Abstract
Long repeated sequences of DNA and their associated secondary structure govern the development and severity of a significant class of neurological diseases. Utilizing the effect of base stacking on fluorescence quantum yield, 2-aminopurine substitutions for adenine previously demonstrated sequestered bases in the stem and exposed bases in the loop for an isolated (CAG)8 sequence. The present studies evaluate (CAG)8 that is incorporated into a duplex, as this three-way junction is a relevant model for intermediates that lead to repeat expansion during DNA replication and repair. From an energetic perspective, thermally-induced denaturation indicates that the duplex arms dictate stability and that secondary structure of the repeated sequence is disrupted. Substitutions with 2-aminopurine probe base exposure throughout this structure, and two conclusions about secondary structure are derived. First, the central region of (CAG)8 is more solvent-exposed than single-stranded DNA, which suggests that hairpin formation in the repeated sequence is disrupted. Second, base stacking becomes compromised in the transition from duplex to (CAG)8, resulting in bases that are most similar to single-stranded DNA at the junction. Thus, an open (CAG)8 loop and exposed bases in the arms indicate that the strand junction profoundly influences repeated sequences within three-way junctions.
Keywords: Trinucleotide Repeats, Three-Way junctions, DNA Hairpins
Repetitive sequences within the human genome are linked with disease, and a question concerns the extent to which their noncanonical secondary structures are involved in and even responsible for cancer and inherited neurological diseases.1,2,3,4 The present studies consider how genomic context influences intrastrand interactions in trinucleotide repeats. A significant advance in understanding a role for DNA structure was first described in 1991, when a common basis of several neurological diseases was shown to be expansion in the number of trinucleotide repeats.3,4,5 Repeat length correlates with manifestation of diseases such as Huntington's disease, for which cognative decline and other neurological effects derive from polyglutamine within huntinton’s protein.6 Across the human population, a range of 6-35 repeats of (CAG) are found in the coding region for this protein. With only 1-4 additional repeats, the risk of contracting this disease increases, and further expansion is associated with the fully developed disease. Central to models for repeat amplification are self-associated DNA strands, such as stem-loop hairpins that are favored by (CNG) repeats, where N is any nucleotide.7 When such strands are not aligned with and extrude from their complementary sequence, mechanisms such as replication and repair can lengthen the original sequence.4,8 For example, in non-dividing cells, single-stranded intermediates can form by oxidative damage.9 Resulting hairpin structures can be protected by proteins, and subsequent lesion repair generates an expanded sequence.
Because they provide the genetic code for ~20 neurological diseases, (CNG) repeats have been the focus of structural studies, and the consensus from a diverse range of techniques is that stem-loop hairpins are favored.10 Intrastrand folding was deduced from rapid electrophoretic mobility relative to unstructured, single-stranded oligonucleotides.11 Stem and loop regions are indicated by chemical and biochemical agents that discern protected and exposed bases, respectively.12,13 NMR studies have established coupling between paired bases and shown that mismatches can be accommodated within the helical stack.14,15 Mismatches in the stem of (CNG) hairpins impact stability in the order (CAG) ~ (CCG) < (CTG) < (CGG), as derived from differential scanning calorimetry and from hyperchromic absorbance changes.13, 16 Via fluorescence studies with the adenine analog 2-aminopurine, heterogeneity in the loop and sequestered mismatches in the stem were highlighted for the (CAG)8 hairpin.17 Another feature of repeated sequences is their conformational heterogeneity, as demonstrated by spectroscopic, calorimetric, and biochemical studies.18,19,20
The present studies consider how structure within repeated sequences is transformed by flanking duplexes. Integrating different secondary structural elements in DNA can alter the properties of the constituent regions to produce junctions and overall structures with distinct features.21,22,23 For three-way junctions with an extruded repeat sequence, their assembly is driven by base pairing in the duplex arms.24 For (CAG) repeats, alternatives to single stem-loop hairpins are indicated by their preferential association with single-strand specific enzymes.24 This issue is explored using 2-aminopurine to evaluate secondary structure in both the repeated and junction regions of three-way junctions (Fig. 1A). Three experiments are discussed: fluorescence intensity changes with position in the three-way junction, fluorescence changes due to thermal denaturation, and efficiency of extrinsic quenching by acrylamide. The results are considered within the context of the stem-loop structure formed by the isolated (CAG)8 sequence. The two important conclusions are that a constrained (CAG)8 loop emanates from the duplex arms and that base pairing is compromised in the duplex. Thus, the strand junction disrupts the ability of the repeated sequence to self-associate, which could impact biochemical mechanisms of repeat expansion.
Figure 1.
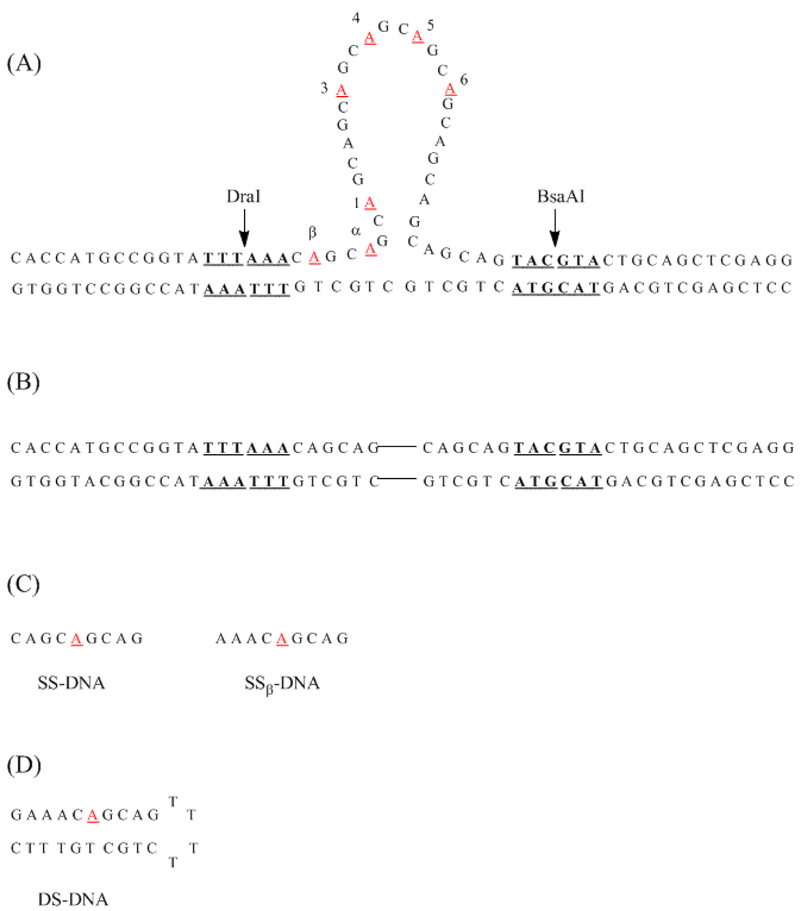
(A) Model structure for the three-way junction formed with (CAG)8. Restriction enzyme sites for DraI and BsaAI are indicated in bold and underlined. The positions of 2-aminopurine substitution are indicated in red and are underlined. (B) Structure of the duplex without (CAG)8. The connecting lines represent the position of the abstracted repeat sequence from Fig. 1A. (C) Structures of single-stranded oligonucleotides with 2-aminopurine. The structure on the right has the same local base sequence as β-CAG. The structure on the left has the same local base sequence as the remaining modified three-way junctions. (D) Structure of double-stranded oligonucleotide with 2-aminopurine.
Materials and Methods
Buffer components (Sigma-Aldrich, St. Louis, MO) and acrylamide (Acros Organics, Belgium) were used as received. All measurements were conducted in a buffer consisting of 10 mM Tris/Tris-H+, 10 mM MgCl2, and 50 mM NaCl at pH 7.9. Oligonucleotides (Integrated DNA Technologies, Coralville, IA) were purified by denaturing 8% PAGE with 7 M urea, TBE buffer at 50 °C (300 V), with subsequent visualizing of the samples by UV light on a TLC plate. Desired bands were removed from the gel, electroeluted in TBE buffer, and passed through a NAP-10 desalting column (GE Healthcare, Piscataway, NJ). Samples were then lyophilized and resuspended in water.
Oligonucleotides, along with their extinction coefficients, are presented in Table 1. Concentrations were determined by extrapolating absorbances at 260 nm of the high-temperature post-transition baselines back to 25 °C. Extinction coefficients of single-stranded oligonucleotides were derived from the nearest neighbor approximation. For modified oligonucleotides, extinction coefficients were evaluated as the sum of the extinction coefficient without 2-aminopurine plus the relatively small contribution from free 2-aminopurine (1000 M−1cm−1 at 260 nm).25 Absorbance was collected at 260 nm as a function temperature on a Cary 300 spectrometer equipped with a multicell holder (Varian, Palo Alto, CA). Given melting temperatures of ~77 °C, experiments were started at 45 °C, and the temperature was increased to 95 °C at a rate of 1 °C/min, allowing 1 minute for equilibration before each reading. Absorbance from buffer was subtracted from the DNA absorbance.
Table 1.
Single-Stranded Oligonucleotides.
| Sequence | Lengtha | εb |
|---|---|---|
| 5’-CACCATGCCGGTA TTTAAA CAG CAG CAG CAG (CAG)8 CAG CAG CAG CAG (CAG)8 CAG CAG TACGTA CTGCAGCTCGAGG-3’ | 74 | 720,500 |
| 5’-CACCATGCC GGT ATT TAA ACAG CAG CAG CAG CAG CAG CAG CAG CAG (CAG)8 CAG CAG TACGTA CTGCAGCTCGAGG-3’ c |
74 | 708,700 |
| 5’-CCTCGAGCTGCAG TACGTA CTGCTG-CTGCTG TTTAAA TACCGGCATGGTG-3’d | 50 | 462,500 |
| 5’-G AAA CAG CAG TTTT CTG CTG TTT C-3’ (DS-CAG) | 24 | 210,900 |
| 5’-AAA CAG CAG-3’ (SSβ-((CAG)8)8) | 9 | 86,300 |
| 5’- CAG CAG CAG-3’ (SS-((CAG)8)8) | 9 | 78,000 |
Lengths in bases
Extinction coefficients (M−1 cm−1) of the unfolded single strands.
Seven variants of this oligonucleotide were used, each with a single substitution of 2- aminpurine, indicated by the red, underlined bases.
Connecting line represents the region corresponding to the (CAG)8.
Three-way junctions were formed by annealing a 74-base oligonucleotide with a central (CAG)8 sequence and flanking 25-base regions that are complementary to a 50-base oligonucleotide (Fig. 1A). Nomenclature for modified structures indicates the position of the 2-aminopurine substitution for adenine. Annealing was performed by heating equimolar amounts of single strands together to 95 °C for 5 minutes with slow cooling to room temperature over a period of more than 12 hours. Purity was verified by 12% polyacrylmide nondenaturing gel electrophoresis.
Fluorescence spectra were collected on a Fluoromax-2 spectrometer (Jobin-Yvon Horiba, Edison, NJ) equipped with DataMax 3.4 software and Neslab RTE-7 circulating bath. The excitation wavelength was 307 nm and the emission wavelength was 370 nm, and the samples were equilibrated for 5 min at each temperature before recording the signal. Fluorescence intensities were acquired using 1 μM concentrations of three-way junctions. For acrylamide studies, standard Stern-Volmer analysis was an adequate model to determine the static quenching constants. Downward curvature of Stern-Volmer plots was observed upon addition of up to 0.5 M concentrations of acrylamide. As correlation of the fitting parameters was noted for more sophisticated models, our studies focused on concentrations below 0.1 M acrylamide where a standard linear Stern-Volmer equation was used. Oligonucleotide concentrations were 0.3-0.5 μM. Averages and standard deviations were calculated from at least three replicate experiments. For both emission and absorbance measurements, melting temperatures were determined as the intersection point between experimental melting curve and the median between lower and upper baselines, which correspond to the folded and denatured forms of the three-way junction, respectively.26 van’t Hoff analyses of melting profiles were used to determine equilibrium constants for conversion of folded structures to single-stranded products as a function of temperature.26 Reaction molecularity was known from the strand stoichiometry of the three-way junction and duplex that was established from gel electrophoresis. Linear fitting of Arrhenius plots were used to calculate ΔH° and ΔS° using data over the range of 20-90% conversion with the assumption of no heat capacity change. Melting was not reversible for these intermolecular complexes.
Formation of annealed three-way junctions was monitored with restriction endonucleases BsaAI and DraI (New England BioLabs, Ipswich, MA) using distinct six base pair sites in the duplex arms. DNA samples with 0.5 μM concentrations in NEB2 buffer were mixed with 10-15 units of an enzyme to the total volume of 20 μL and incubated at 37 °C for at least 3 hours. Samples were mixed with SYBR Gold and the gel loading dye and analyzed on the native 12% polyacrylamide gel electrophoresis in TBE buffer.
Results
Duplex arms dominate stability
To form the model three-way junction, a repeated trinucleotide (CAG)8 that is flanked by two 25-base regions is annealed with a complementary to a 50-base oligonucleotide (Figs. 1A and 2). Integrity of the resulting duplex arms is signified by cleavage at DraI and BsaAI sites centered 16 base pairs from the arm termini (Fig. 2). The factors that influence the stability of the three-way junction are considered using temperature-dependent absorbance changes. First, hyperchromism indicates that base unstacking accompanies thermal denaturation to single-stranded DNA, and two state analyses of their melting profiles indicate that the three-way junction and its component duplex have comparable thermodynamic stabilities (Fig. 3 and 1S). 26 For the duplex, ΔH° = 317 ± 13 kcal/mole and ΔS° = 865 ± 35 cal/(mole K) at 82 °C, while for the three-way junction, ΔH° = 262 ± 24 kcal/mole and ΔS° = 715 ± 68 cal/(mole K) at 79 °C. The errors preclude evaluation of relative stabilities via standard free energy changes at a common temperature. Second, the transition for the three-way junction is monophasic with no observed changes at 67 °C, which is the melting temperature for an isolated (CAG)8. These observations suggest that the duplex arms anchor the three-way junction and that the repeated sequence has a minor thermodynamic contribution to the stability of the three-way junction. To refine this analysis, fluorescence studies were conducted using 2-aminopurine substitutions.
Figure 2.
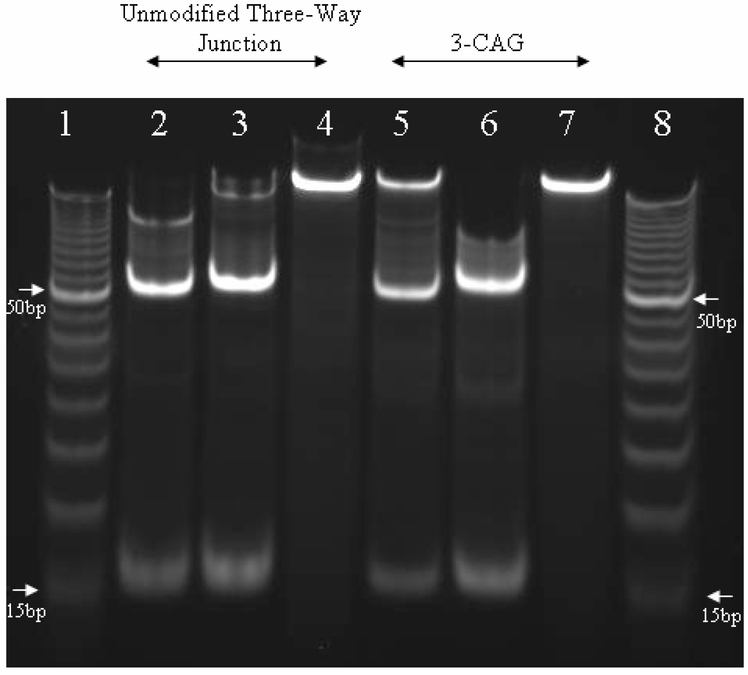
Image of a nondenaturing 12% polyacrylamide gel to assess restriction digestion of three-way junctions. Lanes 1 and 8 contain a 5-base pair ladder with 15 and 50 base pair oligonucleotides marked with arrows. Lanes 2-4 correspond to the unmodified three-way junction without 2-aminopurine. Lanes 2 and 3 represent the products of digestion by DraI and BsaAI, respectively, and the mobilities of the faster bands are between the 15 and 20 base pairs. Lane 4 shows the three-way junction before digestion. Lanes 5-7 correspond to the modified three-way junction 3-CAG, and the mobilities of the faster bands are between the 15 and 20 base pairs. Lanes 5 and 6 represent the products of digestion by DraI and BsaAI, respectively. Lane 7 shows the three-way junction before digestion.
Figure 3.
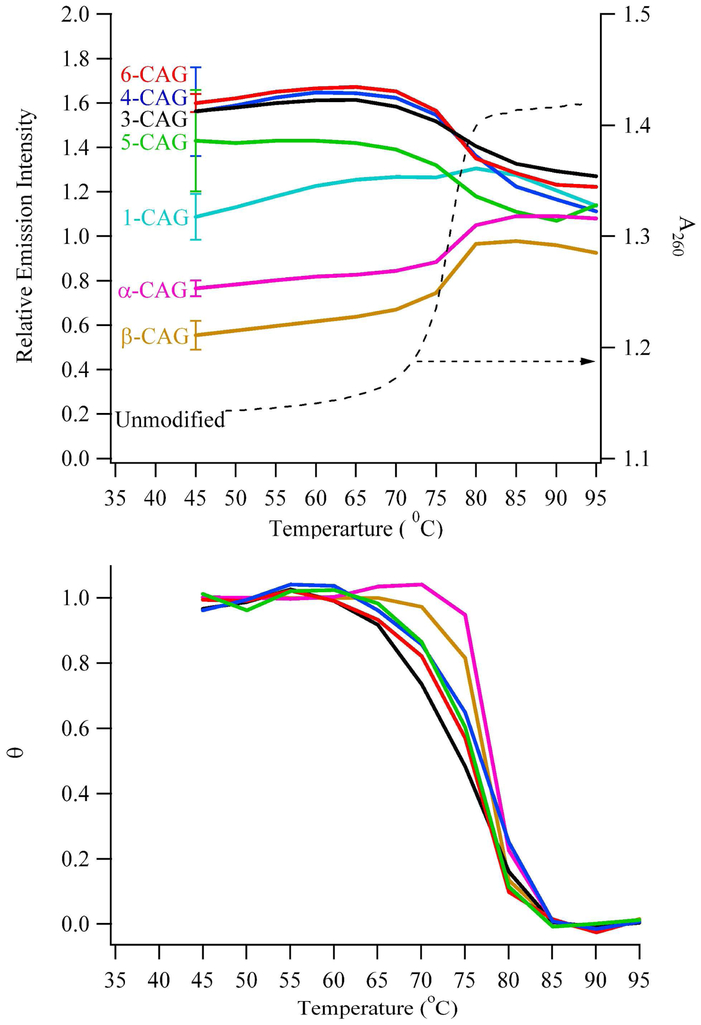
(Top) Emission intensities using λex = 307 nm and λem = 370 nm (left axis) of the modified three-way junctions and absorbance changes at 260 nm (right axis) for the unmodified three-way junction as a function of temperature. Fluorescence intensities were normalized to the emission of single-stranded references. Positions of the substitutions follow notation in Figure 1A. (Bottom) Fractional unfolding (θ) as a function of temperature for the modified three-way junctions. The fluorescence intensities at low and high temperature were used to determine the baseline spectral responses from the folded and unfolded forms, respectively, of the three-way junctions. The color scheme follows that of the top figure.
2-Aminopurine probes secondary structure
To map solvent exposure throughout the three-way junction, seven variants with 2-aminopurine were used, as the fluorescence quantum yield of this isomer of adenine is sensitive to base stacking (Fig. 1A).27,28 To probe the duplex arms, modifications labeled α-CAG and β-CAG were made in the two (CAG) repeats that precede the junction. Within the integrated (CAG)8, five substitutions notated as 1-CAG, 3-CAG, 4-CAG, 5-CAG, and 6-CAG were made in the 1st, 3rd, 4th, 5th, and 6th repeats, respectively. These substitutions were chosen because they represent a range of solvent exposure within the stem and loop of the isolated (CAG)8 hairpin.17 To understand how fluorescence intensities relate to DNA secondary structure, 2-aminopurines in duplex hairpin and short single-stranded oligonucleotides define the range from full to relaxed base stacking, respectively (Fig. 1C and 1D). Conserving local base sequence in these references accounts for base context effects on fluorescence.29,30 Previous studies have established that substitution of adenine with 2-aminopurine retains the conformation and stability of DNA structures, and their effect on the three-way junction is evaluated using thermal denaturation and gel electrophoresis.17,18 Modified and unmodified oligonucleotides have similar melting temperatures, thus indicating that single substitutions of adenine with 2-aminopurine do not significantly alter stability (Table 2). Furthermore, gel mobilities are similar for modified and unmodified structures, indicating that global shapes are similar. Finally, digestions by restriction enzymes yield unmodified and modified fragments with similar mobilities, indicating that duplex integrity is not compromised (Fig. 2).
Table 2.
Melting Temperatures and Acrylamide Quenching Constants.
| Oligonucleotidea | Tm,abs (°C)b | Tm,fluor (°C)c | Kq (M−1)d |
|---|---|---|---|
| β-CAG | 75.9 (0.4) | 77.0 (0.6) | 7.5 (0.2) |
| α-CAG | 77.2 (0.1) | 78.7 (0.3) | 9.8 (0.5) |
| 1-CAG | 76.6 (0.1) | ND | 11.1 (0.2) |
| 3-CAG | 76.8 (0.8) | 77.0 (1.7) | 15.0 (0.2) |
| 4-CAG | 76.4 (0.1) | 77.1 (0.6) | 13.5 (0.7) |
| 5-CAG | 75.9 (0.3) | 77.6 (1.6) | 15.2 (0.7) |
| 6-CAG | 76.4 (0.6) | 76.5 (0.3) | 14.9 (0.4) |
| Unmodified | 77.2 (0.2) | ||
| SS-CAG | 9.6 (1.0) | ||
| DS-CAG | 8.2 (0.6) |
Labels pertain to structure in Fig. 1A.
Melting temperatures derived from hyperchromic absorbance changes at 260 nm. Standard deviations provided in parentheses.
Melting temperatures derived from fluorescence intensity changes of substituted 2- aminopurines. The value for 1-AP was not determined (see Fig. 3). Standard deviations provided in parentheses.
Acrylamide quenching constants.
Junction perturbs arms and repeat sequence
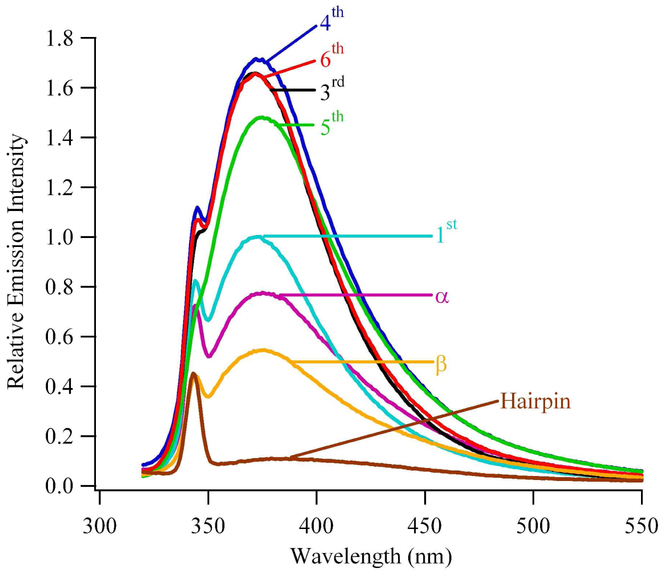
Within the modified three-way junctions, solvent exposure of 2-aminopurine was probed using fluorescence intensity measurements. Fluorescence spectra are consistent with prior studies (Fig. 4).31 When compared with single-stranded DNA, the order of fluorescence intensities is β-CAG < α-CAG < 1-CAG < 3-CAG ~ 4-CAG ~ 5-CAG ~ 6-CAG. This trend suggests that moving from the duplex arm into the (CAG)8 sequence increases base exposure to solvent. Importantly, substitutions in the 3rd, 4th, 5th, and 6th repeats of (CAG)8 have similar fluorescence intensities, indicative of a consistent environment in this central region of the repeated sequence. To evaluate secondary structure within the three-way junction, fluorescence intensities are compared with single-and double-stranded references. The intensity from β-CAG is lower than from single-stranded DNA but is higher than from duplex DNA, and the intensity from α-CAG is still higher. These relative intensities report on solvent exposure and indirectly indicate that base pairing with opposing GTC’s is disrupted by the strand junction, as also inferred from the absorbance-based thermal denaturation studies. Base exposure continues to increase in the 1st repeat of (CAG)8, for which the intensity from 2-aminopurine is comparable with single-stranded DNA. Within the (CAG)8 sequence, higher solvent exposures relative to the single-stranded reference are observed for 3-CAG, 4-CAG, 5-CAG, and 6-CAG. These consistently high intensities for the integrated (CAG)8 depart from the observations of the isolated (CAG)8 hairpin, for which the analogous 3-CAG and 6-CAG have suppressed fluorescence due to stacking in the duplex stem and 4-CAG and 5-CAG have enhanced fluorescence due to their exposure in the loop.17
Figure 4.
Fluorescence spectra of the modified three-way junctions normalized relative to single-stranded references recorded at 45 °C. Positions of the substitutions follow the notation in Figure 1A.
Fluorescence changes that accompany thermal denaturation provide further perspective on the regional conformation within the three-way junction (Fig. 3). For all 2-aminopurine constructs, melting temperatures derived from changes in fluorescence quantum yield are similar, and these are furthermore similar to the value derived from absorbance measurements for the unmodified structure (Fig. 3 and Table 1). Because these modifications probe a range of positions within the three-way junction, similarities in melting temperatures derived from fluorescence and absorbance measurements suggest a two-state melting process in which global unfolding is dictated by the duplex arms. Within experimental repeatability, limiting fluorescence approaches that of single-stranded DNA, and the shapes of the melting profiles describe conformational changes within the three-way junction (Fig. 3). Hyperfluorescent changes for β-CAG and α-CAG indicate that stacked bases within the duplex arms become solvent exposed with denaturation. The intensity variation for 1-CAG is relatively small, which indicates that this position in the three-way junction is similar to a single-stranded oligonucleotide over the entire temperature range. Intensities from 3-CAG, 4-CAG, 5-CAG, and 6-CAG decrease with temperature, which suggests that constraints in the repeat sequence relax with denaturation.
Support from acrylamide quenching
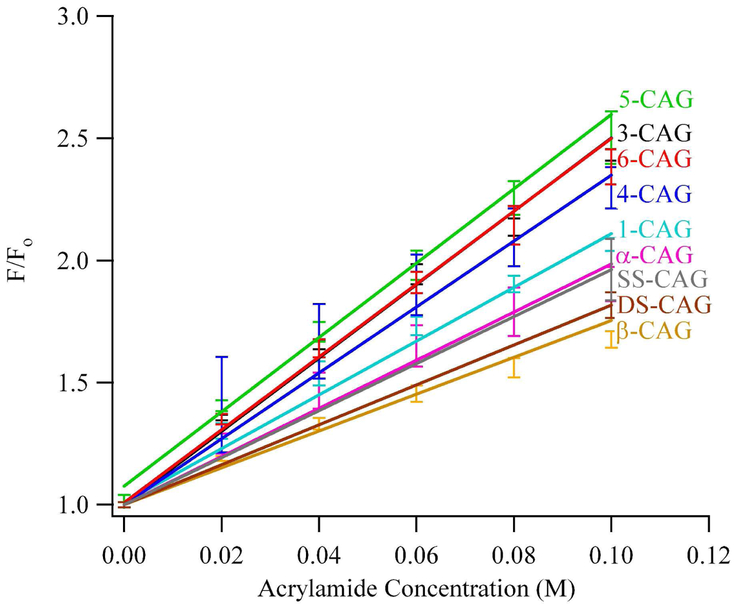
To complement information derived from the inherent fluorescence of 2-aminopurine, extrinsic fluorescence quenching by acrylamide also probed solvent exposure.32 By relating quenching efficiency to secondary structural elements of DNA, structural variations within the three-way junction are inferred.29,30 Relative fluorescence quenching is linear up to 0.1 M acrylamide, and Stern-Volmer analysis was used to extract quenching constants for the 2-aminopurines (Table 2 and Fig. 5). The important conclusions derived from these studies are increased solvent exposure in the junction relative to the duplex arms and a similarly exposed environment in the central region of (CAG)8. Quenching for β-CAG and the hairpin duplex are comparable, which suggests that the β modification is protected from extrinsic quenching by base stacking and pairing. Quenching becomes increasingly efficient proceeding towards the junction to α-CAG and onto 1-CAG. Similar quenching constants are observed for 3-CAG, 4-CAG, 5-CAG, and 6-CAG, and these are consistently higher than for single-stranded DNA references. Furthermore, these values are similar to the value observed for the highly exposed 5th repeat in the loop of the isolated (CAG)8 hairpin.17 This comparison provides further support for an open loop environment in the repeated (CAG)8 sequence in the three-way junction.
Figure 5.
Stern-Volmer plot describing the quenching of modified three-way junctions with acrylamide using λex = 307 nm and λem = 370 nm. The positions of the substitutions follow notation in Figure 1A.
Discussion
The genesis of neurological diseases associated with triplet repeat sequences has been proposed to be hairpin intermediates that alter normal DNA replication and repair processes, and a model three-way junction was formed by incorporating (CAG)8 into double-stranded DNA. This repeated sequence was chosen because the isolated (CAG)8 favors a stem-loop structure, and key structural details were elucidated using 2-aminopurine substitutions for adenine.17 Within the central stem of this folded hairpin, mismatches are sequestered as efficiently as canonical base pairs. Base stacking is assumed to be a major stabilizing force, as prior NMR studies indicate that adenines form self base pairs via a single hydrogen bond.33 In the vicinity of the loop, stem formation is disrupted, as indicated by increasing solvent exposure. The loop is characterized by highly solvent exposed bases, particularly in the 5th (CAG) repeat.
The question of interest in these studies is how intrastrand interactions in (CAG)8 are altered in a 50-base pair duplex, and thermal denaturation studies show that the duplex arms dominate overall stability and that folding of the repeat sequence is perturbed. These conclusions are founded on the comparable stability of the 50 base pair duplex without the repeated sequence and on the apparent monophasic melting profile without a transition corresponding to the isolated hairpin. To provide a higher resolution picture of both the repeated sequence and the strand junction, 2-aminopurine was substituted for adenine. Single- and double-stranded DNA with 2-aminopurine provide structural references for solvent-exposed and -sequestered bases, respectively. The first general conclusion is that the central 3-CAG, 4-CAG, 5-CAG, and 6-CAG in the loop are distinguished by their similarly high level of solvent exposure when compared to single-stranded DNA. These intensities suggest a similarly constrained environment, and fluorescence diminution at high temperature shows that relaxation accompanies denaturation. Acrylamide quenching at these positions is also more efficient relative to single-stranded DNA analogs. As a further point of comparison, quenching efficiency for these central (CAG) repeats is comparable to highly exposed bases in the loop of the isolated (CAG)8 hairpin.17 The second general finding concerns compromised base stacking and pairing at the strand junction. Substitution of 2-aminopurine for adenine results in duplexes maintain two hydrogen bonds with opposing thymines, and the resulting structures have unaltered conformations and are slightly less stable than of DNA.34,35 These prior studies suggest that solvent exposure of 2-aminopurine accurately reports base pairing in the junction region. The intensity from 1-CAG exhibits little variation with temperature and tracks the behavior of single-stranded DNA. In the duplex arm, α-CAG and β-CAG have lower intensities that increase with temperature, indicative of base stacking that is driven by pairing with complementary GTC’s. However, the higher intensity from the most distant β-CAG relative to duplex DNA indicates that base pairing is compromised in the junction region. The intensity trend 1-CAG > α-CAG > β-CAG suggests that base pairing is less perturbed away from the junction, and efficient enzymatic digestion indicates that duplex integrity is fully reestablished at the restriction sites adjacent to the 2 CAG/GTC repeats.
In summary, key structural features of the model three-way junction are an open, constrained (CAG)8 loop and disrupted base interactions at the junction. Enzymatic probing first suggested alternatives to single stem-loop arrangements for repeated sequences that are slipped out from duplexes.24 Uniform digestion of excess CAG repeats by mung bean nuclease indicates that this single-strand specific enzyme could be targeting loops of multiple smaller hairpins or a single open loop. In support of such alternatives secondary structures, electron microscopy shows localization of single-strand binding proteins with three-way junctions that have excess CAG repeats. From an energetic perspective, disrupted base stacking in repeated sequences could be indicated by the similar stability of three-way junctions with and without abasic sites.19 The results could be expected based on the instability of isolated CAG based hairpins relative to other CNG based hairpins, but the constrained conformation of the (CAG)8 indicates that the strand junction has a profound impact on the structure of the repeat. Given the relationship between long repeat lengths and neurological diseases, our current focus is the effect of the junction on longer DNA sequences. Our studies are also considering the strand junction, as tertiary structure is influenced by base pairing at the junction.36,37 Regiospecific studies with 2-aminopurine provide the basis for pursuing these studies.
Conclusion
Expansion in the number of trinucleotide repeats beyond a critical threshold is a significant factor in the development of several neurological diseases, and intrastrand folding of these long sequences diverts normal biochemical processing of DNA. The present studies demonstrate that the context of the repeated sequences influence their secondary structure. Specifically, CAG repeats within a three-way junction adopt an open loop with constrained bases and base pairing at the junction is perturbed by the repeated sequence. These studies provide the foundation for understanding the long-range effect of the junction on longer sequences.
Supplementary Material
Acknowledgements
We thank R. Buscaglia and B. Chaires for helpful discussions regarding 2-aminopurine. We thank S. Story for carefully reading the manuscript.
Funding Information: We thank the National Science Foundation (CHE-0718588 and CBET-0853692), the National Institutes of Health (R15GM071370 and P20 RR-016461 (from the National Center for Research Resource)), and the Henry Dreyfus Teacher-Scholar Awards Program for support.
References
- 1.Wells RD (2007) Non-B DNA conformations, mutagenesis and disease, Trends in Biochemical Sciences 32, 271–278. [DOI] [PubMed] [Google Scholar]
- 2.Vasquez KM, and Hanawalt PC (2009) Intrinsic genomic instability from naturally occurring DNA structures: An introduction to the special issue, Molecular Carcinogenesis 48, 271–272. [DOI] [PubMed] [Google Scholar]
- 3.Kovtun IV, and McMurray CT (2008) Features of trinucleotide repeat instability in vivo, Cell Research 18, 198–213. [DOI] [PubMed] [Google Scholar]
- 4.Mirkin SM (2007) Expandable DNA repeats and human disease, Nature 447, 932–940. [DOI] [PubMed] [Google Scholar]
- 5.(a) Verkerk AJ (1991) Identification of a gene (fmr-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in Fragile X syndrome., Cell 65, 905–914. [DOI] [PubMed] [Google Scholar]; (b) La Spada AR, Wilson EM, Lubahn DB, Harding AE, and Fischbeck KH (1991) Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy, Nature 352, 77–79. [DOI] [PubMed] [Google Scholar]
- 6.Bates GP (2005) History of genetic disease: The molecular genetics of Huntington disease - a history, Nature Reviews Genetics 6, 766–773. [DOI] [PubMed] [Google Scholar]
- 7.Wells RD, Dere R, Hebert ML, Napierala M, and Son LS (2005) Advances in mechanisms of genetic instability related to hereditary neurological diseases, Nucleic Acids Research 33, 3785–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson CE, Edamura KN, and Cleary JD (2005) Repeat instability: Mechanisms of dynamic mutations, Nature Reviews Genetics 6, 729–742. [DOI] [PubMed] [Google Scholar]
- 9.(a) Pearson C, Ewel A, Acharya S, Fishel R, and Sinden R (1997) Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases, Human Molecular Genetics 6, 1117–1123. [DOI] [PubMed] [Google Scholar]; (b) Sinden RR (2001) Neurodegenerative diseases: Origins of instability, Nature 411, 757–758. [DOI] [PubMed] [Google Scholar]
- 10.Mitas M (1997) Trinucleotide repeats associated with human disease, Nucleic Acids Research 25, 2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell JE, Newbury SF, and McClellan JA (1995) Compact structures of d(CNG)n oligonucleotides in solution and their possible relevance to Fragile X and related human genetic diseases, Nucleic Acids Research 23, 1876–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitas M, Yu A, Dill J, Kamp TJ, Chambers EJ, and Haworth IS (1995) Hairpin properties of single-stranded DNA containing a gc-rich triplet repeat: (CTG)15, Nucleic Acids Research 23, 1050–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amrane S, Sacca B, Mills M, Chauhan M, Klump HH, and Mergny J-L (2005) Length-dependent energetics of (CTG)n and (CAG)n trinucleotide repeats, Nucleic Acids Research 33, 4065–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariappan SV, Silks LA 3rd, Chen X, Springer PA, Wu R, Moyzis RK, Bradbury EM, Garcia AE, and Gupta G (1998) Solution structures of the Huntington’s disease DNA triplets, (CAG)n, Journal of Biomolecular Structure & Dynamics 15, 723–44. [DOI] [PubMed] [Google Scholar]
- 15.Zheng M, Huang X, Smith GK, Yang X, and Gao X (1996) Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding. An NMR and UV spectroscopic study, Journal of Molecular Biology 264, 323–36. [DOI] [PubMed] [Google Scholar]
- 16.Paiva AM, and Sheardy RD (2004) Influence of sequence context and length on the structure and stability of triplet repeat DNA oligomers, Biochemistry 43, 14218–27. [DOI] [PubMed] [Google Scholar]
- 17.Degtyareva NN, Reddish MJ, Sengupta B, and Petty JT (2009) Structural studies of a trinucleotide repeat sequence using 2-aminopurine, Biochemistry 48, 2340–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee BJ, Barch M, Castner EW, Völker J, and Breslauer KJ (2007) Structure and dynamics in DNA looped domains: CAG triplet repeat sequence dynamics probed by 2-aminopurine fluorescence, Biochemistry 46, 10756–10766. [DOI] [PubMed] [Google Scholar]
- 19.Volker J, Plum GE, Klump HH, and Breslauer KJ (2009) DNA repair and DNA triplet repeat expansion: The impact of abasic lesions on triplet repeat DNA energetics, Journal of the American Chemical Society 131, 9354–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartenstine MJ, Goodman MF, and Petruska J (2000) Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase, Journal of Biological Chemistry 275, 18382–18390. [DOI] [PubMed] [Google Scholar]
- 21.Early Thomas A. ; Kearns David R.; Burd John F.; Larson Jacquelynn E.; Wells Robert D. (1977) High resolution proton nuclear magnetic resonance investigation of the structural and dynamic properties of d(C15A15)·d(T15G15) Biochemistry 16, 541–551. [DOI] [PubMed] [Google Scholar]
- 22.Ren J, Qu X, Trent JO, and Chaires JB (2002) Tiny telomere DNA, Nucleic Acids Res. 30, 2307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaynutdinov TI, Brown P, Neumann RD, and Panyutin IG (2009) Duplex formation at the 5’ end affects the quadruplex conformation of the human telomeric repeat overhang in sodium but not in potassium, Biochemistry 48, 11169–11177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson CE, Tam M, Wang Y-H, Montgomery SE, Dar AC, Cleary JD, and Nichol K (2002) Slipped-strand DNAs formed by long (CAG)˙(CTG) repeats: Slipped-out repeats and slip-out junctions, Nucleic Acids Research 30, 4534–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox JJ, Wempen I, Hampton A, Doerr IL (1958) Thiation of nucleosides. I. Synthesis of 2-amino-6-mercapto-9-/8-D-ribofuranosylpurine ("thioguanosine") and related purine nucleosides. Journal of the American Chemical Society 80, 1669–1675. [Google Scholar]
- 26.Mergny J-L, and Lacroix L (2003) Analysis of thermal melting curves, Oligonucleotides 13, 515–537. [DOI] [PubMed] [Google Scholar]
- 27.Rachofsky EL, Osman R, and Ross JB (2001) Probing structure and dynamics of DNA with 2-aminopurine: Effects of local environment on fluorescence, Biochemistry 40, 946–56. [DOI] [PubMed] [Google Scholar]
- 28.Xu D, Evans KO, and Nordlund TM (1994) Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption, Biochemistry 33, 9592–9. [DOI] [PubMed] [Google Scholar]
- 29.Ballin JD, Bharill S, Fialcowitz-White EJ, Gryczynski I, Gryczynski Z, and Wilson GM (2007) Site-specific variations in RNA folding thermodynamics visualized by 2-aminopurine fluorescence, Biochemistry 46, 13948–13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballin JD, Prevas JP, Bharill S, Gryczynski I, Gryczynski Z, and Wilson GM (2008) Local RNA conformational dynamics revealed by 2-aminopurine solvent accessibility, Biochemistry 47, 7043–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai P, Cole TD, Thompson E, Millar DP, and Linn S (2003) Steady-state and time-resolved fluorescence studies indicate an unusual conformation of 2-aminopurine within ATAT and TATA duplex DNA sequences, Nucleic Acids Research 31, 2323–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakowicz JR, Principles of Fluorescence Spectroscopy. Plenum Press: New York, 1983. [Google Scholar]
- 33.Mariappan SVS; Chen X; Catasti P; Bradbury EM; Gupta G, Structural Studies on the Unstable Trinucleotide Repeats. Academic Press: New York, 1998. [Google Scholar]
- 34.Xu D; Evans KO; Nordlund TM (1994) Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry 33, 9592–9. [DOI] [PubMed] [Google Scholar]
- 35.Patel N; Berglund H; Nilsson L; Rigler R; McLaughlin LW; Graslund A (1992) Thermodynamics of interaction of a fluorescent DNA oligomer with the anti-tumour drug netropsin. European Journal of Biochemistry 203, 361–367. [DOI] [PubMed] [Google Scholar]
- 36.Lilley DMJ (2000) Structures of helical junctions in nucleic acids. Quarterly Reviews of Biophysics 33, 109–159. [DOI] [PubMed] [Google Scholar]
- 37.Sinden RR, Potaman VN, Oussatcheva EA, Pearson CE, Lyubchenko YL, and Shlyakhtenko LS (2002) Triplet repeat DNA structures and human genetic disease: Dynamic mutations from dynamic DNA, Journal of Biosciences 27, 53–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.