2014;111:4461–4465. doi: 10.1073/pnas.1322003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J, Pazos IM, Gai F. Microscopic insights into the protein-stabilizing effect of trimethylamine N-oxide (TMAO) Proc. Natl. Acad. Sci. USA. 2014;111:8476–8481. doi: 10.1073/pnas.1403224111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvestro A, Pommier J, Pascal MC, Giordano G. The inducible trimethylamine N-oxide reductase of Escherichia coli K12: its localization and inducers. Biochim. Biophys. Acta. 1989;999:208–216. doi: 10.1016/0167-4838(89)90220-3. [DOI] [PubMed] [Google Scholar]
- 20.Proctor LM, Gunsalus RP. Anaerobic respiratory growth of Vibrio harveyi, Vibrio fischeri and Photobacterium leiognathi with trimethylamine N-oxide, nitrate and fumarate: ecological implications. Environ. Microbiol. 2000;2:399–406. doi: 10.1046/j.1462-2920.2000.00121.x. [DOI] [PubMed] [Google Scholar]
- 21.Méjean V, et al. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol. Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 22.Berks BC, Richardson DJ, Reilly A, Willis AC, Ferguson SJ. The napEDABC gene cluster encoding the periplasmic nitrate reductase system of Thiosphaera pantotropha. Biochem. J. 1995;309:983–992. doi: 10.1042/bj3090983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gon S, Giudici-Orticoni MT, Méjean V, Iobbi-Nivol C. Electron transfer and binding of the c-type cytochrome TorC to the trimethylamine N-oxide reductase in Escherichia coli. J. Biol. Chem. 2001;276:11545–11551. doi: 10.1074/jbc.M008875200. [DOI] [PubMed] [Google Scholar]
- 24.Shaw AL, Leimkuhler S, Klipp W, Hanson GR, McEwan AG. Mutational analysis of the dimethylsulfoxide respiratory (dor) operon of Rhodobacter capsulatus. Microbiol. Read. Engl. 1999;145(Pt 6):1409–1420. doi: 10.1099/13500872-145-6-1409. [DOI] [PubMed] [Google Scholar]
- 25.Ilbert M, Méjean V, Giudici-Orticoni M-T, Samama J-P, Iobbi-Nivol C. Involvement of a mate chaperone (TorD) in the maturation pathway of molybdoenzyme TorA. J. Biol. Chem. 2003;278:28787–28792. doi: 10.1074/jbc.M302730200. [DOI] [PubMed] [Google Scholar]
- 26.Genest O, et al. Dedicated metallochaperone connects apoenzyme and molybdenum cofactor biosynthesis components. J. Biol. Chem. 2008;283:21433–21440. doi: 10.1074/jbc.M802954200. [DOI] [PubMed] [Google Scholar]
- 27.Leimkühler S, Iobbi-Nivol C. Bacterial molybdoenzymes: old enzymes for new purposes. FEMS Microbiol. Rev. 2016;40:1–18. doi: 10.1093/femsre/fuv043. [DOI] [PubMed] [Google Scholar]
- 28.Iobbi-Nivol C, Leimkühler S. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim. Biophys. Acta. 2013;1827:1086–1101. doi: 10.1016/j.bbabio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Bjellqvist B, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 30.Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–539. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- 31.Wilkins MR, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. Clifton NJ. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 32.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 33.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 34.Reyes F, Gavira M, Castillo F, Moreno-Vivián C. Periplasmic nitrate-reducing system of the phototrophic bacterium Rhodobacter sphaeroides DSM 158: transcriptional and mutational analysis of the napKEFDABC gene cluster. Biochem. J. 1998;331(Pt 3):897–904. doi: 10.1042/bj3310897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez D, Koch G. Exploring functional membrane microdomains in bacteria: an overview. Curr. Opin. Microbiol. 2017;36:76–84. doi: 10.1016/j.mib.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borrero-de Acuña JM, Timmis KN, Jahn M, Jahn D. Protein complex formation during denitrification by Pseudomonas aeruginosa. Microb. Biotechnol. 2017;10:1523–1534. doi: 10.1111/1751-7915.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberge, F. et al. Dynamic subcellular localization of a respiratory complex controls bacterial respiration. eLife4 (2015). [DOI] [PMC free article] [PubMed]
- 38.Wei H, et al. Functional roles of CymA and NapC in reduction of nitrate and nitrite by Shewanella putrefaciens W3-18-1. Microbiol. Read. Engl. 2016;162:930–941. doi: 10.1099/mic.0.000285. [DOI] [PubMed] [Google Scholar]
- 39.Alves, M. N. et al. Characterization of the periplasmic redox network that sustains the versatile anaerobic metabolism of Shewanella oneidensis MR-1. Front. Microbiol. 6 (2015). [DOI] [PMC free article] [PubMed]
- 40.Schwalb C, Chapman SK, Reid GA. The membrane-bound tetrahaem c-type cytochrome CymA interacts directly with the soluble fumarate reductase in Shewanella. Biochem. Soc. Trans. 2002;30:658–662. doi: 10.1042/bst0300658. [DOI] [PubMed] [Google Scholar]
- 41.Gao H, et al. Impacts of Shewanella oneidensis c-type cytochromes on aerobic and anaerobic respiration. Microb. Biotechnol. 2010;3:455–466. doi: 10.1111/j.1751-7915.2010.00181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iobbi-Nivol C, et al. A reassessment of the range of c-type cytochromes synthesized by Escherichia coli K-12. FEMS Microbiol. Lett. 1994;119:89–94. doi: 10.1111/j.1574-6968.1994.tb06872.x. [DOI] [PubMed] [Google Scholar]
- 43.Shi L-X, Schröder WP. The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochim. Biophys. Acta. 2004;1608:75–96. doi: 10.1016/j.bbabio.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Gassel M, Möllenkamp T, Puppe W, Altendorf K. The KdpF subunit is part of the K( + )-translocating Kdp complex of Escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem. 1999;274:37901–37907. doi: 10.1074/jbc.274.53.37901. [DOI] [PubMed] [Google Scholar]
- 45.Kaufmann P, et al. Modulating the Molybdenum Coordination Sphere of Escherichia coli Trimethylamine N-Oxide Reductase. Biochemistry (Mosc.) 2018;57:1130–1143. doi: 10.1021/acs.biochem.7b01108. [DOI] [PubMed] [Google Scholar]
- 46.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. TIG. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Appleyard RK. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herrero M, de Lorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bordi C, Théraulaz L, Méjean V, Jourlin-Castelli C. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 2003;48:211–223. doi: 10.1046/j.1365-2958.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- 55.Honoré FA, Méjean V, Genest O. Hsp90 Is Essential under Heat Stress in the Bacterium Shewanella oneidensis. Cell Rep. 2017;19:680–687. doi: 10.1016/j.celrep.2017.03.082. [DOI] [PubMed] [Google Scholar]
- 56.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 57.Schägger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 58.Wittig I, Karas M, Schägger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics MCP. 2007;6:1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Boughanemi, S. et al. Microbial oxidative sulfur metabolism: biochemical evidence of the membrane-bound heterodisulfide reductase-like complex of the bacterium Aquifex aeolicus. FEMS Microbiol. Lett. 363 (2016). [DOI] [PubMed]
- 60.Battesti A, Bouveret E. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods San Diego Calif. 2012;58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 61.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller, J. H. Experiments in molecular genetics. (Cold Spring Harbor Laboratory, 1972).
- 63.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 64.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinforma. Oxf. Engl. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bienert S, et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45:D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017;7:10480. doi: 10.1038/s41598-017-09654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodrigues ML, Oliveira TF, Pereira IAC, Archer M. X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J. 2006;25:5951–5960. doi: 10.1038/sj.emboj.7601439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hurwitz N, Schneidman-Duhovny D, Wolfson HJ. Memdock: an α-helical membrane protein docking algorithm. Bioinforma. Oxf. Engl. 2016;32:2444–2450. doi: 10.1093/bioinformatics/btw184. [DOI] [PubMed] [Google Scholar]
- 71.Chang Y-W, et al. Architecture of the type IVa pilus machine. Science. 2016;351:aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arslan E, Schulz H, Zufferey R, Künzler P, Thöny-Meyer L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 1998;251:744–747. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.