Abstract
Sorcin (Soluble resistance related calcium binding protein) is a small soluble penta EF family (PEF) of calcium (Ca2+) binding protein (22,000 Da). It has been reported to play crucial roles in the regulation of calcium homeostasis, apoptosis, vesicle trafficking, cancer development, and multidrug resistance (MDR). Overexpression of sorcin has been reported to be associated with different cancers such as breast cancer, colorectal cancer, gastric cancer, leukemia, lung cancer, nasopharyngeal cancer, ovarian cancer, etc. Essentially, expression of sorcin has been found to be elevated in cancer cells as compared to normal cells, indicating that it has prominent role in cancer. Moreover, sorcin was found to be the regulator of various proteins that has an association with carcinogenesis including NF-κB, STAT3, Akt, ERK1/2, VEGF, MMPs, caspases, etc. Sorcin was also found to regulate apoptosis, as silencing of the same resulted in increased levels of proapoptotic genes and induced mitochondrial apoptotic pathway in cancer. Interestingly, mutations in the sorcin gene have been closely linked with poor overall survival in bladder cancer, brain lower-grade glioma, glioblastoma, glioblastoma multiforme, kidney renal clear cell carcinoma, and stomach adenocarcinoma. Additionally, overexpression of sorcin was also found to induce MDR against different chemotherapeutic drugs. All these findings mark the importance of sorcin in cancer development and MDR. Therefore, there is urgent need to explore the functional mechanism of sorcin and to analyze whether silencing of sorcin would able to chemosensitize MDR cells. The current review summarizes the structure, expression, and functions of sorcin and its importance in the regulation of various malignancies and MDR.
Introduction
Targeting sorcin for cancer treatment
Cancer is one of the exponentially growing health disorders posing a huge thread to humankind. More than any risk factors, improper lifestyle has been found to be the major reason for the development of various cancers [1]. More than half a century of research identified multiple signaling molecules to be associated with cancer, deregulation of which plays crucial role in the onset and progression of disease. These signaling molecules include Akt, ACLY, Erk, IκB kinase (IKK), NF-κB, lipocalins, STAT3, Wnt, TNFα, and TNFα-induced proteins etc. [2], [3], [4], [5], [6], [7], [8], [9], [10]. Several such molecules have also been found to have diagnostic and therapeutic values as numerous drugs targeting these key molecules have been found to exert high therapeutic potential against different cancers [11], [12], [13], [14], [15], [16]. Sorcin (soluble resistance related calcium binding protein) is one such signaling molecule recently gaining more attention in cancer research as it has been found to induce multidrug resistance (MDR) in cancers [17]. It is a 22-kDa, soluble, small, penta EF family (PEF) of calcium (Ca2+) binding protein which has an association with calcium (Ca2+) homeostasis, MDR, and cancer development [18], [19], [20], [21]. SRI, the sorcin encoding gene, was found to be located on human chromosome seven (7q21.12) with 9 exons, and the sorcin protein is composed of 198 amino acid residues [22]. It was reported to be a cytosolic protein, which shows an association with free ribosomes, rough endoplasmic reticulum, mitochondria, microfilament, and perinuclear membranes [23]. Sorcin was first identified in vincristine-resistant Chinese hamster lung cell line DC-3F/VCRd-5 L by Meyers and Biedler and has been shown to increase the drug efflux in MDR cells in a calcium (Ca2+)-dependent manner [17], [24], [25].
Interestingly, analysis of expression pattern of sorcin revealed it to be expressed in most human tissues such as the tissues of bone, heart, brain, kidney, breast, skin, B-lymphocytes, T-lymphocytes, and monocytes. Further, shouting its importance in cancer, sorcin was found to be overexpressed in different cancers including breast cancer, colorectal cancer, gastric cancer, leukemia, lung cancer, nasopharyngeal cancer, and ovarian cancer [26]. It was also shown that sorcin was generally unexpressed in terminally differentiated mature tissues but was highly expressed in majority of tumor tissues, marking it as a potent target for cancer diagnosis and therapy. Many reports show that sorcin may have important role in the progression of cancer by enhancing the various hallmark of cancer such as cell motility, invasion, migration, metastasis, epithelial to mesenchymal transition (EMT), and MDR. Further, sorcin was found to modulate the levels of important cellular proteins, which are involved in the process of tumorigenesis such as NF-κB, CTSZ, STAT3, Akt, ERK1/2, VEGF, MMPs, caspases, etc. [20], [26], [27], [28], [29]. In addition, silencing of this protein resulted in apoptosis and reverted MDR of cancer cells, and additionally, sorcin depletion reduced the levels of various proteins involved in angiogenesis, invasion, and metastasis [27], [29]. Remarkably, overexpression of sorcin was also shown to induce chemoresistance against a variety of chemotherapeutics including 5-fluorouracil, cisplatin, doxorubicin, etoposide, homoharringtonine, paclitaxel, vincristine, etc. [18]. Thus, it is apparent that sorcin indeed has a key role in cancer. However, the exact mechanism(s) of action of sorcin in the initiation and progression of many cancers remains obscure. Presently, a number of studies are being carried out with the aim of exploring the actual cellular functions of sorcin and its involvement in tumorigenesis and development of MDR. Therefore, the current review discusses the collective role and the associated molecular mechanisms of this calcium (Ca2+) binding protein in cancer and MDR summing up the literature available.
Structure of Sorcin
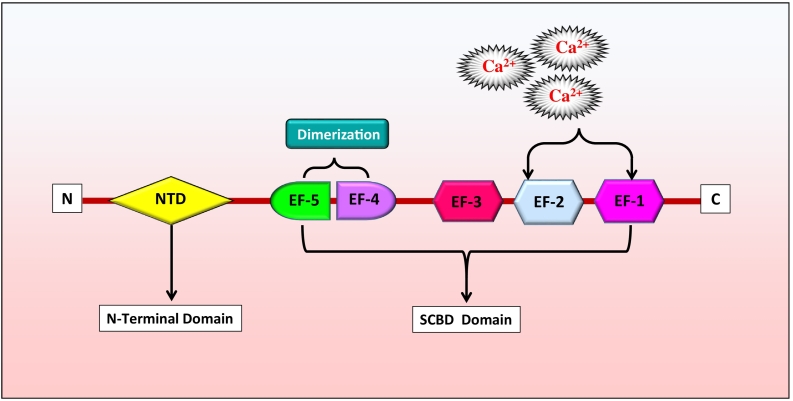
Aforementioned, sorcin, also known as CP-22 and V19, is an acidic, soluble, calcium (Ca2+)-binding protein encoded by SRI gene located at 7q21.12 with nine exons [23], [30]. The SRI gene spans about 21.9 kb in the human genomic DNA and transcribes into five variants, namely, isoform A, B, C, D, and X1 [31]. Among these, isoform A is the most commonly studied variant, which is a 22-kDa protein made of 198 amino acids. Sorcin has been classified into the penta-EF-hand (PEF) protein family as it has five EF-hand motifs. Proteins of the PEF family were known to interact with the membranes through these EF-hand motifs in a Ca2+-dependent manner. Other proteins of this small PEF family are calpain, grancalcin, peflin, and PDCD6 (earlier known as ALG-2) [32], [33], [34], [35]. Sorcin was found to share a high homology with the light chain of calpain protein [36]. X-ray diffraction analysis of sorcin protein crystals revealed sorcin to have a globular shape with an extended N-terminal. It has been shown to form homodimers, and the monomers of these homodimer consist of two domains viz., C-terminal Ca2+-binding domain known as sorcin calcium binding domain and N-terminal glycine-rich domain [35], [37], [38]. The C-terminal of sorcin protein is composed of mainly eight alpha helices connected by loops to form the five EF-hands. These five EF-hands were found to pair with each other (EF1 with EF2 and EF3 with EF4). The free unpaired EF5 hand helps in homodimer formation by pairing with the EF5 of other sorcin. In addition, EF4-EF5 was also shown to participate in the protein dimerization (Figure 1) [35], [38].
Figure 1.
Structure of sorcin protein.
Structural comparison with the other members of PEF family revealed the folds of the sorcin EF motifs, especially the EF1 hand, to be highly conserved. It also suggested that the EF1-EF2 pair has two important Ca2+-binding sites and changes its conformation to a larger extent upon calcium binding, signifying this first EF hand pair to have crucial role in sorcin-calcium signaling (Figure 1) [35]. In line with this, comparison of the crystal structure of apo-Sor and calcium bound sorcin (CaSor) also showed that calcium binding moves the D-helix that joins the EF1-EF2 pair with EF3 and opens the EF1 hand promoting the exposure of hydrophobic surfaces [38]. Sorcin is broadly distributed in vertebrates with highly conserved amino acid sequence. For instance, the protein sequences of mouse and human sorcin differ by only eight amino acids. However, notably, the phosphorylation sites of sorcin were found to differ between different speices [39].
Expression Pattern
Aforementioned, sorcin was found to be expressed in most tissues including the tissues of normal bone, breast, brain, heart, kidney, and skin. Apart from normal tissues, overexpression of sorcin was reported in various cancers such as breast cancer, colorectal cancer, gastric cancer, lung cancer, ovarian cancer, leukemia, and myeloma [20], [26], [27], [40], [41]. In addition, the expression level of sorcin was found to be higher in hepatocellular carcinoma tissues than the neighboring nontumorous and normal liver tissues [28]. Interestingly, the terminally differentiated mature tissues were found to have no sorcin, whereas its expression was very high in majority of tumor tissues, signifying it to be a potent target for cancer [27].
Molecular Targets
Sorcin is a regulatory protein, controlling the expression of various molecules in biological system. Sorcin was found to exert its oncogenic activity by inducing different signaling pathways such as ERK, MAPK/ERK, STAT3, PI3K/Akt, Akt/NF-κB, etc. [26], [28], [42]. It was also shown to induce tumor invasion, migration, and metastasis through modulation of the levels of Cathepsin Z (CTSZ), p-STAT3, and matrix metalloproteinases (MMP-2 and -9) [20]. Further, activation of STAT3 by sorcin was proved to develop chemoresistance and radioresistance in malignant cells through the interaction with transcription factors, including NF-κB (nuclear factor kappa B) [43]. It was also suggested that sorcin might have an important role in EMT and cancer stem cells (CSCs) progression as it modulated the levels of E-cadherin, N-cadherin, fibronectin, α-SMA, vimentin, VEGF, and ERK signaling pathways [28], [29]. Further supporting this, upregulation of sorcin was shown to induce the activity of vimentin (mesenchymal marker) protein, to increase the level of p-ERK1/2, and to downregulate the activity of E-cadherin (epithelial marker) [28]. In addition, overexpression of sorcin was also found to activate PI3K/Akt signaling pathway, which further plays a major role in migration, invasion, and epithelial to mesenchymal phenotype [26]. Adding to its tumorigenic potential, sorcin was also shown to be involved in the regulation of MDR, survival, and cell death associated proteins such as MDR1, MRP1, GST-π, Livin, Src, survivin, c-fos, c-jun, Bax, Bcl-2, caspase-3, caspase-12 and GRP78/BiP (binding immunoglobulin protein), cyclin-D1, c-Myc, p21, and p53 etc. [27], [40], [44], [45]. Besides, sorcin was shown to exert its cytoprotective activity against chemotherapeutic agents by interacting and stabilizing TRAP1 (TNF receptor associated protein 1) against apoptosis in the mitochondria [46]. Similarly, sorcin overexpression was also found to increase the levels of MDR1/P-gp and contribute to the multidrug resistant phenotype by promoting the binding of CREB1 to cAMP response elements (CRE) present in the MDR1/P-gp promotor through increased phosphorylation of CREB1 [21].
Functions of Sorcin
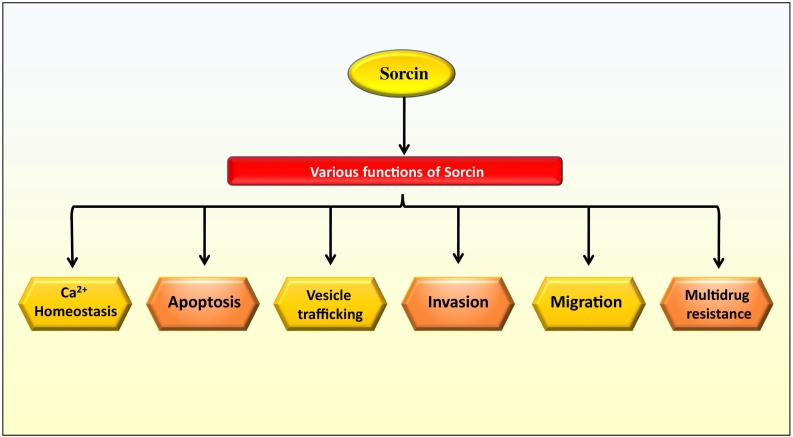
The actual role of sorcin is not fully understood. However, it was observed that sorcin helps in the regulation of homeostasis, apoptosis, vesicles trafficking, and MDR in cells (Figure 2). Sorcin has a significant role in the regulation of calcium (Ca2+) homeostasis in human body. Calcium (Ca2+) plays significant roles in neurons, including synaptic plasticity and apoptosis, and deregulation of this neuronal calcium signaling was known to be one of the central mechanisms of different neurodegenerative diseases such as Alzheimer's disease (AD). Sorcin regulates the calcium homeostasis by two ways such as calcium-dependent binding to calcium channels and calcium binding itself. Sorcin expression enhances the concentration of calcium in endoplasmic reticulum (ER), inhibits ER stress, and induces the resistance to apoptosis. Moreover, the expression level of sorcin was found to be highly upregulated during ER stress [19], [47]. Apart from calcium homeostasis, sorcin was also found to have a key role in the activation of mitosis and cytokinesis as loss of sorcin highly compromised the normal process of mitosis and cytokinesis [47].
Figure 2.
Functions of sorcin in various cellular processes.
It was also shown that sorcin can regulate apoptosis in cancer cells and induce cell cycle progression by calcium-dependent interaction with different kinases such as Polo-like kinase 1 (PLK1, a serine/threonine-protein kinase associated with mitotic spindle poles), Aurora A, and Aurora B. For instance, sorcin was found to interact with PLK1, get phosphorylated, and induce PLK1 autophosphorylation, ultimately regulating its kinase activity [38], [47]. Further, in the heart, sorcin was found to regulate various proteins including cardiac RyR2 (ryanodine receptor), L-type calcium channel, and sodium-calcium exchanger that has an association with excitation and contraction coupling [48]. It was found to inhibit the L-type calcium current (I Ca, L) of the ventricular myocytes (isolated from rabbit) of rabbit [49]. Apart from this, sorcin also targets the sarcolemmal NCX1 (sodium/ calcium exchanger) and induces its expression in the cardiac muscles, and silencing of sorcin by miR-1 helps to regulate the myocardial contraction through calcium signaling [50], [51]. Furthermore, sorcin was also shown to regulate the dimensions and calcium concentration of the ER vesicles through activation of SERCA (sarcoplasmic/endoplasmic reticulum Ca2+-ATPase) and inhibition of RyR (ryanodine receptor) [47]. Altered expression of sorcin was found in the endometrium of women with mysterious infertility during the early-to-mid-secretory phase, suggesting a possible role of sorcin in endometrial receptivity and embryo implantation. Further, it was identified that sorcin regulates the Ca2+-dependent angiogenesis in endometrial cells via activating VEGF/PI3K/Akt pathway and prepares the endometrium for implantation [52].
Roles in Malignancies
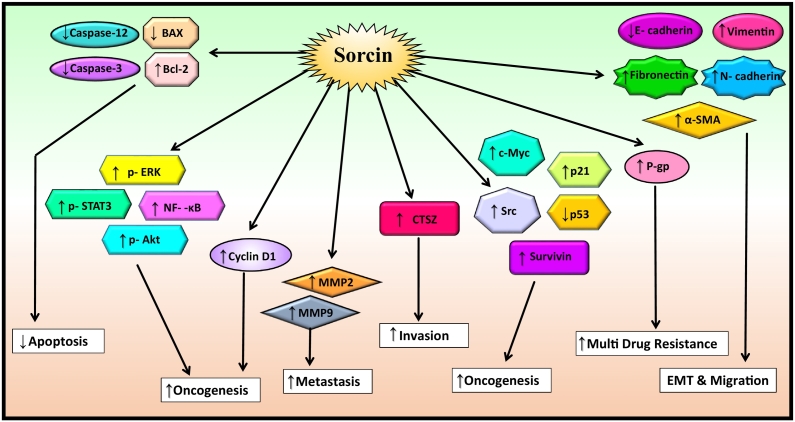
Invasion and migration are the two major manifestations of tumor progression. Numerous studies have shown two patterns of invasion in cancer: individual cell migration and collective cell migration by which tumor cells are able to overcome the hurdle of the extracellular matrix (ECM) and spread into neighboring tissues [53]. Apart from this, cancer also associated with other important processes including inhibition of apoptosis, MDR, epithelial mesenchymal transition, etc., and sorcin has the ability to regulate different oncogenic genes involved in the regulation of these processes such as p-ERK, p-STAT3, NF-κB, p-Akt, survivin, Src, c-myc, etc. It also helps to inhibit apoptosis by inactivating caspase-3 and caspase-12 (Figure 3). Further, sorcin was suggested to promote the invasion and migration of different cancer cells as overexpression of sorcin resulted in an increased level of p-STAT3, MMP2, and MMP9. For instance, overexpression of sorcin was found to enhance the levels of CTSZ, p-STAT3, MMP2, and MMP9 in gastric cancer [20]. Also, the expression of sorcin was found to be 5.4-fold upregulated in gastric cancer cells (SGC7901 cells) than the normal cells [54]. Further, it was reported that upregulation of sorcin in HepG2, hepatocellular carcinoma cells, extensively increased the levels of mesenchymal marker vimentin and phosphorylated ERK1/2 (p-ERK1/2) and considerably reduced the levels of epithelial marker E-cadherin [28]. All these reports describe that sorcin may have an essential role in EMT. Further supporting this, sorcin was shown to stimulate EMT in colorectal cancer cells (HCT116 cells) through the activation of PI3K/Akt signaling pathway; upregulation of N-cadherin, fibronectin, vimentin, and α-SMA; and downregulation of E-cadherin [26].
Figure 3.
Sorcin upregulates the gene involved in cell migration, invasion, oncogenesis, and metastasis and downregulates the gene involved in apoptosis.
↑ = upregulation; ↓ = downregulation.
As observed from the cBioPortal for Cancer Genomics data, several mutations of sorcin protein are associated with different kind of cancers including bladder cancer, colorectal adenocarcinoma, prostate adenocarcinoma, skin cutaneous melanoma, sarcoma, and uterine corpus endometrial carcinoma, etc. (Table 1). In line with this, RNA sequencing analysis of patient samples revealed amplification of SRI gene to be associated with various cancers, which was evident from the TCGA cBioPortal database. Further, TCGA data showed that various cancers possess variable SRI gene amplification frequency. For example, esophagus cancer has the amplification frequency of 8.06% (15 cases), lung squamous cancer 4.11%, ovarian cancer 3.635%, stomach cancer 3.77%, pancreas cancer 2.15%, cholangiocarcinoma 1.96%, cervical cancer 1.29%, melanoma 0.84%, sarcoma 0.75%, breast cancer 0.81%, uterine cancer 0.18%, and testicular germ cell cancer 1.28%. In addition to gene amplification, different mutations of SRI gene were also reported in the TCGA database, and frequency of SRI gene mutation observed in different cancers is as follows: colorectal cancer 0.16%, uterine cancer 0.36%, prostate cancer 0.4%, and sarcoma 0.38%. To further reveal the association of sorcin with survival and prognosis of different cancer patients, the overall survival Kaplan-Meier estimate and disease/progression-free Kaplan-Meier estimate graphs from cBioportal for Cancer Genomics data were analyzed (Tables 1 and 2). As mentioned earlier, according to the TCGA database, sorcin shows different mutations in different cancers such as X84_splice (splice mutation) in bladder cancer, D157N (missense mutation) in colorectal adenocarcinoma, A161T (missense mutation) & Q48⁎(nonsense mutation) in prostate adenocarcinoma, P28L (missense mutation) & C162F (missense mutation) in skin cutaneous melanoma, Y13Tfs ⁎30(FS del mutation) in sarcoma, and A161T (missense mutation) & R106I (missense mutation) in uterine corpus endometrial carcinoma.
Table 1.
Overall Survival by Kaplan-Meier Estimate
| Cancer | Total No. of Samples | Cases with Alteration (No.) |
Cases without Alteration (No.) |
References | |||||
|---|---|---|---|---|---|---|---|---|---|
| Mutation | Total | Deceased | MMS | Total | Deceased | MMS | |||
| Adrenocortical carcinoma | 88 | - | 1 | 0 | NA | 87 | 32 | 79.01 | [64] |
| Bladder cancer | 408 | X84_splice | 6 | 3 | 28.38 | 399 | 174 | 34.95 | [65] |
| Breast invasive cancer | 816 | - | 5 | 0 | NA | 809 | 118 | 129.6 | [66] |
| 482 | - | 1 | 0 | NA | 481 | 60 | 113.74 | [67] | |
| 963 | - | 7 | 0 | NA | 954 | 135 | 129.6 | [68] | |
| Brain lower-grade glioma | 283 | - | 4 | 1 | 25.89 | 278 | 71 | 75.1 | [69] |
| Breast cancer | 2051 | - | 35 | 16 | 163.1 | 1831 | 1075 | 152.9333333 | [70] |
| BUC | 127 | - | 1 | 0 | NA | 126 | 46 | 20.47 | [71] |
| 127 | - | 1 | 0 | NA | 125 | 61 | 23.19 | [72] | |
| Colorectal adenocarcinoma | 212 | D157N | 2 | 0 | NA | 208 | 17 | NA | [73] |
| 220 | D157N | 3 | 1 | NA | 216 | 38 | NA | [74] | |
| Cholangiocarcinoma | 35 | - | 1 | 0 | NA | 32 | 15 | 24.34 | [75] |
| CSCCEA | 191 | X18_splice | 3 | 1 | NA | 188 | 40 | 101.74 | [76] |
| Esophageal carcinoma | 184 | - | 15 | 7 | 44.71 | 169 | 69 | 25.76 | [77] |
| Glioblastoma | 281 | - | 8 | 6 | 10.8 | 238 | 157 | 13.1 | [78] |
| Glioblastoma multiforme | 273 | - | 9 | 7 | 10.81 | 259 | 204 | 13.63 | [79] |
| HNSCC | 279 | - | 7 | 5 | 71.16 | 152 | 64 | 21.75 | [80] |
| 504 | - | 18 | 10 | 30.91 | 484 | 205 | 56.44 | [81] | |
| RPCC | 280 | - | 1 | 0 | NA | 278 | 41 | NA | [82] |
| RCCC | 418 | - | 2 | 1 | 1.94 | 413 | 138 | 75.5 | [83] |
| 448 | - | 3 | 1 | NA | 445 | 151 | 90.41 | [84] | |
| LNDLBL | 48 | - | 1 | 0 | NA | 47 | 9 | 211.07 | [85] |
| OSC | 311 | - | 10 | 8 | 50.33 | 299 | 197 | 44.51 | [86] |
| Pancreatic adenocarcinoma | 149 | - | 4 | 1 | NA | 145 | 81 | 19.65 | [87] |
| Prostate adenocarcinoma | 492 | A161T, Q48⁎ | 5 | 0 | NA | 487 | 9 | NA | [88] |
| Papillary thyroid carcinoma | 399 | - | 1 | 0 | NA | 321 | 13 | NA | [89] |
| Stomach adenocarcinoma | 287 | - | 8 | 1 | 18.33 | 217 | 53 | 30.88 | [90] |
| 393 | - | 17 | 6 | 37.88 | 369 | 133 | 35.97 | [91] | |
| Skin cutaneous melanoma | 287 | P28L, C162F | 5 | 1 | 297.67 | 276 | 159 | 80.62 | [92] |
| Sarcoma | 243 | Y13Tfs ⁎ 30 | 3 | 1 | NA | 240 | 91 | 65.41 | [93] |
| Testicular germ cell cancer | 149 | - | 2 | 0 | NA | 131 | 4 | NA | [94] |
| Thyroid carcinoma | 399 | - | 1 | 0 | NA | 398 | 14 | NA | [95] |
| UCEC | 240 | A161T, R106I | 2 | 0 | NA | 237 | 23 | NA | [96] |
| 242 | A161T, R106I | 3 | 0 | NA | 239 | 32 | NA | [97] | |
Table 2.
Disease/Progression-Free Survival by Kaplan-Meier Estimate.
| Cancer | Total no. of Samples | Cases with Alteration (No.) |
Cases without Alteration (No.) |
References | ||||
|---|---|---|---|---|---|---|---|---|
| Total | Deceased | MMS | Total | Deceased | MMS | |||
| Adrenocortical carcinoma | 88 | 1 | 1 | 12.84 | 78 | 41 | 37.75 | [64] |
| Bladder cancer | 408 | 5 | 3 | 25.23 | 314 | 139 | 32.59 | [65] |
| Breast invasive cancer | 816 | 5 | 1 | NA | 753 | 84 | 214.72 | [66] |
| 963 | 7 | 1 | NA | 872 | 101 | 214.72 | [68] | |
| Brain lower-grade glioma | 283 | 4 | 3 | 10.84 | 256 | 87 | 47.7 | [69] |
| BUC | 127 | 1 | 1 | 25.23 | 98 | 50 | 18.23 | [72] |
| Colorectal adenocarcinoma | 220 | 3 | 1 | NA | 186 | 29 | NA | [74] |
| Cholangiocarcinoma | 35 | 1 | 0 | NA | 30 | 16 | 9.23 | [75] |
| CSCCEA | 191 | 2 | 0 | NA | 171 | 31 | NA | [76] |
| Esophageal carcinoma | 184 | 9 | 5 | 17.58 | 133 | 65 | 21.42 | [77] |
| Glioblastoma | 281 | 8 | 5 | 5.2 | 239 | 125 | 8.5 | [78] |
| Glioblastoma multiforme | 273 | 7 | 6 | 4.6 | 170 | 140 | 7.36 | [79] |
| HNSCC | 504 | 11 | 4 | 21.91 | 368 | 137 | 61.07 | [81] |
| RPCC | 280 | 1 | 0 | NA | 259 | 49 | 106.04 | [82] |
| RCCC | 448 | 2 | 0 | NA | 364 | 114 | 106.77 | [84] |
| LNDLBL | 48 | 1 | 0 | NA | 43 | 12 | 120.53 | [85] |
| OSC | 311 | 7 | 6 | 21.06 | 256 | 204 | 16.1 | [86] |
| Pancreatic adenocarcinoma | 149 | 3 | 3 | 18.23 | 112 | 68 | 14.75 | [87] |
| Prostate adenocarcinoma | 492 | 5 | 2 | 39.98 | 481 | 87 | NA | [88] |
| Papillary thyroid carcinoma | 399 | 1 | 0 | NA | 331 | 23 | NA | [89] |
| Stomach adenocarcinoma | 287 | 7 | 1 | 17.87 | 151 | 29 | 55.06 | [90] |
| 393 | 16 | 6 | 18.86 | 289 | 87 | 55.06 | [91] | |
| Skin cutaneous melanoma | 287 | 4 | 1 | NA | 246 | 192 | 49.21 | [92] |
| Sarcoma | 243 | 2 | 1 | 7.62 | 214 | 119 | 34 | [93] |
| Testicular germ cell cancer | 149 | 2 | 0 | NA | 129 | 33 | 191.43 | [94] |
| Thyroid carcinoma | 399 | 1 | 0 | NA | 385 | 35 | NA | [95] |
| UCEC | 240 | 2 | 0 | NA | 221 | 38 | NA | [96] |
| 242 | 3 | 0 | NA | 225 | 46 | NA | [97] | |
Abbreviations: BUC, bladder urothelial carcinoma; CSCCEA, cervical squamous cell carcinoma and endocervical adenocarcinoma; HNSCC, head & neck squamous cell carcinoma; LNDLBL, lymphoid neoplasm diffuse large B cell lymphoma; MMS, median month survival; OSC, ovarian serous cystadenocarcinoma; RCCC, renal clear cell carcinoma; RPCC, renal papillary cell carcinoma; UCEC, uterine corpus endometrial carcinoma.
Similarly, analysis of the overall survival Kaplan-Meier estimate of different cancers revealed that the median month survival of patients with unaltered sorcin is higher than that of the patients with altered sorcin. The median month survival data for different cancer cases without sorcin gene alteration is as follows: bladder cancer 34.9 months, brain lower-grade glioma 75.1 months, glioblastoma 13.1 months, glioblastoma multiforme 13.63 months, kidney renal clear cell carcinoma 75.5 months, and stomach adenocarcinoma 30.88 months. However, in some of the cancers, patients with altered sorcin showed more median month survival such as breast cancer (163.1 months), esophageal carcinoma (44.71 months), head and neck squamous cell carcinoma (71.16 months), ovarian serous cystadenocarcinoma (50.33 months), and skin cutaneous melanoma (297.67 months) (Table 1). Likewise, the disease/progression-free survival of patients without sorcin alteration was as follows: adrenocortical carcinoma 37.75 months, bladder cancer 32.59 months, breast invasive cancer 214.72 months, brain lower grade glioma 47.7 months, bladder urothelial carcinoma 18.23 months, cholangiocarcinoma 9.23 months, esophageal carcinoma 21.42 months, glioblastoma 8.5 months, glioblastoma multiforme 7.36 months, head and neck squamous cell carcinoma 61.07 months, kidney renal papillary cell carcinoma 106.04 months, kidney renal clear cell carcinoma 106.77 months, lymphoid neoplasm diffuse large B-cell lymphoma 120.53 months, ovarian serous cystadenocarcinoma 16.1 months, pancreatic adenocarcinoma 14.75 months, stomach adenocarcinoma 55.06 months, skin cutaneous melanoma 49.21 months, sarcoma 34 months, and testicular germ cell cancer 191.43 months. And patients with altered sorcin had the disease/progression-free survival as follows: adrenocortical carcinoma 12.84 months, bladder cancer 25.23 months, brain lower-grade glioma 10.84 months, bladder urothelial carcinoma 25.23 months, esophageal carcinoma 17.58 months, glioblastoma 5.2 months, glioblastoma multiforme 4.6 months, head and neck squamous cell carcinoma 21.91 months, ovarian serous cystadenocarcinoma 21.06 months, pancreatic adenocarcinoma 18.23 months, prostate adenocarcinoma 39.98 months, stomach adenocarcinoma 17.87 months, and sarcoma7.62 months (Table 2).
Functions in MDR
MDR has turned out to be a major hurdle for effective cancer chemotherapies. MDR has been defined as the resistance of cancer cells to different chemotherapeutic drugs that may have diverse structures and mechanisms of action. It is the most prominent reason for the failure of most of the chemotherapeutic drugs in cancer treatment as the effect of these drugs decreases when the cancer cells acquired MDR. Cancer cells acquire MDR through ABC transporter family, resistance to apoptosis induction, autophagy, cancer stem cells, miRNA, hypoxia, DNA damage and repair, and epigenetic regulation. Therefore, there is an urgent need to discover MDR associated biomarkers to enhance the success of chemotherapeutic drugs for the treatment of cancer patients [55]. Sorcin is one such protein, which was recently found to be associated with MDR in various cancers. MDR is known to be mediated by different drug resistance genes such as MDR1, MRP1, etc. MDR1, MRP1, and GST-π together form the classical MDR pathway. Thus, these two proteins, MDR1 and MRP1, are well established to have an important role in chemotherapeutic drug efflux and lead to MDR. Interestingly, sorcin was found to regulate the levels of MDR1 and MRP1 along with the expression of various other MDR related genes including GST-π, Livin, Src, survivin, Bcl-2, cyclin-D1, c-myc, p21, and p53. Silencing of sorcin resulted in the downregulation of these genes in addition to p-Akt and NF-κB levels inducing chemoresistance in myeloma cells (Figure 3) [27]. Further, upregulation of sorcin was shown to modulate the activity of different chemotherapeutic drugs such as doxorubicin, paclitaxel, cisplatin, homoharringtonine, vincristine, etc. [25], [56].
Doxorubicin is a well-known chemotherapeutic drug used for treating different cancers including breast cancer, bladder cancer, and blood cancer (leukemia). It was shown that sorcin has the ability to bind with doxorubicin, leading to the decreased levels of drug inside the cells, and also increases its efflux via MDR1 [18]. Likewise, chemoresistance to cisplatin in MDR cells is also associated with the co-amplification of sorcin [57]. Further, it was also reported that sorcin has prognostic value in childhood acute lymphoid leukemia (ALL), which may be related to the upregulation of MDR1/P-gp gene expression which plays an important role in the regulation of drug distribution in different tissues [58]. Similarly, co-amplification of sorcin and MDR1 gene observed in leukemia can be taken as a good indicator of clinical drug resistance and prognosis of the disease [59]. Further proving the importance of sorcin in MDR, overexpression of sorcin in K562 cells by gene transfection led to the increase in drug resistance, from 4.1- to 22.5-fold, to various chemotherapeutic drugs such as doxorubicin, etoposide, homoharringtonine, and vincristine [25]. Moreover, increased expression of this protein in multidrug-resistant cells of various cancers indicates that sorcin may have a pivotal role in the development of resistant phenotype [30], [32], [60]. In contrast to these reports, Parekh et al. (2002) showed the overexpression of sorcin to be associated with reduced paclitaxel resistance in various cancers [56]. Furthermore, it was shown that tetrandrine (Tet) treatment helps in the chomosensitization of K562/A02 cells by downregulating sorcin [61]. Likewise, haishengsu (HSS), a conventional drug from Tegillarca granosa that suppresses the expression of sorcin and P-gp protein was shown to increase the chemosensitivity in leukemic patients and to induce apoptosis in the drug resistant K562/ADM tumors of mice [62], [63].
Conclusion
Taken together, sorcin (22 kDa) is a small penta EF family of soluble calcium (Ca2+) binding protein that shows an association with calcium homeostasis, endometrial receptivity, cancer development, and MDR. It also shows differential expression pattern in malignant cells and MDR cancer cells. It is known to be tightly associated with ribosomes, rough endoplasmic reticulum, mitochondria, and nuclear membranes. In the last few years, sorcin has appeared as one of the most fascinating executive for calcium homeostasis in the cells. However, the expression level of sorcin is much lower than the calmodulin (calcium binding protein) protein. Several reports have implicated that apart from calcium-ion regulation, sorcin may also be involved in maintaining the dimensions of ER vesicles, regulation of cell cycle progression through activation of mitosis and cytokinesis and in regulating the activity of Ca2+-dependent kinases. In addition to this, sorcin also was found to regulate angiogenesis, invasion, and migration of different tumor cells by regulating various key molecules involved in the processes such as NF-κB, CTSZ, STAT3, Akt, ERK1/2, VEGF, MMPs, caspases, and signaling pathways including ERK, MAPK/ERK, and PI3K/Akt. Sorcin was also found to induce metastasis and chemoresistance in malignant cells. Further, the calcium homeostasis, basic function of sorcin, is the important cellular response to stress conditions favoring the drug resistance in tumor progression. Analysis of the hepatocellular carcinoma patient samples revealed sorcin overexpression to be associated with worse prognosis. Upregulation of sorcin in malignant cells significantly induces the cell proliferation, migration, and invasion, and knockdown of the same diminished the proliferation, migration, and invasion of cancer cells, revealing the importance of sorcin in the development and progression of cancer. Adding to its cancer-promoting effects, sorcin was proved to induce MDR against various chemotherapeutic agents through modulation of MDR1, MRP1, NF-κB, apoptotic, antiapoptotic, survival proteins. Downregulation of sorcin may lead to membrane hyperpolarization and reduce calcium content in mitochondria which may further promote the drug-induced apoptosis in malignant cells. TCGA data analysis also showed alteration status of sorcin gene to be significantly associated with survival of cancer patients, suggesting the prognostic value of this protein. Further, the increased levels of sorcin observed in different multidrug-resistant cells implicate the possibility of using it as a potential biomarker for predicting MDR in various cancers. Until now, maximum research on sorcin has focused on the connection between sorcin and diseases but rarely discussed about the regulation of sorcin by different chemotherapeutic drugs. Also, there is an indispensable need for novel therapeutic strategies targeting sorcin for better management of different multidrug resistant cancers. However, further studies are obligatory to unveil the actual role of sorcin in the development of cancers and the multidrug resistant (MDR) phenotype and to define sorcin as a novel diagnostic and therapeutic marker for different cancers.
Acknowledgments
Acknowledgement
This work was supported by BT/529/NE/TBP/2013 awarded to Ajaikumar B Kunnumakkara by Department of Biotechnology (DBT), Government of India. The author Kishore Banik acknowledges the UGC New Delhi for providing him the fellowship.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Gautam Sethi, Email: gautam.sethi@tdt.edu.vn, phcgs@nus.edu.sg.
Ajaikumar B. Kunnumakkara, Email: kunnumakkara@iitg.ernet.in, ajai78@gmail.com.
References
- 1.Anand P, Kunnumakkara AB, Sundaram C, Harikumar KB, Tharakan ST, Lai OS, Sung B, Aggarwal BB. Cancer is a preventable disease that requires major lifestyle changes. Pharm Res. 2008;25(9):2097–2116. doi: 10.1007/s11095-008-9661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy NK, Bordoloi D, Monisha J, Padmavathi G, Kotoky J, Golla R, Kunnumakkara AB. Specific targeting of Akt kinase isoforms: taking the precise path for prevention and treatment of cancer. Curr Drug Targets. 2017;18(4):421–435. doi: 10.2174/1389450117666160307145236. [DOI] [PubMed] [Google Scholar]
- 3.Khwairakpam AD, Shyamananda MS, Sailo BL, Rathnakaram SR, Padmavathi G, Kotoky J, Kunnumakkara AB. ATP citrate lyase (ACLY): a promising target for cancer prevention and treatment. Curr Drug Targets. 2015;16(2):156–163. doi: 10.2174/1389450115666141224125117. [DOI] [PubMed] [Google Scholar]
- 4.Awasthee N, Rai V, Chava S, Nallasamy P, Kunnumakkara AB, Bishayee A, Chauhan SC, Challagundla KB, Gupta SC. Targeting IkappaB kinases for cancer therapy. Semin Cancer Biol. 2018 doi: 10.1016/j.semcancer.2018.02.007. [pii: S1044-579X(17)30046–9] [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monisha J, Padmavathi G, Roy NK, Deka A, Bordoloi D, Anip A, Kunnumakkara AB. NF-kappaB blockers gifted by mother nature: prospectives in cancer cell chemosensitization. Curr Pharm Des. 2016;22(27):4173–4200. doi: 10.2174/1381612822666160609110231. [DOI] [PubMed] [Google Scholar]
- 6.Monisha J, Roy NK, Bordoloi D, Kumar A, Golla R, Kotoky J, Padmavathi G, Kunnumakkara AB. Nuclear factor kappa B: a potential target to persecute head and neck cancer. Curr Drug Targets. 2017;18(2):232–253. doi: 10.2174/1389450117666160201112330. [DOI] [PubMed] [Google Scholar]
- 7.Monisha J, Roy NK, Padmavathi G, Banik K, Bordoloi D, Khwairakpam AD, Arfuso F, Chinnathambi A, Alahmadi TA, Alharbi SA. NGAL is downregulated in oral squamous cell carcinoma and leads to increased survival, proliferation, migration and chemoresistance. Cancers. 2018;10(7) doi: 10.3390/cancers10070228. [pii: E228] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunnumakkara AB, Sailo BL, Banik K, Harsha C, Prasad S, Gupta SC, Bharti AC, Aggarwal BB. Chronic diseases, inflammation, and spices: how are they linked? J Transl Med. 2018;16(1):14. doi: 10.1186/s12967-018-1381-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4(5) doi: 10.1101/cshperspect.a008052. [pii: a008052] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padmavathi G, Banik K, Monisha J, Bordoloi D, Shabnam B, Arfuso F, Sethi G, Fan L, Kunnumakkara AB. Novel tumor necrosis factor-alpha induced protein eight (TNFAIP8/TIPE) family: functions and downstream targets involved in cancer progression. Cancer Lett. 2018;432:260–271. doi: 10.1016/j.canlet.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Bordoloi D, Roy NK, Monisha J, Padmavathi G, Kunnumakkara AB. Multi-targeted agents in cancer cell chemosensitization: what we learnt from curcumin thus far. Recent Pat Anticancer Drug Discov. 2016;11(1):67–97. doi: 10.2174/1574892810666151020101706. [DOI] [PubMed] [Google Scholar]
- 12.Padmavathi G, Rathnakaram SR, Monisha J, Bordoloi D, Roy NK, Kunnumakkara AB. Potential of butein, a tetrahydroxychalcone to obliterate cancer. Phytomedicine. 2015;22(13):1163–1171. doi: 10.1016/j.phymed.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Kunnumakkara AB, Bordoloi D, Padmavathi G, Monisha J, Roy NK, Prasad S, Aggarwal BB. Curcumin, the golden nutraceutical: multitargeting for multiple chronic diseases. Br J Pharmacol. 2017;174(11):1325–1348. doi: 10.1111/bph.13621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sailo BL, Banik K, Padmavathi G, Javadi M, Bordoloi D, Kunnumakkara AB. Tocotrienols: the promising analogues of vitamin E for cancer therapeutics. Pharmacol Res. 2018;130:259–272. doi: 10.1016/j.phrs.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 15.Banik K, Harsha C, Bordoloi D, Lalduhsaki Sailo B, Sethi G, Leong HC, Arfuso F, Mishra S, Wang L, Kumar AP. Therapeutic potential of gambogic acid, a caged xanthone, to target cancer. Cancer Lett. 2018;416:75–86. doi: 10.1016/j.canlet.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Roy NK, Deka A, Bordoloi D, Mishra S, Kumar AP, Sethi G, Kunnumakkara AB. The potential role of boswellic acids in cancer prevention and treatment. Cancer Lett. 2016;377(1):74–86. doi: 10.1016/j.canlet.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Koch G, Smith M, Twentyman P, Wright K. Identification of a novel calcium-binding protein (CP22) in multidrug-resistant murine and hamster cells. FEBS Lett. 1986;195(1-2):275–279. doi: 10.1016/0014-5793(86)80176-4. [DOI] [PubMed] [Google Scholar]
- 18.Genovese I, Fiorillo A, Ilari A, Masciarelli S, Fazi F, Colotti G. Binding of doxorubicin to sorcin impairs cell death and increases drug resistance in cancer cells. Cell Death Dis. 2017;8(7):e2950. doi: 10.1038/cddis.2017.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SI, Lee HJ, Kim SS, Kwon YS, Chun W. Sequestration of sorcin by aberrant forms of tau results in the defective calcium homeostasis. Korean J Physiol Pharmacol. 2016;20(4):387–397. doi: 10.4196/kjpp.2016.20.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuo H, Shu F, She S, Yang M, Zou XQ, Huang J, Hu HD, Hu P, Ren H, Peng SF. Sorcin induces gastric cancer cell migration and invasion contributing to STAT3 activation. Oncotarget. 2017;8(61):104258–104271. doi: 10.18632/oncotarget.22208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamagishi N, Nakao R, Kondo R, Nishitsuji M, Saito Y, Kuga T, Hatayama T, Nakayama Y. Increased expression of sorcin is associated with multidrug resistance in leukemia cells via up-regulation of MDR1 expression through cAMP response element-binding protein. Biochem Biophys Res Commun. 2014;448(4):430–436. doi: 10.1016/j.bbrc.2014.04.125. [DOI] [PubMed] [Google Scholar]
- 22.Wang SL, Tam MF, Ho YS, Pai SH, Kao MC. Isolation and molecular cloning of human sorcin a calcium-binding protein in vincristine-resistant HOB1 lymphoma cells. Biochim Biophys Acta. 1995;1260(3):285–293. doi: 10.1016/0167-4781(94)00206-i. [DOI] [PubMed] [Google Scholar]
- 23.Sugawara I, Mizumoto K, Ohkochi E, Hamada H, Tsuruo T, Mori S. Immunocytochemical identification and localization of the Mr 22,000 calcium-binding protein (sorcin) in an adriamycin-resistant myelogenous leukemia cell line. Jpn J Cancer Res. 1989;80(5):469–474. doi: 10.1111/j.1349-7006.1989.tb02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong Z, Sun P, Chu H, Zhu H, Sun D, Chen J. Overexpression of sorcin in multidrug-resistant human breast cancer. Oncol Lett. 2014;8(6):2393–2398. doi: 10.3892/ol.2014.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Xu Y, Tan Y, Qi J, Xiao Y, Yang C, Zhu Z, Xiong D. Sorcin, an important gene associated with multidrug-resistance in human leukemia cells. Leuk Res. 2006;30(4):469–476. doi: 10.1016/j.leukres.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Tong W, Sun D, Wang Q, Suo J. Sorcin enhances metastasis and promotes epithelial-to-mesenchymal transition of colorectal cancer. Cell Biochem Biophys. 2015;72(2):453–459. doi: 10.1007/s12013-014-0486-3. [DOI] [PubMed] [Google Scholar]
- 27.Xu P, Jiang YF, Wang JH. shRNA-mediated silencing of sorcin increases drug chemosensitivity in myeloma KM3/DDP and U266/ADM cell lines. Int J Clin Exp Pathol. 2015;8(3):2300–2310. [PMC free article] [PubMed] [Google Scholar]
- 28.Lei X, Liang Y, Chen J, Xiao S, Lei J, Li J, Duanmu J, Jiang Q, Liu D, Tang C. Sorcin predicts poor prognosis and promotes metastasis by facilitating epithelial-mesenchymal transition in hepatocellular carcinoma. Sci Rep. 2017;7(1):10049. doi: 10.1038/s41598-017-10365-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Hu Y, Li S, Yang M, Yan C, Fan D, Zhou Y, Zhang Y, Yague E, Xiong D. Sorcin silencing inhibits epithelial-to-mesenchymal transition and suppresses breast cancer metastasis in vivo. Breast Cancer Res Treat. 2014;143(2):287–299. doi: 10.1007/s10549-013-2809-2. [DOI] [PubMed] [Google Scholar]
- 30.Meyers MB, Schneider KA, Spengler BA, Chang TD, Biedler JL. Sorcin (V19), a soluble acidic calcium-binding protein overproduced in multidrug-resistant cells. Identification of the protein by anti-sorcin antibody. Biochem Pharmacol. 1987;36(14):2373–2380. doi: 10.1016/0006-2952(87)90606-x. [DOI] [PubMed] [Google Scholar]
- 31.SRI sorcin [Homo sapiens (human)]. National Center for Biotechnology Information, U.S. National Library of Medicine. https://www.ncbi.nlm.nih.gov/gene?cmd=retrieve&list_uids=6717
- 32.Roberts D, Meyers MB, Biedler JL, Wiggins LG. Association of sorcin with drug resistance in L1210 cells. Cancer Chemother Pharmacol. 1989;23(1):19–25. doi: 10.1007/BF00258452. [DOI] [PubMed] [Google Scholar]
- 33.GenBank: AAA92155.1; sorcin [Homo sapiens]. National Center for Biotechnology Information, U.S. National Library of Medicine. https://www.ncbi.nlm.nih.gov/protein/AAA92155.1
- 34.Lee WP. Purification, cDNA cloning, and expression of human sorcin in vincristine-resistant HOB1 lymphoma cell lines. Arch Biochem Biophys. 1996;325(2):217–226. doi: 10.1006/abbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 35.Xie X, Dwyer MD, Swenson L, Parker MH, Botfield MC. Crystal structure of calcium-free human sorcin: a member of the penta-EF-hand protein family. Protein Sci. 2001;10(12):2419–2425. doi: 10.1110/ps.36701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Bliek AM, Meyers MB, Biedler JL, Hes E, Borst P. A 22-kd protein (sorcin/V19) encoded by an amplified gene in multidrug-resistant cells, is homologous to the calcium-binding light chain of calpain. EMBO J. 1986;5(12):3201–3208. doi: 10.1002/j.1460-2075.1986.tb04630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilari A, Johnson KA, Nastopoulos V, Verzili D, Zamparelli C, Colotti G, Tsernoglou D, Chiancone E. The crystal structure of the sorcin calcium binding domain provides a model of Ca2+-dependent processes in the full-length protein. J Mol Biol. 2002;317(3):447–458. doi: 10.1006/jmbi.2002.5417. [DOI] [PubMed] [Google Scholar]
- 38.Ilari A, Fiorillo A, Poser E, Lalioti VS, Sundell GN, Ivarsson Y, Genovese I, Colotti G. Structural basis of Sorcin-mediated calcium-dependent signal transduction. Sci Rep. 2015;5:16828. doi: 10.1038/srep16828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colotti G, Poser E, Fiorillo A, Genovese I, Chiarini V, Ilari A. Sorcin, a calcium binding protein involved in the multidrug resistance mechanisms in cancer cells. Molecules. 2014;19(9):13976–13989. doi: 10.3390/molecules190913976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi J, Liu N, Zhou Y, Tan Y, Cheng Y, Yang C, Zhu Z, Xiong D. Overexpression of sorcin in multidrug resistant human leukemia cells and its role in regulating cell apoptosis. Biochem Biophys Res Commun. 2006;349(1):303–309. doi: 10.1016/j.bbrc.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 41.Qu Y, Yang Y, Liu B, Xiao W. Comparative proteomic profiling identified sorcin being associated with gemcitabine resistance in non–small cell lung cancer. Med Oncol. 2010;27(4):1303–1308. doi: 10.1007/s12032-009-9379-5. [DOI] [PubMed] [Google Scholar]
- 42.Sun Y, Wang C, Meng Q, Liu Z, Huo X, Sun P, Sun H, Ma X, Peng J, Liu K. Targeting P-glycoprotein and SORCIN: dihydromyricetin strengthens anti-proliferative efficiency of adriamycin via MAPK/ERK and Ca(2+)-mediated apoptosis pathways in MCF-7/ADR and K562/ADR. J Cell Physiol. 2018;233(4):3066–3079. doi: 10.1002/jcp.26087. [DOI] [PubMed] [Google Scholar]
- 43.Aggarwal BB, Kunnumakkara AB, Harikumar KB, Gupta SR, Tharakan ST, Koca C, Dey S, Sung B. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Ann N Y Acad Sci. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maddalena F, Laudiero G, Piscazzi A, Secondo A, Scorziello A, Lombardi V, Matassa DS, Fersini A, Neri V, Esposito F. Sorcin induces a drug-resistant phenotype in human colorectal cancer by modulating Ca(2+) homeostasis. Cancer Res. 2011;71(24):7659–7669. doi: 10.1158/0008-5472.CAN-11-2172. [DOI] [PubMed] [Google Scholar]
- 45.Hu Y, Cheng X, Li S, Zhou Y, Wang J, Cheng T, Yang M, Xiong D. Inhibition of sorcin reverses multidrug resistance of K562/A02 cells and MCF-7/A02 cells via regulating apoptosis-related proteins. Cancer Chemother Pharmacol. 2013;72(4):789–798. doi: 10.1007/s00280-013-2254-2. [DOI] [PubMed] [Google Scholar]
- 46.Landriscina M, Laudiero G, Maddalena F, Amoroso MR, Piscazzi A, Cozzolino F, Monti M, Garbi C, Fersini A, Pucci P. Mitochondrial chaperone Trap1 and the calcium binding protein Sorcin interact and protect cells against apoptosis induced by antiblastic agents. Cancer Res. 2010;70(16):6577–6586. doi: 10.1158/0008-5472.CAN-10-1256. [DOI] [PubMed] [Google Scholar]
- 47.Lalioti VS, Ilari A, O'Connell DJ, Poser E, Sandoval IV, Colotti G. Sorcin links calcium signaling to vesicle trafficking, regulates Polo-like kinase 1 and is necessary for mitosis. PLoS One. 2014;9(1):e85438. doi: 10.1371/journal.pone.0085438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto T, Hisamatsu Y, Ohkusa T, Inoue N, Sato T, Suzuki S, Ikeda Y, Matsuzaki M. Sorcin interacts with sarcoplasmic reticulum Ca(2+)-ATPase and modulates excitation-contraction coupling in the heart. Basic Res Cardiol. 2005;100(3):250–262. doi: 10.1007/s00395-005-0518-7. [DOI] [PubMed] [Google Scholar]
- 49.Fowler MR, Colotti G, Chiancone E, Higuchi Y, Seidler T, Smith GL. Complex modulation of L-type Ca(2+) current inactivation by sorcin in isolated rabbit cardiomyocytes. Pflugers Arch - Eur J Physiol. 2009;457(5):1049–1060. doi: 10.1007/s00424-008-0575-5. [DOI] [PubMed] [Google Scholar]
- 50.Zamparelli C, Macquaide N, Colotti G, Verzili D, Seidler T, Smith GL, Chiancone E. Activation of the cardiac Na(+)-Ca(2+) exchanger by sorcin via the interaction of the respective Ca(2+)-binding domains. J Mol Cell Cardiol. 2010;49(1):132–141. doi: 10.1016/j.yjmcc.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ali R, Huang Y, Maher SE, Kim RW, Giordano FJ, Tellides G, Geirsson A. miR-1 mediated suppression of Sorcin regulates myocardial contractility through modulation of Ca2+ signaling. J Mol Cell Cardiol. 2012;52(5):1027–1037. doi: 10.1016/j.yjmcc.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 52.Gupta K, Sirohi VK, Kumari S, Shukla V, Manohar M, Popli P, Dwivedi A. Sorcin is involved during embryo implantation via activating VEGF/PI3K/Akt pathway in mice. J Mol Endocrinol. 2018;60(2):119–132. doi: 10.1530/JME-17-0153. [DOI] [PubMed] [Google Scholar]
- 53.Krakhmal NV, Zavyalova MV, Denisov EV, Vtorushin SV, Perelmuter VM. Cancer invasion: patterns and mechanisms. Acta Nat. 2015;7(2):17–28. [PMC free article] [PubMed] [Google Scholar]
- 54.Deng L, Su T, Leng A, Zhang X, Xu M, Yan L, Gu H, Zhang G. Upregulation of soluble resistance-related calcium-binding protein (sorcin) in gastric cancer. Med Oncol. 2010;27(4):1102–1108. doi: 10.1007/s12032-009-9342-5. [DOI] [PubMed] [Google Scholar]
- 55.Wu Q, Yang Z, Nie Y, Shi Y, Fan D. Multi-drug resistance in cancer chemotherapeutics: mechanisms and lab approaches. Cancer Lett. 2014;347(2):159–166. doi: 10.1016/j.canlet.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Parekh HK, Deng HB, Choudhary K, Houser SR, Simpkins H. Overexpression of sorcin, a calcium-binding protein, induces a low level of paclitaxel resistance in human ovarian and breast cancer cells. Biochem Pharmacol. 2002;63(6):1149–1158. doi: 10.1016/s0006-2952(02)00850-x. [DOI] [PubMed] [Google Scholar]
- 57.Demidova NS, Ilyinskaya GV, Shiryaeva OA, Chernova OB, Goncharova SA, Kopnin BP. Decreased sensitivity of multidrug-resistant tumor cells to cisplatin is correlated with sorcin gene co-amplification. Neoplasma. 1995;42(4):195–201. [PubMed] [Google Scholar]
- 58.Dabaghi M, Rahgozar S, Moshtaghian J, Moafi A, Abedi M, Pourabutaleb E. Overexpression of SORCIN is a prognostic biomarker for multidrug-resistant pediatric acute lymphoblastic leukemia and correlates with upregulated MDR1/P-gp. Genet Test Mol Biomarkers. 2016;20(9):516–521. doi: 10.1089/gtmb.2016.0031. [DOI] [PubMed] [Google Scholar]
- 59.Li G, Tan Y, Yang C, Zhao C, Zhao H, Wang J, Xue Y, Han M, Qian L, Zhao C. Expression and clinical implications of the soluble drug resistance-related calcium-binding protein (sorcin) gene in leukemia patients. Zhonghua Xue Ye Xue Za Zhi. 2002;23(6):293–296. [PubMed] [Google Scholar]
- 60.Hamada H, Okochi E, Oh-hara T, Tsuruo T. Purification of the Mr 22,000 calcium-binding protein (sorcin) associated with multidrug resistance and its detection with monoclonal antibodies. Cancer Res. 1988;48(11):3173–3178. [PubMed] [Google Scholar]
- 61.Li J, Chen BA, Zhu MS, Gao F, Ding JH, Gao C, Sun YY, Cheng J, Wang J, Zhao G. Influence of tetrandrine on SORCIN gene expression in K562/A02 cell line. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2008;16(1):65–69. [PubMed] [Google Scholar]
- 62.Li GY, Liu JZ, Zhang B, Yang M, Chen SG, Hou M, Wang LX. Tegillarca granosa extract Haishengsu (HSS) suppresses expression of mdr1, BCR/ABL and sorcin in drug-resistant K562/ADM tumors in mice. Adv Med Sci. 2013;58(1):112–117. doi: 10.2478/v10039-012-0069-8. [DOI] [PubMed] [Google Scholar]
- 63.Li GY, Zhang L, Liu JZ, Chen SG, Xiao TW, Liu GZ, Wang JX, Wang LX, Hou M. Marine drug Haishengsu increases chemosensitivity to conventional chemotherapy and improves quality of life in patients with acute leukemia. Biomed Pharmacother. 2016;81:160–165. doi: 10.1016/j.biopha.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Adrenocortical carcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=acc_tcga&Z_SCORE_THRESHOLD=2.0&RPPA_SCORE_THRESHOLD=2.0&data_priority=0&case_set_id=acc_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=acc_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=acc_tcga_gistic
- 65.Bladder cancer (TCGA, Cell 2017). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=blca_tcga_pub_2017&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=blca_tcga_pub_2017_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=blca_tcga_pub_2017_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=blca_tcga_pub_2017_gistic
- 66.Breast invasive carcinoma (TCGA, Cell 2015). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=brca_tcga_pub2015&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=brca_tcga_pub2015_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=brca_tcga_pub2015_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=brca_tcga_pub2015_gistic
- 67.Breast invasive carcinoma (TCGA, Nature 2012). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=brca_tcga_pub&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=brca_tcga_pub_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=brca_tcga_pub_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=brca_tcga_pub_gistic
- 68.Breast invasive carcinoma (TCGA, Provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=brca_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=brca_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=brca_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=brca_tcga_gistic
- 69.Brain lower grade glioma (TCGA, Provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=lgg_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=lgg_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=lgg_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=lgg_tcga_gistic
- 70.Breast cancer (METABRIC, Nature 2012 & Nat Commun 2016). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=brca_metabric&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=brca_metabric_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=brca_metabric_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=brca_metabric_cna
- 71.Bladder urothelial carcinoma (TCGA, Nature 2014). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=blca_tcga_pub&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=blca_tcga_pub_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=blca_tcga_pub_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=blca_tcga_pub_gistic
- 72.Bladder urothelial carcinoma (TCGA, Provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=blca_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=blca_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=blca_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=blca_tcga_gistic
- 73.Colorectal adenocarcinoma (TCGA, Nature 2012). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=coadread_tcga_pub&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=coadread_tcga_pub_cna_seq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=coadread_tcga_pub_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=coadread_tcga_pub_gistic
- 74.Colorectal adenocarcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=coadread_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=coadread_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=coadread_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=coadread_tcga_gistic
- 75.Cholangiocarcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=chol_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=chol_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=chol_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=chol_tcga_gistic
- 76.Cervical squamous cell carcinoma and endocervical adenocarcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=cesc_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=cesc_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=cesc_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=cesc_tcga_gistic
- 77.Esophageal carcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=esca_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=esca_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=esca_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=esca_tcga_gistic
- 78.Glioblastoma (TCGA, Cell 2013). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=gbm_tcga_pub2013&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=gbm_tcga_pub2013_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=gbm_tcga_pub2013_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=gbm_tcga_pub2013_gistic
- 79.Glioblastoma multiforme (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=gbm_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=gbm_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=gbm_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=gbm_tcga_gistic
- 80.Head and neck squamous cell carcinoma (TCGA, Nature 2015) http://www.cbioportal.org/index.do?cancer_study_id=hnsc_tcga_pub&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=hnsc_tcga_pub_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=hnsc_tcga_pub_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=hnsc_tcga_pub_gistic
- 81.Head and neck squamous cell carcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=hnsc_tcga&Z_SCORE_THRESHOLD=2.0&RPPA_SCORE_THRESHOLD=2.0&data_priority=0&case_set_id=hnsc_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=hnsc_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=hnsc_tcga_gistic
- 82.Kidney renal papillary cell carcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=kirp_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=kirp_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=kirp_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=kirp_tcga_gistic
- 83.Kidney renal clear cell carcinoma (TCGA, Nature 2013). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=kirc_tcga_pub&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=kirc_tcga_pub_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=kirc_tcga_pub_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=kirc_tcga_pub_gistic
- 84.Kidney renal clear cell carcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=kirc_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=kirc_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=kirc_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=kirc_tcga_gistic
- 85.Lymphoid neoplasm diffuse large B-cell lymphoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=dlbc_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=dlbc_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=dlbc_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=dlbc_tcga_gistic
- 86.Ovarian serous cystadenocarcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=ov_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=ov_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=ov_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=ov_tcga_gistic
- 87.Pancreatic adenocarcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=paad_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=paad_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=paad_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=paad_tcga_gistic
- 88.Prostate adenocarcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=prad_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=prad_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=prad_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=prad_tcga_gistic
- 89.Papillary thyroid carcinoma (TCGA, Cell 2014). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=thca_tcga_pub&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=thca_tcga_pub_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=thca_tcga_pub_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=thca_tcga_pub_gistic
- 90.Stomach adenocarcinoma (TCGA, Nature 2014). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=stad_tcga_pub&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=stad_tcga_pub_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=stad_tcga_pub_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=stad_tcga_pub_gistic
- 91.Stomach adenocarcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=stad_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=stad_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=stad_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=stad_tcga_gistic
- 92.Skin cutaneous melanoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=skcm_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=skcm_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=skcm_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=skcm_tcga_gistic
- 93.Sarcoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=sarc_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=sarc_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=sarc_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=sarc_tcga_gistic
- 94.Testicular germ cell cancer (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=tgct_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=tgct_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=tgct_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=tgct_tcga_gistic
- 95.Thyroid carcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=thca_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=thca_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=thca_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=thca_tcga_gistic
- 96.Uterine corpus endometrial carcinoma (TCGA, Nature 2013). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=ucec_tcga_pub&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=ucec_tcga_pub_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=ucec_tcga_pub_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=ucec_tcga_pub_gistic
- 97.Uterine corpus endometrial carcinoma (TCGA, provisional). cBioPortal Version 1.15.1-SNAPSHOT. http://www.cbioportal.org/index.do?cancer_study_id=ucec_tcga&Z_SCORE_THRESHOLD=2&RPPA_SCORE_THRESHOLD=2&data_priority=0&case_set_id=ucec_tcga_cnaseq&gene_list=SRI&geneset_list=+&tab_index=tab_visualize&Action=Submit&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=ucec_tcga_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=ucec_tcga_gistic