Abstract
In eukaryotic cells, the γ-monomethylphosphate cap structure has been identified in four small RNAs, namely, U6 , 7SK, B2, and plant U3 RNAs. In this study, we show that in the case of 7SK and B2, as well as in plant U3 RNAs, the 5′ stem-loop followed by a short single-stranded region serves as the capping signal. We previously showed that the nucleotides 1–25 of mouse U6 snRNA, also comprised of a stem-loop followed by a short single-stranded region, function as the capping signal. These data show that capping signals in all four RNAs have common features. The length of the stem-loop among these capped RNAs varied from 20 to 108 nucleotides, with no significant variation in the capping efficiency. In addition to the capping signal, we also observed a minimum RNA length requirement of about 15–25 nucleotides following the stem-loop for efficient capping in vitro. The capping signal in plant U3 snRNA corresponds to the additional 5′ stem-loop found in U3 RNAs from plants and lower eukaryotes but absent in U3 RNA from higher animals. Consistent with this observation, the human U3 RNA that lacks the additional 5′ stem-loop was not a suitable substrate for capping when compared to U6 snRNA.
IN eukaryotes, in addition to mRNAs, several small RNAs contain cap structures on their 5′ ends. These capped small RNAs contain either trimethylguanosine (TMG) or γ-monomethyltriphosphate (methylphosphate) cap structures (Reddy and Busch, 1988; Reddy et al., 1992). In the case of human cells, the TMG-capped RNAs include U1-U5 and U7-U13 RNAs (Reddy and Busch, 1988; Tyc and Steitz, 1989). The TMG cap structures in small RNAs have diverse functional roles, including enhancing the stability (Kleinschmidt and Pederson, 1990) and in some instances transport across the nuclear membrane (Fischer and Luhrmann, 1990; Hamm et al., 1990; Fischer et al., 1991). All TMG-capped RNAs studied to date are transcribed in the nucleus by RNA polymerase II with cotranscriptionally added m7G on their 5′ ends, as is the case with m7G cap formation in mRNAs (Eliceiri, 1980; Skuzeski et al., 1984). In the case of frog U2 snRNA, it was shown that the m7G cap is further methylated to TMG in the cytoplasm (Mattaj, 1986); thus, the formation of TMG cap structure takes place in two discrete steps, the cotranscriptional m7G addition in the nucleus and trimethylation in the cytoplasm. However, m7G cap in TMG-capped RNAs of some organisms, such as Trypanosomes, are formed post-transcriptionally and an in vitro capping system for TMG cap formation has been reported (Zwierzynski and Buck, 1990).
To date, four small RNAs containing the methylphosphate cap structure have been characterized; these are U6 (Singh and Reddy, 1989), 7SK (Gupta et al., 1990a; Shumyatsky et al., 1990), rodent B2 (Shumyatsky et al., 1990), and plant U3 RNAs (Shimba et al., 1992). Although the nuclear U6 snRNA and nucleolar U3 snoRNA are involved in pre-mRNA and pre-rRNA processing, respectively, the functions of nuclear 7SK and cytoplasmic B2 RNAs are not known. Although not yet fully characterized, there is evidence for the existence of additional methylphosphate-capped RNAs (Gupta et al., 1990a; Liu et al., 1992). We studied the function of methylphosphate cap structure using the frog oocyte system, and in the case of U6, 7SK, and B2 RNAs, the methylphosphate-capped RNAs were three to nine times more stable compared to corresponding RNAs containing ppp on their 5′ ends (Shumyatsky et al., 1993). These data show that one of the functions of methylphosphate cap structure is to enhance the half-life of RNAs and as shown earlier, this property is common to other known cap structures such as m7G (Shatkin, 1976, 1985) and TMG cap structures (Kleinschmidt and Pederson, 1990). This may explain the presence of different cap structures in U3 snoRNA from animal and plant cells and suggests that in the case of plant U3 snoRNA, the methylphosphate cap may functionally substitute for TMG cap structure found in U3 snoRNA of animal cells.
Using U6 snRNA as the model substrate, we previously showed that all the three phosphates of the initiation nucleotide incorporated during transcription of the U6 snRNA are retained during cap formation and an RNA sequence-dependent methyltransferase methylates the gamma phosphate of the substrate RNA (Gupta et al., 1990b). In the case of m7G or TMG-capped RNAs, only two of the three (α and β) phosphates of the initiation nucleotide incorporated during transcription are retained in the final RNA. Therefore, the mechanisms of methylphosphate and m7G TMG cap formation are fundamentally different (Gupta et al., 1990b; Reddy et al., 1992). Because U6 (Hamm and Mattaj, 1989) or 7SK snRNA (Shumyatsky et al., 1993) does not leave the nucleus, the methylphosphate cap formation appears to take place in the nucleus. We previously showed that the 1–25 nucleotides of U6 snRNA comprised of a stem-loop followed by a short single-stranded region is necessary and sufficient to direct cap formation in vitro (Singh et al., 1990). In this study, we extend these studies to three other methylphosphate-capped RNAs, and in every case, we found that 5′ stem-loop followed by a short single-stranded region is necessary to direct cap formation in vitro. In addition to this core capping signal, we find additional, apparently sequence-independent, RNA length requirement following these capping signals to direct cap formation in vitro.
MATERIALS AND METHODS
Plasmid
The mouse U6 (Das et al., 1988), human 7SK (Murphy et al., 1987), and mouse B2 (Kramerov et al., 1982) genes were used for generation of plasmids suitable for transcription by T7 RNA polymerase. The T7 promoter sequences have been introduced by means of polymerase chain reaction (PCR) so that the T7 transcripts of the genes started with the first nucleotide of the corresponding RNA. PCR was also used to make shorter versions of U6 and 7SK RNA genes (1–35 nucleotides and 1–115 nucleotides for U6 and 7SK, respectively). B2 RNA gene under T7 promoter was cut with Pf1MI and BglII restriction endonucleases, filled in with T4 DNA polymerase, and ligated using T4 DNA ligase resulting in a deletion corresponding to nucleotides 77 to 105 of B2 DNA sequence.
RNA Synthesis In Vitro With T7 RNA Polymerase
The plasmid DNAs were linearized by cutting at the appropriate restriction sites. Linearized DNA (2 μg each) was incubated in 20 μl of transcription buffer (Epicentre Technologies Corp., Madison, WI), 10 mM DTT, 0.5 mM each of ATP, CTP, UTP, 0.04 mM GTP, 20 U of T7 RNA polymerase, 20 U of RNasin, and 20 μCi [α-32P]GTP (Amersham Corp., Arlington Heights, IL) for 60 min at 37°C. Labeled RNAs were fractionated on 10% denaturing polyacrylamide gels; bands of interest were excised and eluted RNAs were precipitated with 3 vol. of ethanol including 20–40 μg of glycogen or 10 μg of E. coli tRNA as carrier. The purified RNAs were suspended in 10 μl of sterile water and used for further experiments.
Capping In Vitro With HeLa Cell Extracts
The capping reactions were carried out essentially as described earlier by Gupta et al. (1990b). Briefly, approximately 20,000–50,000 cpm of in vitro-transcribed uncapped RNA was incubated with whole HeLa cell extract (∼300 μg of protein) in the capping buffer [50 mM Tris-HCl (pH 8.0), 150 mM KCl, 10 mM MgCl2, and 4 mM AdoMet] at 30°C for various time periods as specified in the figure legends. The reactions were terminated by the addition of SDS/phenol, and the purified RNAs were -digested with nuclease PI and in some cases also with alkaline phosphatase. When the RNA length is different between samples, the amount of radioactivity was adjusted so that approximately the same number of 5′ ends are being analyzed. These digests were fractionated by electrophoresis on DEAE-cellulose paper at pH 3.5 (Singh et al., 1990). The radioactivity was quantified by counting the cut DEAE-cellulose paper in a liquid scintillation counter or using the Betascope 603 blot analyzer system (Betagen, IntelliGenetics Inc., Mountain View, CA). Autoradiography was done at −70°C using Hyperfilm-MP (Amersham) with Lightning Plus screens.
Oocyte Injections
After a low-speed centrifugation of the oocytes to facilitate accurate injection, labeled RNA samples in approximately 20 nl were injected into each oocyte nucleus or 50 nl into cytoplasm. After an overnight incubation, the oocytes were suspended in homomedium [50 mM Tris-HCl (pH 7.5), 5 mM EDTA, 1.5% SDS, 300 mM NaCl], digested with proteinase K, RNAs extracted with phenol-chloroform, chloroform, and precipitated with 3 vol. of ethanol. Extracted RNAs were analyzed on denaturing 10% polyacrylamide gels, and usually three to five oocyte equivalents of RNA per lane were loaded on polyacrylamide gels.
RESULTS
The Methylphosphate Cap Formation Can Be Uncoupled From Transcription In Vivo
We showed previously that U6 snRNA can be transcribed in vitro in the presence of AdoHcys, which is a competitive inhibitor of methylation reactions including the methylphosphate cap formation (Singh et al., 1990; Gupta et al., 1990b). In addition, U6 snRNA synthesized under T7 promoter with ppp on its 5′ end can be accurately capped in vitro by the HeLa cell extract (Gupta et al., 1990b). These data indicated that transcription and capping can be uncoupled in vitro. We wanted to see whether methylphosphate cap formation and transcription are actually uncoupled in vivo. To achieve this, we injected U6, 7SK, 5S, and 7–2 RNA genes into the nuclei of frog oocytes and the synthesized RNAs were analyzed for the presence of methylphosphate cap structure.
In the case of U6 (Fig. 1, lane 1) and 7SK RNAs (Fig. 1, lane 3), only 15–30% of the RNAs were capped with mpppG on their 5′ ends and the rest was found to be pppG. Because 5S RNA and 7–2 RNAs do not contain the cap structure, as expected, both 5S and 7–2 RNAs were not capped (Fig. 1, lanes 2 and 5, respectively). An aliquot of the 7SK RNA (lane 4) and 5S RNA (lane 6) was also treated with alkaline phosphatase after nuclease PI digestion. As expected, the pppG in these two cases was completely digested and mpppG cap structure formed in 7SK snRNA was resistant to digestion by alkaline phosphatase (lane 4). These data demonstrate that the synthesis of 106 nucleotide-long U6 RNA, as well as 331 nucleotide-long 7SK RNA, could be completed in vivo without an obligate requirement for capping. These data also show that capping in the case of frog oocytes is rather slow and rate limiting compared to transcription of these RNAs.
FIG. 1.
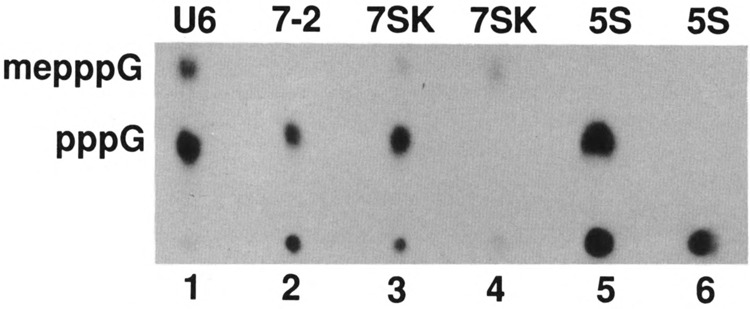
Analysis of cap structure of small RNAs synthesized in frog oocytes. Approximately 20 nl containing [α-32P]GTP and 4 ng of different small RNA genes were injected into each frog oocyte nucleus, incubated for 16 h, and the labeled RNAs were purified by electrophoresis on a polyacrylamide gel. The RNAs indicated on top of each lane were isolated, purified, and digested with nuclease PI (lanes 1–3 and 5), or with nuclease PI followed by alkaline phosphatase (lanes 4 and 6), electrophoresed on DEAE-cellulose paper, and subjected to auto-radiography. me, methyl.
Kinetics of Capping In Vitro
To study the efficiency of capping of different RNA substrates in vitro, we needed conditions where the RNA substrate is not limiting. We used labeled U6, 7SK, and B2 RNAs for in vitro capping reactions and quantified the capped and uncapped forms of added substrate RNAs at different intervals ranging from 2 to 30 min. Although there are no apparent primary sequence homologies between 7SK, U6 and B2 RNA sequences, all these three RNAs were capped with similar efficiencies (Fig. 2). To compare the efficiency of capping of different RNAs that vary in length as well as in sequence, we chose a 10-min time period where approximately 50% of the added RNAs are still in uncapped form (Fig. 2) and used these conditions to determine the capping efficiency of different RNAs. In some experiments, the quantity of cell extract added to the capping reactions was reduced to obtain 50% capping in about a 30-min time period.
FIG. 2.
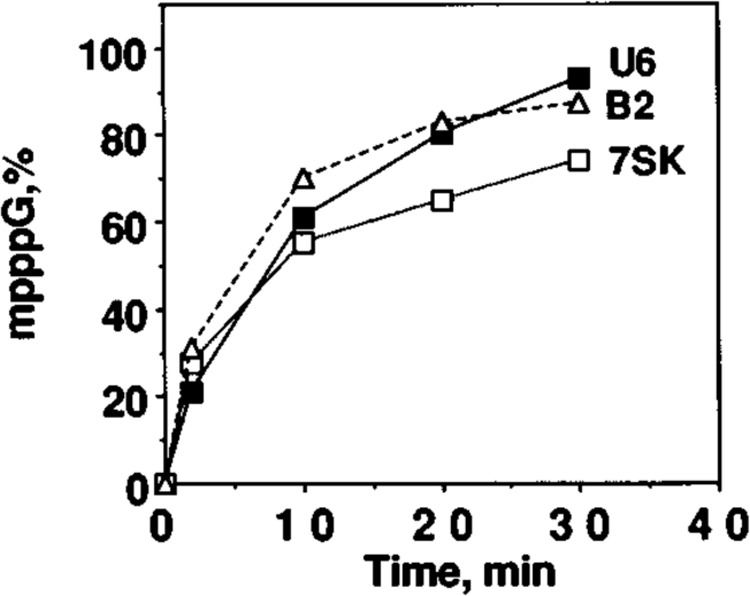
Kinetics of methylphosphate cap formation in vitro with different RNAs as substrates. U6, 7SK, and B2 small RNAs were transcribed in vitro with T7 RNA polymerase in the presence of [α-32P]GTP as the labeled nucleotide, and equal moles of each RNA were used as substrate in an in vitro capping reaction. After various periods of incubation, RNAs were purified, digested with nuclease P1, electrophoresed on DEAE-cellulose paper, and subjected to autoradiography. The radioactivity in pppG and mpppG spots was quantified using a Betascope blot analyzer, and the radioactivity in pppG and mpppG was taken as 100%. Conversion of ppp to mpppG with increasing period of incubation is presented.
5′ End Sequences From 7SK, B2, and Plant U3 RNAs Can Direct Capping In Vitro
Plasmid DNAs containing U6, 7SK, B2, and Plant U3 RNA genes were used as templates, and RNAs were synthesized using T7 RNA polymerase. All the resulting RNAs contained 5′ ends identical to the corresponding cellular RNAs but differed on their 3′ ends; in addition, these RNAs also did not contain any sequences corresponding to the vector following the RNA sequences. These RNAs were incubated with HeLa cell extract and the fraction of 5′ ppp converted to methylphosphate cap was quantified (Table 1). First, we used U6 snRNA as a substrate and U61-46 RNA (Fig. 3A, lane 2) was capped as efficiently as the full-length U6 RNA (Fig. 3A, lane 1); however, U61-35 RNA was capped with only 10% efficiency (lane 3). We also used U61-31 and U61-41 RNA, and these RNAs were capped very inefficiently compared to U6 snRNA (Fig. 5, lanes 2 and 3; Table 1). The 5′ stem-loop in U6 snRNA was destroyed by site-directed mu-tagenesis in two linker-scan mutants. These two mutants were no longer capped in vitro (Fig. 3, lanes 5 and 6) compared to wild-type U6 snRNA (lane 4). These data confirm our earlier data (Singh et al., 1990) that the integrity of the 5′ stem-loop is essential for capping.
TABLE 1.
CAPPING EFFICIENCIES OF VARIOUS snRNAs AND snRNA MUTANTS
| U6 RNA | |
| 1–106 (Dra I) | 100 |
| 1–46 (Mbo II) | 95 |
| 1–41 (Oligos) | 15 |
| 1–35 (Oligos) | <5 |
| 1–35 + 26 vector | 90 |
| 7SK RNA | |
| 1–331 (DraI) | 100 |
| 1–126 (Alu I) | 90 |
| 1–108 (Nla III) | <10 |
| 1–47 (ScrF I) | <5 |
| B2 RNA | |
| 1–180 (DraI) | 100 |
| 1–104 (Bgl II) | 100 |
| 1–83 (Fok I) | 100 |
| 1–77 (NlaHI) | 10 |
| 1–77 + poly A | 60 |
| 1–74 (PflmMI) | 7 |
| 1–74 + poly A | 65 |
| 1–66 (ScrF I) | <5 |
| 1–50 (Ava II) | <5 |
| 1–16 (Dde I) | <5 |
| U3 RNA (human) | |
| 1–215 (RsaI) | 40 |
| 1–107 (DdeI) | 10 |
| 1–91 (MnlI) | 7 |
| 1–80 (MnlI) | <5 |
| 1–64 (HgiAI) | <5 |
| U3 RNA (plant) | |
| 1–230 (BamHI) | 100 |
| 1–97 (HaeHI) | 90 |
| 1–55 (EcoRI) | <5 |
| 1–15 + 35 vector (HindIII) | 90 |
| U2 RNA (human) | |
| 1–188 (SmaI) | <5 |
Capping efficiencies of truncated and/or modified RNAs were calculated, taking the capping efficiency of the corresponding wild-type snRNA as 100%. The results presented are averages of three independent experiments.
FIG. 3.
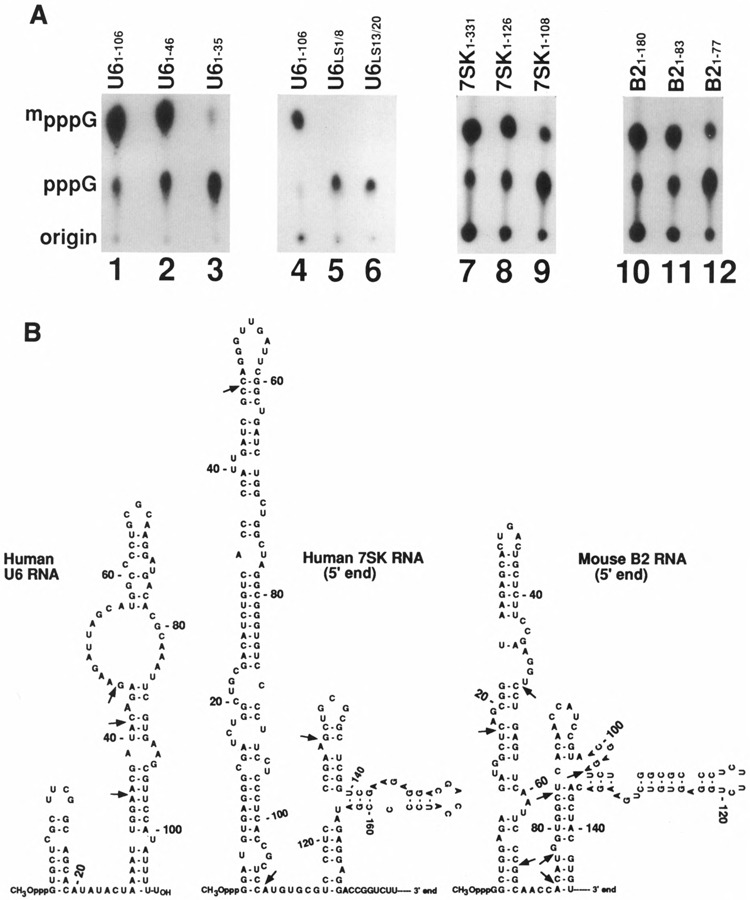
(A) Effect of 3′ deletions on capping efficiency of U6, 7SK, and B2 RNAs. The U6, 7SK, and B2 small RNA genes under T7, promoter were truncated, and full-length or truncated versions with 3′ deletions were incubated under in vitro capping conditions. The U6 gene linker-scan mutants described in Singh et al. (1990) were transcribed in the presence of AdoHcys and labeled RNAs were assayed for capping. The numbers above indicate the length of the RNA starting from initiation nucleotide 1. The U6 LS1/8 contained GGTACC between nucleotides 1 and 8; LS13/20 also contained GGTACC between nucleotides 1 and 6. After the capping reaction, the RNAs were purified, digested with nuclease PI, electrophoresed on DEAE-cellulose paper, and subjected to autoradiography. (B) Secondary structures of U6, 7SK, and B2 small RNAs. The secondary structure of U6 snRNA is from Epstein et al. (1980). The 7SK and B2 RNA secondary structures are from Wassarman and Steitz (1991) and Blandon et al. (1990), respectively. The arrows indicate the sites where RNAs were truncated and used for capping in vitro.
Fig 5.
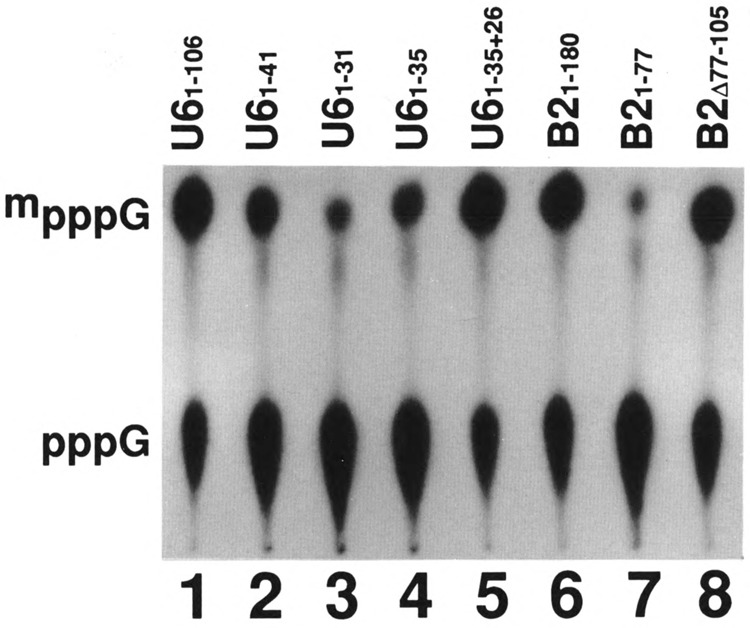
Effect of unrelated sequences 3′ to the capping signal on the efficiency of capping in vitro. Labeled U6 and B2 small RNAs with different 3′ truncations and alterations, as indicated on the top of each lane, were prepared in vitro and subjected to capping reaction in vitro. The labeled RNAs were purified, digested with nuclease P1, electrophoresed on DEAE-cellulose paper, and subjected to autoradiography. U61-35-26 RNA is 61 nucleotides long and has 26 nucleotides of vector sequence following the U61-35 sequence.
In the case of 7SK1-126 RNA (lane 8), the capping efficiency was similar to full-length 7SK RNA (lane 7), and 7SK1-108 RNA was capped with less than 10% efficiency (Fig. 3A, lane 9) compared to full-length 7SK RNA. B21-83 RNA was capped as efficiently (lane 11) as the full-length B2 RNA (lane 10); however, B21-77 RNA was capped with less than 10% efficiency (lane 12). Figure 3B shows the secondary structure of human U6 and secondary structures corresponding to the 5′ ends of 7SK and B2 RNAs. It is interesting to note that the secondary structures of 7SK (Wassarman and Steitz, 1991) and B2 RNAs (Blandon et al., 1990) contain a stem-loop on their 5′ ends, and both B2 RNA1-83 with 14 nucleotides after the stem-loop and 7SK RNA1-126 with 18 nucleotides following the stemloop were capped nearly as efficiently as the corresponding full-length RNAs. In contrast, RNA with 11 nucleotides after the stem-loop, 7SK1-108 RNA with only the 5′ stem-loop and B21-77 with eight nucleotides after the stem-loop were not good substrates for capping in vitro (Fig. 3A, B, and Table 1). These data show that in addition to the U6 snRNA, the 5′ ends of 7SK and B2 RNAs are capable of directing the methylphosphate cap formation. These data also suggest that it is not the total length of the RNA starting from the 5′ end that is important for capping; rather, it is some minimum length of the RNA following the stem-loop that is important in determining the capping efficiency.
5′ End Sequences of 7SK and B2 RNAs Are Required for Capping In Vitro
Nucleotides 1–25 comprise part of the first stemloop of the human U6 snRNA (Fig. 3B). We previously demonstrated that U6Δ2-25 snRNA lacking 2–25 nucleotides does not serve as a substrate for capping (Singh et a l., 1990). We synthesized U6Δ1-25 RNA in the presence of AdoHcys so that this RNA does not contain the methylphosphate cap structure. When this RNA was used as a substrate for capping, the capping efficiency was found to be <5% (Fig. 4, lane 2). We also prepared constructs where the 5′ end sequences corresponding to a portion of the stem-loop of 7SK and B2 RNAs are deleted and these RNAs were tested for their ability to direct capping in vitro. Both 7SKΔ2-48 (lane 4) and B2Δ2-38 (lane 6) were not capable of directing cap formation in vitro. As expected, the U6 (lane 1), 7SK (lane 3), and B2 RNA (lane 5) were capped efficiently under the same conditions. These data demonstrate that, just like in U6 snRNA, the 5′ end sequences that contribute to the stem-loop structures in the case of 7SK and B2 RNAs (see Fig. 3B) are also required for directing cap formation in vitro.
FIG. 4.
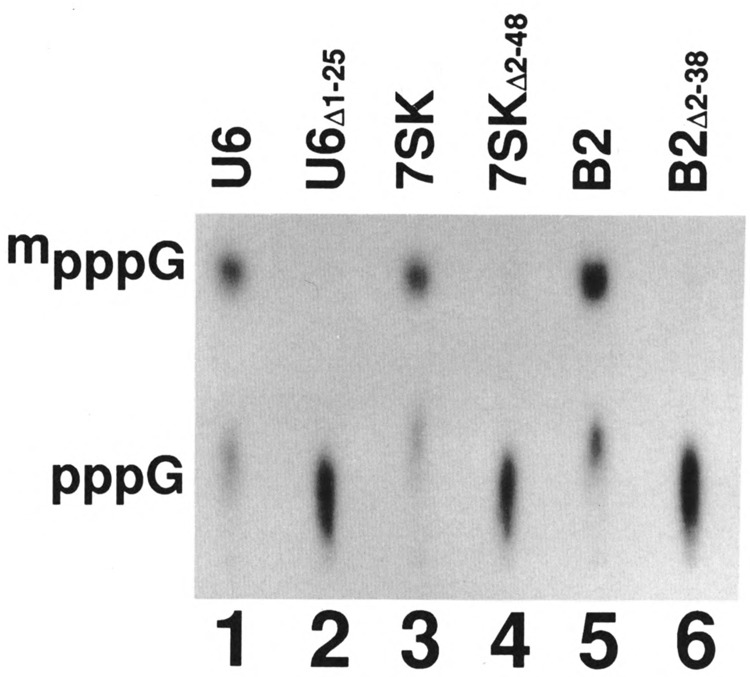
Effect of 5′ deletions on the capping efficiency of U6, 7SK, and B2 RNAs. U6, 7SK, and B2 small RNAs, where sequences near the 5′ end were deleted, were prepared with 5′ ppp by transcription in vitro in the presence of [α-32P]GTP. Full-length and the corresponding RNAs with deletions near the 5′ end were subjected to capping reaction in vitro; the labeled RNAs were purified, digested with nuclease P I, electrophoresed on DEAE-cellulose paper, and subjected to autoradiography.
Role of Random RNA Sequences 3′ to the Capping Signal
Our earlier studies on the identification of capping signal in U6 snRNA were carried out by transcription of altered U6 snRNA genes in vitro using the HeLa cell extract (Singh et a l., 1990). In many of these experiments, the transcription was driven by the U6 gene promoter and the transcription termination occurred at a cluster o f T residues in the vector sequence. Consequently, the transcripts contained portions of the U6 RNA sequence on the 5′ end and sequences derived from the vector on the 3′ end. We wanted to see whether capping signal corresponding to the 5′ end of U6 RNA without the vector sequence on the 3′ end is sufficient to direct cap formation.
We tested U6 RNA with different lengths, all of which contained the same 5′ end, and only U61-46 RNA (Fig. 3, lane 2) or U6 RNA longer than 46 nucleotides (data not shown) were capped as efficiently as the wild-type U6 RNA. U61-41 was capped with 35% efficiency (Fig. 5, lane 2), and U61-31 and U61-35 RNA were capped with an efficiency of 10% or less (Fig. 5, lanes 3 and 4, respectively; Table 1). These data indicated that although U61-25 is the capping signal (Singh et al., 1990), this signal is not functional unless some additional sequence is present on the 3′ end of the capping signal. Because U61-25 snRNA with vector sequence on its 3′ end was as good as the sequence from the U6 snRNA (Singh et al., 1990), there appears to be no specific sequence requirement 3′ to the core capping signal. We directly tested this possibility by testing U61-35 RNA containing 25 nucleotides of the vector on its 3′ end for capping efficiency. This RNA was capped as efficiently as the wild-type U6 RNA (compare lanes 1 and 5). These data further support the suggestion that there is a length requirement following the essential 5′ stem-loop.
We also tested B21-77 RNA, which does not get capped by itself (Fig. 5, lane 7), and added an unrelated sequence on its 3′ end. This modified B2 RNA again was capped (Fig. 5, lane 8) as efficiently as the wild-type RNAs (Fig. 5, lane 6). These data are consistent with the results obtained with U6 snRNA and confirm that random sequences are sufficient to fulfill the length requirement following the core capping signal.
Poly A Sequence Following B21-77 RNA Is Sufficient to Direct Cap Formation In Vitro
We also tested whether a simple homopolymer like poly A can substitute for this length requirement 3′ to the capping signal. B21-77 RNA, which by itself does not get capped in vitro, was polyadenylated with poly A polymerase and this RNA was tested for capping in vitro. As expected, B21-77 RNA, as well as B21-74 RNA, was capped with <5% efficiency (Fig. 6, lanes 2 and 3, respectively). B21-77 RNA (lane 4), as well as B21-74 (lane 5) with poly A on its 3′ end, was capped with approximately 60% efficiency compared to wild-type B2 RNA (lane 1). These data show that the capping enzyme prefers a heteropolymer; however, a homopolymer like poly A can, to a large extent, substitute for a heteropolymer.
FIG. 6.
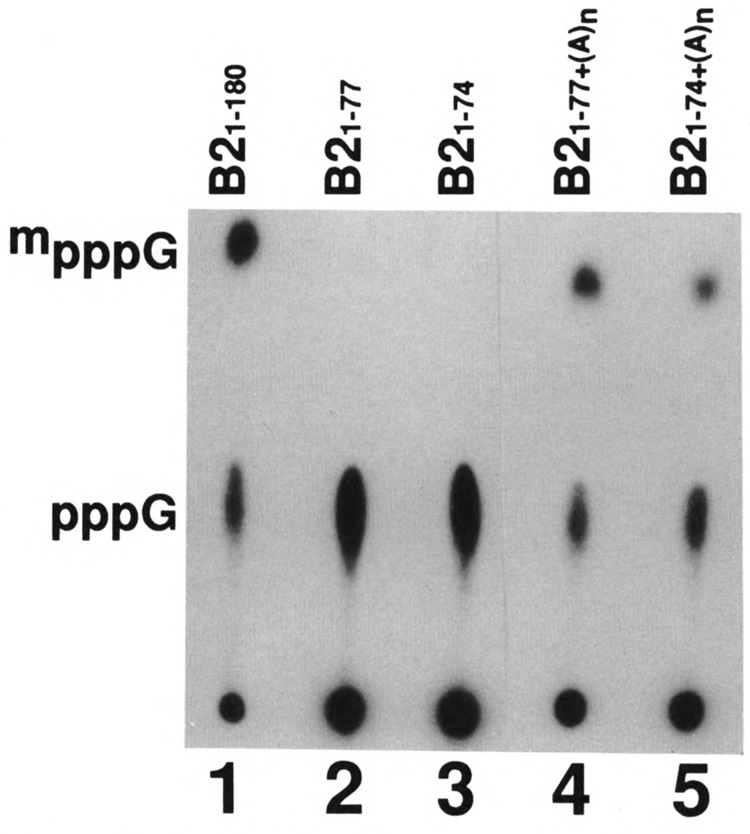
Effect of poly A sequences on the capping efficiency. Labeled B21-74 and B21-77 RNAs were polyadenylated with poly A polymerase and the average length of poly A added was approximately 150 A residues. These poly A+ B2 RNAs were subjected to capping reaction in vitro. In addition, the full-length B2 RNA and B21-74 and B21-77 RNAs without poly A were also used as controls. The labeled RNAs, after the capping reaction, were purified, digested with nuclease P1, electrophoresed on DEAE-cellulose paper, and subjected to autoradiography.
Any Stem-Loop Followed by a Single-Stranded Region Is Not Sufficient for Capping In Vitro
Although the four methylphosphate-capped small RNAs have similar structural requirements (i.e., a stem-loop followed by a single-stranded region), there is no obvious primary sequence common to these four RNAs. In addition, there are very few nucleotides conserved among capping signals in the 5′ stem-loop structure of U6 RNA from various yeasts (Roiha et al., 1989). These observations raised the possibility that any stem-loop structure followed by some single-stranded region may be sufficient to direct capping in vitro. The transcripts containing the human immunodeficiency virus type 1 (HIV-1) TAR element and the RNA from the coding region of gene 10 of the T7 phage (pET-3) can form a stable stem-loop at their 5′ ends (Fig. 7A).
FIG. 7.
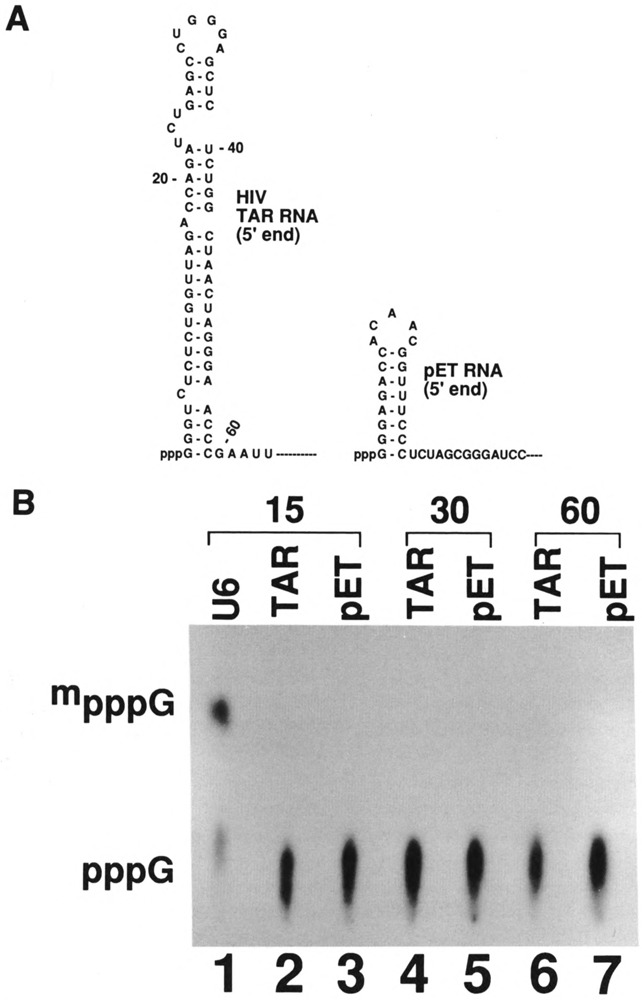
(A) Secondary structures of the 5′ end of the HIV-1 TAR RNA and T7 phase DNA. TAR RNA sequence corresponds to nucleotides 1–59 of HIV-1 RNA (Muesing et al., 1987) and pET sequence corresponds to nucleotides 1–26 of bacteriophage T7 DNA (Rosenberg et al., 1987). Remainder of the sequence is from the plasmid vector. (B) Capping of unrelated stem-loop structures in vitro. Plasmids containing the T7 TAR element and T7 pET-3 sequence were linearized with Pvu II and EcoR V, respectively, transcribed by T7 RNA polymerase, and the transcripts were purified on polyacrylamide gels. The T7 TAR construct gave a 170 nucleotide-long RNA that has a 5′ stem-loop of 59 nucleotides. The T7 pET-3 construct gave a transcript of 174 nucleotides that includes a 5′ stem-loop of 21 nucleotides. Lane 1: U6 RNA as a control; 15, 30, or 60 shown on top are the period (min) of incubation. These transcripts were incubated with HeLa cell extract for capping for various time periods; RNAs were purified, digested with nuclease P1, electrophoresed on DEAE-cellulose paper, and subjected to autoradiography.
To test this possibility, we prepared these two RNAs and incubated them in vitro under conditions where capping of U6 snRNA is approximately 50% complete (Fig. 7B, lane 1). There was no detectable capping in the case of pET-3 RNA after 10 min (lane 3), 20 min (lane 5), or 1 h (lane 7) of incubation. In the case of TAR RNA, there was no detectable capping after 10 min (lane 2) or 20 min (lane 4) of incubation. However, there was some detectable low level of capping of TAR RNA transcripts after incubation for 1 h (lane 6) or longer (data not shown). These data show that any stem-loop followed by a single-stranded region will not serve as a substrate for capping in vitro.
Capping Signal in Plant and Human U3 RNA
The plant U3 snoRNAs are unique in having methylphosphate cap structures whereas U3 snoRNAs form animal cells and lower eukaryotes, including yeasts, contain the TMG cap structure (Shimba et al., 1992). In addition, the 5′ end of human U3 snoRNA secondary structure contains one stem-loop, whereas the 5′ end sequence of plant U3 snoRNAs contains two stem-loops (Fig. 8A). We prepared plant U3 RNAs of different sizes and tested them for capping efficiency. Again, full-length plant U3 RNA and U31-90 RNA, which contains 50 nucleotides after the stem-loop, were capped; however, U31-55 RNA, which has only 15 nucleotides after the stem-loop, was not capped (Fig. 8B, lane 4 and Table 1). When we used U31-45 RNA with 35 nucleotides of vector sequence on the 3′ end, this RNA was capped as efficiently (Fig. 8B, lane3) as the wild-type U3 snoRNA (Fig. 8B, lane 1). These data show that methylphosphate cap containing plant ID snoRNA has similar structural requirements as the U6, 7SK, and B2 RNAs for directing cap formation in vitro.
FIG. 8.
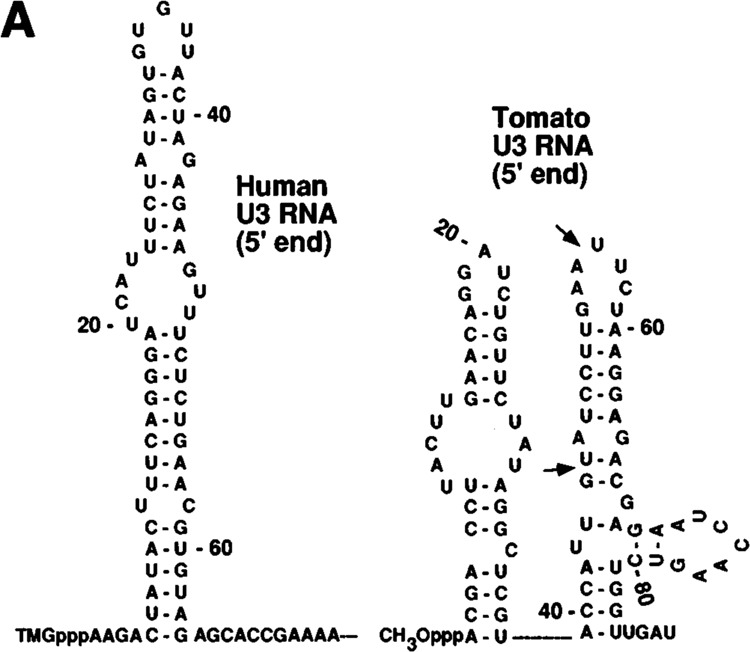
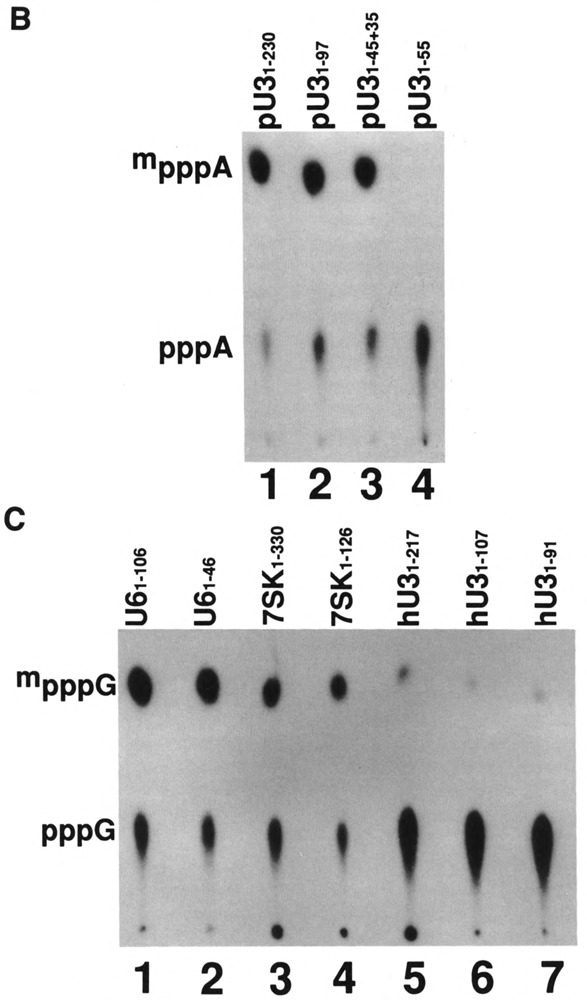
(A) Secondary structures of human and plant U3 snoRNAs. The secondary structures corresponding to the 5′ portions of human U3 snoRNA (Parker and Steitz, 1987) and tomato U3 snoRNA (Kiss et al., 1991) are shown. The arrows indicate the sites where RNAs were truncated and used for capping in vitro. (B) Capping of tomato U3 RNA in vitro. Labeled tomato U3 RNA and 3′-truncated or altered tomato U3 RNAs, as indicated on the top of each lane, were prepared in vitro and incubated under capping conditions. The purified RNAs were digested with nuclease P1, electrophoresed on DEAE-cellulose paper, and subjected to autoradiography. (C) Capping of human U3 RNAs in vitro. Full-length human U3 snoRNA and two 3′-truncated U3 RNAs and other RNAs, as indicated on the top of each lane, were synthesized in vitro and incubated under capping conditions. The RNAs at the end of the incubation were purified, digested with nuclease P1, electrophoresed on DEAE-cellulose paper, and subjected to autoradiography.
Plant U3 snoRNA genes are normally transcribed by RNA polymerase III (Kiss et al., 1991). These investigators modified the tomato U3 snoRNA gene so that it could be transcribed by RNA polymerase II and acquired the TMG cap (Kiss et al., 1991). These data showed that plant U3 snoRNAs have methylphosphate capping signal as well as TMG formation signal. Therefore, we wanted to test whether human U3 snoRNA has methylphosphate capping signal in addition to the signal for TMG formation. Wild-type human U3 snoRNA and two shorter versions were prepared and their capping efficiency was compared with U6 and 7SK snRNAs (Fig. 8C). The human U3 snoRNA was capped with about 10% efficiency (lane 5) compared to U6 or 7SK RNA (lanes 1 and 3, respectively). In addition, although the 5′ ends of U6 (lane 2) or 7SK (lane 4) snRNAs alone were capped efficiently, the human U31-107 RNA (lane 6) and IB^ RNA (lane 7) containing the 5′ stem-loop followed by 43 and 27 nucleotides, respectively, were capped with less than 5% efficiency. These data show that human U3 snoRNA does not contain a capping signal found in other methylphosphate-capped small RNAs.
DISCUSSION
One of the main observations made in this study is that in the four methylphosphate cap-containing small RNAs that have been studied thus far, the capping signals correspond to their 5′ ends consisting of a stem-loop followed by a short, single-stranded region. The size of the stem-loop among these RNAs varied significantly, from 20 nucle-requirement in this portion of the capping signal.
We also looked for common sequences/features between capping signals among various methylphosphate-capped RNAs. In most of the U6 snRNAs, a G-C pair four nucleotides starting from 5′ end, and ACAU near the base of the stem-loop were found otides in the case of human U6 snRNA to 108 nucleotides in the case of human 7SK snRNA, with no significant difference in the capping efficiency (Fig. 2). These data suggest that although the stem-loop is required for the capping, the size of this stem-loop is not very critical. In addition, phylogenetic comparison of U6 snRNA from different organisms has shown that the stem-loop corresponding to the capping signal in U6 snRNA from different yeasts varies in size from 11 nucleotides to 46 nucleotides (Roiha et al., 1989). These data also support the suggestion that the size of the stem-loop can vary substantially and still act as an efficient capping signal.
The second observation is that in addition to the core capping signal of 25 nucleotides at the 5′ end of human U6 snRNA, there is an RNA length requirement for capping in vitro. The U61-35 RNA encompassing the core capping signal was not capable of directing capping in vitro. The same RNA length requirement following the stem-loop was found in the case of 7SK, B2, and plant U3 small RNAs. However, this length requirement can be satisfied by vector sequence or RNA sequence from another portion of the RNA. Even a poly A sequence was largely able to substitute for this random sequence. These results suggest that there is no strict sequence to be conserved in animal and yeast RNAs (Roiha et al., 1989). However, neither human 7SK nor mouse B2 RNA contained these sequence features, although these RNAs are capped as efficiently as the U6 snRNA (Fig. 2). Our data indicate that all these RNAs are capped by a common capping enzyme (Shimba and Reddy, 1994). These results raise the interesting question as to what this capping enzyme is recognizing in these different RNAs. At this stage, only two things seem to be common and required for capping. These are: 1) the 5′ stem-loop followed by a short, single-stranded region; 2) approximately a 10–20 nucleotide-long random RNA sequence following this core capping signal. The 5′ stem-loop in TAR RNA is very stable with a stability number of −37.6 kcal/mol1 (Muesing et al., 1987) compared to −10.8 kcal/moll for 5′ stem-loop in human U6 snRNA (Singh et al., 1990). In spite of this, stable stem-loops followed by long, single-stranded regions in TAR or pET RNAs could not direct cap formation (Fig. 7A and B), suggesting that there are critical features in these capping signals that are yet to be identified.
The secondary structure of nucleolar U3 RNA from different organisms has been studied by different investigators. All the available information supports the notion that U3 RNA from animal cells contain two stem-loops designated I and II (Parker and Steitz, 1987). However, the U3 snoRNA from lower eukaryotes like yeast (Porter et al., 1988; Segault et al., 1992; Selinger et al., 1992) and plants (Kiss and Solymosy, 1990) contain three stem-loops designated IA, IB, and II. Although the 3′ stem-loop (II) is common and conserved in all known U3 snoRNAs, the stem-loop IA, which we showed to be the methylphosphate capping signal in plant cells (Shimba et al., 1992; this study), is not present in animal cell U3 snoRNA. It appears that these sequences corresponding to the capping signal present in lower eukaryotes were lost during evolution of higher animals. These observations are consistent with the possibility that an ancestral U3 snoRNA transcribed by RNA polymerase III underwent conversion into the U3 gene that is transcribed by RNA polymerase II and the methylphosphate capping signal in animal U3 snoRNA genes was subsequently lost.
ACKNOWLEDGEMENTS
We thank Drs. S. Baserga, S. Murphy, and W. Harper for human U3, 7SK, and TAR/pET clones. These studies were supported by a grant from NIH-GM38320.
REFERENCES
- Blandon T. S., Fregean C. J., McBuzney M. W. (1990), Mol Cell Biol 10, 4058–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G., Henning D., Wright D., and Reddy R. (1988), EMBO J 7, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliceiri G. L. (1980), J Cell Physiol 102, 199–207. [DOI] [PubMed] [Google Scholar]
- Epstein P., Reddy R., Henning D., and Reddy R. (1980), J Biol Chem 255, 8901–8906. [PubMed] [Google Scholar]
- Fischer U., and Luhrmann R. (1990), Science 249, 786–790. [DOI] [PubMed] [Google Scholar]
- Fischer U., Darzynkiewicz E., Tahara S., Dathan N. A., Lührmann R., and Mattaj I. W. (1991), J Cell Biol 113, 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Singh R., and Reddy R. (1990b), J Biol Chem 265, 9491–9495. [PubMed] [Google Scholar]
- Gupta S., Busch R. K., Singh R., and Reddy R. (1990a), J Biol Chem 265, 19137–19142. [PubMed] [Google Scholar]
- Hamm J., and Mattaj I. W. (1989), EMBO J 8, 4179–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Darzynkiewicz E., Tahara S. M., and Mattaj I. W. (1990), Cell 62, 569–577. [DOI] [PubMed] [Google Scholar]
- Kiss T., Marshallsay C., and Filipowicz W. (1991), Cell 65, 517–526. [DOI] [PubMed] [Google Scholar]
- Kiss T., and Solymosy F. (1990), Nucleic Acids Res 19, 1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt A. M. and Pederson T. (1990), Proc Natl Acad Sci USA 87, 1283–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D. A., Lekakh I. V., Samarina O. P., and Ryskov A. P. (1982), Nucleic Acids Res 10, 7477–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M-H., Busch R., Buckley B., and Reddy R. (1992), Nucleic Acids Res 20, 4299–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. (1986), Cell 46, 905–911. [DOI] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., and Capon D. J. (1987), Cell 48, 691–701. [DOI] [PubMed] [Google Scholar]
- Murphy S., DiLiegro C., and Melli M. (1987), Cell 51, 81–87. [DOI] [PubMed] [Google Scholar]
- Parker K., and Steitz J. A. (1987), Mol Cell Biol 7, 2899–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter G. L., Brennwald P. J., Holm K. A., and Wise J. A. (1988), Nucleic Acids Res 16, 10131–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., and Busch H. (1988), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles (Birnsteil M. L., ed.), Springer-Verlag, Berlin, pp. 1–37. [Google Scholar]
- Reddy R., Singh R., and Shimba S. (1992), Pharmacol Ther 54, 249–267. [DOI] [PubMed] [Google Scholar]
- Roiha H., Shuster E. O., Brow D. A., and Guthrie C. (1989), Gene 82, 137–144. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D-S., Lin S-W., Dunn J. J., and Studier F. W. (1987), Gene 56, 125–135. [DOI] [PubMed] [Google Scholar]
- Segault V., Mougin A., Georgiev A., Baroques A., and Branlant C. (1992), Nucleic Acids Res 20, 3443–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger D. A., Porter G. L., Brennwald P. J., and Wise J. A. (1992), Mol Biol Evol 9, 297–308. [DOI] [PubMed] [Google Scholar]
- Shatkin A. J. (1976), Cell 9, 645–653. [DOI] [PubMed] [Google Scholar]
- Shatkin A. J. (1985), Cell 40, 223–224. [DOI] [PubMed] [Google Scholar]
- Shimba S., Buckley B., Reddy R., Kiss T., and Filipowicz W. (1992), J Biol Chem 267, 13772–13777. [PubMed] [Google Scholar]
- Shimba S., and Reddy R. (1994), J Biol Chem 269, 12419–12423. [PubMed] [Google Scholar]
- Shumyatsky G. P., Tillib S. V., and Kramerov D. A. (1990), Nucleic Acids Res 18, 6347–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumyatsky G., Wright D., and Reddy R. (1993), Nucleic Acids Res 21, 4756–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Gupta S., and Reddy R. (1990), Mol Cell Biol 10, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., and Reddy R. (1989), Proc Natl Acad Sci USA 86, 8280–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski J. M., Lund E., Murphy J. T., Steinberg T. H., Burgess R. R., and Dahlberg J. E. (1984), 259, 8345–8352. [PubMed] [Google Scholar]
- Tyc K., and Steitz J. A. (1989), EMBO J 8, 3133–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., and Steitz J. A. (1991), Mol Cell Biol 11, 3432–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwierzynsky T. A., and Buck G. A. (1990), Nucleic Acids Res 18, 4197–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]


