Abstract
Cancer cells and embryonic tissues share a number of cellular and molecular properties, suggesting that induced pluripotent stem cells (iPSCs) may be harnessed to elicit anti-tumor responses in cancer vaccines. RNA-sequencing revealed that human and murine iPSCs express tumor-associated antigens, and we show here a proof-of principle for using irradiated iPSCs in autologous anti-tumor vaccines. In a prophylactic setting, iPSC vaccines prevent tumor growth in syngeneic murine breast cancer, mesothelioma, and melanoma models. As an adjuvant, the iPSC vaccine inhibited melanoma recurrence at the resection site and reduced metastatic tumor load, which was associated with fewer Th17 cells and increased CD11b+GR1hi myeloid cells. Adoptive transfer of T cells isolated from vaccine treated-tumor-bearing mice inhibited tumor growth in unvaccinated recipients, indicating that the iPSC vaccine promotes an antigen-specific anti-tumor T cell response. Our data suggest a generalizable strategy for multiple types of cancer that could prove highly valuable in clinical immunotherapy.
Keywords: Pluripotent stem cells, immunotherapy, prophylactic vaccination, adjuvant therapy, shared epitopes, breast cancer, melanoma, mesothelioma, resection area, metastases, immune profiling
Introduction
Nearly a century ago, researchers observed that immunization with embryonic materials led to the rejection of transplanted tumors (Brewer et al., 2009). More recently, studies identified shared transcriptome profiles and antigens on various tumor cells and embryonic cells (Ben-Porath et al., 2008; Ghosh et al., 2011). This has led to the hypothesis that embryonic stem cells (ESCs) could be used as immunization agents to promote an anti-tumor response. A major advantage of whole cell vaccination over traditional vaccines, which consist of inactivated organisms or protein products, is that a broad range of antigens can be presented to T-cells, including unknown antigens (Palena et al., 2006; Yaddanapudi et al., 2012). However, the use of fetal and embryonic materials as vaccines to induce anti-tumor immunity has not yet advanced beyond animal models, owing largely to ethical challenges surrounding these therapies.
Since the discovery of induced pluripotent stem cells (iPSCs) (Takahashi et al., 2007; Takahashi and Yamanaka, 2006), pluripotent cells from a patient’s own tissues can be created that share nearly identical gene expression and surface markers profiles with ESCs (Bock et al., 2011; Mallon et al., 2013; Mallon et al., 2014; Soldner et al., 2009), circumventing a major ethical roadblock. Additionally, the tumorigenic (Kooreman and Wu, 2010; Lee et al., 2013) and immunogenic (de Almeida et al., 2014; Zhao et al., 2011) properties of iPSCs with autologous transplantation suggest potential efficacy in cancer vaccination. Importantly, autologous iPSCs may provide a more accurate and representative panel of patient’s tumor immunogens than non-autologously-derived ESCs. Here we test the hypothesis that iPSCs may work as a whole cell-based vaccine that presents T-cells with a broad heterogeneity in cancer-related epitopes.
Results
Human and murine iPSCs express tumor-specific and tumor-associated antigens
We first performed RNA sequencing on 11 different human iPSC clones to compare expression profiles from a selected cancer-related gene list to human ESCs (hESCs), cancer tissues, and healthy tissues (Figure S1A). Based on this gene list, we found human iPSCs cluster with hESCs and the cancer tissues, revealing important gene expression overlap in cancer genes between different cancer types and iPSCs. The upregulation of a subset of these genes was then also validated in murine iPSCs and ESCs (Figure S1B). These findings suggest the possibility of using iPSCs in different species to prime the host in developing immunity against known and perhaps, also unknown tumor-specific antigens (TSA) and tumor-associated antigens (TAA).
iPSC-vaccine primed mice mount strong B- and T-cell responses against breast cancer in vitro and in vivo
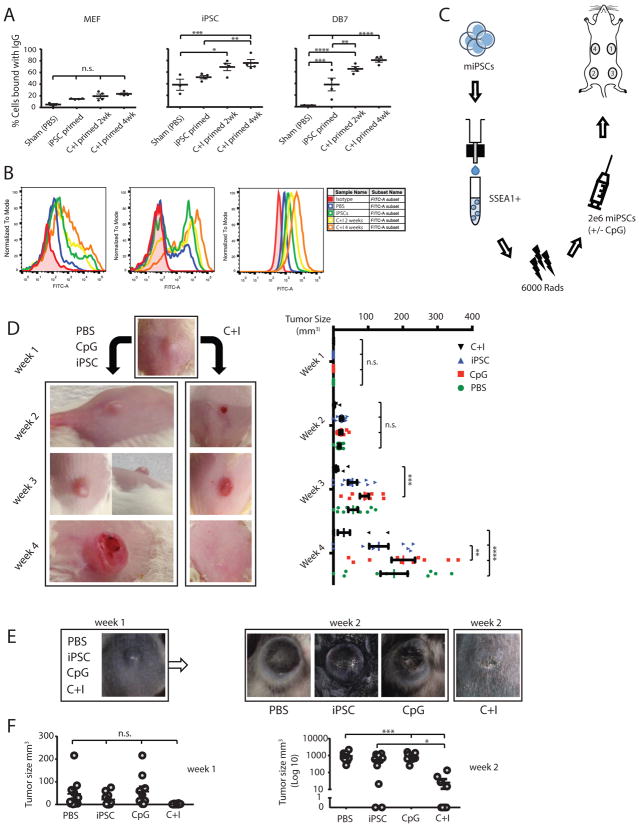
Using FVB strain iPSCs (Figure S2A, D) and the adjuvant CpG, proven to be successful in tumor vaccination (Gilkeson et al., 1998; Goldstein et al., 2011; Mor et al., 1997; Mukherjee et al., 2007), we observed an effective immune response to a murine breast cancer (DB7) with a CpG and iPSCs (C+I) combination. In brief, we first established the effect of CpG and an optimal vaccination schedule. We primed FVB mice with iPSCs or C+I for two weeks or four weeks and found the strongest in vitro T-cell responses to DB7 tumor lysate in the C+I four-week group (Figure S2E, F). In addition, a vaccination schedule of four weeks with the C+I combination resulted in the highest IgG binding (80.0 ± 3.4%) to DB7 and was therefore used for subsequent vaccination rounds (Figure 1A, B). After optimizing the schedule (Figure 1C), we proceeded with the vaccination of 40 FVB mice, divided into four groups: 1) PBS, 2) CpG only, 3) iPSCs only, and 4) C+I. After four once-weekly vaccinations, 5x104 DB7 cancer cells were injected subcutaneously and tumor size was monitored using caliper measurement. After one week, all mice presented with a similar lesion at the injection site that regressed in 7 out of 10 C+I treated mice and progressed to larger tumors in the other groups (Figure 1D; Figure S3A, B). Four weeks after tumor inoculation, five mice per group were sacrificed to analyze the immune profiles in blood, spleen, and draining lymph nodes (dLNs). The other five mice per group were used for long-term survival studies for up to one year. Most were sacrificed in the first two weeks after the end of the experiment when their tumor exceeded 1 cm3. However, two mice in the C+I treatment group survived one year and had antibody titers against iPSCs and DB7 similar to the start of the experiment and were able to fully reject 5x104 cancer cells upon reintroduction (Figure S3C, D). The control mice in this experiment, primed with iPSC-derived endothelial cells, were unable to mount IgG responses to the DB7 cell line, thereby ruling out the possibility that the culturing conditions with FBS-containing media could be responsible for the cross-reactivity or endogenous murine leukemia viral antigens.
Figure 1. Assessing the optimal vaccination schedule, followed by successful prophylactic treatment of breast cancer and melanoma in mice.
(A) Optimal vaccination was set to C+I vaccination for four weeks, as assessed by % IgG binding to DB7, without a significant increase in non-specific MEF binding (n=3 control animals, n=4 iPSC primed animals, n=4 C+I primed 2 week, and n=4 C+I primed 4 week animals, mean±s.e.m., ANOVA with Tukey’s multiple comparison test). (B) Representative FACS plot of serum IgG binding of PBS 4-week, iPSC 4-week, C+I 2-week, or C+I 4-week vaccinated mice to embryonic fibroblasts, iPSCs, and DB7 cancer cells. As a control sample for differentiated cells, a partly differentiated cell culture was included in the analysis. This is shown by IgG positive and negative cells, indicating that the IgG binding is specific to the undifferentiated portion of the analyzed cells. C+I 4-week vaccinated mice showed the best IgG binding to DB7 breast cancer cells. (C) Schema showing vaccine preparation consists of sorting murine iPSCs for pluripotency, irradiation, resuspension in adjuvant solution, and subcutaneous injection in the flank, sites 1 to 4. (D) Vaccination of FVB mice with the C+I vaccine resulted in a complete rejection of the cancer cells in 7 out of 10 mice by four weeks and overall reductions in DB7 tumor size (n=10 per group; representative images; left panel). Quantification of the data presented in right panel. (E) Vaccination of C57BL/6 mice with the C+I vaccine resulted in significant reduction of melanoma sizes initiated by the aggressive B16F0 melanoma cell line by week two (n=8 PBS, n=9 iPSC primed, n=10 CpG primed, and n=9 C+I primed). (F) Quantification of the tumor size data presented in panel E. Data in D and F expressed as mean±s.e.m., ANOVA with Tukey’s multiple comparison test, *p<0.05, **p<0.001, ***p<0.001, ****p<0.0001).
C+I vaccination provides breast cancer and melanoma immunity by upregulating antigen presentation and T-helper/cytotoxic T-cell activity
To test the effectiveness of our vaccine in targeting multiple cancer types, an additional experiment was performed using the melanoma cell line B16F0, which is syngeneic to the C57BL/6 mouse strain. C57BL/6 iPSCs were generated (Figure S2B, D) and 40 mice were again divided into PBS, CpG, iPSCs, and C+I groups and treated for four weeks. Following this, 5x104 B16F0 cells were subcutaneously injected in the lower back. Tumor growth assessment by caliper measurement showed significantly lower tumor progression by week 2 in the C+I group (Figure 1E, F; Figure S3E, F). Due to large tumor sizes in the control groups, the mice were sacrificed two weeks after tumor injection. Afterwards, the immune cell profiles in blood, dLNs, and spleens were analyzed using flow cytometry. Cytometric analysis showed a significant decrease in CD4+CD25+ FoxP3+ regulatory T-cells (T-regs) in blood and an increase in effector/memory helper T-cells in dLNs two weeks after tumor injections in C57BL/6 mice (Figure 2A, B), as well as increased percentages of mature antigen presenting cells (APCs) (Figure 2C).
Figure 2. Prophylactic vaccination leads to increased antigen presentation in dLNs and subsequent effector/memory T-cell responses in dLNs and spleen.
(A) Two weeks after B16F0 introduction, iPSC and C+I vaccinated mice showed a significant reduction in percentages of regulatory T-cells (CD4+CD25+FoxP3+) and an increase in effector/memory helper T-cells (CD4+CD44+) in the peripheral blood of C+I vaccinated mice. At that point, only limited upregulation of effector/memory cytotoxic T-cells (CD8+CD44+) was seen. (B) The dLNs in the C+I group had significantly higher percentages of effector/memory helper T-cells and (C) increased antigen presentation by mature antigen presenting cells (APCs) such as macrophages (CD11b+F4/80+MHC-II+CD86+) and dendritic cells (CD11c+MHC-II+CD86+). (D) C+I vaccinated FVB mice showed increased percentages of activated cytotoxic T-cells (CD8+Granzyme-B+) in spleens four weeks after DB7 introduction. (E) dLNs of these mice revealed an increased frequency of mature antigen-presenting macrophages as well as (F) effector/memory helper T-cells and cytotoxic T-cells. (n=5 per group, mean±s.e.m., ANOVA with Tukey’s multiple comparison test, *p<0.05, **p<0.001, ***p<0.001, ****p<0.0001).
At four weeks, FVB mice in the C+I vaccinated group had significant increases in the effector/memory cytotoxic T-cells in the spleen and dLNs (Figure 2D, F). The tumor-specificity of these cytotoxic T-cells was further confirmed by increased secretion of IFN-γ by splenocytes isolated from C+I vaccinated mice in response to DB7 tumor lysate (Figure 3A, B, Figure S4A, B). As with the C57BL/6 mice, upregulation of mature APCs and helper T-cells was also seen in dLNs of FVB mice (Figure 2E, F). Both mouse strains remained healthy throughout the study and showed no signs of autoimmune responses due to the vaccine in serum and in tissues (Figure S4C–F). Lastly, the effectiveness of the C+I vaccine was assessed in the more clinically relevant orthotopic model of breast cancer. Significant tumor size differences were seen as early as one week after orthotopic transfer of cancer cells in C+I vaccinated mice compared to vehicle control, followed by further tumor reduction over the course of three weeks (Figure 3C, E). Using an additional group of orthotopic breast cancer mice, in vivo tumor specificity was tested by adoptively transferring splenocytes from C+I vaccinated or vehicle (PBS+CpG) vaccinated mice into these tumor-bearing mice (Figure 3D). This resulted in a significant reduction of tumor sizes in the C+I vaccinated group compared to the vehicle-vaccinated group (Figure 3F).
Figure 3. Tumor specific properties of C+I vaccine in-vitro as well as in-vivo in an orthotopic tumor model of breast cancer.
(A) Dual ELISPOT assay (red: granzyme-β, blue: IFN-γ) for immune cell activation of splenocytes in the C+I vaccinated group (iPSC vaccinated; n=6) compared to CpG alone (vehicle; n=4) group upon exposure to iPSC lysate and DB7 lysate (also see Figure S4A, B). (B) Significant increase of number of IFN-γ spots in C+I vaccinated group compared to the vehicle group. (Spots calculated by Adobe Photoshop software based on color differences. ***p<0.001, Student’s t-test). (C) Representative images of tumor volume in C+I vaccinated mice compared to vehicle mice in an orthotopic tumor model of breast cancer at three weeks after tumor inoculation. (D) Representative images of tumor volume in tumor bearing mice after receiving adoptive transfer of splenocytes from C+I vaccinated mice compared to vehicle mice in an orthotopic tumor model of breast cancer at three weeks after adoptive transfer. (E) Quantification of the results from panel C shows a significant reduction of tumor volume in C+I vaccinated mice compared to vehicle mice in an orthotopic tumor model of breast cancer over the course of three weeks. (F) Significant reduction of tumor volume in tumor-bearing mice from panel D over the course of three weeks after adoptive transfer of splenocytes from C+I vaccinated mice (n=7) compared to mice receiving splenocytes from vehicle vaccinated mice (n=8). (***p<0.001, one way ANOVA).
Tumor immunity in C+I vaccinated mice is the result of shared epitopes between iPSCs and cancer cells
To test whether the C+I vaccine provides immunity against shared epitopes between iPSCs and cancer cells, we performed additional experiments to assess two-way immunity by demonstrating (1) cancer immunity by C+I primed T-cells and (2) iPSC immunity by tumor-experienced lymphocytes (TELs). For the first experiment, isolated T-cells from C+I vaccinated or vehicle (PBS+CpG) vaccinated mice were adoptively transferred to a group of tumor-bearing orthotopic breast cancer mice (n=7 per group) and tumor growth was measured over the course of four weeks (Figure 4A). This resulted in a significant reduction of tumor sizes in the C+I vaccinated group compared to the vehicle vaccinated group as early as one week after the adoptive transfer (Figure 4B). For the second experiment, another batch of C+I (n=10) or vehicle (n=10) vaccinated mice were inoculated with breast cancer cells and tumor growth was measured at one week (Figure 4C, D). Afterwards, we extracted TELs from the dLNs near the tumor site (Torcellan et al., 2017). These TELs were then adoptively transferred to iPSC-inoculated NOD-SCID mice (5x106 TELs per mouse; n=4 per group) and teratoma development was measured for four weeks. Significant reduction in teratoma sizes was seen at four weeks in the NOD-SCID mice receiving TELs from C+I animals that were able to reject the DB7 tumor cells, whereas mice receiving TELs from vehicle vaccinated animals developed large teratomas (Figure 4E, F).
Figure 4. Shared epitopes between cancer cells and iPSCs provide T-cells with two-way immunity.
(A) Representative images from tumor-bearing mice four weeks after receiving T-cells from either vehicle or C+I vaccinated mice. (B) Quantification of the tumor sizes of tumor-bearing mice in Figure 4A over the course of four weeks after receiving T-cells from vehicle or C+I vaccinated mice, as measured by caliper. Significant reduction of tumor sizes was seen as early as 1 week after the adoptive transfer of T-cells from C+I vaccinated mice and remained significantly reduced during the course of the experiment (** p<0.01, *** p<0.001; Student T-test). (C) Representative images of vehicle (CpG only; n=10) and C+I vaccinated mice (n=10) at one week after orthotopic tumor inoculation. (D) Quantification of the tumor sizes displayed in Figure 4C shows robust rejection of the DB7 breast cancer cells (*** p<0.001; Student T-test). (E) Representative images of NOD-SCID mice receiving TELs from the dLNs from vehicle or C+I vaccinated mice from the experiment in Figure 4C (n=4 per group). (F) Images from the teratomas isolated from mice in Figure 4E (top panel), and quantification reveals a significant reduction in teratoma sizes (bottom panel) from the C+I immunized group (* p<0.05; Student T-test).
C+I vaccination in a mesothelioma model elicits a pro-inflammatory profile for tumor infiltrating lymphocytes (TILs)
As an alternative model for prophylactic treatment, we selected the mesothelioma cell line AC29, syngeneic to CBA/J mice. Again, CBA/J iPSCs were created (Figure S2C, D) and mice were vaccinated for four weeks with PBS (P), CpG and iPSCs (C+I), or CpG with irradiated AC29 cancer cells (C+A) as a positive control. Afterwards, 2x106 AC29 cells (A) or 2x106 iPSCs (I) were injected subcutaneously, and after one week the TILs were analyzed for their immune profile and TCR sequences. Immune profiling was performed with cytometry by time of flight (CyTOF) analysis using a phenotype and intracellular staining kit, which revealed an increased presence of effector/memory CD4+ (24.0%) and CD8+ T-cells (22.4%), with a reduction in T-regs in the C+I/A group (1.9%) compared to P/A control (21.1%, 14.2% and 3.0%, respectively) (Figure 5A). Using Citrus (cluster identification, characterization and regression) analysis (Bruggner et al., 2014), B-cells and T-cells expressing IL-2, IL-4, and IL-5 were found to be predictive of tumor regression in C+I vaccinated mice compared to the PBS control group (Figure 5B; Figure S5A–B, D). Interestingly, systemic cytokine levels were significantly lower in the vaccinated group and were found to correlate with the positive control mice showing tumor rejection (C+I/iPSC; C+A/AC29) (Figure 6A; Figure S6A–B). TCR sequencing in the PBS control group revealed an overlap in T-cell clones that are commonly present in thymus and spleen (Figure S6C). In contrast, the TCRs in the C+I group were more diverse among different mice. In addition, there was a generally lower frequency of the clones in the thymus and more similar frequencies in the spleen, likely because of mouse-specific responses to the C+I vaccine (Figure 6B; Figure S6D). Interestingly, there was one TCR clone that was shared by 4 of 5 mice in the C+I group, but was not present in any of the other groups; this clone was also extremely rare in naïve mice.
Figure 5. TILs show a pro-inflammatory phenotype with B-cell and CD4+ T-cell anti-tumor responses.
(A) One week after 2x106 AC29 (A) mesothelioma cells were injected in CpG+iPSC (C+I) vaccinated mice (n=5), TILs in this C+I/A group showed an increase in the frequency of effector/memory CD4+ and CD8+ cells and a reduction in T-reg numbers, compared to PBS (P) vaccinated mice (n=5; P/A group), as assessed by SPADE analysis of CyTOF data. The positive control groups, C+I vaccinated and CpG+AC29 (C+A) vaccinated mice, fully rejected iPSCs (n=5; C+I/I) and AC29 cells (n=5; C+A/A), respectively, with a subsequently enhanced presence of monocytes and macrophages and stromal cells. (B) Citrus analysis of CyTOF data revealed that higher levels of IL-2, IL-4, and IL-5 in B-cell and helper T-cell clusters in the C+I mice are responsible for the intra-tumoral immune response.
Figure 6. C+I vaccination leads to a systemic immune profile similar to positive control groups of tumor rejection and upregulation of vaccine-specific T-cell clones.
(A) Luminex analysis of serum from the different treatment groups at one week after tumor cell introduction reveals a significantly lower presence of systemic cytokines in the positive control mice (C+I/iPSC, C+A/AC29) compared to PBS control mice (PBS/AC29). The C+I/AC29 group follows a similar trend as the positive control samples (C+I/iPSC and C+A/AC29, ANOVA with Tukey’s multiple comparison test, *p<0.05, **p<0.001, ***p<0.001). (B) Among C+I vaccinated mice (C+I1 through C+I5/AC29), there was greater unique vaccine-associated variance within the TILs, whereas PBS-vaccinated mice (PBS1 through 5/AC29) demonstrated a higher uniformity among T-cells that are commonly present in lymphoid organs (Figure S6C-D).
C+I adjuvant therapy after tumor resection leads to decreased tumor load in resection areas and dLNs
To assess the effectiveness of the vaccine as an adjuvant therapy after tumor resection, we next injected 5x104 B16F0 tumor cells subcutaneously in the lower back of C57BL/6 mice and R2- or R1-resected mice after two weeks. R2-resected mice had no visible recurrence of melanoma in the resection area (RA) after receiving two adjuvant rounds of C+I vaccine, whereas PBS control vaccinated mice had visible tumors within the RAs (Figure S7A). R1-resected mice were vaccinated for four weeks with the C+I vaccine (n=10), CpG (n=10), and PBS (n=8) (Figure 7A), after which dLNs and RAs were analyzed using a tumor-specific primer designed to detect and quantify the B16F0 melanoma line (Figure S7B–G). Tumor load in the dLNs was reduced in both CpG only and the C+I vaccine groups, indicating that CpG acted as a potent adjuvant to induce tumor degradation upon near-tumor injection (Figure S7H). Interestingly, in areas more distant from the vaccination sites, only the C+I vaccinated group had significantly lower tumor recurrence in the RA (Figure 7B). Systemically, this is explained by reactivation of the immune system (Chung et al., 2013; Dolcetti et al., 2010; Numasaki et al., 2003), as well as a reduction of B16 melanoma-promoting Th17 cells (He et al., 2010) compared to the control groups (Figure 7C; Figure S5C, E).
Figure 7. Adjuvant vaccination after tumor resection leads to clean RAs and reactivation of the immune system to target cancer cells.
(A) B16F0 tumor-bearing mice underwent R1 tumor resection, were randomized into different treatment groups, and were vaccinated with either C+I, CpG, or PBS for four weeks. (B) DNA from skin biopsies (*) in resection areas (RAs) showed a significant reduction in the percentage of tumor cells after four vaccination rounds with the C+I vaccine, as assessed by ddPCR. (C) Vaccination post-tumor resection led to a reduction of Th17 cells (CD4+CD62L+TCR-b+(IL-2/IL-17A); CD4+CD62L+CD44+TCR-b+(IL-17A)) and an increased presence of TNF-α expressing myeloid cells (CD11b+CD44+GR1hi(TNF-a)) and IL-4 expressing CD19+CD62L+CD44+ B-cells (n=8 PBS, n=10 CpG, n=10 C+I, mean±s.e.m., ANOVA with Tukey’s multiple comparison test, *p<0.05). SQ: subcutaneous injection.
Discussion
Tumor establishment and progression involve highly proliferative hypoimmunogenic cells that evade the surveillance of the immune system. Therefore, new avenues within the field of cancer treatment are being pursued to target cancer by reactivating the immune system. One way researchers are trying to achieve this is by using chimeric antigen receptors (CARs) with promising results (Lee et al., 2015; Maude et al., 2014; Maus et al., 2014). The idea behind this therapy is to create a cancer-specific antigen receptor and couple this to an effector cell (e.g., T-cell), with newer generations of CARs that might even incorporate the co-stimulatory pathways. However, thus far results have been mixed with some patients relapsing, possibly due to loss of expression of the targeted antigen (Grupp et al., 2013; Maude et al., 2014). One way to circumvent this would be to identify new tumor-specific antigens, but large numbers of tumor antigens are still unknown.
Pluripotent cells and tissues share known and likely also unknown TSAs and TAAs with cancer cells, and therefore could be a potential agent to prime an immune system to target cancer. This modified cell would then function as a surrogate cell type that resembles the targeted cancer type. A few groups have pursued the use of embryonic cells for priming the immune system in targeting cancer, but thus far without showing efficacy and safety for the treatment of various types of cancer (Li et al., 2009; Yaddanapudi et al., 2012). In addition, they still rely on the use of ethically concerning ESCs and a genetically modified cell line as an adjuvant (Yaddanapudi et al., 2012), making these treatments less suitable for personalized clinical translation.
In this study, we showed that prophylactic immunization of several mouse strains with an iPSC-based vaccine produces an effective immune response to multiple cancer types by upregulation of mature APCs in the dLNs with a subsequent increase in helper T-cells and cytotoxic T-cells locally, and later on, systemically. Interestingly, this led to a systemically favorable T-effector/T-reg ratio, which has been found to reduce tolerizing conditions (Zou, 2005). With our adoptive transfer data on transplantation of C+I primed splenocytes into tumor-bearing mice, we demonstrated the tumor-specificity of our iPSC vaccine, which based on our in vitro data, was likely the result of IFN-γ+ effector T-cells. The lifespan of these IFN- γ+ effector T-cells (8–10 days) would also explain why there was tumor regression after the adoptive transfer of C+I primed splenocytes in the orthotopic model of breast cancer for the first two weeks, after which a small increase in tumor size was seen (Dooms and Abbas, 2002). To test if the immunity created by the vaccine is the result of shared epitopes between iPSCs and cancer cells, we performed adoptive transfer of C+I-primed T-cells to breast cancer bearing mice and adoptive transfer of TELs to iPSC-inoculated NOD-SCID mice. With these experiments, we were able show that C+I-primed T-cells rejected the DB7 breast cancer cells and that the primed TELs were able to reduce teratoma size or stop teratoma formation altogether. This "two-way immunity" demonstrates shared epitopes between iPSCs and cancer cells.
Looking into the early intra-tumor immune response, we found mainly B-cells and T-cells expressing IL-2, IL-4, and IL-5 with a switch from common T-cell clones to rarer vaccine-associated T-cell clones. Most of these high frequency clones vary between the vaccinated mice, suggesting that each mouse mounts a cross-reactive immune response based on different epitopes from the iPSCs. This provides further evidence that iPSCs share a larger repertoire of cancer-related epitopes, indicating that this surrogate cell type could be a potential candidate to limit the chances of immune evasion by the cancer cells as has occasionally reported in CAR therapy (Grupp et al., 2013; Maude et al., 2014).
Another issue with CAR therapy is organ toxicity from cytokine storms upon transfusion of CAR T-cells (Morgan et al., 2010). As we showed in our CBA/J mouse data using the Luminex assay, systemic cytokine levels are low, and instead there is a localized immune response within the tumor similar to the positive control group of tumor rejection. In addition, tissue analysis of our mice at different time points after vaccination did not show any increases in immune cells within heart and kidney tissues compared to negative control groups, nor were elevated levels of anti-nuclear antigen (ANA) IgG seen in serum from C+I vaccinated mice.
As a therapy for established melanomas, the C+I vaccine was not effective in reducing tumor growth, which is likely due to an established immunosuppressive tumor microenvironment that could potentially be remedied by combining the C+I vaccine with checkpoint blockade treatment (Le et al., 2015). However, as an adjuvant therapy after R1 resection of melanoma, we found that the C+I vaccine reactivated the immune system in rejecting remnant melanoma cells by the systemic upregulation of IL-4 expressing B-cells, TNF-α-expressing CD11b+GR1hi myeloid cells, as well as a reduction of tumor-promoting Th17 cells. In this setting, the cancer epitope heterogeneity of iPSCs, combined with the ease of their generation, may make this therapy readily available as adjuvant immunotherapy for multiple cancer types within weeks after diagnosis.
This last point is crucial for immunotherapy, because it is commonly known that that the tumor microenvironment could limit effectiveness of tumor immunity by suppressive immune cells residing within the tumor. After debulking of the tumor and disrupting the tumor microenvironment to create an “inflamed” tumor site, immunotherapy should be more effective (Gajewski et al., 2013). This is demonstrated in our R1 resected melanoma model, which again emphasizes the need for a multi TSA- and TAA-based vaccine to be readily available at time of tumor resection. Having a surrogate whole cell vaccine with multiple known and, likely also unknown, TSAs and TAAs available at such a short time after diagnosis would allow the priming of the immune system to target large numbers of cancer-specific antigens at a time when cancer cells are most vulnerable.
Even though an overlap was seen in murine and human TAA genes, it is important to note the differences in murine and human immunology before extrapolation above-mentioned data to humans (Mestas and Hughes, 2004). Further testing of the C+I vaccine on human samples ex vivo should therefore be performed to show efficacy in humans.
Taken together, our data show the feasibility of creating broad tumor immunity against multiple cancer types using an iPSC-based vaccine that presents the immune system with large quantities of tumor antigens. Compared to current immunotherapy strategies, our iPSC vaccine is capable of reactivating the immune system to target established cancers without therapy-associated adverse effects and can be created within a few weeks after diagnosis. These beneficial properties make this iPSC vaccine a potential option for personalized adjuvant immunotherapy shortly after conventional primary treatment of cancer.
Star Methods
Experimental Models and Subject Details
Animal models
Young adult female FVB, C57BL/6J, and CBA/J mice (6–8 weeks old) were used. Animals were randomly assigned to the different treatment groups. Tumor-bearing mice were excluded from the experiment if their physical condition required euthanasia before the experimental deadline, due to criteria such as tumor sizes exceeding 1 cm3, visible distress, pain, or illness. All experiments were approved by the Stanford University Administrative Panel of Laboratory Animal Care (APLAC).
Generation of murine iPSCs from fibroblasts
Fibroblasts from FVB, C57BL/6J, and CBA/J mice (The Jackson Laboratory, Bar Harbor, Maine) were grown in DMEM Glutamax (ThermoFisher Scientific, Waltham, MA, USA) with 20% fetal bovine serum (FBS) and 1x NEAA (ThermoFisher Scientific). Fibroblasts were dissociated using TrypLE Express (ThermoFisher Scientific) and 1x106 fibroblasts were resuspended in electroporation buffer (Neon system, ThermoFisher Scientific). Cells were transfected with a codon-optimized mini-intronic plasmid (coMIP) containing the four reprogramming factors Oct4, Sox2, c-Myc, and KLF4 (Diecke et al., 2015). After transfection, cells were plated on irradiated mouse embryonic feeder (MEF) cells and cultured in DMEM with 15% FBS, 1x NEAA, and 10 ng/ml murine leukemia inhibiting factor (mLIF; EMD Millipore, MA, USA). After iPSC colonies started to appear, they were manually picked and transferred to a fresh feeder layer. The iPSC colonies were grown for a few passages and then transferred to 0.2% gelatin-coated plates to be sorted for SSEA-1 using magnetic bead sorting (Miltenyi, Germany) to keep a pure undifferentiated population. For characterization, iPSCs were stained for Oct4, Nanog, Sox2 (Santa Cruz, CA, USA), SSEA1, and c-Myc (EMD Millipore) to assess pluripotency. In addition, a teratoma assay was performed on all iPSC lines by transplantation of 1x106 iPSCs in the hindlimb of NOD-SCID mice (The Jackson Laboratory). All cell lines were tested for mycoplasma contamination and found to be negative.
Cancer cell lines and implantation
The breast cancer line DB7 was a gift from Dr. Joe Smith (University of Utah, USA). It was derived from FVB mice and is a non-metastatic cell line. The B16F0 melanoma cell line was purchased from ATCC (Manassas, VA, USA) and is syngeneic to C57Bl/6 mice. It has low-grade lymphoid metastatic potential to the lungs. The AC29 mesothelioma cancer line was purchased from Sigma-Aldrich (St. Louis, MO, USA). The cancer lines were grown in DMEM, 10% FBS under normal culture conditions. For the C57BL/6 and FVB mice, 5x104 cancer cells were resuspended in 100 μl PBS and injected subcutaneously in the lower back of the mice. The CBA/J mice were injected with 2x106 cancer cells. Tumor growth was assessed weekly by caliper measurement. At the end of the study, tumors were explanted and gross examination of draining lymph nodes and lung tissue was performed for any metastases.
Method Details
CpG + iPSC vaccine preparation and immunization
For each mouse, 2x106 SSEA-1-sorted syngeneic murine iPSCs were irradiated at 6,000 rads prior to injection. Cells were suspended in 100 μl of 5 μM CpG (Invivogen, San Diego, USA), dissolved in PBS, and loaded into ¼ cc insulin syringes (Terumo). Mice were placed in an induction chamber and anesthetized with 2% isoflurane (Isothesia, Butler Schein) in 100% oxygen with a delivery rate of 2μl/min until the loss of righting reflex, as per APLAC guidelines at Stanford University. Immunization was performed by subcutaneous injection of the vaccine in the flanks of the mice, with the injection site changing every week. Mice were monitored weekly for early signs of auto-reactivity to the vaccine by weight measurements and gross examination of overall appearance. Vaccination preparation and dosage were the same for the prophylactic and adjuvant treatment experiments. The prophylactic vaccination studies were replicated several times in the same mouse strain, different mouse strains and by different investigators. The investigator analyzing the tumor sizes and data from the adjuvant treatment experiment was blinded for the different treatment groups.
Mixed lymphocyte reaction (MLR)
Spleens were isolated, minced, and filtered through a 70 μm strainer. After multiple washes with glucose-containing RPMI, the pellet was resuspended in ACK lysis buffer for removal of red blood cells. CFSE-labeled (ThermoFisher Scientific) splenocytes from C+I vaccinated mice were then plated at a density of 1x105 cells per 100 ul in a 96-well plate and incubated for 72 hr with another 100 μl solution of DB7 tumor lysate, ranging from 1–10 μg. After 72 hr, the plate was spun down and the supernatant isolated for cytokine analysis using the mouse Th1/Th2/Th17 Cytokines Multi-Analyte ELISArray kit (Qiagen, Hilden, Germany), and the cell pellet was analyzed with the LSR-II Flow Cytometer to assess T-cell proliferation.
IgG binding assay
Cells were washed multiple times with PBS and resuspended in 100 μl FACS buffer with the addition of 2 μl of serum from the vaccinated mice and incubated for 30 minutes on 4°C. Following this, cells were washed multiple times and incubated with an anti-IgG FITC secondary antibody (ThermoFisher Scientific) for another 20 min on 4°C. As an isotype control an IgG antibody, pre-adsorbed for murine IgG and IgM, was included. The cells were then analyzed using the LSR-II Flow Cytometer.
Histopathology of explanted organs
At time of sacrifice, the heart and kidneys were explanted from vaccinated mice and processed for histopathology. Briefly, the organs were fixed overnight in 4% paraformaldehyde and transferred to 70% ethanol for 24 hr. Fixed samples were embedded in paraffin and 5 μm sections were cut and stained with hematoxylin and eosin (H&E) for histological analysis by a pathologist.
Isolation of inflammatory cells and serum from blood, spleen, tumor, and dLNs
FVB, C57BL/6, and CBA/J experimental mice were sacrificed at 4, 2, and 1 week(s), respectively, after tumor inoculation. Tissues were isolated from the mice and placed in a digestion buffer containing RPMI, FBS, collagenase, DNAse, trypsin inhibitor and HEPES, then minced and placed in a shaker at 37°C for 45 min. Samples were than filtered through a 70 μm strainer, spun down, and resuspended in ACK lysis buffer to remove any red blood cells. After lysis, the cell suspension was washed with PBS and used for subsequent analyses. Additionally, dissociated tumors were passed through a Percoll gradient to remove non-immune cells and isolate tumor-infiltrating leukocytes (TILs). Blood was collected in two separate tubes per mouse for PBMC (EDTA containing tube) and serum isolation (uncoated tube).
Staining of inflammatory cells for FACS analysis
Inflammatory cells isolated from blood and tissues were resuspended in 200 μl FACS buffer (DPBS, 2% FBS and 200 μM EDTA), blocked with a FcR-blocking Reagent (BD Pharmingen, San Diego, CA, USA), and divided into 2 tubes. One tube was stained with a surface marker panel, containing CD3, CD4, CD25 (eBioscience), CD8a, CD44, CD45 (Biolegend), and the intracellular markers Granzyme-B (eBioscience) and FoxP3 (Biolegend). The second tube was stained for F4/80, MHC-II (eBioscience), CD86 (BD Biosciences), CD11b, CD11c, NK1.1, Ly6-G, and CD45 (Biolegend). A rat IgG2b κ isotype control was included for CD44, FoxP3 (Biolegend), and MHC-II (eBioscience). A rat IgG2a κ isotype was included for the Granzyme-B (eBioscience) and CD86 (BD Biosciences) staining. For CD25 staining, the IgG1 κ isotype (eBioscience) was included. In both panels, the fixable viability dye 780 (Invitrogen) was added to exclude dead cells from the analysis. Extracellular staining was performed prior to fixing and permeabilizing the samples for staining with intracellular markers. Samples were analyzed on the LSR-II Flow Cytometer analyzer in the Beckmann FACS facility (Stanford University).
Teratoma formation
Teratoma formation was performed as previously described (Nelakanti et al., 2015), with the exception of site of injection. For this manuscript, a flank injection was preferred over a hindlimb injection to ensure easier access for over-time measurements of teratoma size. In brief, 1x106 iPSCs were resuspended in growth factor reduced Matrigel™ (50 μl per injection) and injected in the flank of immunodeficient mice (NOD-SCID IL2Rgammanull;NSG). Teratoma sizes at site of injection were measured over time using a caliper. After four weeks, mice were sacrificed and the teratomas harvested for final measurements.
Generation of tumor lysate
1x107 tumor cells were used from in vitro culture and resuspended in 1 ml of PBS. The cell suspension was frozen to -80°C for 45 min and then thawed on 37°C for 30 min. This process was repeated for a total of three times. Afterwards, the suspension was spun down and the supernatant, containing tumor lysate, was isolated for protein concentration measurements using the Pierce BCA Protein Assay Kit (ThermoFisher Scientific).
Luminex multiplex cytokine assay
Production of various cytokines was measured in cell culture supernatant and serum samples using a multiplex-Luminex platform (LabMap200 System; Luminex) in conjunction with Panomics antibodies at the Human Immune Monitoring Center at Stanford University.
ELISPOT assay
Splenocytes (5x105) were isolated as described above and co-cultured with either iPSC or DB7 lysate (35 μg) for the duration of 37 hr, after which the secretion of granzyme-β and IFN-γ was measured by Enzyme-Linked ImmunoSpot (ELISPOT) according to the manufacturer’s instructions (cat# ELD5819, R&D Systems, Diaclone). Adobe Photoshop CS6 software was used for the calculation of size and number of IFN-γ positive spots.
Adoptive transfer of splenocytes and T-cells
C+I vaccinated and vehicle vaccinated mice were sacrificed and their splenocytes isolated, as previously described (Galvan et al., 2015; Naas et al., 2010; Sodhi et al., 1985). In brief, the spleens were digested and passed through a 70 μm strainer. Afterwards red blood cells were lysed with ACK lysis buffer (cat# 118-156-101, Quality biology, INC.) and the remaining splenocytes washed with PBS. The splenocytes were then dissolved in 200 μl PBS solution and intravenously injected in an orthotopic model of breast cancer by tail vein injection. For the adoptive transfer of T-cells, the procedure is as described above, with the addition of a magnetic bead sorting using the Pan T-cell isolation kit to acquire CD3+ T-cells (#130-095-130, Miltenyi, Germany) after the final washing step.
Orthotopic tumor model
FVB mice were injected with 2x106 DB7 tumor cells directly into the mammary fat pad tissue, as previously described (Kocaturk and Versteeg, 2015). The range of cancer cell number was based on previous reports (Chen et al., 2011; Evans et al., 2014) and was set at 2x106 DB7 cancer cells after validating the model and achieving a tumor incidence of 100%.
Isolation of TELs from draining lymph nodes
After sacrificing the C+I and vehicle-vaccinated mice, their dLNs were isolated, minced and passed through a 70 μm strainer. After washing the cells with PBS, the T-cell portion of the TELs were isolated using the Pan T-cell isolation kit to acquire CD3+ T-cells (#130-095-130, Miltenyi, Germany).
Anti-nuclear antibody (ANA) ELISA
Murine blood was collected from PBS, CpG only, or CpG-iPSCs vaccinated mice and the plasma was separated from the blood via centrifugation for 15 min at 1000g. The plasma samples were diluted at 1:200 with sample dilution. The concentrations of anti-nuclear antibodies (IgG) were determined using an ELISA kit, according to the manufacturer's instructions (Antibodies-Online; antibodies-online.com). For this experiment, four biological replicates per group were used and for each biological replicate three technical replicates were included.
Cytometry by Time of Flight (CyTOF)
Immune cells were isolated from explanted tissues according to aforementioned methods. Cells were stained with the Mouse Spleen/Lymph Node Phenotyping kit, the Mouse Intracellular Cytokine I Panel kit, and the viability dye Cisplatin (Fluidigm, South San Francisco, CA). Cells were resuspended in MaxPar water at a concentration of 1x105–1x108 cells per ml with the addition of normalization beads and ran on a CyTOF2 (Fluidigm) machine. Following this, the data were normalized using the normalization beads. The data were analyzed using the Cytobank online software for spanning tree progression analysis of density-normalized events (SPADE) (Qiu et al., 2011).
PCR detection of the large genomic deletion in CDKN2A
Primers were designed to detect the junction of the large deletion in CDKN2A of the B16 melanoma cell line (Figure S7B). Each 25 μl PCR reaction solution contained 1.25 units of PrimeSTAR® GXL DNA Polymerase (Clontech) and 50–100 ng of genomic DNA extracted by DNeasy Blood & Tissue Kit (Qiagen) (Figure S7C). PCR products were then analyzed by Sanger sequencing and aligned with the gene database in NCBI (Figure S7D).
T-cell Receptor (TCR) sequencing
The DNA from the TILs infiltrating the AC29 tumors was isolated using the DNeasy Blood & Tissue kit (Qiagen). Samples were submitted to Adaptive Biotechnologies (Seattle, WA) for a survey level TCR sequencing. The minimum DNA content from the submitted samples was 150 ng per sample with DNA quality A260/280 between 1.8 to 2.0. Data analysis as well as assessment of TCR clonality between samples were performed in collaboration with Adaptive Biotechnologies. In brief, a list of TCR clones within each sample and their frequencies within the DNA sample were provided. For the T-cell overlap search, the amino acid sequences of the clones appearing in 4 or 5 of the samples in the two sample groups were compared. Data from the CI treatment group and the PBS control group were ruled comparable with similar average productive unique values (PBS: 3582.2, CI: 3005.4).
Quantification and Statistical Analysis
All values are expressed as mean ± s.d. or mean ± s.e.m. as indicated. Intergroup differences were appropriately assessed by either unpaired two-tailed Student’s t-test or one-way/two-way analysis of variance (ANOVA) with Tukey’s posthoc test using PRISM GraphPad software. * P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Cluster identification, characterization and regression (Citrus)
In brief, based on hierarchical clustering and a regularized regression model, Citrus generates a list of stratifying clusters and behaviors from multidimensional data. In addition, it can describe their features (e.g., intracellular cytokines) and provide a predictive model for newly acquired data or validation samples. The stratifying features from these clusters were plotted as median expression on the x-axis (Figure 5B, 7C; Figure S5B). CyTOF data were analyzed using Cytobank and gated for viable single cells, after which the FCS files were uploaded in the GUI from Citrus 0.8 and the script was run in R (version 3.0.3). For the analysis of the splenocytes exposed to B16F0 tumor lysate, Citrus analysis was performed with 10,000 sampling events with 0.2% (567 events) minimum clustering. For the TILs, Citrus analysis was based on 1,000 sampling events with 500 events minimum clustering. Clustering features were found to be of interest with a cv.min and cv.fdr.constrained of less than 25.
Quantification of tumor load for melanoma by digital droplet PCR (ddPCR)
Primers and probe were designed to detect 3 SNPs (colored in red) that are specific to the B16 melanoma cell line. DNA was extracted from the tumor resection area and dLNs of C57BL/6 mice four weeks after R1 tumor resection using the DNeasy Blood & Tissue Kit (Qiagen). Each ddPCR reaction solution was reconstituted to a final volume of 20 μL using 40 to 50 ng of DNA template and ddPCR™ Supermix for Probes, without dUTP (BioRad). Each sample was quantified by using 2 probes: MT probe to assess the tumor load, and TaqMan®Copy Number TFRC probe (Mm00000692_cn, ThermoFisher) to assess the cell amount (Figure S7E, F). The final primer and probe concentrations were 900 nM and 250 nM, respectively. Droplet formation was carried out using a QX100 droplet generator with 20 μL of PCR reaction solution. A rubber gasket was placed over the cartridge and loaded into the droplet generator. The emulsion (~35 μl in volume) was then slowly transferred using a multichannel pipette to a 96-Well twin.tec™ PCR Plates (Eppendorf). The plate was then heat-sealed with foil and the emulsion was cycled to end point per the manufacturer's protocol with annealing temperature at 62.5°C. The samples were then read using a BioRad QX100 reader. The standard curve was created for different amounts of tumor load, including 0%, 1%, 5%, 10%, 25%, 50%, 75%, 90%, 95%, 99%, and 100%, and linear regression equation was utilized to quantify the tumor load for each DNA sample (Figure S7G). Following are the sequences of the primers and probes for detecting tumor load:
Forward primer, 5’ACTAGCCAGAGGATCTTAAAGACT3’;
Reverse primer, 5’GCCATCACTGGAAAGAGAGGC3’;
Mutant Probe,
 (Blackhole Quencher)3’; (red indicating mutant-specific
alleles).
(Blackhole Quencher)3’; (red indicating mutant-specific
alleles).
Analysis of RNA-sequencing data
The pair-end human RNA-seq data from normal and cancer cell lines were downloaded from the GEO repository of ENCODE project (http://www.genome.gov/encode/) (Consortium, 2011). The RNA-seq of iPSCs reprogrammed by different methods were generated from previous publication (Churko et al., 2017). The fastq files of the sequencing were aligned to the human genome (hg19) by Hisat (https://ccb.jhu.edu/software/hisat/index.shtml) (Kim et al., 2015). The aligned reads were assigned by HT-seq (http://www-huber.embl.de/users/anders/HTSeq) (Anders et al., 2015) to the gene annotation (version 19) provided by the GenCode project (http://www.gencodegenes.org/) (Harrow et al., 2012). The gene expression levels were estimated and normalized by DESeq (https://bioconductor.org/packages/release/bioc/html/DESeq.html) (Anders and Huber, 2010; Anders et al., 2013). The gene expression levels were extracted from a curated selection of cancer-related genes described in seven datasets (allOnco; Bushman Lab, University of Pennsylvania) after which a heatmap was generated using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E).
Analysis of murine RNA sequencing data
The pair-end/single-end RNA-seq data from tissues were downloaded from the mouse ENCODE project (http://mouseencode.org/). The mouse iPSC data were downloaded from GSE36294 (Chang et al., 2014). The fastq files of the sequencing were aligned to the mouse genome (mm10) by Hisat (https://ccb.jhu.edu/software/hisat/index.shtml). The aligned reads were assigned by HT-seq (http://www-huber.embl.de/users/anders/HTSeq) to the gene annotation (version M3) provided by the GenCode project (http://www.gencodegenes.org/). The gene expression levels were estimated and normalized by DESeq (https://bioconductor.org/packages/release/bioc/html/DESeq.html). The heatmap was generated using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E).
Data and Software Availability
T-cell receptor sequencing data have been deposited in the immuneACCESS platform under the following accession number: doi:10.21417/B7B648
KEY RESOURCES TABLE
The table highlights the genetically modified organisms and strains, cell lines, reagents, software, and source data essential to reproduce results presented in the manuscript. Depending on the nature of the study, this may include standard laboratory materials (i.e., food chow for metabolism studies), but the Table is not meant to be comprehensive list of all materials and resources used (e.g., essential chemicals such as SDS, sucrose, or standard culture media don’t need to be listed in the Table). Items in the Table must also be reported in the Method Details section within the context of their use. The number of primers and RNA sequences that may be listed in the Table is restricted to no more than ten each. If there are more than ten primers or RNA sequences to report, please provide this information as a supplementary document and reference this file (e.g., See Table S1 for XX) in the Key Resources Table.
Please note that ALL references cited in the Key Resources Table must be included in the References list. Please report the information as follows:
REAGENT or RESOURCE: Provide full descriptive name of the item so that it can be identified and linked with its description in the manuscript (e.g., provide version number for software, host source for antibody, strain name). In the Experimental Models section, please include all models used in the paper and describe each line/strain as: model organism: name used for strain/line in paper: genotype. (i.e., Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J). In the Biological Samples section, please list all samples obtained from commercial sources or biological repositories. Please note that software mentioned in the Methods Details or Data and Software Availability section needs to be also included in the table. See the sample Table at the end of this document for examples of how to report reagents.
SOURCE: Report the company, manufacturer, or individual that provided the item or where the item can obtained (e.g., stock center or repository). For materials distributed by Addgene, please cite the article describing the plasmid and include “Addgene” as part of the identifier. If an item is from another lab, please include the name of the principal investigator and a citation if it has been previously published. If the material is being reported for the first time in the current paper, please indicate as “this paper.” For software, please provide the company name if it is commercially available or cite the paper in which it has been initially described.
-
IDENTIFIER: Include catalog numbers (entered in the column as “Cat#” followed by the number, e.g., Cat#3879S). Where available, please include unique entities such as RRIDs, Model Organism Database numbers, accession numbers, and PDB or CAS IDs. For antibodies, if applicable and available, please also include the lot number or clone identity. For software or data resources, please include the URL where the resource can be downloaded. Please ensure accuracy of the identifiers, as they are essential for generation of hyperlinks to external sources when available. Please see the Elsevier list of Data Repositories with automated bidirectional linking for details. When listing more than one identifier for the same item, use semicolons to separate them (e.g. Cat#3879S; RRID: AB_2255011). If an identifier is not available, please enter “N/A” in the column.
A NOTE ABOUT RRIDs: We highly recommend using RRIDs as the identifier (in particular for antibodies and organisms, but also for software tools and databases). For more details on how to obtain or generate an RRID for existing or newly generated resources, please visit the RII or search for RRIDs.
Please use the empty table that follows to organize the information in the sections defined by the subheading, skipping sections not relevant to your study. Please do not add subheadings. To add a row, place the cursor at the end of the row above where you would like to add the row, just outside the right border of the table. Then press the ENTER key to add the row. Please delete empty rows. Each entry must be on a separate row; do not list multiple items in a single table cell. Please see the sample table at the end of this document for examples of how reagents should be cited.
TABLE FOR AUTHOR TO COMPLETE
Please upload the completed table as a separate document. Please do not add subheadings to the Key Resources Table. If you wish to make an entry that does not fall into one of the subheadings below, please contact your handling editor. (NOTE: For authors publishing in Current Biology, please note that references within the KRT should be in numbered style, rather than Harvard.)
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | ||||
|---|---|---|---|---|---|---|
| Antibodies | ||||||
| PE/Cy7 anti-mouse CD3 | eBioscience | 25-0031-82; RRID: AB_469572 | ||||
| APC anti-mouse CD4 | BD Bioscience | 553051 | ||||
| PE anti-mouse CD4 | eBioscience | 12-0042-83; RRID: AB_465510 | ||||
| PerCP/Cy5.5 anti-mouse CD8 | Biolegend | 100734; RRID: AB_2075238 | ||||
| PE/Cy7 anti-mouse CD11b | Biolegend | 101216; RRID: AB_312799 | ||||
| Pacific Blue anti-mouse/human CD44 | Biolegend | 103020; RRID: AB_493683 | ||||
| APC anti-mouse CD11c | Biolegend | 117310; RRID: AB_313779 | ||||
| FITC anti-mouse CD86 | BD Bioscience | 553691 | ||||
| eFluor 450 anti-mouse F4/80 | eBioscience | 48-4801-82; RRID: AB_1548747 | ||||
| Alexa Fluor 488 anti-mouse FoxP3 | Biolegend | 126406; RRID: AB_1089113 | ||||
| PE anti-mouse Granzyme-B | eBioscience | 12-8898-82; RRID: AB_10870787 | ||||
| Brilliant Violet 510 anti-mouse CD45 | eBioscience | 103137; RRID: AB_2561392 | ||||
| Alexa Fluor 700 anti-mouse CD25 | eBioscience | 56-0251-82; RRID: AB_891422 | ||||
| eFluor 780 Fixable Viability Dye | eBioscience | 65-0865-14 | ||||
| Alexa Fluor 700 anti-mouse GR-1 Ly6 | Biolegend | 108422; RRID: AB_2137487 | ||||
| PerCP/Cy5.5 anti-mouse NK1.1 | Biolegend | 108728; RRID: AB_2132705 | ||||
| PerCP/Cy5.5 anti-mouse DX-5 | Biolegend | 108916; RRID: AB_2129358 | ||||
| PE anti-mouse MHC-II | eBioscience | 12-5321-81; RRID: AB_465928 | ||||
| Alexa Fluor 488 anti-mouse IgG | Thermo Fisher Scientific | A-11001; RRID: | ||||
| Anti-rabbit IgG Cross-adsorbed | Thermo Fisher Scientific | AB_253406931213; RRID: AB_228376 | ||||
| Pacific Blue Rat IgG2b κ isotype control | Biolegend | 400627 | ||||
| Alexa Fluor 488 Rat IgG2b κ isotype control | Biolegend | 400625 | ||||
| PE Rat IgG2b κ isotype control | Thermo-Fisher Scientific | 12-4031-82; RRID: AB_470042 | ||||
| FITC Rat IgG2a κ isotype control | BD Biosciences | 553929 | ||||
| PE Rat IgG2a κ isotype control | eBioscience | 12-4321-80; RRID: AB_1834380 | ||||
| Alexa Fluor 700 Rat IgG1 κ isotype control | eBioscience | 56-4301-80; RRID: AB_494017 | ||||
| Anti SSEA-1 microbeads | Miltenyi Biotec | 130-094-530 | ||||
| Oct 3/4 anti-mouse | Santa Cruz Biotechnology | sc-5279 | ||||
| c-Myc anti-mouse | EMD Millipore | 06-340 | ||||
| SSEA-1 anti-mouse | EMD Millipore | MAB4301 | ||||
| Nanog anti-mouse | Santa Cruz Biotechnology | sc-33760 | ||||
| Sox2 anti-mouse | Santa Cruz Biotechnology | sc-365823 | ||||
| Mouse on Mouse (M.O.M.) Basic Kit | Vector Laboratories | BMK-2202 | ||||
| Fc Block anti-mouse | BD Biosciences | 553141 | ||||
| Bacterial and Virus Strains | ||||||
| Biological Samples | ||||||
| Chemicals, Peptides, and Recombinant Proteins | ||||||
| DMEM, high glucose, GlutaMAX | Gibco | 10569-010 | ||||
| Fetal Bovine Serum | Life Technologies | 26140079 | ||||
| DPBS, no calcium, no magnesium | Gibco | 14190250 | ||||
| MEM Non-Essential Amino Acids | Gibco | 11140050 | ||||
| TrypLE Express | Gibco | 12605-036 | ||||
| CpG ODN 1826 | Invivogen | tlrl-1826-1 | ||||
| Murine Leukemia Inhibitory Factor (mLIF) | EMD Millipore | ESG1106 | ||||
| Gelatin | Sigma Aldrich | G1393-100ML | ||||
| Isothesia | Henry Schein | 029405 | ||||
| Matrigel™ matrix growth factor reduced | BD Biosciences | 356231 | ||||
| RPMI medium 1640 | Life Technologies | 11875-119 | ||||
| ACK lysis buffer | Quality Biological | 118-156-101 | ||||
| Collagenase D | Roche | 11088882001 | ||||
| Deoxyribonuclease I from bovine pancreas | Sigma Aldrich | D4263-5VL | ||||
| Trypsin Inhibitor | Sigma Aldrich | T9003-1G | ||||
| HEPES 1M | Sigma Aldrich | H3662 | ||||
| Percoll GE Density Gradient Media | VWR | 89428-524 | ||||
| UltraPure 0.5M EDTA | Life Technologies | 15575-020 | ||||
| PrimeStar GXL DNA Polymerase | Clontech | R050A | ||||
| ddPCR Supermix for Probes | Bio-Rad | 1863024 | ||||
| Critical Commercial Assays | ||||||
| Neon Transfection System 100μl Kit | Thermo Fisher Scientific | MPK10025 | ||||
| MycoAlert Detection Kit | Lonza | LT07-318 | ||||
| MycoAlert Assay Control Set | Lonza | LT07-518 | ||||
| CFSE Cell Trace staining | Thermo Fisher Scientific | C34554 | ||||
| Mouse Th1/Th2/Th17 Cytokine Multi-Analyte ELISArray | Qiagen | MEM-003A | ||||
| MaxPar Mouse Spleen/Lymph Node Phenotyping kit | Fluidigm | 201306 | ||||
| MaxPar Mouse Intracellular Cytokine I Panel kit | Fluidigm | 201310 | ||||
| Cisplatin viability dye | Fluidigm | 201062 | ||||
| Cytofix/Cytoperm Permeabilization Solution kit | BD Biosciences | BDB554714 | ||||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 | ||||
| Mouse IFN-gamma/Granzyme B Dual-Color ELISpot Kit | R&D Systems | ELD5819 | ||||
| Pan T-cell Isolation Kit II, mouse | Miltenyi Biotec | 130-095-130 | ||||
| Anti-nuclear Antigen (IgG) mouse ELISA Kit | Antibodies-Online | ABIN366290 | ||||
| DNeasy Blood & Tissue kit | Qiagen | 69504 | ||||
| Deposited Data | ||||||
| TCRβ sequencing tumor-infiltrating lymphocytes | This paper | immunoACCESS: doi:10.21417/B7B648 | ||||
| RNA-seq from human somatic/cancer cell lines | ENCODE project | http://www.genome.gov/encode/ | ||||
| RNA-seq from human iPS cells | Churko et al., 2017 | |||||
| RNA-seq form murine somatic cell lines | ENCODE project | http://mouseencode.org/ | ||||
| RNA-seq from murine iPS cells | Chang et al., 2014 | N/A | ||||
| Experimental Models: Cell Lines | ||||||
| EmbryoMax® Primary Mouse Embryo Fibroblasts | EMD Millipore | PMEF-N | ||||
| DB7 breast cancer cells | The University of Utah; Dr. Joe Smith | |||||
| B16F0 melanoma cells | ATCC | CRL-6322 | ||||
| AC29 mesothelioma cells | Sigma Aldrich | 10092308 | ||||
| Experimental Models: Organisms/Strains | ||||||
| Mouse: FVB/NJ | The Jackson Laboratory | 001800 | ||||
| Mouse: C57BL/6J | The Jackson Laboratory | 000664 | ||||
| Mouse: CBA/J | The Jackson Laboratory | 000656 | ||||
| NOD-SCID IL2Rgammanull (NSG) | The Jackson Laboratory | 005557 | ||||
| Oligonucleotides | ||||||
| TaqMan®Copy Number TFRC probe (Mm00000692_cn) | Thermo Fisher Scientific | 4400291 | ||||
| 5’ACTAGCCAGAGGATCTTAAAGACT3’ | This paper | N/A | ||||
| 5’GCCATCACTGGAAAGAGAGGC3’ | This paper | N/A | ||||
|
|
This paper | N/A | ||||
| Recombinant DNA | ||||||
| Codon-optimized mini-intronic plasmid (coMIP) | Diecke et al., 2015 | N/A | ||||
| Software and Algorithms | ||||||
| ImmunoSeq Analyzer | Adaptive Biotechnologies | https://clients.adaptivebiotech.com/ | ||||
| Prism GraphPad 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ | ||||
| Cytobank | Cytobank, Inc. | https://stanford.cytobank.org/cytobank/login | ||||
| Citrus 0.8 | Nolan lab; Stanford, USA. | https://github.com/nolanlab/citrus/wiki/Installing-Citrus | ||||
| R; version 3.0.3 | The R Project | https://www.r-project.org/ | ||||
| Adobe Photoshop CS6 | Adobe | http://www.adobe.com/nl/products/photoshop.html | ||||
| Other | ||||||
| 29 Gauge x ” ½ needle 3/10cc | Terumo Medical | SS30M2913 | ||||
| Multiplex-Luminex platform (LabMap200 System) | HIMC; Stanford University, USA | N/A | ||||
TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and Virus Strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological Samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children's Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124-87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical Commercial Assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; and Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental Models: Cell Lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy- 18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; | Bloomington Drosophila | BDSC:34393; |
| P{TRiP.HMS00609}attP2 | Stock Center | FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein- Volhard, 2000 | ZFIN: ZDB-GENO-060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino:
MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT |
Gene Tools | ZFIN: ZDB- MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand:
UAGGGACUUAGGGUUCUCUCUAGGGACUUAG GGUUCUCUCUAGGGA |
This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and Algorithms | ||
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal. | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
Contact for Reagents and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph C. Wu (joewu@stanford.edu).
Supplementary Material
Figure S1. (Related to Figure 1) Heatmap analysis of RNA sequencing data from human and murine iPSCs, healthy tissues, and cancer tissues. (A) RNA sequencing from 11 human iPSC clones generated with different reprogramming methods (C, E, M, L, U, and S) reveals grouping with hESCs and different cancer types (Encode database) based on a selected list of cancer-related genes described in seven datasets (allOnco; Bushman Lab, University of Pennsylvania). Minicircle (C1, C2); Episomal (E1, E2); mRNA (M1, M2); Lentivirus (L1); microRNA (U1, U2); Sendai virus (S1, S2); hESCs (H7.1); Hepatocellular Carcinoma (HepG2); Pancreatic Carcinoma (PANC1); Cervical Carcinoma (Hela.S3); Glioblastoma/Astrocytoma (U87); fibroblasts (Fib); skin fibroblasts (BJ); lung fibroblasts (Nhlf); and Skeletal Muscle Myoblasts (Hsmm). (B) Based on more than 2,000 human tumor-associated antigens (TAA) and tumor-specific antigens (TSA) RNA, we were able to identify the presence 116 of these tumor antigens in murine iPSCs (MEF.iPSC and APC.iPSC) and ESCs. Similar to the human data, these are highly expressed in the pluripotent populations but only lowly or not expressed in the somatic tissues. MEF-derived iPSCs (MEF.iPSC); APC-derived iPSCs (APC.iPSC); murine ESCs (R1.ESC); murine ESCs (ES.Bruce4); brain tissue (brain_W10); central nervous system (CNS_E18); colon tissue (colon_W08); heart tissue (heart_W10); kidney tissue (kidney_W10); large intestines (large.intestine_W8); limb tissue (limb_E14.5); liver tissue (liver_W10); lung tissue (lung_W10); mammary gland tissue (mammary.gland_W8); and small intestines (small.intestine_W8).
Figure S2. (Related to Figure 1–2, 5) Characterization of iPSCs for pluripotency and in vitro optimization of the vaccination schedule using the adjuvant CpG. Pluripotency staining of (A) FVB, (B) C57BL/6, and (C) CBA/J murine iPSCs with pluripotency markers Oct4, c-Myc, Nanog, and SSEA-1. (D) All three cell lines were able to form teratomas in vivo upon transplantation of 1x106 iPSCs in the hindlimb of NOD/SCID mice. (E) Using the adjuvant (CpG), most effective proliferation of CFSE-labeled splenocytes in response to DB7 lysate was seen in the cytotoxic T-cells in the C+I 4-week vaccination group. (F) This was also the only group with increased cytokine release in the supernatant with a higher dose of DB7 tumor lysate (U=unstimulated splenocytes; S=stimulated splenocytes with allogeneic C57BL/6 splenocytes).
Figure S3. (Related to Figure 1) Tumor growth over time in FVB and C57BL/6 tumor-bearing mice. (A) Representative images of PBS, CpG only, iPSCs only, and CpG+iPSC (C+I) vaccinated mice one week after subcutaneous injection of 5x104 DB7 cancer cells. (B) Images of five mice per group at four weeks after DB7 breast cancer cell injection that were selected for immune profile analysis of spleen, dLNs, and blood. (C) One year after the experiment in Figure 1D, C+I treated mice (n=2) were reintroduced with 5x104 DB7 breast cancer cells and fully rejected the cancer cells by four weeks. In contrast, mice vaccinated with iPSC-derived endothelial cells (iECs; n=4) were unable to reject the cancer cells, which proliferated into large tumors. (D) One year after vaccination and full rejection of DB7 breast cancer cells, C+I primed animals still have high antibody titers against iPSCs and DB7, unlike mice that were vaccinated with iECs. (E) Images of all C57BL/6 mice in the four treatment groups two weeks after B16F0 melanoma cell injection. Several mice in the C+I group showed a regression of tumor sites or inflammation as indicated by local swelling, redness, and, in some cases spontaneous bleeding. (F) Representative images showing mice in the C+I group that were used for immune profile analysis of spleens, dLNs, and blood at time of sacrifice.
Figure S4. (Related to Figure 1, 3–4) C+I vaccination leads to tumor specificity without causing auto-inflammatory responses. Cell culture images from splenocytes isolated from CpG only (vehicle; n=4) and C+I vaccinated mice (n=6) incubated with (A) DB7 tumor lysate or (B) iPSC lysate. Blue spots represent IFNγ secretion by splenocytes and were identified with Adobe Photoshop software in the bottom row images for each condition. (C) Serum from PBS (n=4), CpG (n=4), and CpG+iPSC (n=4) vaccinated mice was isolated after four rounds of vaccination. No significant differences were found in anti-nuclear antibody titers among the different groups (total samples: n=12 with three technical replicates for each sample) (one way ANOVA; p=0.29, ns). (D) For the duration of the experiment, FVB mice (left panel) and C57Bl/6 mice (right panel) were thriving with a healthy appearance and weight increase over time. (E) Representative images of heart (left panel) and kidney samples (right panel) from FVB mice in the three treatment groups revealed no increases in the numbers of inflammatory cells in any of the groups compared to PBS control. (F) Similar findings were seen in the heart (left panel) and kidney tissues (right panel) from C57BL/6 mice.
Figure S5. (Related to Figure 5) Citrus cluster description and features from TILs (CBA/J) and splenocytes from tumor-resected mice (C57BL/6). (A) Clustering analysis of the CyTOF data from Figure 5B using Citrus with their surface marker profile and (B) predictive features of the cytokine profiles from these clusters in the C+I vaccinated group compared to PBS control. (C) Citrus cluster description from Figure 7C. (D) Cross-validation error rate from Citrus analysis in Figure 5B. (E) Cross-validation error rate from Citrus analysis in Figure 7C.
Figure S6. (Related to Figure 6) C+I vaccination in CBA/J mice leads to reduced tumor establishment and a reduction of commonly shared intra-tumor T-cell clones in favor of vaccine-specific clones. (A) Representative image at day 2 of CBA/J mice vaccinated with PBS, CpG+iPSC (C+I), or CpG+AC29 (C+A) and then introduced with either 2x106 AC29 or 2x106 iPSCs. (B) Image (left panel) and tumor sizes (right panel) of all mice at week 1 post-tumor injection, revealing a complete rejection of the iPSCs and cancer cells in the positive control groups (C+I/iPSC = CpG+iPSC vaccinated mice introduced with iPSCs, and C+A/AC29 = CpG+AC29 vaccinated mice introduced with AC29 cells). The last three mice in the C+I/AC29 already showed tumor regression, whereas the first two had severe erythema and swelling at the tumor sites. (C) Frequency of T-cell clones with their receptor sequences in the PBS+AC29 control group if they were shared by 4 or 5 out of 5 mice. TILs had 28 clones that are present in multiple mouse strains in spleen (n=3) as well as thymus (n=3). The larger public clone repertoire within the PBS group therefore likely consisted of shared TCR clones on the basis of chance. (D) Frequency of T-cell clones with their receptor sequences in the C+I/AC29 treatment group if they were shared by 4 or 5 out of 5 mice. TILs only showed six clones that were present in multiple mouse strains in spleen (n=3) as well as thymus (n=3). In these cases, the generally lower frequency of the clones in the thymus and more similar frequencies of the clones in the spleen suggest that the majority of the commonly shared clones in the vaccine group may be more important than public clones. In addition, clone CGARDAGGQDTQYF is extremely rare and was only present in 1 out of 3 spleens in the C57BL/6 mouse strain and was not present in the PBS, C+I/iPSC, or C+A/AC29 groups, indicating it was likely C+I vaccine-specific.
Figure S7. (Related to Figure 7) Adjuvant C+I vaccination prevents the recurrence of tumor after R2 resection of B16F0 melanoma and reduced tumor load in dLNs. (A) B16F0 tumor-bearing mice (n=6) with a tumor size ~1 cm3 received a R2 resection where a small piece of tumor (blue arrow) was left behind in the resection area (RA). C+I vaccinated mice (n=3) had no visible tumor recurrence in the RAs after two rounds of vaccination, whereas the PBS control group (n=3) had visible and palpable tumors. (B) PCR/ddPCR primer development based on a large deletion in CDKN2A in B16 melanoma cells (Fw1-Rv1, Fw2-Rv2). (C) B16 melanoma cell primers were designed to show the deletion by PCR in the melanoma samples without amplification in C57BL/6 wild type samples. (D) PCR products were then validated and analyzed by Sanger sequencing and aligned with the gene database in NCBI. (E–F) Using ddPCR with a mutant probe based on the CDKN2A deletion and a TaqMan®Copy Number probe the quantity of tumor cells within each DNA sample could be assessed. (G) The standard curve was plotted using different concentrations of mutant DNA, revealing a good correlation between % of mutant DNA and the readout from the BioRad QX100 reader. (H) Using ddPCR, a reduction of average tumor load in dLNs of R1 resected mice in the CpG and C+I groups was seen compared to PBS control. If dLNs were positive (>2% threshold), the C+I vaccinated group had the lowest percentage of tumor cells in dLNs.
Acknowledgments
We thank Dr. A. Connolly for the histopathology analysis of the hearts and kidneys; G. Busque for assistance with the caliper measurements of the tumors; and S. Carree and S. Limb for assistance with the graphic design of the cartoons; and J. Churko for providing the RNA sequencing data from the different human iPSC clones. This work was supported in part by grants from the Korean R&D grant HI14C3417 (Y.K.), California Institute of Regenerative Medicine (CIRM) DR2A-05394 and RT3-07798, National Institute of Health (NIH) R01 AI085575, R01 HL113006, R01 HL133272 (J.C.W.), and U19 AI057229 (M.M.D.).
Footnotes
Author Contributions
N.G.K. developed the experimental design, performed and interpreted experiments and wrote the manuscript; Y.K. performed and interpreted experiments; P.E.deA performed preliminary experiments and assisted with the experimental design; V.T. developed the ddPCR primers and performed the ddPCR; S.D. assisted with iPSC generation; N-Y.S. performed biostatistical analyses on the RNA-sequencing data; T-T.W. established the orthotopic breast cancer model and performed the adoptive transfer of splenocytes; H.Y. assisted with iPSC culture; D.D. assisted with the preliminary in vitro experiments; R.N. and T.B. assisted with the vaccinations, tumor measurements, harvest, and processing of tissues for downstream analyses; D.T.P assisted with splenocyte isolation for adoptive transfer and analyzing the RNA-sequencing data; I.B. assisted with the optimization of the vaccination schedule; A.H. provided experimental advice with the T-cell receptor sequencing; P.H.A.Q., J.F.H., R.L., and M.M.D. provided experimental advice and manuscript writing; J.C.W. provided experimental advice and design, manuscript writing, and funding support.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature Genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BG, Mitchell RA, Harandi A, Eaton JW. Embryonic vaccines against cancer: an early history. Exp Mol Pathol. 2009;86:192–197. doi: 10.1016/j.yexmp.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. PNAS. 2014;111:E2770–2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang G, Gao S, Hou X, Xu Z, Liu Y, Kang L, Tao Y, Liu W, Huang B, Kou X, et al. High-throughput sequencing reveals the disruption of methylation of imprinted gene in induced pluripotent stem cells. Cell Res. 2014;24:293–306. doi: 10.1038/cr.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Tang YA, Huang SM, Juan HF, Wu LW, Sun YC, Wang SC, Wu KW, Balraj G, Chang TT, et al. A novel sialyltransferase inhibitor suppresses FAK/paxillin signaling and cancer angiogenesis and metastasis pathways. Cancer Res. 2011;71:473–483. doi: 10.1158/0008-5472.CAN-10-1303. [DOI] [PubMed] [Google Scholar]
- Chung AS, Wu X, Zhuang G, Ngu H, Kasman I, Zhang J, Vernes JM, Jiang Z, Meng YG, Peale FV, et al. An interleukin-17-mediated paracrine network promotes tumor resistance to anti-angiogenic therapy. Nature Medicine. 2013;19:1114–1123. doi: 10.1038/nm.3291. [DOI] [PubMed] [Google Scholar]
- Churko J, Lee J, Ameen M, Gu M, Venkaasubramanian M, Diecke S, Sallam K, Im H, Wang G, Gold JD, et al. Transcriptomic and epigenomic differences in human induced pluripotent stem cells generated from six reprogramming methods. Nat Biomed Eng. 2017;1:826–837. doi: 10.1038/s41551-017-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida PE, Meyer EH, Kooreman NG, Diecke S, Dey D, Sanchez-Freire V, Hu S, Ebert A, Odegaard J, Mordwinkin NM, et al. Transplanted terminally differentiated induced pluripotent stem cells are accepted by immune mechanisms similar to self-tolerance. Nature Communications. 2014;5:3903. doi: 10.1038/ncomms4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diecke S, Lu J, Lee J, Termglinchan V, Kooreman NG, Burridge PW, Ebert AD, Churko JM, Sharma A, Kay MA, et al. Novel codon-optimized mini-intronic plasmid for efficient, inexpensive, and xeno-free induction of pluripotency. Sci Rep. 2015;5:8081. doi: 10.1038/srep08081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolcetti L, Peranzoni E, Ugel S, Marigo I, Fernandez Gomez A, Mesa C, Geilich M, Winkels G, Traggiai E, Casati A, et al. Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF. European Journal of Immunology. 2010;40:22–35. doi: 10.1002/eji.200939903. [DOI] [PubMed] [Google Scholar]
- Dooms H, Abbas AK. Life and death in effector T cells. Nat Immunol. 2002;3:797–798. doi: 10.1038/ni0902-797. [DOI] [PubMed] [Google Scholar]
- Evans MS, Chaurette JP, Adams ST, Jr, Reddy GR, Paley MA, Aronin N, Prescher JA, Miller SC. A synthetic luciferin improves bioluminescence imaging in live mice. Nat Methods. 2014;11:393–395. doi: 10.1038/nmeth.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan DL, O'Neil RT, Foster AE, Huye L, Bear A, Rooney CM, Wilson MH. Anti-tumor effects after adoptive transfer of IL-12 transposon-modified murine splenocytes in the OT-I-melanoma mouse Model. PLoS One. 2015;10:e0140744. doi: 10.1371/journal.pone.0140744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh Z, Huang M, Hu S, Wilson KD, Dey D, Wu JC. Dissecting the oncogenic and tumorigenic potential of differentiated human induced pluripotent stem cells and human embryonic stem cells. Cancer Research. 2011;71:5030–5039. doi: 10.1158/0008-5472.CAN-10-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkeson GS, Conover J, Halpern M, Pisetsky DS, Feagin A, Klinman DM. Effects of bacterial DNA on cytokine production by (NZB/NZW)F1 mice. J Immunol. 1998;161:3890–3895. [PubMed] [Google Scholar]
- Goldstein MJ, Varghese B, Brody JD, Rajapaksa R, Kohrt H, Czerwinski DK, Levy S, Levy R. A CpG-loaded tumor cell vaccine induces antitumor CD4+ T cells that are effective in adoptive therapy for large and established tumors. Blood. 2011;117:118–127. doi: 10.1182/blood-2010-06-288456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Li H, Yusuf N, Elmets CA, Li J, Mountz JD, Xu H. IL-17 promotes tumor development through the induction of tumor promoting microenvironments at tumor sites and myeloid-derived suppressor cells. J Immunol. 2010;184:2281–2288. doi: 10.4049/jimmunol.0902574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocaturk B, Versteeg HH. Orthotopic injection of breast cancer cells into the mammary fat pad of mice to study tumor growth. J Vis Exp. 2015;(96) doi: 10.3791/51967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooreman NG, Wu JC. Tumorigenicity of pluripotent stem cells: biological insights from molecular imaging. J R Soc Interface. 2010;7(Suppl 6):S753–763. doi: 10.1098/rsif.2010.0353.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zeng H, Xu RH, Liu B, Li Z. Vaccination with human pluripotent stem cells generates a broad spectrum of immunological and clinical responses against colon cancer. Stem Cells. 2009;27:3103–3111. doi: 10.1002/stem.234. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Chenoweth JG, Johnson KR, Hamilton RS, Tesar PJ, Yavatkar AS, Tyson LJ, Park K, Chen KG, Fann YC, et al. StemCellDB: the human pluripotent stem cell database at the National Institutes of Health. Stem Cell Research. 2013;10:57–66. doi: 10.1016/j.scr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T, Ghorbani M, Soare C, Scherling N, Muller R, Ghorbani P, Diaz-Mitoma F. Adoptive transfer of splenocytes to study cell-mediated immune responses in hepatitis C infection using HCV transgenic mice. Comparative Hepatology. 2010;9:7. doi: 10.1186/1476-5926-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelakanti RV, Kooreman NG, Wu JC. Teratoma formation: a tool for monitoring pluripotency in stem cell research. Curr Protoc Stem Cell Biol. 2015;32:4A.8.1–4A.8.17. doi: 10.1002/9780470151808.sc04a08s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon BS, Hamilton RS, Kozhich OA, Johnson KR, Fann YC, Rao MS, Robey PG. Comparison of the molecular profiles of human embryonic and induced pluripotent stem cells of isogenic origin. Stem Cell Research. 2014;12:376–386. doi: 10.1016/j.scr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maus MV, Grupp SA, Porter DL, June CH. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood. 2014;123:2625–2635. doi: 10.1182/blood-2013-11-492231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Mor G, Singla M, Steinberg AD, Hoffman SL, Okuda K, Klinman DM. Do DNA vaccines induce autoimmune disease? Human Gene Therapy. 1997;8:293–300. doi: 10.1089/hum.1997.8.3-293. [DOI] [PubMed] [Google Scholar]
- Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Pathangey LB, Bradley JB, Tinder TL, Basu GD, Akporiaye ET, Gendler SJ. MUC1-specific immune therapy generates a strong anti-tumor response in a MUC1-tolerant colon cancer model. Vaccine. 2007;25:1607–1618. doi: 10.1016/j.vaccine.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numasaki M, Fukushi J, Ono M, Narula SK, Zavodny PJ, Kudo T, Robbins PD, Tahara H, Lotze MT. Interleukin-17 promotes angiogenesis and tumor growth. Blood. 2003;101:2620–2627. doi: 10.1182/blood-2002-05-1461. [DOI] [PubMed] [Google Scholar]
- Palena C, Abrams SI, Schlom J, Hodge JW. Cancer vaccines: preclinical studies and novel strategies. Adv Cancer Res. 2006;95:115–145. doi: 10.1016/S0065-230X(06)95004-0. [DOI] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nature Biotechnology. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi A, Tandon P, Sarna S. Adoptive transfer of immunity against solid fibrosarcoma in mice with splenocytes and peritoneal exudate cells obtained after in vitro sensitization and in vivo immunization with cis-dichlorodiamine platinum(II) treated fibrosarcoma cells. Arch Geschwulstforsch. 1985;55:47–61. [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Torcellan T, Hampton HR, Bailey J, Tomura M, Brink R, Chtanova T. In vivo photolabeling of tumor-infiltrating cells reveals highly regulated egress of T-cell subsets from tumors. PNAS. 2017;114:5677–5682. doi: 10.1073/pnas.1618446114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaddanapudi K, Mitchell RA, Putty K, Willer S, Sharma RK, Yan J, Bodduluri H, Eaton JW. Vaccination with embryonic stem cells protects against lung cancer: is a broad-spectrum prophylactic vaccine against cancer possible? PLoS One. 2012;7:e42289. doi: 10.1371/journal.pone.0042289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (Related to Figure 1) Heatmap analysis of RNA sequencing data from human and murine iPSCs, healthy tissues, and cancer tissues. (A) RNA sequencing from 11 human iPSC clones generated with different reprogramming methods (C, E, M, L, U, and S) reveals grouping with hESCs and different cancer types (Encode database) based on a selected list of cancer-related genes described in seven datasets (allOnco; Bushman Lab, University of Pennsylvania). Minicircle (C1, C2); Episomal (E1, E2); mRNA (M1, M2); Lentivirus (L1); microRNA (U1, U2); Sendai virus (S1, S2); hESCs (H7.1); Hepatocellular Carcinoma (HepG2); Pancreatic Carcinoma (PANC1); Cervical Carcinoma (Hela.S3); Glioblastoma/Astrocytoma (U87); fibroblasts (Fib); skin fibroblasts (BJ); lung fibroblasts (Nhlf); and Skeletal Muscle Myoblasts (Hsmm). (B) Based on more than 2,000 human tumor-associated antigens (TAA) and tumor-specific antigens (TSA) RNA, we were able to identify the presence 116 of these tumor antigens in murine iPSCs (MEF.iPSC and APC.iPSC) and ESCs. Similar to the human data, these are highly expressed in the pluripotent populations but only lowly or not expressed in the somatic tissues. MEF-derived iPSCs (MEF.iPSC); APC-derived iPSCs (APC.iPSC); murine ESCs (R1.ESC); murine ESCs (ES.Bruce4); brain tissue (brain_W10); central nervous system (CNS_E18); colon tissue (colon_W08); heart tissue (heart_W10); kidney tissue (kidney_W10); large intestines (large.intestine_W8); limb tissue (limb_E14.5); liver tissue (liver_W10); lung tissue (lung_W10); mammary gland tissue (mammary.gland_W8); and small intestines (small.intestine_W8).
Figure S2. (Related to Figure 1–2, 5) Characterization of iPSCs for pluripotency and in vitro optimization of the vaccination schedule using the adjuvant CpG. Pluripotency staining of (A) FVB, (B) C57BL/6, and (C) CBA/J murine iPSCs with pluripotency markers Oct4, c-Myc, Nanog, and SSEA-1. (D) All three cell lines were able to form teratomas in vivo upon transplantation of 1x106 iPSCs in the hindlimb of NOD/SCID mice. (E) Using the adjuvant (CpG), most effective proliferation of CFSE-labeled splenocytes in response to DB7 lysate was seen in the cytotoxic T-cells in the C+I 4-week vaccination group. (F) This was also the only group with increased cytokine release in the supernatant with a higher dose of DB7 tumor lysate (U=unstimulated splenocytes; S=stimulated splenocytes with allogeneic C57BL/6 splenocytes).
Figure S3. (Related to Figure 1) Tumor growth over time in FVB and C57BL/6 tumor-bearing mice. (A) Representative images of PBS, CpG only, iPSCs only, and CpG+iPSC (C+I) vaccinated mice one week after subcutaneous injection of 5x104 DB7 cancer cells. (B) Images of five mice per group at four weeks after DB7 breast cancer cell injection that were selected for immune profile analysis of spleen, dLNs, and blood. (C) One year after the experiment in Figure 1D, C+I treated mice (n=2) were reintroduced with 5x104 DB7 breast cancer cells and fully rejected the cancer cells by four weeks. In contrast, mice vaccinated with iPSC-derived endothelial cells (iECs; n=4) were unable to reject the cancer cells, which proliferated into large tumors. (D) One year after vaccination and full rejection of DB7 breast cancer cells, C+I primed animals still have high antibody titers against iPSCs and DB7, unlike mice that were vaccinated with iECs. (E) Images of all C57BL/6 mice in the four treatment groups two weeks after B16F0 melanoma cell injection. Several mice in the C+I group showed a regression of tumor sites or inflammation as indicated by local swelling, redness, and, in some cases spontaneous bleeding. (F) Representative images showing mice in the C+I group that were used for immune profile analysis of spleens, dLNs, and blood at time of sacrifice.
Figure S4. (Related to Figure 1, 3–4) C+I vaccination leads to tumor specificity without causing auto-inflammatory responses. Cell culture images from splenocytes isolated from CpG only (vehicle; n=4) and C+I vaccinated mice (n=6) incubated with (A) DB7 tumor lysate or (B) iPSC lysate. Blue spots represent IFNγ secretion by splenocytes and were identified with Adobe Photoshop software in the bottom row images for each condition. (C) Serum from PBS (n=4), CpG (n=4), and CpG+iPSC (n=4) vaccinated mice was isolated after four rounds of vaccination. No significant differences were found in anti-nuclear antibody titers among the different groups (total samples: n=12 with three technical replicates for each sample) (one way ANOVA; p=0.29, ns). (D) For the duration of the experiment, FVB mice (left panel) and C57Bl/6 mice (right panel) were thriving with a healthy appearance and weight increase over time. (E) Representative images of heart (left panel) and kidney samples (right panel) from FVB mice in the three treatment groups revealed no increases in the numbers of inflammatory cells in any of the groups compared to PBS control. (F) Similar findings were seen in the heart (left panel) and kidney tissues (right panel) from C57BL/6 mice.
Figure S5. (Related to Figure 5) Citrus cluster description and features from TILs (CBA/J) and splenocytes from tumor-resected mice (C57BL/6). (A) Clustering analysis of the CyTOF data from Figure 5B using Citrus with their surface marker profile and (B) predictive features of the cytokine profiles from these clusters in the C+I vaccinated group compared to PBS control. (C) Citrus cluster description from Figure 7C. (D) Cross-validation error rate from Citrus analysis in Figure 5B. (E) Cross-validation error rate from Citrus analysis in Figure 7C.
Figure S6. (Related to Figure 6) C+I vaccination in CBA/J mice leads to reduced tumor establishment and a reduction of commonly shared intra-tumor T-cell clones in favor of vaccine-specific clones. (A) Representative image at day 2 of CBA/J mice vaccinated with PBS, CpG+iPSC (C+I), or CpG+AC29 (C+A) and then introduced with either 2x106 AC29 or 2x106 iPSCs. (B) Image (left panel) and tumor sizes (right panel) of all mice at week 1 post-tumor injection, revealing a complete rejection of the iPSCs and cancer cells in the positive control groups (C+I/iPSC = CpG+iPSC vaccinated mice introduced with iPSCs, and C+A/AC29 = CpG+AC29 vaccinated mice introduced with AC29 cells). The last three mice in the C+I/AC29 already showed tumor regression, whereas the first two had severe erythema and swelling at the tumor sites. (C) Frequency of T-cell clones with their receptor sequences in the PBS+AC29 control group if they were shared by 4 or 5 out of 5 mice. TILs had 28 clones that are present in multiple mouse strains in spleen (n=3) as well as thymus (n=3). The larger public clone repertoire within the PBS group therefore likely consisted of shared TCR clones on the basis of chance. (D) Frequency of T-cell clones with their receptor sequences in the C+I/AC29 treatment group if they were shared by 4 or 5 out of 5 mice. TILs only showed six clones that were present in multiple mouse strains in spleen (n=3) as well as thymus (n=3). In these cases, the generally lower frequency of the clones in the thymus and more similar frequencies of the clones in the spleen suggest that the majority of the commonly shared clones in the vaccine group may be more important than public clones. In addition, clone CGARDAGGQDTQYF is extremely rare and was only present in 1 out of 3 spleens in the C57BL/6 mouse strain and was not present in the PBS, C+I/iPSC, or C+A/AC29 groups, indicating it was likely C+I vaccine-specific.
Figure S7. (Related to Figure 7) Adjuvant C+I vaccination prevents the recurrence of tumor after R2 resection of B16F0 melanoma and reduced tumor load in dLNs. (A) B16F0 tumor-bearing mice (n=6) with a tumor size ~1 cm3 received a R2 resection where a small piece of tumor (blue arrow) was left behind in the resection area (RA). C+I vaccinated mice (n=3) had no visible tumor recurrence in the RAs after two rounds of vaccination, whereas the PBS control group (n=3) had visible and palpable tumors. (B) PCR/ddPCR primer development based on a large deletion in CDKN2A in B16 melanoma cells (Fw1-Rv1, Fw2-Rv2). (C) B16 melanoma cell primers were designed to show the deletion by PCR in the melanoma samples without amplification in C57BL/6 wild type samples. (D) PCR products were then validated and analyzed by Sanger sequencing and aligned with the gene database in NCBI. (E–F) Using ddPCR with a mutant probe based on the CDKN2A deletion and a TaqMan®Copy Number probe the quantity of tumor cells within each DNA sample could be assessed. (G) The standard curve was plotted using different concentrations of mutant DNA, revealing a good correlation between % of mutant DNA and the readout from the BioRad QX100 reader. (H) Using ddPCR, a reduction of average tumor load in dLNs of R1 resected mice in the CpG and C+I groups was seen compared to PBS control. If dLNs were positive (>2% threshold), the C+I vaccinated group had the lowest percentage of tumor cells in dLNs.