Abstract
Background
Recent studies showed preliminary data on associations of MRI-based imaging phenotypes of breast tumours with breast cancer molecular, genomic, and related characteristics. In this study, we present a comprehensive analysis of this relationship.
Methods
We analysed a set of 922 patients with invasive breast cancer and pre-operative MRI. The MRIs were analysed by a computer algorithm to extract 529 features of the tumour and the surrounding tissue. Machine-learning-based models based on the imaging features were trained using a portion of the data (461 patients) to predict the following molecular, genomic, and proliferation characteristics: tumour surrogate molecular subtype, oestrogen receptor, progesterone receptor and human epidermal growth factor status, as well as a tumour proliferation marker (Ki-67). Trained models were evaluated on the set of the remaining 461 patients.
Results
Multivariate models were predictive of Luminal A subtype with AUC = 0.697 (95% CI: 0.647–0.746, p < .0001), triple negative breast cancer with AUC = 0.654 (95% CI: 0.589–0.727, p < .0001), ER status with AUC = 0.649 (95% CI: 0.591–0.705, p < .001), and PR status with AUC = 0.622 (95% CI: 0.569–0.674, p < .0001). Associations between individual features and subtypes we also found.
Conclusions
There is a moderate association between tumour molecular biomarkers and algorithmically assessed imaging features.
Subject terms: Breast cancer, Predictive markers
Introduction
Radiogenomic1 (a.k.a. imaging-genomic) analysis of breast cancer, which investigates the relationship between breast tumour imaging characteristics and tumour molecular, genomic, proliferation, and related features, has been gaining significant interest in recent years.2–18 Establishing a strong relationship between tumour imaging phenotypes and molecular markers could provide a non-invasive surrogate means of genomic analysis. This could be done by using a non-invasive imaging signature instead of a genomic signature that requires invasive tissue sampling. Otherwise, these relationships could help identify groups of patients that may benefit from additional genomic analysis.
Previous studies2,3,5,7–16,18 have demonstrated the potential for radiogenomic associations in breast cancer, predominantly associations of MR imaging features with molecular subtype or gene assays to predict cancer recurrence. However, as shown in Table 1, the majority of the prior studies on breast radiogenomics have used moderate sample sizes (most studies to date used <100 subjects) and small numbers of imaging features (typically <100). Also, only two prior studies have used an independent validation set to study the predictive ability of their imaging features.3,18 Consequently, the reported strengths of the imaging associations have varied widely. For example, for discriminating triple-negative breast cancer versus other subtypes, area under the curve (AUC) values of 0.92 in a study,3 0.79 in another study,18 0.78 in a different study,11 and 0.67 in another study17 were reported. Furthermore, each study uses a different, often very limited, set of imaging features which renders the comparison of specific results infeasible.
Table 1.
Prior studies reporting association of breast MR imaging features and genomic characteristics
| First author, year and reference | Number of imaging features from MR | Number of patients (dataset information: S for single and M—for multiple institutions) | Breast cancer related principal research question |
|---|---|---|---|
| Uematsu et al.9 | 9 | 176 (S) | Correlation of Imaging features and pathologic findings in TNBC and non-TNBC |
| Costantini et al.14 | 14 | 225 (S) | Comparison of imaging features of TNBC and non-TNBC |
| Yamamoto et al.7 | 26 | 10 (M) | Association of imaging features and interferon breast cancer subtype |
| Sung et al.8 | 7 | 321 (S) | Comparison of imaging features of TNBC and non-TNBC |
| Agner et al.13 | 120 | 65 (S) | Imaging features for predicting TNBC and other cancers |
| Mazurowski et al.15 | 23 | 48 (M) | Association of imaging features and molecular subtypes |
| Blaschke et al.10 | 6 | 112 (S) | Association of imaging features and molecular subtypes |
| Grimm et al.6 | 56 | 275 (S) | Association of imaging features and molecular subtypes |
| Guo et al.12 | 38 | 91 (M) | Integrated radiomics and genomics data to predict clinical phenotypes |
| Wang et al.11 | 85 | 84 (M) | Association of imaging features of background parenchymal enhancement and TNBC |
| Yamaguchi et al.16 | 5 | 186 (S) | Association of imaging features and molecular subtypes |
| Li et al.17 | 37 | 91 (M) | Association of imaging features and molecular subtypes |
| Fan et al.3 | 88 | 96 (S) | Association of imaging features and molecular subtypes |
| Wu et al.18 | 35 | 210 (M) | Association of imaging features and molecular subtypes |
TNBC triple negative breast cancer
In this study, we address this issue by presenting a comprehensive study of associations of tumour surrogate molecular subtype, receptor status, and proliferation of a set of 922 patients and 529 MR imaging features. The dataset’s heterogeneous imaging parameters (manufacturer, magnetic field strength, acquisition parameters) ensure that the results are not limited to a very specific MR setting. The set of features used in this study was constructed to represent a wide range of imaging characteristics including size, shape, texture, and enhancement of both the tumour and the surrounding tissue. We included both features published in the literature as well as those developed in our laboratory and developed machine learning-based multivariate models to test the effectiveness of these features.
Materials and methods
Patient population
In this local institutional board approved study, we identified 1150 consecutive female patients, from 1 January 2000 to 23 March 2014 with invasive breast cancer and available pre-operative MRI at our institution. A waiver for informed consent was also secured for this study. Patients with prior breast surgery, history of breast cancer, or neoadjuvant therapy prior to the MRI acquisition were excluded. From these patients, 922 patients were selected for our study using the specific criteria shown in Fig. 1. These 922 patients included 271 patients that were used in our earlier much more limited analyses6,19 which in addition to analysing a notably smaller cohort, did not investigate the associations of MR imaging features with ER, PR, and HER2 status, and proliferation marker (Ki-67). Our previous studies with the subset of this cohort also did not validate the findings on an independent test set.
Fig. 1.
Flowchart of inclusion and exclusion criteria for patients
Pathology data
Pathology results from the first immunohistochemistry (IHC) analysis, or the clinician’s note if not available (n = 65), were reviewed for the ER, PR, and H2 status. An Allred score from the IHC greater than or equal to 3+ was considered positive for ER and PR. For the determination of HER2 status, an IHC HER2 score of 3+, or a score of 2+ with an additional condition of amplification of HER2 gene by FISH (PathVysion Her2 DNA Probe kit, Abbott Laboratories, Chicago, IL)6 was considered positive. Following the criteria described in earlier publications,20,21 the surrogate molecular subtype was determined as: Luminal A (ER and/or PR positive, HER2 negative), Luminal B (ER and/or PR positive, HER2 positive), HER2 (ER and PR negative, HER2 positive), triple-negative (ER, PR, and HER2 negative). We also recorded the proliferation marker (Ki-67) values from the initial IHC data, when available. Ki-67 was considered high if it was greater than 14 as in refs.22,23. Additionally, the majority (84%) of tumours were ductal, 10% were lobular, 4% belonged to other categories, and 4% did not have this data available.
Imaging data
For the patients included in our study, we collected axial breast MRIs that were acquired by 1.5T or 3T scanners in the prone position. Scanner related details and MR acquisition parameters can be found in Supplementary material (Tables S1 and S2, respectively). The following MRI sequences were available: a non-fat saturated T1-weighted sequence, a fat-saturated gradient echo T1-weighted pre-contrast sequence, and typically four post-contrast T1-weighted sequences acquired after the IV administration of contrast agent (using a weight-based protocol of 0.2 mL/kg). In our cohort, three types of contrast agents were used as follows: gadobutrol (Gadavist, Bayer Healthcare, Berlin, Germany) for 2 (0.2%) patients, gadopentetate dimeglumine (Magnevist, Bayer Healthcare, Berlin, Germany) for 560 (60.8%) patients, and gadobenate dimeglumine (Multihance, Bracco, Milan, Italy) for 263 (28.5%) patients. The specific name of the contrast agent used was not available for 97 (10.5%) patients. The median acquisition time between a pair of post-contrast sequences was 131 s.
Each case was annotated by one of eight fellowship-trained breast imagers (1–22 years of post-fellowship experience, 3–32% of cases annotated by different readers). For each patient, a graphical user interface developed in our laboratory displayed to the reader the following MR sequences: (a) pre-contrast, (b) first post-contrast, and (c) subtracted (obtained by subtracting the pre-contrast from the first post-contrast). Tumours were delineated by three-dimensional boxes provided by the reader.
Image segmentation
Using a reader’s annotation (3D box), we applied a fuzzy C-means automatic segmentation24 to obtain the tumour mask. The breast and fibroglandular tissue masks were automatically extracted from the N4-corrected25 T1-non-fat saturated (T1-NFS) images and first post-contrast sequences. Thus, we had four masks extracted for each patient: (a) tumour (semi-automatic) (b) breast mask (automatic) (c) fibroglandular tissue (FGT) mask from T1-NFS (automatic) (d) FGT mask from post-contrast sequence (automatic). We removed any tumour voxels overlapping with the FGT masks to arrive at the final FGT masks used for feature extraction.
Imaging feature extraction and organisation
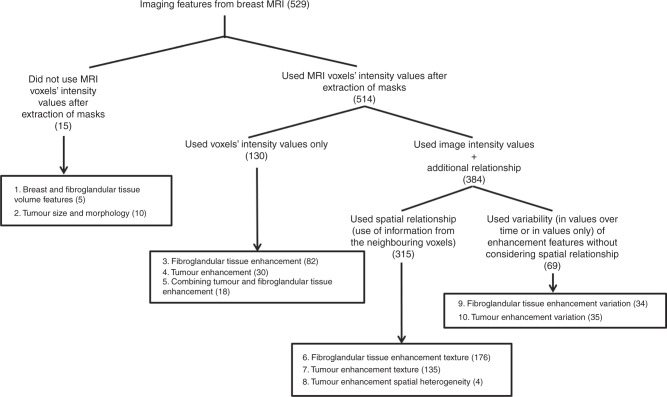
A review of the literature on breast MR image processing, computer-aided diagnosis, and radiomics guided our feature selection. The goal was to compile a comprehensive set of features that have been shown to be effective predictors and could quantify characteristics of the breast, tumour, and FGT. Based on the source and the nature of image processing represented by a feature, we categorised all features as shown in Fig. 2. Based on the type of the feature extracted, different subsets of the available MR sequences were used (all of them if necessary).
Fig. 2.
Feature distribution as per the different groups
Features were derived from the following sources: (a) features that capture the properties of breast as a whole are in category 1, (b) categories 2, 4, 7, 8 and 10 quantify characteristics of the tumour, (c) characteristics of FGT are expressed by features included in categories 1, 3, 6 and 9, and (d) category 5 represents features that capture the properties of tumour and FGT enhancement simultaneously. As per the image processing, (a) features in categories 1 and 2 do not require the voxel intensity values after the extraction of the corresponding masks (b) features in categories 3, 4 and 5 use voxel intensity values for capturing tumour/tissue enhancement but the spatial relationship of the voxels are not explored, (c) features in categories 6, 7 and 8 exploit the spatial relationship of the voxels while quantifying enhancement (d) features in categories 9 and 10 use variation in enhancement over time or in values but does not use the spatial relationship.
The list of the features are available in the Appendix (Supplementary Material A) of our previous publication.26
Training and test sets
Half (461) of the available 922 patients were included in our training set and the remaining 461 were included in our test set. The training set included 271 patients that were used in our earlier analyses6 to minimise the potential bias that could arise by including such cases in the test set since we have worked with this subset of data previously. A random split of the remaining data was used to arrive at the equal split between the training set and the test set.
Model development and statistical analysis
The primary goal of this study was to evaluate whether machine learning-based multivariate models based on imaging features can distinguish between different tumour subtypes and can predict molecular, genomic, and proliferation markers such as ER status, PR status, HER2 status, and proliferation (Ki-67). We considered eight-specific prediction tasks corresponding to eight radiogenomics associations where an imaging-based model aims to distinguish: (1) Luminal A versus other subtypes, (2) Luminal B versus other subtypes, (3) HER2 versus other subtypes, (4) TNBC versus other subtypes, (5) ER positivity versus OR negativity, (6) PR positivity versus PR negativity, (7) HER2 positivity versus HER2 negativity, and (8) high Ki-67 versus low Ki-67.
A multivariate model for each of these 8 tasks was developed using the training set in a following manner. First, N features with the highest area under the receiver operating characteristic (ROC) curve (AUC) were selected using the corresponding feature as the independent variable and the binary label pertinent to the task as the dependent variable. Then features with high absolute value of correlation (>c) with other features were removed from the feature set to form a pre-selected feature set. Then a random forest classifier was trained using the pre-selected set of features. The parameters of the random forest classifier (number of features that are selected for each tree and number of cases included in each tree leaf) as well as the parameters N and c described above were selected through a cross-validation experiment within the training set in which the training and evaluation were repeated multiple times for a range of values of the four parameters. The parameters that provided the highest cross-validation AUC were selected. This procedure was repeated separately for each of the 8 classification tasks resulting in one final model trained using the training set for each of the tasks. Once the parameters were optimised and the models were developed using the training set, they were evaluated on the independent test set using area under the ROC curve metric. Confidence intervals were estimated using the DeLong method27 and the significance of the association was established using a logistic regression model using the classifier output as the covariate. A p-value <0.00625 (0.05/8 models) was considered significant. An additional analysis was conducted in subgroups formed in the test sets based on the scanner manufacturers (GE and Siemens), race (white and other races), and menopausal status (pre and post). The grouping within the race category was done in order to have a sufficient number of cases in each subgroup (some races have a very low representation in our cohort).
Furthermore, we conducted an exploratory analysis to establish which individual features showed the highest association with the molecular markers. Specifically, for all features, we computed the AUC for the eight prediction tasks on the training set. We sorted the features in descending order according to AUC and removed highly correlated features (|r| > 0.5) to select the top (N = 10) features for each of the eight prediction tasks. For tasks (1–4), we removed repeated occurrences of any feature to arrive at a set of 37 unique features and calculated the AUCs and confidence intervals for these selected features on the test set. For each of tasks (5–8) we computed AUCs and confidence intervals for 10 features.
All analysis, except for the computation of confidence interval was conducted in MATLAB, 2016b (MathWorks, Natick, Mass). The confidence intervals were computed in R, version 3.4.0 (http://www.r-project.org/).
Results
Table 2 shows the patient clinical characteristics. The distribution of patients according to molecular biomarkers, in the training and test set shown in Table 3 are very similar.
Table 2.
Clinicopathological characteristics of the overall patient population, by molecular subtypes, receptor status positivity and Ki-67 availability
| Patient characteristics | Entire cohort | Luminal A | Luminal B | HER2 | TNBC | ER positive | PR positive | HER2 positive | Ki-67 |
|---|---|---|---|---|---|---|---|---|---|
| Number of patients | 922 (100%) | 595 (64.53%) | 104 (11.27%) | 59 (6.39%) | 164 (17.79%) | 686 (74.40%) | 598 (64.86%) | 163 (17.68%) | 450 (48.81%) |
| Median age (age range) in years | 52.25 (21.75–89.49) | 53.61 (25.7–89.49) | 46.54 (29.78–79.52) | 51.92 (27.14–79.08) | 50.4 (21.75–80.70) | 52.82 (25.7–89.49) | 52.42 (25.7–89.49) | 48.38 (27.1–79.52) | 52.5 (23.98–80.46) |
| Race | |||||||||
| White | 651 (70.61%) | 442 (74.29%) | 75 (72.12%) | 36 (61.02%) | 98 (59.76%) | 510 (74.34%) | 453 (75.75%) | 111 (68.10%) | 332 (73.38%) |
| Black | 203 (22.02%) | 107 (17.98%) | 21 (20.19%) | 15 (25.42%) | 60 (36.59%) | 123 (17.93%) | 103 (17.22%) | 36 (22.09%) | 88 (19.56%) |
| Others* | 49 (5.31%) | 30 (5.04%) | 8 (7.69%) | 6 (10.17%) | 5 (3.05%) | 38 (5.54%) | 27 (4.52%) | 14 (8.59%) | 21 (4.67%) |
| Not available | 19 (2.06%) | 16 (2.69%) | 0 | 2 (3.39%) | 1 (0.61%) | 15 (2.19%) | 15 (2.51%) | 2 (1.23%) | 9 (2.00%) |
| Menopausal status | |||||||||
| Pre | 407 (44.14%) | 240 (40.34%) | 59 (56.73%) | 23 (38.98%) | 85 (51.83%) | 293 (42.71%) | 263 (43.98%) | 82 (50.31%) | 198 (44.00%) |
| Post | 499 (54.12%) | 344 (57.82%) | 43 (41.35%) | 36 (61.02%) | 76 (46.34%) | 380 (55.39%) | 323 (54.01%) | 79 (48.47%) | 249 (55.33%) |
| Not available | 16 (1.74%) | 11 (1.85%) | 2 (1.92%) | 0 | 3 (1.83%) | 13 (1.90%) | 12 (2.01%) | 2 (1.23%) | 3 (0.67%) |
| Tumour staging (size) 1 | |||||||||
| T1 | 409 (44.36%) | 289 (48.57%) | 41 (39.42%) | 16 (27.12%) | 63 (38.41%) | 327 (47.67%) | 292 (48.83%) | 57 (34.97%) | 194 (43.11%) |
| T2 | 395 (42.84%) | 234 (39.33%) | 51 (49.04%) | 32 (54.24%) | 78 (47.56%) | 278 (40.52%) | 244 (40.80%) | 83 (50.92%) | 194 (43.11%) |
| T3 | 90 (9.76%) | 57 (9.58%) | 8 (7.69%) | 9 (15.25%) | 16 (9.76%) | 63 (9.18%) | 47 (7.86%) | 17 (10.43%) | 50 (11.11%) |
| T4 | 22 (2.39%) | 11 (1.85%) | 4 (3.85%) | 0 | 7 (4.27%) | 14 (2.04%) | 12 (2.01%) | 4 (2.45%) | 9 (2.00%) |
| Not available | 6 (0.65%) | 4 (0.67%) | 0 | 2 (3.39%) | 0 | 4 (0.58%) | 3 (0.50%) | 2 (1.23%) | 3 (0.67%) |
ER oestrogen receptor, HER2 human epidermal growth factor, PR progesterone receptor, TNBC triple negative breast cancer. *Includes Asian, Native, Hispanic, Multi, Hawaiian, and American Indian
Table 3.
Distribution of patients in the training and test sets as per the molecular subtype, receptor status, and Ki-67 values
| Molecular marker | Details | Count in training set | Count in test set |
|---|---|---|---|
| Molecular subtype (NTRAIN = 461 NTEST = 461) | Luminal A | 305 (66.16%) | 290 (62.91%) |
| Luminal B | 47 (10.20) | 57 (12.36%) | |
| HER2 | 27 (5.86%) | 32 (6.94%) | |
| TNBC | 82 (17.79%) | 82 (17.79%) | |
| ER status (NTRAIN = 461 NTEST = 461) | Positive | 341 (73.97%) | 345 (74.84%) |
| Negative | 120 (26.03%) | 116 (25.16%) | |
| PR status (NTRAIN = 461 NTEST = 461) | Positive | 306 (66.38%) | 292 (63.34%) |
| Negative | 155 (33.62%) | 169 (36.66%) | |
| HER2 status (NTRAIN = 461 NTEST = 461) | Positive | 74 (16.05%) | 89 (19.31%) |
| Negative | 387 (83.95%) | 372 (80.69%) | |
| Ki-67 (NTRAIN = 246 NTEST = 204) | High | 153 (62.20%) | 155 (75.98%) |
| Low | 93 (37.80%) | 49 (24.02%) |
ER oestrogen receptor, HER2 human epidermal growth factor, PR progesterone receptor
The results from the multivariate models are presented in Table 4. Regarding molecular subtypes, the highest performance was obtained for the models distinguishing Luminal A from other subtypes with AUC = 0.697 (95% CI: 0.647–0.746, p < 1.24e−11) and TNBC from the other subtypes AUC = 0.654 (95% CI: 0.589–0.720, p < 1.42e−05). The performances for distinguishing HER2 from other subtypes and for Luminal B from other subtypes were somewhat lower and did not reach statistical significance (p = 0.03 and p = 0.13, respectively). Regarding individual molecular markers, the models showed significant prognostic value for distinguishing ER+ from ER− patients (p < 4.2e−06), PR+ from PR− patients (p < 1.93e−04). The model for predicting high vs low proliferation (Ki-67) showed AUC = 0.624 with a p-value on the margin of significance (p = 0.01).
Table 4.
AUC, CI, and p-values obtained for the trained multivariate models in the test set
| Name of the Task | AUC in test set with 95% CI | p-Value for the model in test set |
|---|---|---|
| Luminal A vs other subtypes | 0.697 (0.647–0.746) | <0.0001* |
| Luminal B vs other subtypes | 0.566 (0.494–0.638) | 0.13 |
| HER2 vs other subtypes | 0.633 (0.539–0.727) | 0.03 |
| TNBC vs other subtypes | 0.654 (0.589–0.720) | <0.0001* |
| ER positivity vs ER negativity | 0.649 (0.591–0.705) | <0.0001* |
| PR positivity vs PR negativity | 0.622 (0.569–0.674) | <0.001* |
| HER2 positivity vs HER2 negativity | 0.500 (0.433–0.567) | 0.81 |
| High Ki-67 vs low Ki-67 | 0.624 (0.531–0.718) | 0.01 |
AUC area under the curve, CI confidence interval, ER oestrogen receptor, HER2 human epidermal growth factor, PR progesterone receptor. *Statistically significant p < 0.00625
The results for the subgroup wise analysis using the test set are shown in Table 5. No notable and systematic differences in the performance of the trained models were observed in the subgroups for a majority of the tasks. A minor difference was found in the task of discriminating TNBC patients from other subtypes between the pre and post-menopausal cohorts. Some differences were observed in the discrimination of high Ki-67 from low Ki-67 for different scanner manufacturer, races, and menopausal status.
Table 5.
AUC and 95% CI obtained for the trained multivariate models in the test set divided into subsets by scanner manufacturer, race, and menopausal status
| Scanner manufacturer | Race | Menopausal status | ||||
|---|---|---|---|---|---|---|
| GE (N = 284) | Siemens (N = 177) | White (N = 322) | Other declared races (N = 129) | Pre (N = 216) | Post (N = 235) | |
| Luminal A vs other Subtypes | 0.701 (0.638–0.763) | 0.685 (0.605–0.765) | 0.708 (0.648–0.769) | 0.646 (0.55–0.741) | 0.652 (0.579–0.725) | 0.737 (0.668–0.807) |
| Luminal B vs other Subtypes | 0.579 (0.476–0.683) | 0.521 (0.41–0.632) | 0.560 (0.467–0.653) | 0.543 (0.427–0.659) | 0.585 (0.487–0.682) | 0.529 (0.417–0.640) |
| HER2 vs other Subtypes | 0.651 (0.534–0.768) | 0.625 (0.467–0.783) | 0.652 (0.527–0.776) | 0.620 (0.472–0.767) | 0.612 (0.469–0.548) | 0.660 (0.535–0.847) |
| TNBC vs other Subtypes | 0.671 (0.586–0.756) | 0.623 (0.52–0.727) | 0.658 (0.571–0.746) | 0.619 (0.511–0.726) | 0.667 (0.582–0.753) | 0.612 (0.496–0.727) |
| ER positivity vs ER negativity | 0.638 (0.565–0.712) | 0.670 (0.577–0.762) | 0.616 (0.542–0.691) | 0.669 (0.574–0.765) | 0.644 (0.564–0.724) | 0.644 (0.559–0.730) |
| PR positivity vs PR negativity | 0.616 (0.549–0.683) | 0.633 (0.549–0.717) | 0.611 (0.545–0.678) | 0.616 (0.516–0.717) | 0.627 (0.551–0.702) | 0.616 (0.542–0.690) |
| HER2 positivity vs HER2 negativity | 0.500 (0.408–0.593) | 0.495 (0.394–0.596) | 0.514 (0.428–0.599) | 0.505 (0.392–0.617) | 0.508 (0.412–0.604) | 0.500 (0.405–0.596) |
| High Ki-67 vs Low Ki-67* | 0.648 (0.536–0.760) | 0.590 (0.408–0.772) | 0.601 (0.495–0.708) | 0.725 (0.527–0.922) | 0.576 (0.417–0.734) | 0.640 (0.520–0.760) |
*Number of cases in each category is less than the mentioned counts in second row due to missing Ki-67 values
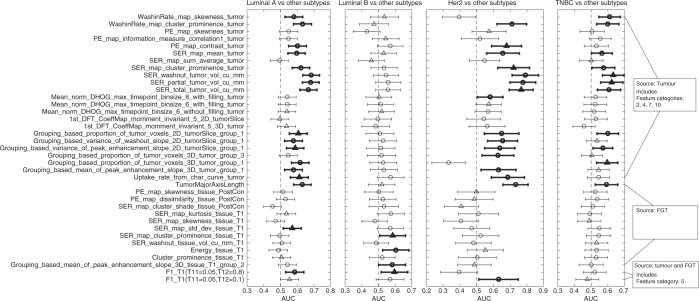
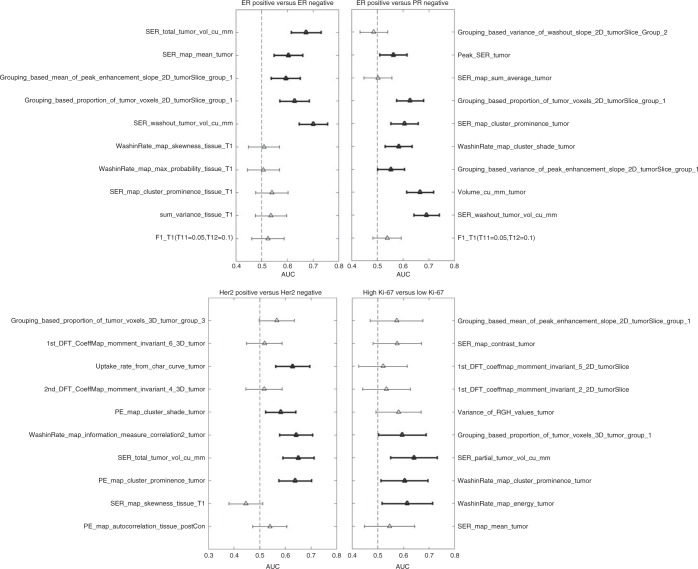
The results for the exploratory univariate analysis are presented in Figs. 3 and 4. The number of features with AUC confidence interval not overlapping with 0.5 was 18, 4, 18, and 11 for the tasks of distinguishing Luminal A, Luminal B, HER2, and TNBC from other subtypes, respectively. All but two of these features maintained their directionality as obtained from the training set. Except for 4 features, all of these features were extracted from the tumour only. Among these 4 features, 2 were extracted from FGT and 2 were extracted using both tumour and tissue, a result consistent with.15 The higher values of AUCs were obtained for discriminating HER2 molecular subtype versus others and Luminal A versus other subtypes.
Fig. 3.
The AUC (indicated by a circular or triangular marker) and confidence intervals (indicated by the endpoints of the lines cutting the marker) values for 37 selected features for prediction tasks 1–4 (indicated at the top of each column). The triangular marker indicates that the corresponding feature was among the 10 features selected from the training set for predicting the corresponding subtype versus others, otherwise the marker is circular. The bold lines indicate that the lower bound of the confidence interval is greater than an AUC of 0.5 and the feature maintained its directionality from the training set
Fig. 4.
The AUC (indicated by a triangular marker) and confidence intervals (indicated by the endpoints of the lines cutting the marker) values for prediction tasks 5–8 (indicated at the top of each column). The bold lines indicate that the lower bound of the confidence interval is >0.5 and the feature maintained its directionality from the training set
For ER positivity versus ER negativity, 5 features were found to have AUC higher than 0.5 for the lower bound of the confidence interval of AUC. This condition was met by 7, 5, and 4 features for PR positivity versus PR negativity, HER2 positivity versus negativity, and high Ki-67 versus low Ki-67, respectively. All of these features were extracted from tumours.
Discussion
In this study, we conducted a comprehensive radiogenomic analysis of breast cancer in the context of dynamic contrast enhancement MRI using a machine-learning-based approach. Motivated by the recent surge in the development of new features in breast MRI radiomics and a strong interest in breast cancer radiogenomics, we looked at the effectiveness and generalisability of the features available in the literature as well as features proposed by our group to predict receptor status, proliferation and, surrogate molecular subtypes in a heterogeneous cohort of 922 patients.
Our study demonstrated that there were associations between characteristics of tumours and FGT in dynamic contrast-enhanced MRI and tumour molecular composition. The associations using multivariate models were, however, only of moderate strength with the highest AUC of 0.697 for distinguishing Luminal A from other subtypes. Overall, we demonstrated the strongest associations for OR and PR which suggests that among the imaging features tested, there is differential expression in the imaging phenotype for OR and PR status. Lower levels of association were found for HER2 and Ki-67 which did not reach the level of statistical significance after accounting for multiple hypothesis testing (p < 0.03 and p < 0.01, respectively). HER2 is a vascular growth factor receptor responsible in part for tumoural angiogenesis. Positive HER2 status has been associated with an increased incidence of multifocal and multicentric disease, increased apparent diffusion coefficient (ADC) scores, and more rapid early enhancement.28–32 Higher ADC values have also been associated with lower Ki-67 scores, but diffusion weighted imaging is not typically performed in routine breast MRI and was not included in this study.33 If prediction of HER2 and Ki-67 with imaging features is of value, then the inclusion of ADC measurement may be of additional help.
Our results indicate that computer-extracted features might be helpful in identifying biological characteristics of the tumours which help plan patient therapy. However, given the performance of the models, MR imaging features alone could not be used as a non-invasive surrogate of the molecular markers evaluated in this study. The moderate association between MRI features and subtypes demonstrates the promise of such features as part of a composite marker that might include additional clinical variables and imaging features from other modalities to determine tumour genomics. Such a composite biomarker might provide additional, heretofore unknown, benefits to treatment planning that allows for more personalised care. Finally, understanding the relationship between tumour biology and the corresponding radiological phenotype furthers the overall understanding of breast cancer as it informs about specific phenotypical expression of different underlying genomic composition. Our detailed analysis of individual imaging features showed that it is mostly the imaging characteristics of the tumour and less of the normal breast parenchyma that show associations with genomics. However, some features that quantify the relationship between the tumour and normal breast parenchyma enhancement6 had radiogenomic associations. Among these tumour features, it was predominantly those that capture enhancement dynamics that showed the highest association with genomics and particularly those related to signal enhancement ratio, which quantifies the relationship between the uptake and the washout times.
A strength of our study is the use of an independent test set which validates that the relationships identified in the training cohort can be generalised to a larger population. The use of a large number of evaluated imaging features and sophisticated machine learning-based multivariate models may result in finding relationships as the result of chance or the result of overfitting the models to the training data. Therefore, the use of a test set to validate the process is of utmost importance. To our knowledge, only two prior investigators have incorporated this important step into their analysis.3,18
In this study, we analysed a highly heterogeneous cohort of imaging and patient parameters: (a) age range of women from 21 to 89 years and seven races (b) tumours of all TNM sizes, nuclear grade, OR, PR, and HER2 status (c) 10 different combinations of magnetic field strengths and scanner manufacturers (d) 3 different types of contrast agents were used for the patients (e) range of values applied for image acquisition in terms of slice thickness, repetition times, echo times, acquisition matrices, flip angles and FOVs (e) eight different expert radiologists served as readers. Our additional analysis on subgroups formed using different scanner manufacturers, races, and menopausal status of the patients did not demonstrate major differences in the performance of the trained models the majority of the tasks.
This study had some limitations. While the heterogeneous cohort used in this study allows for more generalisable conclusions, it is likely that that variability in imaging acquisition parameters were a significant source of noise in our analysis and stronger associations could be found in a cohort with uniform imaging parameters. Future analysis could be conducted to evaluate this issue when a larger number of patients scanned in a uniform manner are available.
A future study could also evaluate a variety of image preprocessing techniques that could alleviate the variability in image acquisition. Furthermore, this study relied on surrogate molecular subtypes defined from ER, PR, and HER2 which are not as robustly predictive of outcomes as formal genetic analysis.34
In summary, we evaluated associations of imaging variables with the following molecular, genomic, and proliferation characteristics: tumour surrogate molecular subtype, ER, PR, and HER2 status, and the tumour proliferation Ki-67 marker in an independent dataset. We showed moderate associations of imaging features with Luminal A subtype, TNBC, ER, and PR status. This shows a potential for extending the usage of imaging in oncology. However, this needs to be done with caution and likely in conjunction with other variables.
Electronic supplementary material
Acknowledgements
Study design: A.S., M.A.M.; provision of study materials or patients: A.S., M.R.H., L.J.G., M.A.M.; data processing, analysis, and interpretation: all authors.; manuscript writing and editing: all authors.
Competing interests
The authors declare no competing interests.
Funding
The authors acknowledge funding from the North Carolina Biotechnology Center (2016-BIG-6520) and National Institutes of Health (1R01EB021360).
Data availabity
The materials might be made available upon request, some restrictions will apply.
Ethics approval and consent to participate
Ethics approval for the study was provided by the Duke University Health System Institutional Review Board. A waiver for informed consent was also secured from the same review board.
Electronic supplementary material
Supplementary Information is available for this paper at 10.1038/s41416-018-0185-8.
References
- 1.Mazurowski MA. Radiogenomics: what it is and why it is important. J. Am. Coll. Radiol. 2015;12:862–866. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 2.Li H, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016;281:382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan M, et al. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS ONE. 2017;12:e0171683. doi: 10.1371/journal.pone.0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan T, et al. A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting oncotypeDX risk scores. Sci. Rep. 2016;6:21394. doi: 10.1038/srep21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashraf AB, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology. 2014;272:374–384. doi: 10.1148/radiol.14131375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J. Magn. Reson. Imaging. 2015;42:902–907. doi: 10.1002/jmri.24879. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. Am. J. Roentgenol. 2012;199:654–663. doi: 10.2214/AJR.11.7824. [DOI] [PubMed] [Google Scholar]
- 8.Sung JS, et al. MR imaging features of triple-negative breast cancers. Breast J. 2013;19:643–649. doi: 10.1111/tbj.12182. [DOI] [PubMed] [Google Scholar]
- 9.Uematsu T, Kasami M, Yuen S. Triple-negative breast cancer: correlation between MR imaging and pathologic findings. Radiology. 2009;250:638–647. doi: 10.1148/radiol.2503081054. [DOI] [PubMed] [Google Scholar]
- 10.Blaschke E, Abe H. MRI phenotype of breast cancer: kinetic assessment for molecular subtypes. J. Magn. Reson. Imaging. 2015;42:920–924. doi: 10.1002/jmri.24884. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, et al. Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: a pilot radiomics study. PLoS ONE. 2015;10:e0143308. doi: 10.1371/journal.pone.0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo W, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. JMIOBU. 2015;2:041007. doi: 10.1117/1.JMI.2.4.041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agner SC, et al. Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study. Radiology. 2014;272:91–99. doi: 10.1148/radiol.14121031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costantini M, et al. Magnetic resonance imaging features in triple-negative breast cancer: comparison with luminal and HER2-overexpressing tumors. Clin. Breast Cancer. 2012;12:331–339. doi: 10.1016/j.clbc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: Luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273:365–372. doi: 10.1148/radiol.14132641. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi K, et al. Intratumoral heterogeneity of the distribution of kinetic parameters in breast cancer: comparison based on the molecular subtypes of invasive breast cancer. Breast Cancer. 2015;22:496–502. doi: 10.1007/s12282-013-0512-0. [DOI] [PubMed] [Google Scholar]
- 17.Li H, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. Npj Breast Cancer. 2016;2:16012. doi: 10.1038/npjbcancer.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, et al. Identifying relations between imaging phenotypes and molecular subtypes of breast cancer: model discovery and external validation. J. Magn. Reson. Imaging. 2017;46:1017–1027. doi: 10.1002/jmri.25661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazurowski MA, et al. Recurrence-free survival in breast cancer is associated with MRI tumor enhancement dynamics quantified using computer algorithms. Eur. J. Radiol. 2015;84:2117–2122. doi: 10.1016/j.ejrad.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Huber KE, Carey LA, Wazer DE. Breast cancer molecular subtypes in patients with locally advanced disease: impact on prognosis, patterns of recurrence, and response to therapy. Semin. Radiat. Oncol. 2009;19:204–210. doi: 10.1016/j.semradonc.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi Y, et al. Analysis of complete response by MRI following neoadjuvant chemotherapy predicts pathological tumor responses differently for molecular subtypes of breast cancer. Oncol. Lett. 2013;5:83–89. doi: 10.3892/ol.2012.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veronese SM, Maisano C, Scibilia J. Comparative prognostic value of Ki-67 and MIB-1 proliferation indices in breast cancer. Anticancer Res. 1995;15(6B):2717–2722. [PubMed] [Google Scholar]
- 23.Cheang MCU, et al. Ki67 Index, HER2 Status, and prognosis of patients with luminal B breast cancer. J. Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bezdek, J. C. Pattern Recognition with Fuzzy Objective Function Algorithms: Kluwer Academic Publishers; Norwell, MA, USA 1981. 256 p.
- 25.Tustison NJ, et al. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha A, Yu X, Sahoo D, Mazurowski MA. Effects of MRI scanner parameters on breast cancer radiomics. Expert Syst. Appl. 2017;87:384–391. doi: 10.1016/j.eswa.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLong, E. R., DeLong, D. M., Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics44, 837–845 (1988). [PubMed]
- 28.Grimm LJ, Johnson KS, Marcom PK, Baker JA, Soo MS. Can breast cancer molecular subtype help to select patients for preoperative MR imaging? Radiology. 2015;274:352–358. doi: 10.1148/radiol.14140594. [DOI] [PubMed] [Google Scholar]
- 29.Ha R, et al. Breast cancer molecular subtype as a predictor of the utility of preoperative MRI. Am. J. Roentgenol. 2015;204:1354–1360. doi: 10.2214/AJR.14.13666. [DOI] [PubMed] [Google Scholar]
- 30.Kim, E. J. et al. Histogram analysis of apparent diffusion coefficient at 3.0t: correlation with prognostic factors and subtypes of invasive ductal carcinoma. J. Magn. Reson. Imaging. 2015. [DOI] [PubMed]
- 31.Martincich L, et al. Correlations between diffusion-weighted imaging and breast cancer biomarkers. Eur. Radiol. 2012;22:1519–1528. doi: 10.1007/s00330-012-2403-8. [DOI] [PubMed] [Google Scholar]
- 32.Blaschke E, Abe H. MRI phenotype of breast cancer: kinetic assessment for molecular subtypes. J. Magn. Reson. Imaging. 2015;42:920–924. doi: 10.1002/jmri.24884. [DOI] [PubMed] [Google Scholar]
- 33.Mori N, et al. Luminal-type breast cancer: correlation of apparent diffusion coefficients with the Ki-67 labeling index. Radiology. 2015;274:66–73. doi: 10.1148/radiol.14140283. [DOI] [PubMed] [Google Scholar]
- 34.Guiu S, et al. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Ann. Oncol. 2012;23:2997–3006. doi: 10.1093/annonc/mds586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.