Abstract
Contextual functional interpretation of -omics data derived from clinical samples is a classical and difficult problem in computational systems biology. The measurement of thousands of data points on single samples has become routine but relating ‘big data’ datasets to the complexities of human pathobiology is an area of ongoing research. Complicating this is the fact that many publically available datasets use bulk transcriptomics data from complex tissues like blood. The most prevalent analytic approaches derive molecular ‘signatures’ of disease states or apply modular analysis frameworks to the data. Here we describe ANIMA (association network integration for multiscale analysis), a network-based data integration method using clinical phenotype and microarray data as inputs. ANIMA is implemented in R and Neo4j and runs in Docker containers. In short, the build algorithm iterates over one or more transcriptomics datasets to generate a large, multipartite association network by executing multiple independent analytic steps (differential expression, deconvolution, modular analysis based on co-expression, pathway analysis) and integrating the results. Once the network is built, it can be queried directly using Cypher, or via custom functions that communicate with the graph database via language-specific APIs. We developed a web application using Shiny, which provides fully interactive, multiscale views of the data. Using our approach, we show that we can reconstruct multiple features of disease states at various scales of organization, from transcript abundance patterns of individual genes through co-expression patterns of groups of genes to patterns of cellular behaviour in whole blood samples, both in single experiments as well as in a meta-analysis of multiple datasets.
Keywords: Transcriptomics, complex networks, graph databases, data integration
Introduction
The human immune system can be regarded as a complex adaptive system 1, even though it is integrated with the more complex system of the whole organism. A true systems view 2 of the immune system needs to account for the various aspects that characterize complex adaptive systems generally. This includes emergence, non-linearity, self-organization, noise, scaling, heterogeneity, a network architecture and preservation of context for individual observations 3. Whole blood is a “window” into the immune system 4, allowing a reasonably detailed assessment of the overall state of the immune system based on analysis of easily obtained blood samples. Human diseases can be conceived of as complex state vectors embedded in high dimensional state space.
Here we present ANIMA, Association Network Integration for Multiscale Analysis, a framework for discovery of high order, high complexity and low dimensional state space vectors from high dimensional low order, low complexity data (i.e. non-normalised microarray data and clinical/ sample phenotype data) by producing and interrogating a multiscale association network, which allows summary and visualization of different, but simultaneously valid views of the state of the immune system under different conditions and at multiple scales.
To showcase this, we have chosen three publically available datasets which were generated using whole blood from human subjects with one of three infectious diseases (acute HIV infection, malaria and respiratory viral infections).
Method outline
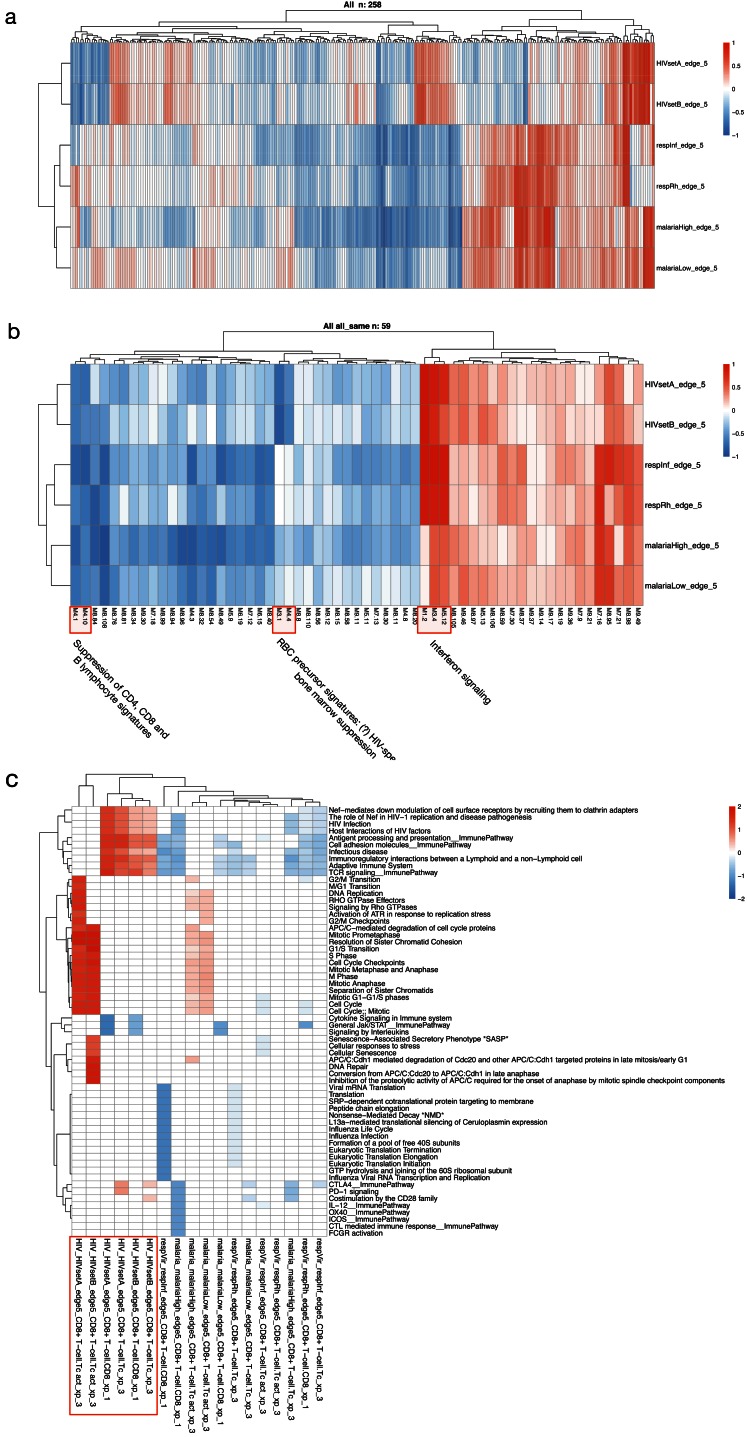
ANIMA generates multiscale association network components from multiple data types (expression data, clinical data and annotation data, e.g. biological pathways databases), and merges all components into a single network ( Figure 1).
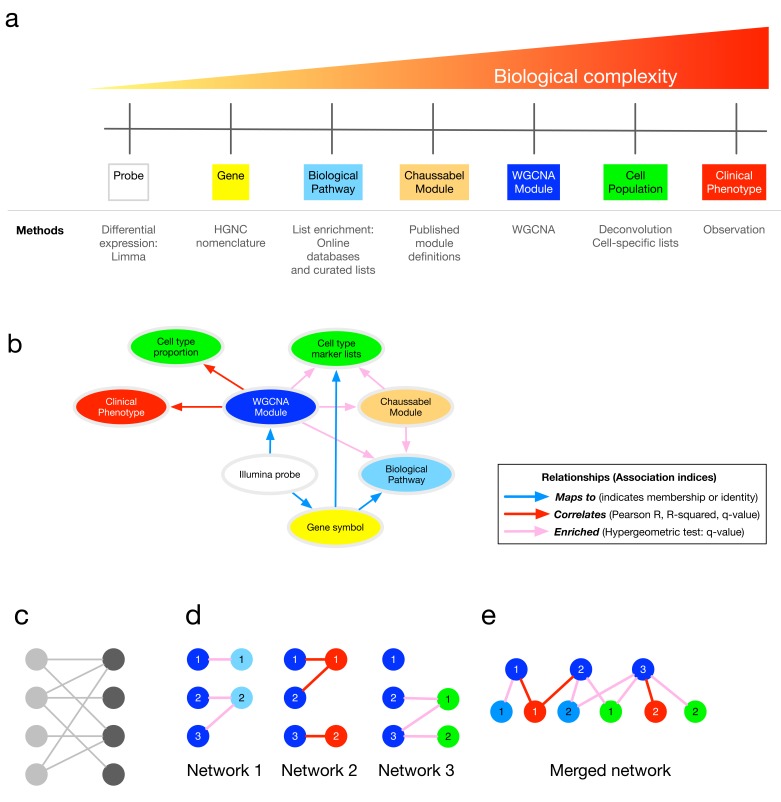
Figure 1. Method overview.
( A) Analytical approaches and biological complexity. ( B) Relationships between output types ( C) A bipartite graph, with two classes of nodes connected by edges. ( D) The separate bipartite graphs, with one node type in common. ( E) Multipartite graph obtained after merging the three graphs in ( D). Abbreviations: HGNC, HUGO Gene Nomenclature Committee; WGCNA, weighted gene co-expression network analysis.
First, the algorithm constructs twenty-nine bipartite networks from a multi-scale analytic pipeline ( Figure 1A, C), combining the output, and the relationships between different classes of output, of various analytic approaches ( Table 1). Each network contains the associations between two distinct data types ( Figure 1B, D, Table 2). The final association network is a result of graph union, merging all bipartite networks on shared node types while retaining all edges ( Figure 1E). This results in a data structure that exposes the relationships between modular gene expression and higher-level phenomena, while retaining key probe- and gene-level information. Three types of association (with their respective association indices) are utilised, resulting in three distinct edge types in the final data structure (See Figure 1B, 1E, Table 2 and supplementary methods; Supplementary File 1). Weighted gene co-expression network analysis 5 (WGCNA) is the core analytic method in ANIMA as this is used to discover biological processes in the system of interest.
Table 1. List of analytic approaches used in constructing the ANIMA database.
| Approach | Method | R package/implementation | Reference |
|---|---|---|---|
| Array data import and
normalisation |
variance stabilising transformation,
quantile normalisation |
lumi | 9 |
| Probe filter | quality filter to remove non-informative
probes from analysis |
ReMoat, ReAnnotator | 10, 11 |
| Differential expression | linear model/moderated t-test | limma | 12 |
| Estimate of cell-type
proportions |
Deconvolution by least-squares fitting | CellMix | 13 |
| Chaussabel module
expression |
Published module definitions and custom
R code |
14, 15 | |
| Chaussabel module
differential expression |
linear model/moderated t-test | limma | 12 |
| WGCNA module detection | Clustering of topological overlap matrix
and dynamic tree cutting |
WGCNA | 5 |
| Chaussabel and WGCNA
module annotation |
List enrichment testing by
hypergeometric test and multiple testing correction |
WGCNA
(UserListEnrichment); ReactomePA |
5, 16 |
| WGCNA module metrics | Differential expression, module AUC,
signature enrichment |
Custom code | |
| Module eigengene
correlations |
Pearson or Spearman rank correlation | WGCNA, custom code | 5 |
| Construction of bipartite
graphs |
Bipartite graphs from adjacency lists
(based on list enrichment) or incidence matrices (based on correlation) |
igraph | 17 |
| Merging of all bipartite
graphs |
Graph union | Neo4j: cypher command
merge |
18 |
Table 2. Types of associations used in the ANIMA database.
| Association
type |
Association index | Intermediate
result |
Multiple testing
correction |
|---|---|---|---|
| Correlation | Pearson R or
Spearman rho |
Incidence
matrix |
Yes |
| List
enrichment |
hypergeometric
index |
Adjacency
list |
Yes |
| Mapping | simple mapping | Adjacency
list |
Not applicable |
Network construction combines three processes. Firstly, mathematical operations on data are performed, independent of prior knowledge. This aspect of the approach is completely unsupervised. The second process involves the testing of hypotheses, to determine differential transcript abundance and differential co-expression of transcripts, requiring knowledge of phenotype/ trait classes for the samples. Finally, the results of the first two processes are integrated in various ways with prior knowledge. The result is a collection of statistically robust analytic results and various associations between them and known biology, in the form of a large, multipartite graph. This large graph is stored in a Neo4j graph database (referred to as the ANIMA database), making the overall network structure as well as the individual nodes and edges accessible for further analysis (see supplementary methods; Supplementary File 1).
After network construction, information in the graph is accessible and utilized to expose new information not present in any of the individual steps. The key to making the ANIMA database useful lies in the use of functions and web applications (see supplementary methods; Supplementary File 1) that query this large multipartite graph and return visualization of relationships or tables of nodes and/ or links (associations between nodes). This has been implemented in several R functions which underpin a Shiny web application called ANIMA REGO.
The novelty and value in ANIMA lies in its three broad approaches to data interpretation: Firstly, it allows detailed, multiscale investigation of a single dataset with a focused research question where two phenotype classes are compared (multiscale class comparison). Secondly, as datasets are stratified by default using two variables (typically one identifying two disease classes, and the other either a potential confounder (e.g. sex), or a second biologic variable of interest, (like co-infection with a second pathogen), we can examine the interaction of the second variable with the first using a factorial study design (factorial analysis). Finally, multiple datasets, and multiple conditions can be meaningfully compared and contrasted to identify similarities and differences (meta-analysis), both at cellular and modular level.
Results
Validation study: data sets
We analysed three publically available microarray datasets using the ANIMA pipeline and toolset (ArrayExpress/ GEO identifiers: E-GEOD-29429, E-GEOD-34404/ GSE34404, E-GEOD-68310/ GSE68310). The first compares a cohort of subjects with acute HIV infection to healthy controls 6, the second compares symptomatic malaria to asymptomatic controls in children from a malaria-endemic region 7, and the last compares the host response in early symptomatic viral respiratory infections 8. Where the datasets contained samples from multiple timepoints, we restricted the analysis to healthy controls and the first disease timepoint. The respiratory virus infection dataset contained samples from subjects with infections other than influenza or rhinovirus; we excluded those from this analysis. Figure S1 (Supplementary File 1) shows the experimental design for the factorial analysis in limma for each of the datasets, together with the numbers of samples in each of the individual groups. Crucially, each of the datasets also included clinical data, which was integrated in the analysis.
The ANIMA network
The result of the script that builds the ANIMA network is a large, mostly connected, multipartite graph. Figure S2 ( Supplementary File 1) shows a subgraph of this network (for dataset HIVsetB, edge 5).
Searching the ANIMA network using network paths and filtering on node and edge properties
In the most direct approach, the large ANIMA graph in the neo4j database can be searched directly from a web browser using the Cypher Query Language (CQL).
For instance, the query
MATCH (ph:pheno)-[r1]-(n:wgcna {square:'HIVsetB',edge:5})-[r2]-(p:PROBE)-[r3]-(s:SYMBOL) WHERE r1.weight > 0.6 AND p.logfc > 2 RETURN *
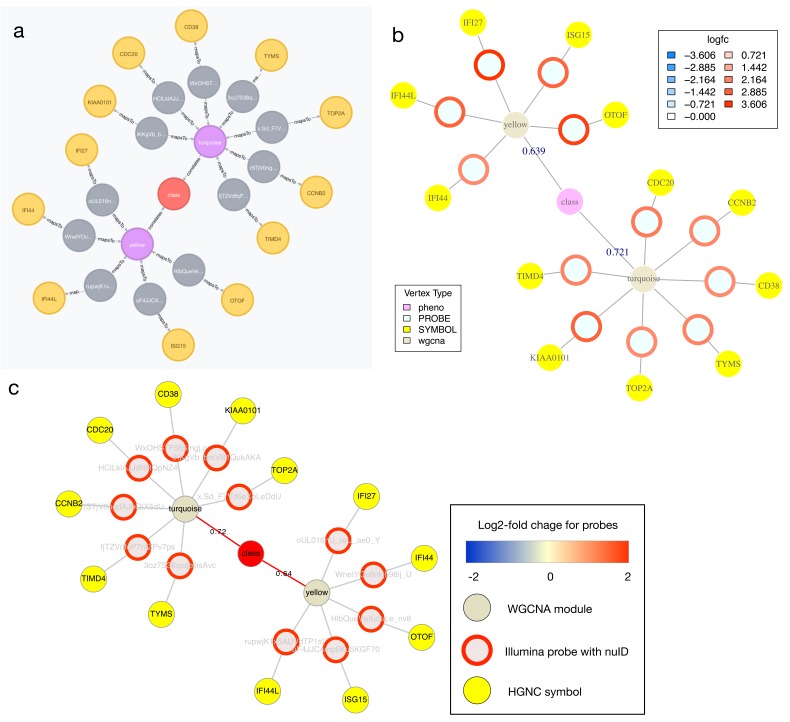
returns WGCNA modules for the HIV positive vs control comparison in the HIVsetB dataset whose module eigengene (ME) is strongly correlated with a clinical variable (Pearson R > .6), and it also returns the genes mapping to probes within those modules that have high levels of over-abundance in HIV-1 infected individuals. In addition to the standard web browser interface for neo4j, into which the above query can be directly entered and returned in the browser window ( Figure 2A), we also provide a function in R ( igraph_plotter) that returns the network found, and plots this within R ( Figure 2B), exports the network as node and edge lists for import in other software like Cytoscape ( Figure 2C), or returns the result as an igraph 17 object for further manipulation within R.
Figure 2. Visualising Cypher query results.
Relationships between nodes extracted from the ANIMA database using a Cypher query applied to the HIVsetB data (N HIV=30, N Controls=17, see Figure S1; Supplementary File 1). Shown are two WGCNA modules that contain probes with increased transcript abundance in acute HIV infection and whose module eigengene is positively correlated with disease class (an ordinal variable). ( A) Result from native browser interface for Neo4j. ( B) Result plotted from within an R session connected to the ANIMA database, using the igraph_plotter function. Log 2-fold change values for the individual probes are shown by coloured rings; values are shown in the legend. ( C) The same result, visualized in Cytoscape, taking advantage of the igraph_plotter function to export node and edge lists for easy import into Cytoscape. Links/ edges are annotated with Pearson correlation coefficients where applicable.
Approach 1: Multiscale class comparison
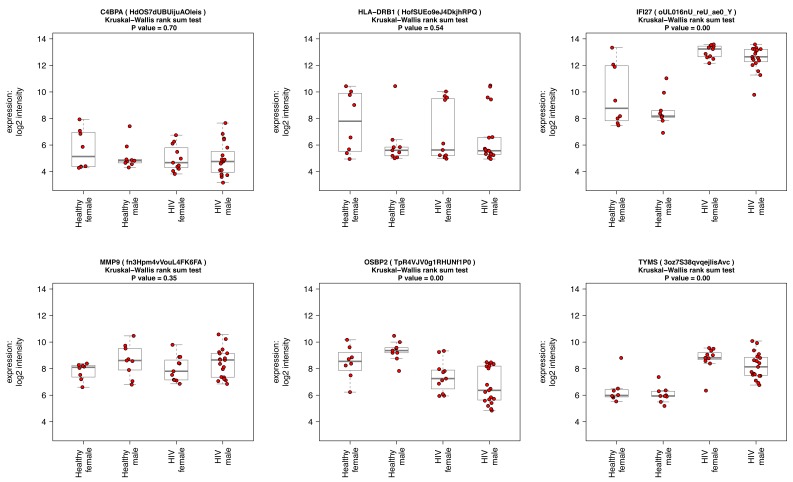
Accessing individual transcript abundance levels in multiple conditions. It is useful to view transcript abundance patterns of specified probesets, for instance to compare microarray data to RT-PCR validation data, or to investigate the behaviour of groups of biologically related genes in various conditions. We provide an interface for this in ANIMA. The user can submit a search string (in the form of a regular expression) containing gene names (HUGO Gene Nomenclature Committee (HGNC) symbols), and box-and whisker plots for the results are returned. In the original paper on acute HIV infection the authors discuss a gene set of six conserved genes that appear at multiple timepoints in an inferred regulatory network of viral set point 6. We show the normalized expression data stratified by HIV status and sex in the two datasets included in the HIV analysis for these six genes ( Figure 3). In the paper on acute viral respiratory infection, IFI27 and PI3 are identified to differ between acute influenza A and human rhinovirus infections. In influenza, IFI27 is upregulated and PI3 downregulated relative to human rhinovirus. The malaria study replicated prior knowledge of differential transcript abundance for C1QB, MMP9, C3AR1, IL18R and HMOX1; we show similar results for these transcripts. Supplementary Table 1 lists the results of differential expression analysis for the above transcripts in the three conditions, providing validation of data at individual transcript level.
Figure 3. Visualising individual probe-level expression data.
Box-and-whisker plots showing normalized, log 2-transformed probe-level expression data for six selected genes, obtained by a custom function in R in four groups: Healthy female, N = 8, Healthy male, N = 9, acute HIV female, N = 11, acute HIV male, N = 19; data from HIVsetB dataset. Gene (and probe nuIDs for disambiguation) are given for reference; the y-axis shows log2 scale normalized intensity values. Box and whisker plots show median, interquartile range, and range. Outliers are defined as values that lie beyond the whiskers, which extend to maximally 1.5 X the length of the box. Individual datapoints are superimposed in red on the box-and-whisker plots. The four groups are compared using Kruskal Wallis rank sum test and the P-value for the comparison is shown in the plot title. Results for individual pairwise comparisons are not shown.
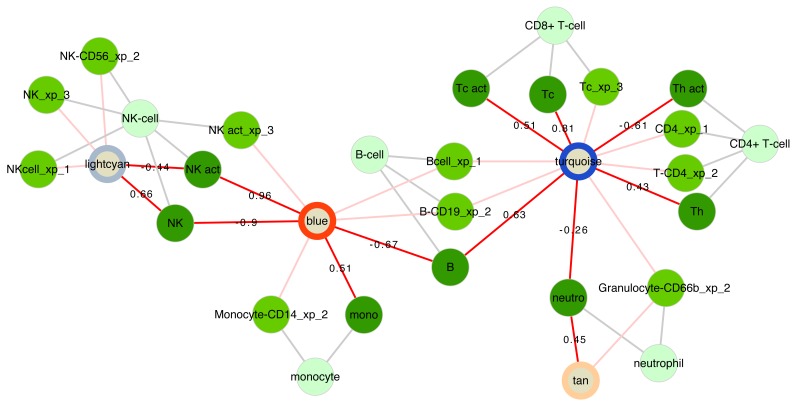
Functional annotation of WGCNA modules. An important question is what do WGCNA modules represent, given that these are groups of genes that co-vary across samples, but that can differ dramatically in size. Instead of searching only for evidence of co-regulation of expression by transcription factors, one should consider other causes of this co-variance. We propose the notion of WGCNA modules representing biological processes that may be regulated at different scales. For instance, a group of transcripts will co-vary across samples if these transcripts are expressed (predominantly) in a single cell type, and the proportions of this cell type varies between samples ( Figure 4). Another group of transcripts may be expressed in multiple cell types, but represents a concerted transcriptional program executed in response to a specific stimulus, such as interferon-alpha stimulating the expression of a specific group of interferon-regulated genes (see below). Generally, we determine the function of WGCNA modules based on all statistically significant associations with pathways and cells.
Figure 4. Cell associations of WGCNA modules.
Relationships of WGCNA modules and different cell types in the respInf dataset (Day 0 acute influenza, N = 46 vs baseline healthy samples, N = 48, see Figure S1; Supplementary File 1). Shown are WGCNA modules whose expression correlates with specific cell-type proportions (dark green, edges annotated with Pearson correlation coefficient R) and that are enriched for the genes specific to that cell type (medium green, suffixes xp_1-3 indicate the respective gene list on which the cell assignments were based, see Supplementary methods). The classes of cells are indicated in light green. The modules are annotated with coloured rings representing the difference in median eigengene values between cases and controls (diffME, see Supplementary methods); blue indicates modules which are under-expressed, and red indicates modules that are over-expressed in cases relative to controls.
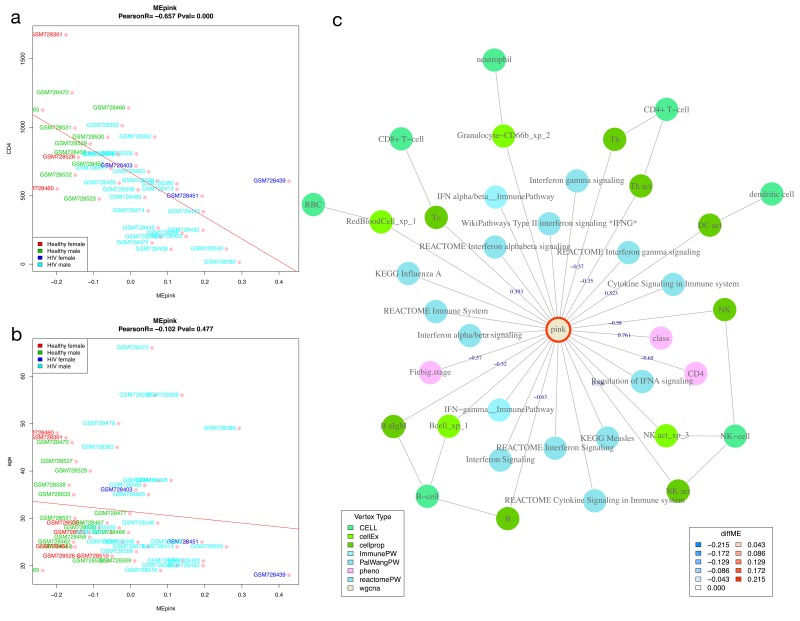
Relationship of modules to clinical variables. We performed Pearson correlation of WGCNA module eigengenes (ME) and clinical variables (described in supplementary methods). Figure 5 shows correlation of the pink ME with age and CD4 count in the HIVsetA data set. Clearly, the pink module is significantly associated with CD4 count, an association that is independent of age. Further investigation shows that this module is linked to interferon signalling, and probably expressed in a variety of cells. This agrees with our understanding of acute HIV infection, which is associated with a robust type-I interferon response and an acute drop in CD4 count 19.
Figure 5. Correlation of module eigengenes with clinical variables.
Shown is the Pearson correlation of the pink module eigengene with CD4 count (cells/microlitre) ( A) and with age (years) ( B) in acute HIV (N = 28) vs healthy controls (N = 23) in the HIVsetA dataset. Study subject IDs are used as point labels, and coloured as indicated in the legend. Plot titles show the Pearson coefficient R and the associated P-value. ( C) WGCNA module annotation obtained from the Neo4j database for the pink module. Edges are labelled with the correlation coefficient (R) where applicable. Note that the same coefficient is obtained for CD4 count as in panel A. Legends are shown for vertex type and diffME (a measure of differential co-expression (see Supplementary methods), i.e. the extent that the module eigengene median varies between two classes). Abbreviations: diffME, differential module eigengene.
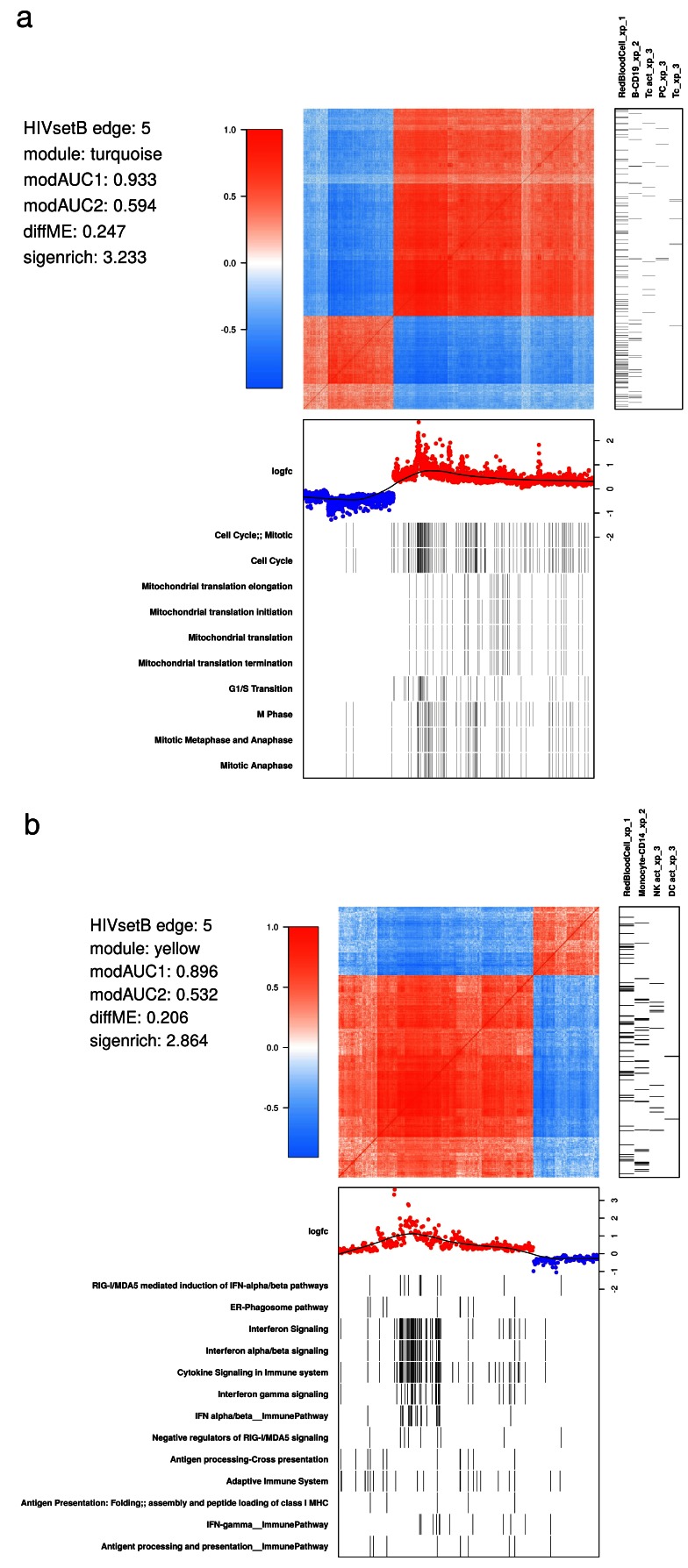
Investigating the structure of WGCNA modules. WGCNA modules are groups of co-expressed transcripts. Demonstrating the extent and direction of correlation of their constituent probes, and their relationships to biological pathways requires sophisticated visualization ( Figure 6). We showcase the example using the HIVsetB dataset and focus on two modules. The turquoise module shows the coordinated action of genes involved in cell division, relating to the cell proliferation in the lymphoid compartment. The yellow module is clearly related to interferon signalling; of interest here is that within this module there is a group of transcripts highly correlated with each other, all related to interferon signalling, suggesting that these transcripts may all be downstream of a single regulatory factor. Also clear from the figure is that this module is not limited to a single cell type, but rather to innate immune cells in general.
Figure 6. WGCNA module structure.
( A) Correlation matrix of all probes in the turquoise module in the HIVsetB dataset (N HIV=30, N Controls=17, see Figure S1; Supplementary File 1). Colours in the heatmap represent Pearson correlation coefficients, ranging from -1 to 1, as indicated by the legend. The module is enriched for lymphocyte-specific genes (right annotation panel) as well as cell cycle/mitosis associated genes, suggesting that various lymphocyte subsets in acute HIV infection are actively proliferating. (bottom annotation panel). Log 2-fold change values refer to differential transcript abundance in acute HIV relative to healthy controls. ( B) Correlation matrix of all probes in the yellow module in the HIVsetB dataset. It is enriched for innate cell genes as well as interferon signaling, suggesting that innate immune cells are in an interferon-induced state. Additional annotation information is provided to the left of the heatmap. The parameters modAUC1, modAUC2, diffME and sigenrich are defined in Supplementary methods. The plot is generated using a custom R function ( mwat).
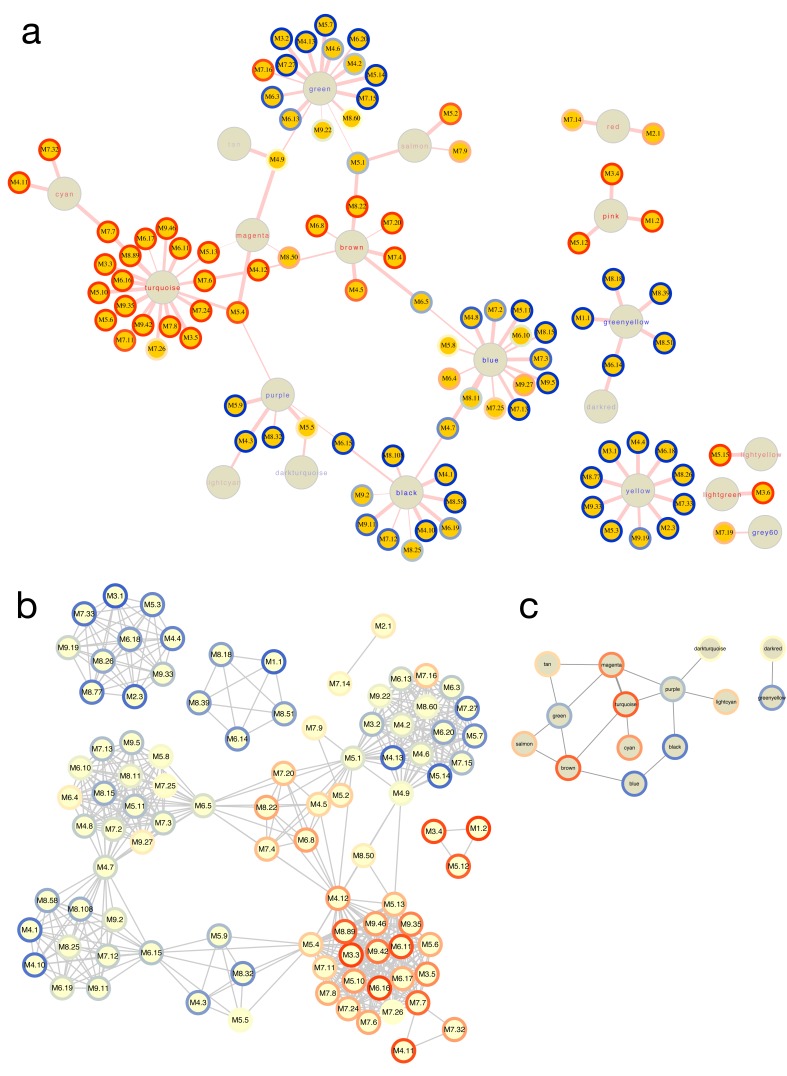
Relationships between module-based approaches. Both WGCNA and the modular approach pioneered by Chaussabel, et al. 11 rely on clustering of similarity matrices to derive modules. A key difference in these methods in ANIMA is that the Chaussabel modules are pre-defined, whereas the WGCNA modules are derived from the transcriptional data under study. It is therefore interesting to discover the relationships between these two approaches. Figure 7A shows the bipartite network of WGCNA and Chaussabel modules derived from the HIVsetA dataset, as defined by the hypergeometric index. Figures 7B and 7C show the two projections of the bipartite graph. The hypergeometric index is not the only way associations between the two module types can be demonstrated; for instance, a more indirect association can be inferred when a WGCNA and Chaussabel module map to the same biological pathway. It is clear from the plots that only a subset of the list of Chaussabel modules associates with WGCNA modules in any given condition.
Figure 7. Relationships between WGCNA and Chaussabel modules.
( A) Bipartite graph of the two module types based on the hypergeometric association index in the HIVsetA dataset (acute HIV, N = 28 vs healthy controls, N = 23). Strikingly, Chaussabel modules tend to have the same direction of differential expression (indicated by the rim colour of the Chaussabel modules, red indicating up-regulation in acute HIV, and blue indicating downregulation) as WGCNA modules they map to, indicated by the label colour of the module. ( B) Projection 1 of ( A), showing relationships between Chaussabel modules based on shared WGCNA modules; dense cliques of modules are observed. ( C) Projection 2 of ( A), showing relationships between WGCNA modules based on shared Chaussabel modules. All associations (hypergeometric test) shown are corrected for multiple testing, BH-corrected P-value < 0.05. All outputs were generated using the igraph_plotter function, exporting vertex and edge tables of the bipartite graph and the two projections and importing these into Cytoscape.
Deconvolution. An important question in analysing transcription data from complex tissues is whether differences in transcript abundance are attributable to transcriptional regulation in one or more cell types, or to changes in the composition of the overall leukocyte populations.
Figure S3 (Supplementary File 1) shows the results for the HIVsetB dataset, and Table S2 (Supplementary File 1) shows the results of non-parametric statistical testing for differences in median cell-type proportions, per cell-type and class comparison with uncorrected P values as well as P-values corrected for multiple testing using the Benjamini-Hochberg procedure; these were encoded as parameters diffP and diffQ of the cellprop node type in the ANIMA database. Neutrophils were the most abundant cell-type estimated from the array data. The proportions of activated NK cells and CD8+ T cells were significantly elevated in acute HIV infection, and proportions of B cells and CD4+ T cells were reduced, in line with published observations 20.
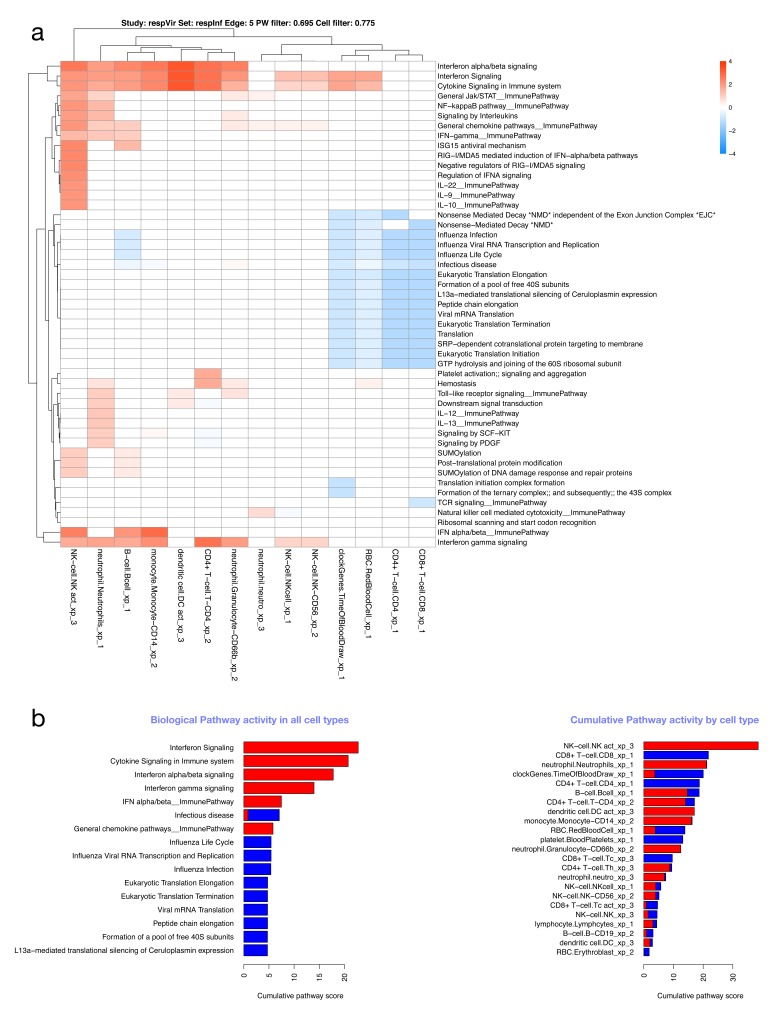
Virtual cells: an estimate of functional phenotypes of different immune cell types. Given that the immune response is mediated by different types of cell, we attempted to re-create “virtual cells” based on the assumption the genes that co-vary in terms of transcript abundance across samples with that of cell-type specific genes are expressed in that particular cell type. With these relationships, we generated virtual cells (probe co-expression matrices annotated with biological pathways and probe differential expression data). Figure S4 (Supplementary File 1) shows two cells (B-cells and neutrophils) in acute HIV infection; both characterized by interferon signalling. Given the “virtual cells” and their functions we can compare pathway-level transcript abundance in different cell types by creating a matrix of cell types and pathways, with each entry representing the “pathway activity” for a given cell and pathway combination. To illustrate this, we show replication of the finding of NK-cell activation in acute influenza infection ( respInf dataset, Figure 8 A,B), and in addition provide more detail on which pathways are probably up- or downregulated in these and multiple other cells.
Figure 8. Cell/pathway activity matrix.
( A) Cell/pathway activity matrix for all cell-types for the respInf dataset (Day 0 acute influenza, N = 46 vs baseline healthy samples, N = 48, see Figure S1). The clustered heatmap shows pathway activity scores representing the mean log-2 fold change for all probes in the pathway for a particular cell type (see Supplementary methods). There is a clear interferon response in multiple cell types, as well as down-regulation of other pathways associated with translation. ( B) Barplots highlighting the most highly differentially regulated pathways (left panel, determined by row sums of matrix in A), and cells with highest levels of differential expression (right panel, determined by column sums of matrix in A). In all cases, up- and downregulated pathway scores are kept separate.
Approach 2: Factorial analysis
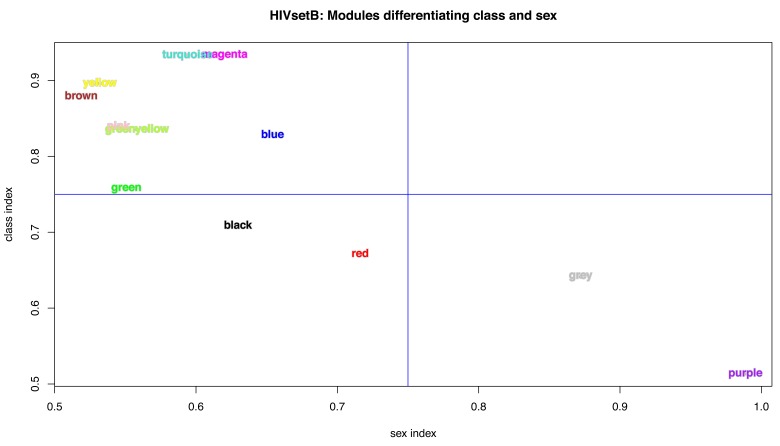
Modules driven by sample class or sex. Our main interest in investigating transcriptomic datasets is to identify molecular and cellular processes that drive, or at least are associated with, specific phenotypic traits of the samples. Therefore, the experimental design for the differential abundance analysis and the probe filtering steps prior to WGCNA module discovery were both designed to highlight processes associated with two factors: disease class and sex; the former because this is the basis of the research question for the three studies, and the latter because sex has a distinct influence on immune system function 21. We developed a novel approach to quantify the association of the WGCNA module expression with these two factors (See supplementary methods; Supplementary File 1). Figure 9 shows that, in the case of acute HIV infection, most modules are associated with the disease process ( HIVsetB dataset), but that one module ( purple) is strongly associated with sex. Figure S5 ( Supplementary File 1) shows the module eigengenes for the yellow and purple modules, demonstrating differential class associations for WGCNA modules; this finding would have been missed had the data not been stratified by both HIV infection status and sex. Table S4 shows the module statistics for this dataset.
Figure 9. WGCNA module indices.
Plot of module indices representing the area-under-the-ROC curve for the two classes for all WGCNA modules in the HIVsetB dataset (N HIV=30, N Controls=17, see Figure S1; Supplementary File 1). The indices are named per the variable they aim to differentiate (disease class or sex). The class index corresponds to the modAUC1 variable and the sex index corresponds to modAUC2. These indices are calculated form the module eigengenes and given class assignments using functions from the rocr package. See text and Supplementary methods for details.
Additional modules were identified that associated with neither sample class nor sex. These represent biologic processes that manifest in heterogeneity of the sampled population. Figure S6 ( Supplementary File 1) plots the study subjects in the HIV infection data set ( HIVsetA) on two axes represented by two module eigengenes.
Approach 3: Meta-analysis of multiple datasets
Module-level meta-analysis. Meta-analysis of multiple related expression datasets can lead to insights not available from analysis of any single datasets, and can highlight common patterns of transcript abundance across different conditions, or meaningful differences across highly common conditions. We implemented the approach pioneered 14 and refined 15 by Chaussabel, et al. to perform modular transcriptional repertoire analysis 22 on the six datasets. This approach is particularly suited to meta-analysis, as the composition of the modules is always identical. Figure 10 shows modular patterns for the six datasets in a clustered heatmap as well as the subset of modules with similar expression patterns across the six data sets, demonstrating universal patterns in the immune response to infection. Supplementary Table S3 ( Supplementary File 2) lists module functions based on all significant enrichment associations for the modules in Figure 10B. Universal upregulation of interferon-related modules particularly stand out, as does the suppression of modules associated with CD4+ T cells, CD8 T cells and B cells.
Figure 10. Meta-analysis of transcriptional and cellular patterns.
( A) All 258 Chaussabel modules plotted as a heatmap in all six datasets. ( B) The subset of modules all expressed in the same direction. Three module groups of interest are identified. ( C) Cell/pathway activity matrix for a single cell type (CD8 + T cell) based on three celltype-gene lists (xp1, xp2, xp3, see Supplementary methods) in all three conditions. Activity in CD8+ T cells in HIV all cluster together, and differ from both malaria and respiratory infections. Cell labels are constructed by [condition]_[dataset]_[comparison]_[cell class]_[cell type]_[gene list].
Meta-analysis at the cell-type level. A second approach to meta-analysis was implemented using virtual cells based on WGCNA modules. Here we compared the pathway scores in a single or several cell types across multiple conditions. For instance, comparing the CD8 T-cell response in acute HIV, acute viral respiratory infection, and symptomatic malaria, we find that proliferation of activated CD8+ T cells characterizes acute HIV infection, and to a lesser extent symptomatic malaria infection. In contrast, there is a suppression of CD8 T cell activity in blood in other conditions, due in part to a reduction in CD8+ T-cell proportions in whole blood ( Figure 10C).
Discussion
Systems immunology aims to understand the complex web of relationships between immune system components (cells, cytokines, effector molecules and other mediators) in immune-mediated disease states. Much progress has been made in single-cell techniques that yield large amounts of information, e.g. single-cell RNAseq and mass cytometry. These approaches are expensive, and not easy to apply in incompletely characterized disease systems, as they require a certain amount of focus (e.g. selection of one or a few cell types for RNAseq, and marker selection for CyTOF). In contrast, RNAseq or microarray analysis of complex tissue samples (like blood) in principle contain information on the transcriptomic state of all cells present in the sample and is thus an unbiased approach. The main difficulty with this lies in the interpretation of the data, and in many cases a complexity-reducing analysis approach is employed, where the focus is placed on differentially expressed genes. Other approaches based on co-expression analysis often fail to explain the drivers of the co-expression patterns.
We demonstrate, using a novel method of aggregating information obtained from clinical and microarray data, an ability to reconstruct many aspects of the immune response, and to discuss this not in the language of probes, genes and signatures, but rather as coordinated biological processes and the cellular context for these processes, allowing the generation of hypotheses at multiple scales. Our multiscale class comparison approach can be used to validate findings from individual papers, for instance the intense NK-cell activation described in influenza 8.
Using our factorial approach, we can begin to dissect inter individual heterogeneity in transcriptional patterns from transcriptional patterns that are in a causal relationship with defined factors. Application of the two meta-analysis approaches allows comparison of arbitrary datasets to detect similarities and differences at modular and cellular levels. An interesting and somewhat unexpected finding is that acute symptomatic malaria and acute respiratory viral illnesses are more similar to each other than to acute HIV infection, another viral illness. Despite these differences, we demonstrate that, at least for these three rather different infections, a broadly similar pattern of transcriptional module activity can be described.
In summary, ANIMA is both a robust implementation of various well-regarded analytic paradigms in microarray analysis, as well as a framework for integrating these various methods to expose relationships at multiple scales and render these computationally accessible.
Software availability
Source code (R code, dockerfiles) are available at GitHub at https://github.com/adeffur/anima under a CC BY-NC 4.0 license
An archive of source code releases linked to github is available on Zenodo ( http://doi.org/10.5281/zenodo.1163398 23)
The docker image used to implement ANIMA is available at Docker hub ( https://hub.docker.com/r/animatest/anima/) with the tag v3.3.3
The same image has been archived and is available on Zenodo ( https://doi.org/10.5281/zenodo.1161475 24)
System requirements
-
1.
Minimum system requirements for installing and running Docker must be met. Docker is freely available for macOS, Microsoft Windows and various Linux distributions. See https://docs.docker.com/install/ for further information
-
2.
To run the full ANIMA pipeline, including the build phase, Docker must be provisioned with at least 50GB RAM, and 6–8 cores
-
3.
To run the ANIMA web application, at least 8GB of system RAM is required, but 16GB is preferable.
Data availability
Array Express: Whole Blood Transcriptional Response to Early Acute HIV, E-GEOD-29429
Gene Expression Omnibus: The genomic architecture of host whole blood transcriptional response to malaria infection, GSE34404
Gene Expression Omnibus: Host transcriptional response to influenza and other acute respiratory viral infections – a prospective cohort study, GSE68310
A single zip archive will all data, and clinical and other metadata has been archived on Zenodo; for reproducibility purposes it is advised that this archive is used as described in the manual ( Supplementary File 3) ( https://doi.org/10.5281/zenodo.1161380 25).
Funding Statement
This work is supported by the Wellcome Trust [104803, 203135] This study was also supported by the Medical Research Council of South Africa, Hasso Plattner Foundation, and Newton Fund [NA150173]
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved with reservations]
Supplementary material
-
1.
Supplementary File 1: ANIMA_method_SUPP_submission_WOR.7z
This document contains supplementary methods, additional figures and tables, implementation and replication information.
Click here for additional data file. (3.8MB, tgz) -
2.
Supplementary File 2: Table S3 New.xlsx
Supplementary Table S3 in Excel format.
Click here for additional data file. (36.1KB, tgz) -
3.
Supplementary File 3: ANIMA_manual_v1.0.0.pdf
This document contains full instructions to replicate the ANIMA pipeline and web application, as well as screenshots describing the functionality of the web application.
Click here for additional data file. (7.4MB, tgz)
Pre-print
A preprint of this work has been submitted to biorxiv (doi: https://doi.org/10.1101/257642 26)
References
- 1. Ahmed E, Hashish AH: On modelling the immune system as a complex system. Theory Biosci. 2006;124(3–4):413–418. 10.1016/j.thbio.2005.07.001 [DOI] [PubMed] [Google Scholar]
- 2. Benoist C, Germain RN, Mathis D: A plaidoyer for 'systems immunology'. Immunol Rev. 2006;210:229–234. 10.1111/j.0105-2896.2006.00374.x [DOI] [PubMed] [Google Scholar]
- 3. Mazzocchi F: Complexity in biology. Exceeding the limits of reductionism and determinism using complexity theory. EMBO Rep. 2008;9(1):10–14. 10.1038/sj.embor.7401147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaussabel D, Pascual V, Banchereau J: Assessing the human immune system through blood transcriptomics. BMC Biol. 2010;8:84. 10.1186/1741-7007-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Langfelder P, Horvath S: WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang HH, Soderberg K, Skinner JA, et al. : Transcriptional network predicts viral set point during acute HIV-1 infection. J Am Med Inform Assoc. 2012;19(6):1103–1109. 10.1136/amiajnl-2012-000867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Idaghdour Y, Quinlan J, Goulet JP, et al. : Evidence for additive and interaction effects of host genotype and infection in malaria. Proc Natl Acad Sci U S A. 2012;109(42):16786–16793. 10.1073/pnas.1204945109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhai Y, Franco LM, Atmar RL, et al. : Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections--A Prospective Cohort Study. PLoS Pathog. 2015;11(6):e1004869. 10.1371/journal.ppat.1004869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Du P, Kibbe WA, Lin SM: lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. 10.1093/bioinformatics/btn224 [DOI] [PubMed] [Google Scholar]
- 10. Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, et al. : A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38(3):e17. 10.1093/nar/gkp942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arloth J, Bader DM, Röh S, et al. : Re-Annotator: Annotation Pipeline for Microarray Probe Sequences. PLoS One. 2015;10(10):e0139516. 10.1371/journal.pone.0139516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smyth GK: Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1): Article3, 1–25. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 13. Gaujoux R, Seoighe C: CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013;29(17):2211–2212. 10.1093/bioinformatics/btt351 [DOI] [PubMed] [Google Scholar]
- 14. Chaussabel D, Quinn C, Shen J, et al. : A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29(1):150–164. 10.1016/j.immuni.2008.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Obermoser G, Presnell S, Domico K, et al. : Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38(4):831–844. 10.1016/j.immuni.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu G, He QY: ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2015;12(2):477–479. 10.1039/c5mb00663e [DOI] [PubMed] [Google Scholar]
- 17. Csardi G, Nepusz T: The igraph software package for complex network research. InterJournal. 2006. Reference Source [Google Scholar]
- 18. Neo4j's Graph Query Language: An Introduction to Cypher. (Accessed: 3rd January 2017). Reference Source [Google Scholar]
- 19. Hardy GA, Sieg S, Rodriguez B, et al. : Interferon-α is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS One. 2013;8(2):e56527. 10.1371/journal.pone.0056527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McMichael AJ, Borrow P, Tomaras GD, et al. : The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10(1):11–23. 10.1038/nri2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klein SL, Flanagan KL: Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626–638. 10.1038/nri.2016.90 [DOI] [PubMed] [Google Scholar]
- 22. Chaussabel D, Baldwin N: Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014;14(4):271–280. 10.1038/nri3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deffur A: adeffur/ANIMA: ANIMA Source code (Version v1.0.0). Zenodo. 2018. 10.5281/zenodo.1163398 [DOI] [Google Scholar]
- 24. Deffur A, Wilkinson RJ, Mayosi BM, et al. : ANIMA: Association Network Integration for Multiscale Analysis TAR archive of anima Docker image used in the publication) (Version 3.3.3). Zenodo. 2018. 10.5281/zenodo.1161476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deffur A, Wilkinson RJ, Mayosi BM, et al. : ANIMA: Association Network Integration for Multiscale Analysis source data, metadata and scripts) (Version 1.0.0). Zenodo. 2018. 10.5281/zenodo.1161381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deffur A, Wilkinson RJ, Mayosi BM, et al. : ANIMA: Association Network Integration for Multiscale Analysis. bioRxiv. 257642 10.1101/257642 [DOI] [PMC free article] [PubMed] [Google Scholar]