Abstract
There are two gene sets for U2 snRNA in the sea urchin L. variegatus, the U2E gene, which is expressed in oogenesis and early embryogenesis and then silenced, and the U2L gene, which is expressed constitutively. There are four major promoter elements found in both U2 snRNA genes; an essential TATA box at −25 to −30, a proximal element (PSE) at −55 required for expression, an element at −100 necessary for maximal expression, and an upstream activating sequence (UAS) necessary for maximal expression. The elements of the U2E and U2L promoters were interchanged and the effect on the temporal pattern of expression determined after microinjection of the genes into sea urchin zygotes. When the U2E PSE element was introduced into the U2L gene, the temporal pattern of expression of the gene was changed to that of the U2E gene. Converting the U2L gene proximal element into the early U2 gene proximal element by altering 14 nucleotides in the promoter also changed the temporal pattern of expression of the U2L gene. Factors that interact with the U2E PSE, detected by a gel mobility shift assay and DNasel footprinting, were present in blastula but not late gastrula embryos. In contrast, interchanging the – 100 element did not greatly affect the temporal pattern of expression, and factors that interact with the U2E gene – 100 box were present in both late gastrula and blastula embryos.
THE snRNAs are critical molecules necessary for the processing of mRNA precursors (Maniatis and Reed, 1987; Sharp, 1987; Lührmann et al., 1990). During early embryonic development in the sea urchin, there is rapid synthesis of snRNAs, starting at the 16–32 cell stage (Nijhawan and Marzluff, 1979). The majority of the U1 snRNAs synthesized in early development are transcribed from a tandemly repeated gene set (Nash et al., 1989). After hatching, the rate of synthesis of snRNAs drops and the tandemly repeated gene set is silenced (Santiago and Marzluff, 1989). The snRNAs synthesized later in development and in adult cells are derived from a gene set present in low copy number. There is a similar temporal pattern of expression of the sea urchin U2 snRNAs; a tandemly repeated gene set expressed in oogenesis and early embryogenesis and a low copy number set expressed later in embryogenesis and in adult cells (Stefanovic et al., 1991). A similar temporal regulation of snRNA expression in early development has been described for frog U1 and U4 snRNAs (Forbes et al., 1984; Lund and Dahlberg, 1987), and there are developmentally regulated variants of mouse U1 snRNAs (Lund et al., 1985; Lobo et al., 1988) and chicken U4 snRNAs (Korf et al., 1988).
snRNA promoters differ from the promoters of other genes transcribed by RNA polymerase II. In vertebrates, there is an essential proximal element, the PSE, located at about −50 to −60, which determines the start site. There is also a distal sequence element, the DSE, that has many properties of an enhancer (Dahlberg and Lund, 1989; Parry et al., 1989). All vertebrate snRNA genes, including genes from mammals, frogs, and birds (Korf and Stumph, 1986) isolated thus far, and including the poorly expressed U7 (Phillips and Turner, 1991; Gruber et al., 1991) and U11 snRNA genes (Suter-Crazzolara and Keller, 1991), have common PSE and DSE sequences. The sequences involved in differential expression of different snRNA genes have yet to be identified.
The sea urchin snRNA gene promoters have a similar spatial structure to the vertebrate promoters. However, the sea urchin snRNA genes are not expressed in Xenopus oocytes (Strub and Birnstiel, 1986), and hence the sea urchin snRNA promoter elements are not recognized by vertebrate transcription factors. There is an essential element in the sea urchin U2 (Stefanovic and Marzluff, 1992) and U1 (Weldelburg and Marzluff, 1992) snRNA genes located 50 to 60 nts 5′ to the start of transcription. However, this element is not conserved among the sea urchin U1, U2 (Stefanovic et al., 1991), and U7 snRNA (Southgate and Busslinger, 1989) genes. The sea urchin U2 snRNA genes differ from the vertebrate snRNA genes transcribed by RNA polymerase II in that they also contain a TATA box at −25 to −30 that is required for expression (Stefanovic and Marzluff, 1992).
We have previously described the isolation of two U2 snRNA genes from the sea urchin L. variegatus, the U2E, which is tandemly repeated and expressed early in embryogenesis, and the U2L gene, which is a single-copy gene expressed constitutively during embryogenesis (Stefanovic et al., 1991). Both genes have similar promoter structures, with a TATA box, a PSE at −60, an element at −100 required for complete expression, and an upstream activating sequence (Stefanovic and Marzluff, 1992). Other than the TATA box, the promoter elements of the U2E and U2L genes do not share any common sequence elements. We describe experiments demonstrating that the PSE element is an important determinant of temporal expression of the U2E gene, whereas the −100 box and upstream sequences play, at most, a minor role in temporal regulation.
MATERIALS AND METHODS
Construction of Clones
The U2E and U2L U2-U1 hybrid genes (U2EH and U2LH) containing the respective U2 promoters and Ul 3′ ends have been described previously (Stefanovic et al., 1991). The LEPL gene that contains nucleotides −27 to −118 of the U2E promoter inserted into the U2L promoter has also been described previously. These genes are shown in Fig. 1. The creation of the site-specific mutants and swapping of the −60 and − 100 elements has been previously described (Stefanovic and Marzluff, 1992). The substitution of the U2E gene PSE element into the U2L gene (LEpseL) was done by site-directed mutagenesis using the method of Kunkel (1985). The riboprobe used for mapping of transcripts was an antisense RNA extending from the nt +330 to nt − 152 of the U2LH template, synthesized in vitro using SP6 polymerase.
FIG. 1.
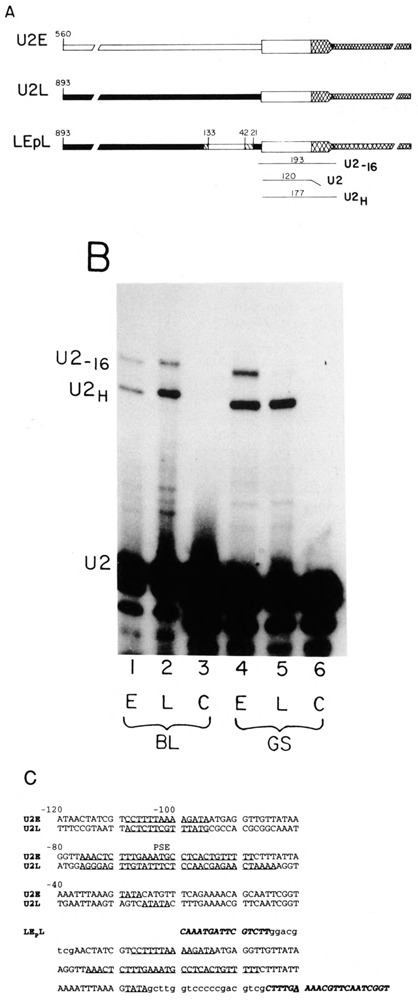
U2 gene constructs and the riboprobe protection assay. (A) Hybrid genes were constructed by fusing the U2E or U2L gene with the LvU1.1 gene as described previously (Stefanovic et al., 1991; Stefanovic and Marzluff, 1992). These two genes give rise to identical transcripts driven by two different U2 promoters. The LEPL gene has the U2E promoter sequence from −27 to −118 including the TATA box, the −60 sequence, and the −100 sequence, introduced into the U2L hybrid gene. The numbers on the figure indicate the distance of the sequence from the first nucleotide of U2 snRNA. The solid boxes are sequence from the U2L gene and the open boxes from the U2E gene. The hatched regions are the sequences introduced during construction of the clone. The sequence of the LEPL promoter is given in (C). The transcript from this gene starts 16 nt 5′ of the normal U2 start site because the U2E promoter sequences have been displaced from the normal U2 start site by 16 nucleotides. This gene was used as an internal control in all of the experiments described. Below the genes is a schematic of the riboprobe protection assay used to analyze the expression of the injected genes. Using a single probe, transcripts from the endogenous U2 genes, the U2-U1 hybrid genes, and the LEPL gene can be distinguished. (B) Expression of the U2 genes during development. The U2EH (lanes 1 and 4) and U2LH (lanes 2 and 5) genes were injected into sea urchin zygotes together with the LEPL gene. The embryos were harvested at the blastula (20 h, lanes 1–3) or late gastrula (66 h, lanes 4–6) stage, and the RNAs were assayed using the riboprobe procedure outlined in (A). Lanes 3 and 6 are uninjected control embryos. This figure is the same analysis presented previously (Stefanovic et al., 1991). The protected fragments are U2: 120 nts protection by the endogenous U2 RNAs; U2H: 177 nts protection by the transcripts from the U2EH and U2LH; U2_16: 193 nts protection by the transcripts from the LEPL gene. (C) The sequences of the proximal promoter region of the U2E and U2L genes are given. The −100 box, the PSE elements, and the TATA boxes are underlined. These elements have been defined previously (Stefanovic and Marzluff, 1992). The sequence of the region of the promoter of the LEPL gene is also given. In italics are the sequences from the U2L gene and in lower case are the polylinker sequences introduced during cloning. The −100, PSE, and TATA box of the U2E gene are underlined. The start site of transcription is the A, which is underlined.
Microinjection Into Sea Urchin Zygotes
Microinjection was performed by the method of Davidson and coworkers (McMahon et al., 1985), modified as described previously (Colin et al., 1988; Stefanovic et al., 1991; Stefanovic and Marzluff, 1992). Briefly, DNA from the test gene and the internal standard LEPL gene were linearized with a restriction enzyme that cuts 3′ of the gene, adjusted to a concentration of 50 μg/ml, mixed together in a 1:1 ratio and injected into L. pictus fertilized eggs immediately after formation of the fertilization membrane. Embryos were grown at 15°C until the hatching blastula stage (20 h) or until the late gastrula-early prism stage (66–72 h). About 100–200 embryos were collected per experiment; the RNA was extracted and analyzed by an RNase protection assay, as described previously (Stefanovic et al., 1991). The protected fragments were resolved by polyacrylamide gel electrophoresis and detected by autoradiography. Quantitation was done using a Betascope or by densitometric scanning of the X-ray film.
Preparation of Embryonic Nuclear Extracts
L. variegatus embryos were grown at 23°C to the blastula (8 h) or gastrula (24 h) stage. Highly purified nuclei were prepared as described (Morris and Marzluff, 1985). Starting with 2 ml of purified nuclei (109/ml), nuclear extracts were made by the method of Morris (Morris et al., 1986) and dialyzed against buffer C (20 mM Hepes, pH 7.8, 40 mM KC1, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, and 20% glycerol) for 4 h. Total blastula nuclear extract was used in all binding experiments except those in Fig. 5B. The extract from gastrula nuclei (and blastula nuclei for Fig. 5B) was fractionated over a 1 ml heparin-agarose column to enrich for DNA binding proteins. After loading the extract, the column was washed with 5 volumes of TM (100 mM Tris, pH 7.8, 12.5 mM MgCl2 1 mM EDTA, 1 mM DTT, and 20% glycerol) + 0.1 M KC1 and the proteins eluted with 2 volumes of TM + 0.4 M KC1. The eluate was dialyzed against buffer C for 4 h. All of the −100 box and PSE binding activity present in blastula extracts was present in the 0.4 M KC1 fraction. The protein concentration was estimated by the Bradford assay and was 1.5 μg/|μl for the blastula extract and 0.3 μg/μl for 0.4 M KC1 fraction from the heparin-agarose column of the blastula and gastrula extract.
FIG. 5.
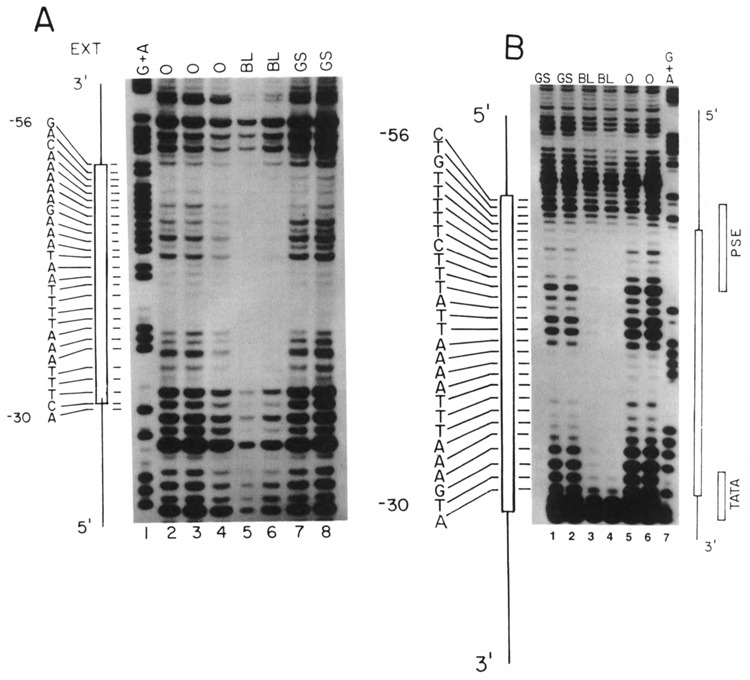
Footprinting analysis of the U2E proximal element. (A) A restriction fragment extending from − 119 to −25 nts 5′ of the U2E gene was labeled at the 5′ end at the EcoRI site and incubated with DNase in the absence (lanes 2–4) or presence of 4.8 μg of crude nuclear protein from blastula embryos (lanes 5,6) or with 1.8 μg of heparin-agarose fractionated gastrula nuclear protein (lanes 7,8). The gastrula nuclear extract was the same extract used in Fig. 4. Lane 1 is a G + A sequencing reaction. The sequence of the protected region is shown on the left. (B) The restriction fragment used in (A) was labeled on the 3′ end at the EcoRI site and incubated with DNase in the absence (lanes 5,6) or presence of 4.8 μg of nuclear protein from blastula embryos (lanes 3,4) or 2.4 μg of heparin-agarose fractionated nuclear protein from gastrula embryos. The gastrula nuclear extract was the same extract used in Figs. 4 and 7A. The sequence of the protected region is shown on the left. Lane 7 is a G + A sequencing reaction. (C) The same fragment used in (A) was analyzed by DNase I footprinting, in the presence and absence of competitor oligonucleotides. Lane 1 is a G + A sequencing reaction. Lane 2 is the DNA fragment incubated in the absence of extract. In lanes 3–7 the DNA was incubated with 4.8 μg of protein extracted from nuclei from blastula stage embryos. In lanes 4 and 5, a 1000- and 2000-fold excess of a nonspecific competitor oligonucleotide was included and in lanes 6 and 7 a 1000- or 2000-fold excess of a synthetic double-stranded oligonucleotide containing nucleotides −47 to −67 of the U2E gene, which contains the U2E PSE sequence, was included. The sequence of the protected region is shown on the left.
Gel Mobility Shift and Footprint Experiments
For gel retardation experiments the early gene PSE probe was prepared by digesting the U2ER60 DNA with PstI and Sail to release a 41 nt fragment containing the PSE. The fragment was labeled at the 3′ end using the Klenow fragment of DNA polymerase I and [α-32P]dCTP and/or [α-32P]dATP. In the competition experiments a 50–100-fold molar excess of the same restriction fragment was used as a specific competitor. The restriction fragment corresponding to the PSE element of the U2L gene PSE was also used as a competitor, and an oligonucleotide with the sequence of part of the coding region of a mouse histone H3 gene was used as a nonspecific competitor. The U2E probe for the −100 box experiments was prepared by digestion of the U2ER100 gene with the Sail and SacII and filling in the Sail site with the Klenow fragment of DNA poll and [α-32P]dCTP. The U2L −100 box probe was prepared similarly starting with the U2LR100 gene. The same unlabeled restriction fragments were used as competitor DNA. The labeled DNA (2 ng) was incubated with nuclear proteins in the presence of 2 μg of dI/dC (Pharmacia) and the indicated competitors, in a total volume of 15 μl of 20 mM Hepes, pH 7.8, 60 mM KC1, 5 mM MgCl2, 1 mM DTT, 8% glycerol. Complexes formed were resolved on a 4% nondenaturing polyacrylamide gel in 40 mM Tris-glycine, pH 8.3.
The probes used for DNasel footprinting were derived from the U2E gene promoter fragment extending from nt −119 to nt −25, which was cloned into pUC18. For footprinting of the −100 box, the plasmid was linearized with HinDIII (near nt − 119), labeled on the 3′ end as described above or on the 5′ end with polynucleotide kinase and [γ-32P]ATP following removal of the 5′ terminal phosphate with calf intestinal phosphatase. The labeled fragment was released by digestion with EcoRI. For footprinting the PSE element the plasmid was linearized with EcoRI, end-labeled as described above, and the labeled fragment released with Hindlll and the labeled DNA fragment purified by preparative gel electrophoresis.
The nuclear proteins were preincubated in 20 mM Hepes, pH 7.8, 60 mM KC1, 5 mM MgCl2, 1 mM DTT, 8% glycerol plus 0.1% NP-40 for 15 min on ice. Aliquots of 8 μ1 were incubated with 2 ng of probe and 1 μg of dI/dC in a total volume of 10 μl for 30 min at room temperature (in the case of 3′ end-labeled probes) or on ice (in the case of 5′ end-labeled probes). The samples contained 4.8 μg of total blastula nuclear proteins and 1.8 μg of the partially purified gastrula nuclear proteins, unless otherwise indicated. Following the incubation, 10 μl of 5 mM CaCl2, 10 mM MgCl2 was added and samples were digested with the indicated amounts of DNase for 1 min on ice. The digestion was stopped by the addition of EDTA and 1% SDS; the DNA was extracted with phenol/chloroform and precipitated with ethanol. The DNA fragments were resolved on 7 M urea-8% polyacrylamide gels. For the competition experiments, 1000- and 2000-fold excess of double-stranded oligonucleotides were included in the incubation. The sequence of the specific competitor was nts 46–67 of the U2E PSE and the nonspecific competitor was GATCTTCA-TGAATATTCACATC.
RESULTS
To study the sequences responsible for regulation of developmental expression of the two sea urchin U2 and snRNA genes, we constructed two chimeric genes (U2EH and U2LH) that have the promoter and first 120 nts of the U2 snRNA genes and the 3′ end of the L. variegatus U1 snRNA gene. These two genes express identical RNAs and the transcript from these genes is readily distinguished from the endogenous snRNAs. We have used these genes to map the important elements in the U2 snRNA promoter (Stefanovic and Marzluff, 1992) and have demonstrated that these two genes are temporally regulated when they are injected into L. pictus embryos (Stefanovic et al., 1991). The U2Lh gene is expressed at a higher rate than the U2EH gene at the late gastrula (prism) stage than at the blastula stage. This is likely due to the inactivation of the U2E gene in the later stages of embryogenesis (Stefanovic et al., 1991). We describe experiments defining the sequences responsible for the temporal regulation of the U2E promoter.
The U2EH and U2LH genes are shown in Fig. 1A. Also shown in Fig. 1A is the LEPL gene, which we used as an internal control gene in most of these studies. This gene contains a fragment of U2E promoter extending from nt −27 to nt −118 introduced in place of the sequence in the U2L promoter. The three important elements necessary for expression of the U2E gene, the TATA box, the PSE, and the −100 box, are all contained on this fragment.
The U2L gene upstream activating sequence is also present in the LEPL gene. In constructing the LEPL gene, a 16 nt fragment of the polylinker was inserted between the TATA box and the start of the U2 RNA. As a result, the LEPL hybrid promoter initiates transcription about nt upstream of the normal start site, allowing us to distinguish this transcript from the transcript of the U2H genes (Fig. 1B).
The sequences of the 120 nts prior to the start site containing the critical elements for expression of the U2E and U2L genes are shown in Fig. 1C. The sequence of the same region of the LEPL gene is also presented.
Sequences Between −27 and −118 Determine the Temporal Expression of the U2E Gene
To demonstrate that the LEPL gene was temporally regulated like the U2E gene, equal amounts of the U2Eh or U2LH genes and the LEPL gene were microinjected into L. pictus zygotes. L. pictus is closely related to L. variegatus and the LvU2 genes are expressed well in L. pictus. Embryos were grown to hatching blastula stage (24 h) and late gastrula (prism) stage (66–72 h), and the embryos that developed normally (100–200 per experiment) were harvested and cellular RNA was extracted. The riboprobe assay used to map the different transcripts is also shown in Fig. 1A. Using as a probe the U2Lh gene, we can map the endogenous U2 RNA, the transcripts from the hybrid genes, and the transcripts from the LEPL gene as separate protected fragments.
Because we are measuring the accumulation of an RNA from the time of injection to the time of harvesting, it is important that the test gene transcripts and control gene transcript have a comparable stability. Because the LEPL gene produces an RNA that has only 16 additional nt at the 5′ end, and the rest of the sequence is identical to the U2H genes, these two RNAs are likely to have similar stabilities. This assumption is supported by the fact that the LEPL and U2EH RNAs accumulate to similar amounts after microinjection.
Figure 1B shows the result of an experiment in which the U2LH and U2EH genes were separately coinjected along with the LEPL gene and embryos harvested at the blastula and late gastrula stages. The ratio of expression of the U2EH and the LEPL gene was similar at the two stages (Fig. 1B, lanes 1 and 4). In contrast, the ratio of expression of the U2Lh to the LEPL gene was fourfold higher at the late gastrula than at the blastula stage (Fig. 1B, lanes 2 and 5), indicating that the LEPL gene has the same temporal pattern of expression as the U2E gene. This is the same magnitude of change seen when the intact U2LH and U2EH genes are compared (Stefanovic et al., 1991). Thus, the sequences required for temporal expression lie between nucleotides −27 and − 118. We used the LEPL gene as an internal control for the early pattern of temporal expression.
The choice of an internal control gene for expression in early development is not a simple one. All the genes that have been studied from the sea urchin change transcription rates during development. Thus, we have chosen to use a gene that has the same temporal expression pattern as one of the U2 genes, the U2E gene, as a control in subsequent experiments. Because there is variation in the absolute amount of expression of the microinjected genes between different batches of embryos, it is essential to compare the expression of two genes that have been injected into the same embryos simultaneously. The LEpL gene meets the criteria for a control gene that is easily distinguished from the test genes, is expressed in similar amounts as the test genes, and is readily measured in a single assay, allowing direct comparison of the same RNA samples with a single probe.
The PSE Element Is Sufficient for the Temporal Regulation of the U2E Gene
Three different experiments demonstrate that the U2E PSE sequence is sufficient to confer proper temporal regulation. We exchanged 25 nt containing the PSE element between the U2E and the U2L genes. We created unique restriction enzyme sites (PstI and SalI) within the 10 nts flanking the PSE elements of both genes. These double mutants, U2ER60 and U2LR60, were the reference genes (Fig. 2A). The EL60E and LE60L clones were constructed by exchanging the Pstl-Sall fragments between the genes and are also shown in Fig. 2A. Expression of the reference genes U2ER60 and U2LR60 and the EL60E and LE60L genes, was estimated relative to the internal standard LEPL gene at the blastula and the late gastrula stage. The reference genes showed the expected temporal pattern of expression (Fig. 2B, lanes 4–7; Table 1). The U2ER60 gene was expressed in a constant ratio to the LEPL gene at the blastula and the gastrula stage (Fig. 2B, lanes 4 and 5), demonstrating that it was expressed like the U2E gene. The U2LR60 gene showed a six- to eightfold higher expression at the late gastrula stage compared with the LEPL gene (Fig. 2B, lanes 6 and 7), as expected for a U2L gene.
FIG. 2.
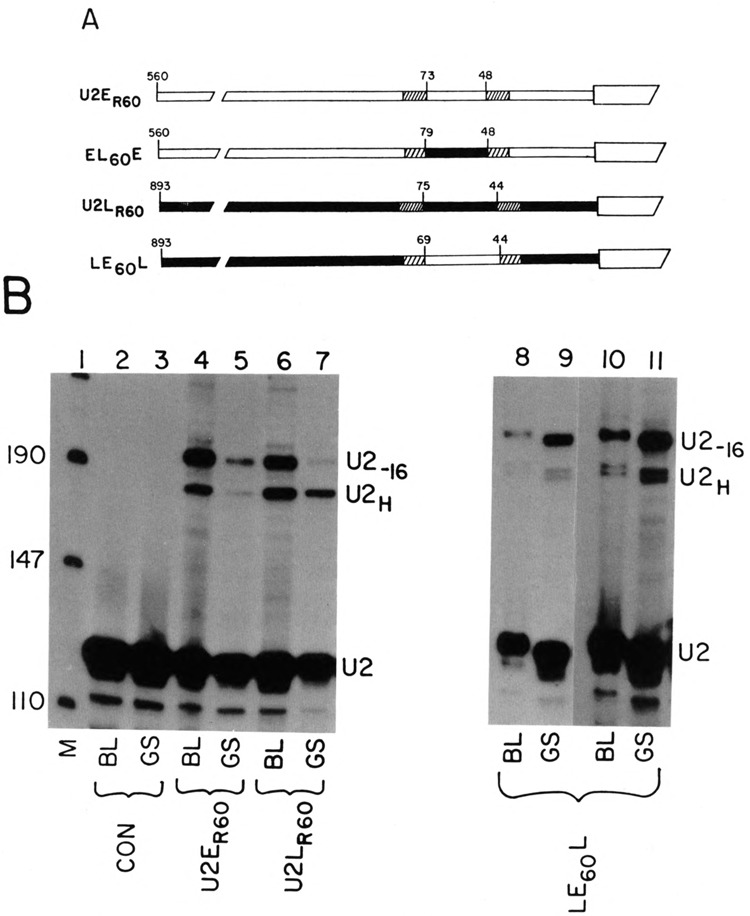
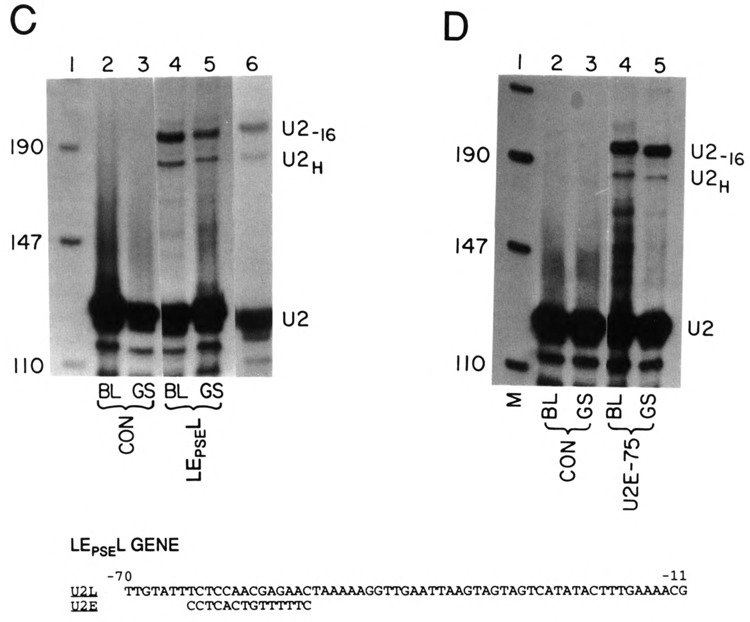
Role of the PSE sequence in developmental regulation of the U2 gene. (A) The U2ER60 and U2LR60 genes have substitutions in nonessential regions that result in introduction of restriction enzyme sites that were used in interchanging the − 60 sequences. The solid boxes are sequence from the U2L gene and the open boxes from the U2E gene. The hatched regions are the alterations made by site-specific mutation to introduce the restriction enzyme sites used for swapping the sequences. The sequences of the site-specific mutations have been described previously and the sequences of the swapped regions are in Fig. 1C (Stefanovic and Marzluff, 1992). The EL60E gene has the −60 region of the U2L gene introduced into the U2E gene. The LE60L gene has the −60 region of the U2E gene introduced into the U2L gene. The numbers indicate the position in the promoter relative to the start of the U2 snRNA in the gene from which the fragment was derived. (B) The genes in (A) were injected together with the LE60L gene into sea urchin zygotes and the embryos harvested at the blastula (20 h, lanes 2,4,6,8,10) or the late gastrula stage (62 h, lanes 3,5,7,9,11). Total cell RNA was prepared and analyzed using the riboprobe protection assay. The protected fragments are described in the legend to Fig. 1B. Lane 1 is pUC18 digested with HpaII. Lanes 2 and 3 are uninjected control embryos. The genes used are indicated below each lane. Lanes 10 and 11 are a darker exposure of lanes 8 and 9. The two protected fragments from the LE60L gene are due to heterogeneous start sites from this gene, which have been described previously (Stefanovic and Marzluff, 1992). (C) The LE60L gene has the sequence from −63 to −49 from the U2L gene replaced with the corresponding sequence from the U2E gene. The sequence shown is of the U2L gene and the 14 nts introduced to give the LEPSEL gene are shown below the figure. This gene was injected into sea urchin zygotes together with the LEPL gene and the embryos were harvested at either the blastula (lane 4) or late gastrula (lane 5) stage. Lanes 2 and 3 are uninjected embryos harvested at the blastula and late gastrula stage, respectively. Lane 6 is a lighter exposure of lane 4. Lane 1 is pUC 18 digested with HpaII. (D) Sea urchin zygotes were injected with the LEPL gene together with the U2E-75 gene, which contains only 75 nts of 5′ flanking sequence. Embryos were harvested at either the blastula (lanes 2 and 4) or late gastrula (lanes 3 and 5) stage. Total cell RNA was prepared and assayed using the riboprobe protection assay. The protected fragments are labeled as in Fig. 2. Lanes 2 and 3 are control uninjected embryos. Lane 1 is pUC18 digested with HpaII.
TABLE 1.
DEVELOPMENTAL EXPRESSION OF THE U2 snRNA PROMOTERS
| Relative Expression | ||
|---|---|---|
| Blastula | Gastrula | |
| LEPL | 1.0 | 0.95 |
| U2L | 3.1 (1.0) | 11 (3.5) |
| LEPSEL | 0.33(1.0) | 0.27 (0.8) |
| LE60L | 0.25 (1.0) | 0.31 (1.2) |
| E-75 | 0.09(1.0) | 0.08 (0.9) |
| E60R | 0.33(1.0) | 0.23 (0.67) |
| L60R | 1.3 (1.0) | 8.5 (6.2) |
| EL100E | 0.16(1.0) | 0.15(0.9) |
| LE100L | 0.67(1.0) | 1.25(1.8) |
| E100r | 0.8 (1.0) | 0.7 (0.9) |
| L100r | 3.0 (1.0) | 11 (3.7) |
The relative expression of the various promoter mutations of the U2L and U2E genes was determined by laser densitometry or by scanning the gels in a Betascope. The expression of the LEPL gene was calculated relative to the U2EH gene and was 50% of the expression of the U2E gene in the blastula stage. Expression of all the other genes was calculated relative to the LEPL gene. The degree of temporal regulation (expression at blastula divided by the expression at gastrula) is given in parentheses. The results are the average of at least two independent assays of the genes, which differed by less than 20%.
The LE60L gene gives rise to two transcripts with different start sites of transcription (Stefanovic and Marzluff, 1992). This is probably due to the fact that the U2E PSE sequence is four nucleotides closer to the first nucleotide of the U2 snRNA in the LEgoL gene and as a result about 50% of the transcripts initiate at nucleotide 4 in the U2 snRNA sequence (Stefanovic and Marzluff, 1992). The relative amounts of the two transcripts did not change between the blastula and the late gastrula stage, and we expressed the amount of expression of the LE60L gene as the sum of these two transcripts. The LE60L gene is expressed equally well relative to the control gene at both the blastula and late gastrula stages (Fig. 3B, lanes 12–15). Two exposures of the gel (Fig. 2B, lanes 8–11) are shown for better comparison of the relative intensities. This is the characteristic temporal expression of the U2E gene and it is in a sharp contrast to the fourfold change in the ratio of test gene to internal standard gene observed for the wild-type late U2LH gene (Fig. 1B) and the reference gene U2LR60 (Fig. 2B, lanes 6 and 7). Thus, simply changing 32 nts, including the PSE, converts temporal expression of the U2L gene to that of the U2E gene.
FIG. 3.
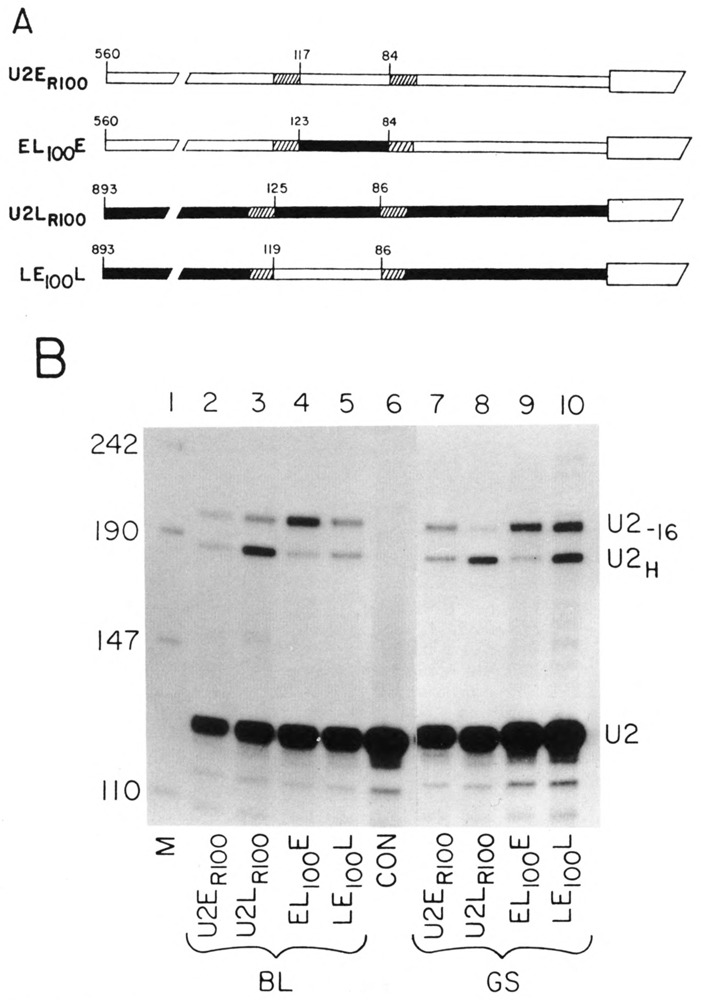
Role of the − 100 sequence in temporally regulated expression. (A) The genes with the −100 sequences interchanged between the two genes. The solid boxes are sequence from the U2L gene and the open boxes from the U2E gene. The hatched regions are the alterations made by site-specific mutation to introduce the restriction enzyme sites used for swapping the sequences. The sequences of the site-specific mutations have been described previously (Stefanovic and Marzluff, 1992) and the sequences of the swapped regions are in Fig. 1C. The U2ER100 and U2LR100 genes differ from the parent U2L and U2E genes by replacements in nonessential regions to introduce restriction enzyme sites useful for swapping the promoter regions. The EL100E gene has the −100 region from the U2L gene introduced into the U2E gene. The LE100L gene has the −100 region from the U2E gene introduced into the U2L gene. The numbers indicate the position in the promoter relative to the start of the U2 snRNA in the gene from which the fragment was derived. (B) The genes shown in (A) were injected together with the LEPL gene into sea urchin zygotes and the embryos were cultured until the blastula (lanes 2–6, 20 h) or late gastrula (lanes 7–10) stage. Total RNA was prepared and analyzed by the riboprobe protection assay. The protected fragments are described in Fig. 2. Lane 1 is pUC18 digested with HpaII. The genes used are indicated below each lane.
To demonstrate the change in the temporal pattern of expression was due precisely to the PSE sequence, we converted the U2L PSE element into the U2E PSE element by changing 14 nucleotides of the promoter. The nucleotides chosen were the critical nucleotides required for expression of the gene (Stefanovic and Marzluff, 1992). The sequence of the PSE element in the LEPSEL gene is shown at the bottom of Fig. 2C. The LEPSEL gene was expressed when it was injected into L. pictus zygotes. A single transcript, initiated at the first nucleotide of U2 snRNA, was produced from the LEPSEL gene, because the PSE element is now positioned at the “normal” place relative to the start site (Fig. 2C, lanes 4–6). The temporal pattern of expression of the LEPSEL gene was identical to the control LEPL gene (Fig. 2C, cf. lanes 4 and 6 with lane 5). Lane 6 is a shorter exposure of lane 4 to allow better comparison of band intensities. Thus, by substitution of just 14 nt of the U2E gene PSE element with a sequence of the U2L PSE, we were able to alter the temporal expression of the U2L gene to that of the U2E gene.
If the PSE element is responsible for temporal regulation, then the U2E gene deletion mutant (U2E-75) containing only 75 nt of 5′ flanking sequence (including the PSE and the TATA box) should retain the regulated temporal expression characteristic of the U2E gene. The U2E-75 gene is expressed at 20% of the wild-type level (Stefanovic and Marzluff, 1992). Figure 2D shows the expression of the U2E-75 gene at the blastula (lane 4) and late gastrula stage (lane 5) compared with the expression of the LEPL gene. The expression of the U2E-75 gene parallels the expression of the LEPL gene during development. Thus, only 75 nucleotides of 5′ flanking sequence, including the PSE element, is sufficient to confer proper temporal expression on the U2E gene. Taken together, the above results allow us to conclude that the U2E PSE element, the sequence of which is completely different from the U2L PSE, is a major cis-acting sequence responsible for temporal expression of the U2E genes in embryonic development.
The EL60E gene was expressed very poorly, apparently due to the inability of the PSE element of the U2L gene to interact with the other promoter elements of the U2E gene, and hence the temporal regulation of this gene could not be determined (Stefanovic and Marzluff, 1992; data not shown).
The results of the expression experiments are summarized in Table 1. The absolute expression of the test gene relative to the control LEPL gene is given and the relative expression of each test gene at the blastula versus the gastrula stage is given in parentheses. The LE60L and LEPSEL and the E-75 gene are expressed in parallel with the LEPL gene at both the blastula and the gastrula stage.
The −100 Box Is Not Required for Temporal Regulation of the U2 Genes
To test if the −100 element is involved in temporal regulation of the early U2 gene expression, we again created two unique restriction enzyme sites within unimportant sequences flanking the −100 element of both genes to give the genes U2ER100 and U2Lr100 (Fig. 3A). These double mutants served as controls for proper temporal expression and have been described previously (Stefanovic and Marzluff, 1992). We then swapped an approximately 50 nt region between the restriction sites creating the EL100E and LE100L genes. These genes are also shown in Fig. 3A. All four of these genes were injected together with the LEpL gene, and the expression of the genes at the blastula and late gastrula stages was compared. Figure 3B (lanes 2–6) shows the relative expression of the test and control genes at the blastula stage and lanes 7–10 show the expression at the late gastrula stage. A gene that is expressed in a temporal pattern similar to the U2E gene will have an identical ratio with the control gene at the two stages, whereas a gene that is expressed like the U2L gene will show higher relative expression to the control gene at the late gastrula compared with the blastula stage. The changes in the genes affected the absolute expression of the RNAs because swapping the −100 elements did reduce the expression of the EL100E and LE100L genes (Fig. 3B, cf. lanes 2 with 4 and 3 with 5). As expected, the reference genes, U2ER100 and U2LR100, showed a similar pattern of expression as the intact genes (Fig. 1B) at the two developmental stages (Fig. 3B, cf. lanes 2 and 7, lanes 3 and 8; Table 1). The EL100E gene (Fig. 3B, lanes 4 and 9) showed the same expression compared with the LEPL gene at both developmental stages, indicating that it was expressed like the U2E gene. The LE100L gene showed a 1.8-fold increased expression at the late gastrula stage compared with the blastula stage relative to the LEPL gene (Fig. 3B, lanes 5 and 10), although the increase was smaller than the fourfold increase observed with the intact gene. Switching the −100 elements did not affect the temporal expression of the U2E gene, although there was a small effect on the temporal expression of the U2L gene.
Factors That Bind the U2E PSE Element Are Present in Blastula But Not Gastrula Nuclear Extracts
The PSE element plays a major role in the expression, and as we showed above, in the temporal regulation of the sea urchin U2 genes. We demonstrate, by gel retardation and DNAse I footprinting experiments, that a factor or factors associate with the early gene PSE element and that these factors are active in blastula but not in gastrula nuclei. Figure 4A shows a specific protein-DNA complex was formed with a 25 nt restriction fragment containing the U2E PSE sequence. Complex formation was sequence specific because it was abolished by addition of excess of the specific unlabeled DNA fragment (Fig. 4A, lane 3), but not with the same molar excess of two nonspecific competitors (Fig. 4A, lanes 4 and 5). One of the competitors was an unrelated oligonucleotide (Fig. 4A, lane 5) and the other was an oligonucleotide with the U2L PSE sequence (Fig. 4A, lane 4). This result is consistent with the U2E and U2L PSE elements binding different factors, as suggested by their sequence difference. The faster migrating complex detected in this experiment (NS) is nonspecific because its formation was not affected by addition of various competitors, and the nonspecific binding activity was removed by heparin-agarose chromatography (see Fig. 4B).
FIG. 4.
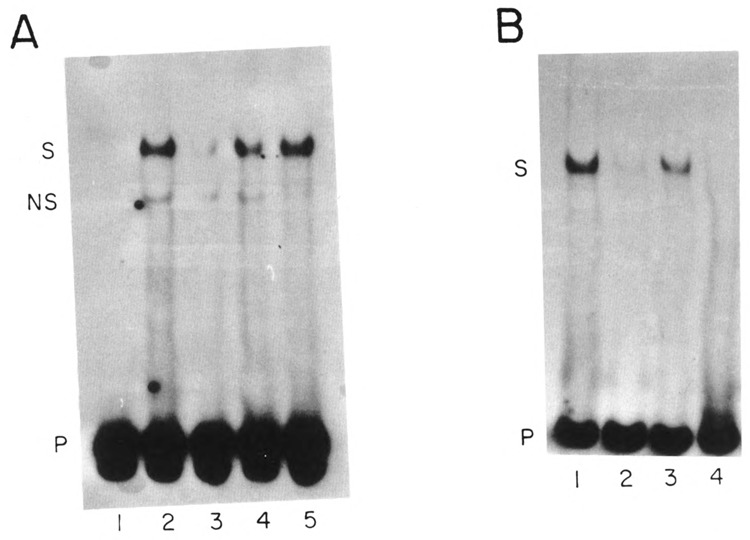
Complexes formed on the U2E proximal element. (A) A restriction fragment containing nts −69 to −47 from the U2E gene was incubated in buffer (lane 1) or with 3.5 μg of nuclear protein from blastula embryos (lanes 2–5) and the protein-DNA complexes analyzed by native gel electrophoresis. In lane 3 a 50-fold excess of unlabeled homologous competitor was included. In lane 4 a 50-fold excess of unlabeled fragment containing the PSE from the U2L gene and in lane 5 a 50-fold excess of an unlabeled fragment from a mouse histone gene was included. The band labeled S is the specific complex and the band labeled NS is a nonspecific complex. (B) The restriction fragment containing the U2E PSE used in (A) was incubated with 2.6 μg of heparin-agarose purified nuclear protein from blastula embryos (lanes 1–3) or 2.6 μg of heparin-agarose purified nuclear protein from gastrula embryos (lane 4) and the protein-DNA complexes analyzed by gel electrophoresis. In lane 2 a 100-fold excess of homologous competitor DNA was included and in lane 3 a 100-fold excess of a fragment containing the PSE from the U2L was included. The band labeled S is the specific DNA-protein complex.
The mobility-shift experiment was repeated using a heparin-agarose fractionated blastula extract and an extract from gastrula embryos, which was also fractionated on heparin-agarose to enrich for DNA binding proteins. The PSE binding activity in the blastula nuclear extract was quantitatively bound to heparin-agarose (not shown). A specific complex was formed using the U2 PSE oligonucleotide (Fig. 4B, lane 1), which was competed by the unlabeled U2E PSE oligonucleotide (Fig. 4B, lane 2), but not by the unlabeled U2L PSE oligonucleotide (Fig. 4B, lane 3). There was no complex formation observed with the heparin-agarose fractionated gastrula extract (Fig. 4B, lane 4). There was also no complex formation observed with the crude gastrula extract, or with the material that did not bind to heparin-agarose (data not shown). As shown below, this gastrula extract does contain factors that bind the U2E − 100 box (see below, Fig. 6). This suggests that the PSE binding activity for the U2E gene is not present in gastrula embryos.
FIG. 6.
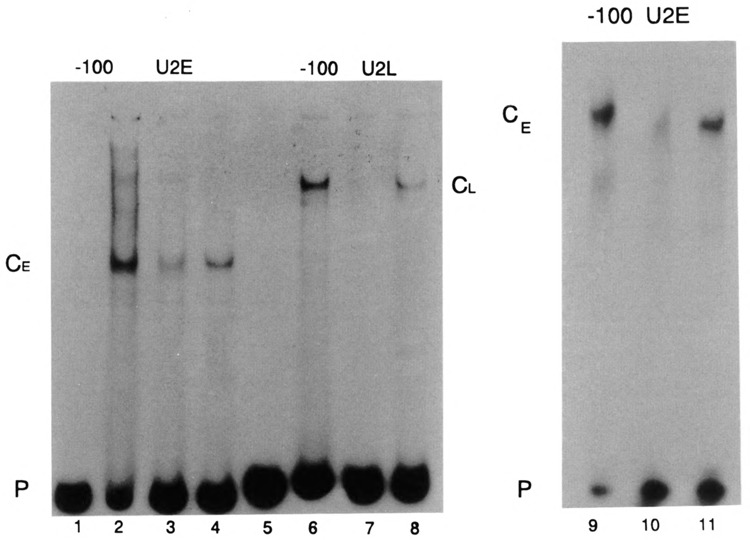
Different factors interact with the −100 box of the U2E and LVU2L genes. (A) The DNA restriction fragments containing the U2E and U2L − 100 boxes were labeled using the Klenow fragment of DNA polymerase I to fill in the 5′ overhangs. These fragments are shown in lanes 1 and 5. The labeled fragments were incubated with 8 μg of nuclear protein from blastula nuclei and the complexes were resolved by native gel electrophoresis on the same gel. In lanes 3 and 8 a 20-fold excess of the restriction fragment containing the U2E −100 box was included in the incubation and in lanes 4 and 7 a 20-fold excess of the U2L − 100 box restriction fragment was included in the incubation. In lanes 9–11 the U2E −100 box restriction fragment was incubated with 6.4 μg of a different extract of nuclear proteins from blastula embryos. A 100-fold excess of unlabeled U2E −100 box fragment was included in lane 10 and a 100-fold excess of unlabeled U2L −100 box fragment was included in lane 11. The complexes formed on the early and late −100 boxes are denoted CE and CL, respectively. B and P is the labeled restriction fragment.
DNase I footprinting experiments were also done with the U2E PSE. There was protection from DNase I cleavage of the lower (noncoding) DNA strand between nucleotides −30 and −56 (Fig. 5A, lanes 5 and 6) by the blastula nuclear extract. There is complete protection of about 24 nt encompassing the 3′ end of the PSE and extending 3′ of the PSE towards the TATA box. There is protection of the first two nucleotides of the TATA box and partial protection of the rest of the TATA box. There is also partial protection of the 5′ end of the PSE. In contrast, there was no footprint observed upon incubation with the heparin-agarose fractionated gastrula extract (lanes 7 and 8), consistent with the lack of PSE binding activity in this extract. There was also a clear footprint of the upper strand (Fig. 5B) encompassing the core of the PSE element and extending 3′ over approximately the same region of DNA from −30 to −56 (Fig. 5B, lanes 3 and 4). Thus, there is a complex that binds and protects both DNA strands. The 3′ border of the footprint extends into the first two nucleotides of the TATA box on both strands.
The large size of the footprint suggests that there may be several proteins interacting with the fragment containing the PSE sequence. The PSE sequence is absolutely required for the formation of the footprint, because the footprint observed with the blastula nuclear extract was abolished when an excess of a synthetic oligonucleotide containing just the PSE sequence was included in the reaction (Fig. 5C, lanes 6 and 7). There was no competition of the footprint by a nonspecific oligonucleotide (Fig. 5C, lanes 4 and 5). This result indicates that formation of the large footprint was dependent on the PSE element. Both the PSE element and the TATA box are indispensable for transcription, although the nucleotides between the two are dispensable (Stefanovic and Marzluff, 1992).
We were unable to detect binding activity to the U2L PSE element either by mobility-shift assays or DNAse I footprinting (not shown), despite using up to 10 times more protein in the reaction.
Factors Interacting With the U2E −100 Box Are Not Temporally Regulated
Two types of experiments demonstrate that the gastrula extracts contained functional DNA binding proteins; the gastrula extracts will footprint the − 100 elements and the gastrula extracts also contain un-degraded TBP. We have shown previously that the − 100 element of both the U2E and U2L genes is a distinct cis-acting sequence that is not necessary for basal transcription, but rather is required for coupling the distal elements in the promoter to the basal promoter (Stefanovic and Marzluff, 1992). Restriction fragments containing the −100 box sequence of the U2E and U2L gene were incubated with a nuclear extract prepared from hatched blastula embryos and the resulting complexes analyzed by gel electrophoresis under nondenaturing conditions. Specific complexes were formed on each of the restriction fragments containing the U2E and U2L −100 boxes, and these complexes had different mobilities, suggesting that they contained different components (Fig. 6, lanes 2 and 6). Competition experiments showed that the homologous probes competed for binding (Fig. 6, lanes 3 and 7, 10), whereas the U2L −100 box sequence did not compete with the U2E −100 box probe (Fig. 6, lanes 4, 11) and the U2E −100 box sequence did not compete with the U2L −100 box probe (Fig. 6, lane 8). The competition for the U2E − 100 box is shown more clearly in Fig. 6, lane 9–11, where larger amounts of competitors were used. This experiment was done with a different blastula nuclear extract than was used in Fig. 6 (lanes 1–8). When a 100-fold excess of specific competitor was used there was complete competition of complex formation (Fig. 6, lane 10), and there was no competition by a 100-fold excess of the U2L − 100 box competitor (Fig. 6, lane 11).
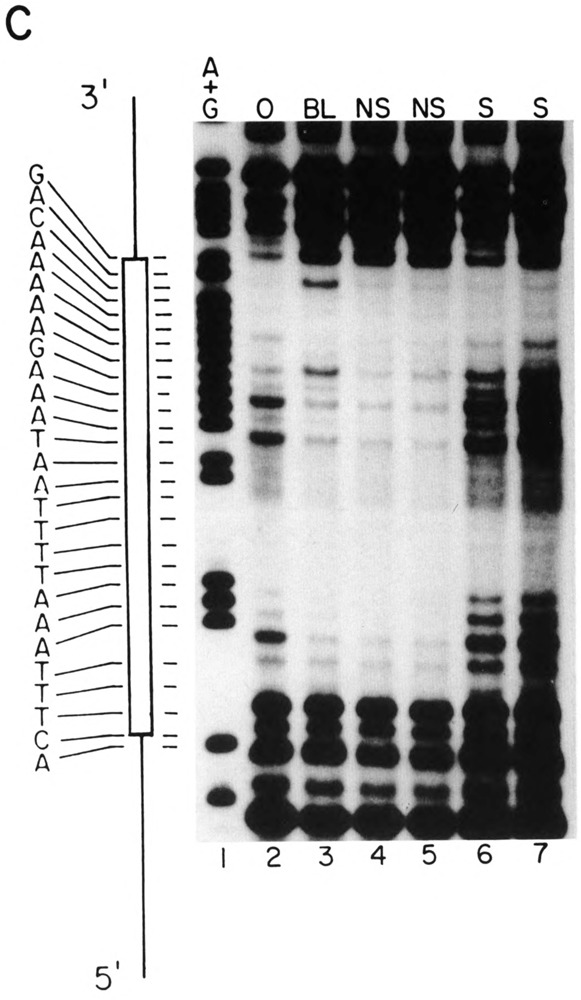
We also used DNase I footprinting to detect factors that interact with the − 100 region of the U2E promoter. Figure 7 presents a footprint of the region of the U2E promoter that contains the −100 element, using a blastula nuclear extract (lanes 5 and 6) or a gastrula nuclear extract that had been fractionated on heparin-agarose to enrich for DNA binding proteins (Fig. 7A, lanes 7 and 8). This is the same gastrula extract that was used for the footprint analysis of the PSE sequence in Fig. 5. All of the − 100 box binding activity is present in the heparin-agarose fraction (not shown). There is a clear protected region extending from −86 to −103 on the coding (upper) strand. Identical footprints were obtained with both the blastula and gastrula extracts; 2.5-fold less protein was necessary for protection by the gastrula nuclear extract because the DNA binding proteins had been enriched by heparin-agarose chromatography. The region protected overlaps with the −100 box element (−107 to −98) identified by microinjection experiments (Stefanovic and Marzluff, 1992). As shown above, the −100 box does not play a major role in temporal regulation of the U2E gene, in keeping with the observation that extracts from both gastrula and blastula stages contain a factor that protects the −100 sequence from DNase I.
FIG. 7.
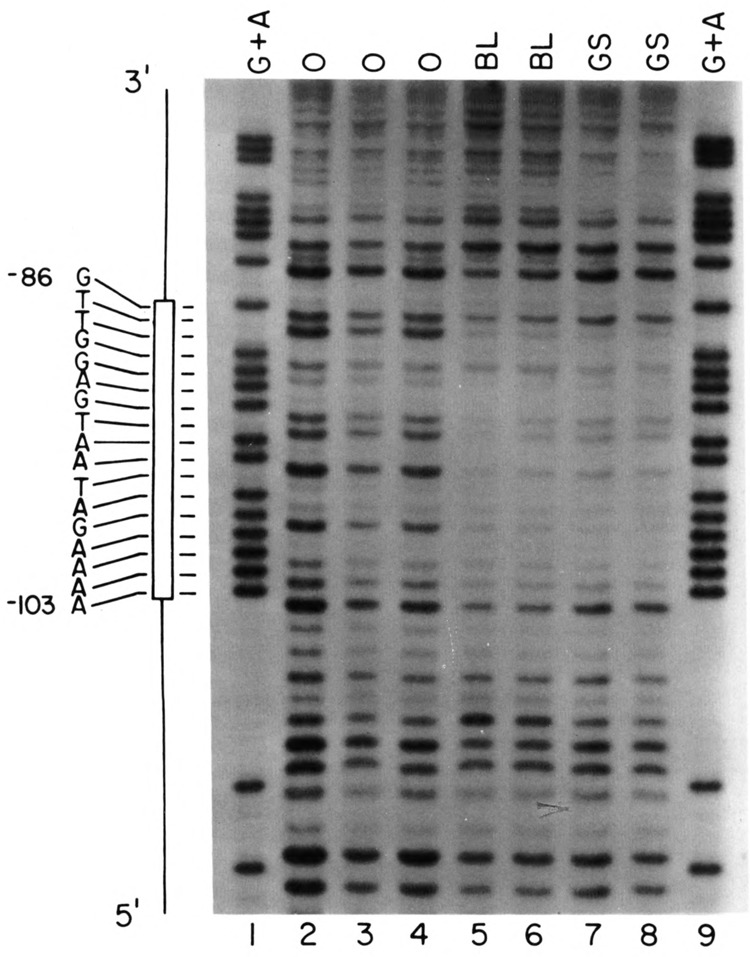
Specific complexes formed on the −100 sequences analyzed by DNasel footprinting. (A) A 94 nt DNA fragment containing the −100 sequence of the U2E gene was labeled at the 5′ end of the HindIII site and treated with DNase in the absence (lanes 2–4) or presence of 4.8 μg of crude nuclear protein from blastula embryos (lanes 5,6) or 1.8 μg of heparin-agarose fractionated gastrula proteins (lanes 7,8) and then analyzed by electrophoresis in a 7 M urea-8% polyacrylamide gel and the DNA fragments detected by autoradiography. Lanes 1 and 9 are G + A sequencing reactions. The sequence of the protected region is shown on the left.
To demonstrate that there was not significant proteolysis in the gastrula extract that might have resulted in loss of the PSE binding protein and that essential transcription factors were present in the gastrula extract, we assayed the amount of the TATA binding protein (TBP) using monoclonal antibodies directed against the conserved portion of TBP from Drosophila (Hoey et al., 1993; a generous gift of Drs. Weinzierl and Tijan). These antibodies detect a single 27 kDa protein in sea urchin nuclear extracts from both blastula and gastrula stages (Fig.8). There was no evidence of proteolysis in either extract.
FIG. 8.
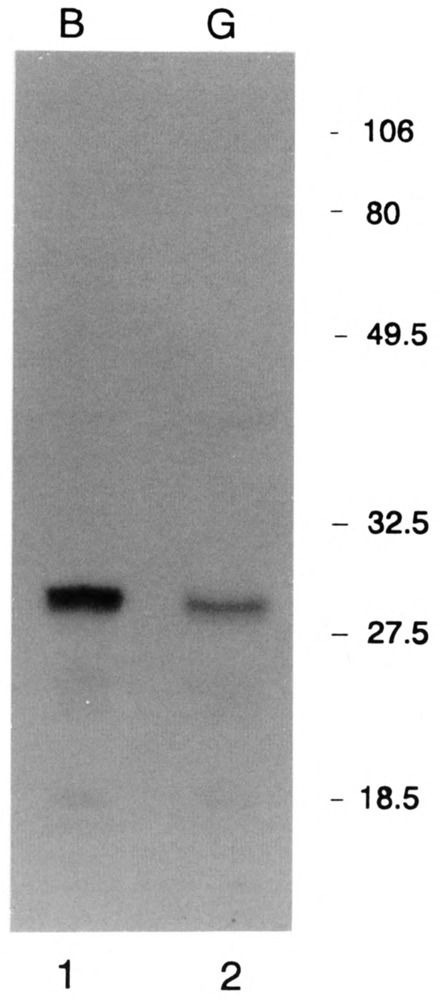
Presence of intact TBP in both the blastula and gastrula nuclear extracts. 52 μg of total nuclear protein from blastula (lane 1) and 36 μg of total nuclear protein from gastrula (lane 2) embryos were resolved by SDS-polyacrylamide gel electrophoresis and the protein was transferred to nitrocellulose by electroblotting. The filter was incubated in a mixture of monoclonal antibodies 16E8 and 58C9 to Drosophila TBP (Hoey et al., 1993; a gift of Drs. Weinzierl and Tjian) and developed using rabbit anti-mouse antibody coupled to alakaline phosphatase.
From the above results we conclude that the blastula nuclear extract contains a factor (or factors) that specifically binds to the U2E proximal promoter, and that this binding activity is absent (or greatly reduced) in gastrula embryos. The same gastrula extract, on the other hand, contains factors that interact with the −100 elements of both U2 genes, as well as factors that interact with multiple elements in the U1 promoter (Stevenson et al., 1992). They also contained intact TBP, but did not contain any detectable PSE binding activity, suggesting that the PSE binding activity is developmentally regulated. These data are totally consistent with the suggestion that the U2E PSE element is the primary element that determines the temporal expression pattern of the U2E gene.
DISCUSSION
There is a large accumulation of snRNAs in the nucleus of the developing sea urchin oocyte. Following maturation of the egg, the snRNAs are located in the cytoplasm, where they remain for the first four cell divisions (Nash et al., 1987). At about the 16–32 cell stage the synthesis of the snRNAs is activated (Nijhawan and Marzluff, 1979), resulting in a three- to fourfold increase in the total amount of U1 and U2 snRNAs by the hatching blastula stage (Nash et al., 1987; Stefanovic et al., 1991). Subsequently there is a dramatic decrease in the rate of snRNA synthesis between the hatching blastula and the late gastrula stage (Nijhawan and Marzluff, 1979). There are sequence variants of U1 (Nash et al., 1989), U2 (Stefanovic et al., 1991), and U6 RNA (S. Sakallah and W. Marzluff, unpublished results) present in sea urchin oocytes and embryos. There are separate tandemly repeated gene sets encoding U1 and U2 snRNAs that are expressed only in oogenesis and early embryogenesis. The tandemly repeated U1 and U2 snRNA genes are not expressed in adult sea urchins (Nash et al., 1989; Stefanovic et al., 1991), and their expression is greatly reduced between the blastula and late gastrula stage in the sea urchin L. variegatus (Santiago and Marzluff, 1989; Stefanovicetal., 1991).
The adult snRNA variants are present in small amounts in sea urchin oocytes and early embryos, suggesting that they are constitutively expressed (Nash et al., 1989; Stefanovic et al., 1991). Because the genes encoding these variants are present in low copy number, the RNAs are present in much lower levels than the transcripts from the tandemly repeated genes. In early embryogenesis the levels of both the adult and embryonic snRNAs increase (Nash et al., 1989; Stefanovic et al., 1991), indicating that both gene sets are actively transcribed in early embryos. The major regulatory step is the silencing of the tandemly repeated genes (Santiago and Marzluff, 1989), although we cannot rule out the possibility that the expression of the adult genes is also stimulated at later embryonic stages. This temporal pattern of regulation is similar to that found for the two sets of sea urchin histone genes (Maxson et al., 1983).
We have previously identified four positive cis-acting elements located in similar sites in the U2E and U2L promoters. These include the distal UAS, which has a fourfold effect on expression; the −100 box, which couples the UAS to the proximal promoter elements; the PSE element at −60; and the −30 TATA box, both of which are essential for expression (Stefanovic and Marzluff, 1992). The promoter sequences of the U2E and U2L genes share no common motifs except for the TATA box. We present evidence that the PSE element is a sequence involved in the temporal regulation of transcription of the early genes, based on several lines of evidence.
A gene containing nts 27 to 118 (including the TATA box, the PSE, and the −100 box) of the U2E promoter replacing those elements in the U2L promoter is expressed with the same temporal pattern as the U2E gene.
Interchanging 26 nts, including the PSE elements of the two genes, resulted in converting the expression of the U2L gene to the temporal pattern of expression seen for the U2E gene.
Substituting 14 nts into the U2L PSE to convert it to the U2E PSE altered the temporal pattern of expression of the U2L gene.
The factor(s) that bind the U2E PSE are present in nuclei from blastula embryos but not in nuclei from gastrula embryos, whereas factors that bind the U2E −100 box are present in nuclei from both late gastrula and blastula embryos.
Microinjected snRNA Genes Show Proper Developmental Regulation
Upon microinjection into sea urchin zygotes, transcripts from the tandemly repeated U2E gene and the U2L gene accumulated in comparable amounts at the blastula stage. Note that in the microinjection experiments the two genes are present in similar copy number, unlike the situation with the endogenous U2 genes. However, by the late gastrula stage transcripts from the U2L single copy gene are four times more prevalent than transcripts from the U2E gene [(Stefanovic et al., 1991) and Fig. 1B]. Accumulation of an RNA is a balance between the rate of synthesis and rate of degradation. Because the snRNAs are quite stable during embryonic development (Nijhawan and Marzluff, 1979), the assay almost certainly underestimates the change that occurs in the expression of the microinjected genes during development. Because the two hybrid U2H transcripts in our microinjection experiments are identical, their stability must be the same and, consequently, the different level of accumulation implies a temporally regulated synthesis. Our results strongly support the notion that there is an element within each individual gene that is primarily responsible for developmental expression. Similar results have been found when the expression of histone genes was studied; individual genes contain much, if not all, of the information for proper temporal expression (Colin et al., 1988; DiLiberto et al., 1989).
The PSE Element Is Responsible for Temporally Regulated Expression of the U2E Gene
The PSE elements are located at about position −55 in both U2 gene promoters, a similar location to the essential PSE elements found in vertebrate snRNA genes. The U2E and U2L PSE elements do not share sequence homology with each other or with the vertebrate sequence. The PSE elements in the sea urchin U2 snRNA genes are indispensable for transcription and in combination with the TATA motif are sufficient for correct initiation of transcription (Stefanovic and Marzluff, 1992). Three different observations support the contention that the U2E PSE is responsible for the major component of the temporal regulation. Deletion of all but 75 nts of the U2E promoter reduces expression 80% but the expression is still temporally regulated (Fig. 2D). The only remaining promoter elements are the PSE and the TATA box. Substitution of 32 nts of the U2E promoter into the U2L promoter, including the PSE, changes the temporal expression of the U2L gene to the U2E gene (Fig. 2B). Changing just 14 nt to convert the U2L PSE sequence to the U2E PSE sequence also changes the temporal expression of the U2L gene (Fig. 2C).
Mobility-shift experiments using a DNA fragment containing the U2E PSE demonstrate the presence of a factor specific for the U2E PSE that is present in nuclear extracts from blastula but not gastrula embryos (Fig. 4). The U2L PSE does not compete for formation of this complex, suggesting that separate factors bind to the two different PSE elements. DNase I footprinting demonstrates that a complex is formed that protects the region from the U2E PSE from the PSE element to the TATA box. This complex is formed in nuclear extracts from blastula embryos and not with nuclear extracts from gastrula embryos (Figs. 4,5). Both DNA strands are protected, and protection extends significantly beyond the core PSE sequence, all the way to the TATA box. Mutation of the region between the PSE element and the TATA box has only a small effect on expression (Stefanovic and Marzluff, 1992), suggesting that the factors protecting this region are not critical to expression. The entire large footprint is competed by incubation with the oligonucleotide containing only the PSE, supporting the contention that the PSE element is directing the formation of the complex (Fig. 5C). It is quite possible that a specific factor(s) primarily contacts DNA at the PSE and TATA sequence and initiates assembly of a transcription complex, which is defined by these two exacting elements. The critical requirements for formation of this complex are a core PSE sequence and a TATA box positioned at the proper distance (our unpublished results). The lack of a footprint over any of this region in the gastrula stage is likely due to the destruction of the PSE binding activity between the blastula and the gastrula stage, because the entire footprint can be completed by an oligonucleotide containing only the PSE sequence (Fig. 5C).
The −100 Box Is Not Critical for Proper Temporal Control
The −100 element was identified as an element necessary for coupling the upstream activating sequences with the proximal promoter elements (Stefanovic and Marzluff, 1992). The U2E gene element shares a strong homology with the similarly positioned sequence in the S. purpuratus U2E gene (13 out of 14 nt are invariant), but no similarity with the −100 box in the U2L gene (Stefanovic et al., 1991; Stefanovic and Marzluff, 1992). Swapping of the −100 elements between U2E and U2L genes (Fig. 3B) did not have a major effect on temporal regulation. In addition, the expression of the U2E promoter with the − 100 element deleted is properly temporally regulated (Fig. 3C). The factor(s) that bind the U2E and U2L −100 boxes are distinct (Fig. 6) and the factor that binds the U2E − 100 box is present in extracts of nuclei from both blastula and gastrula embryos (Fig. 7). The U2E −100 factor may be involved in expression of other genes, which are not temporally regulated, accounting for its persistence in gastrula embryos. The vertebrate snRNA genes use some of the common transcription factors used in transcription of other genes by RNA polymerase II (Korf and Stumph, 1986; Parry et al., 1989; Tanaka et al., 1988) and the sea urchin snRNA genes may do the same.
Temporal Regulation of the Sea Urchin snRNA Genes
The data reported here clearly identify the PSE element as the critical sequence involved in temporal regulation of the U2E gene. In vertebrates, all the snRNAs share a common PSE element, which presumably interacts with a common factor required for transcription of all of the vertebrate snRNA genes (Gunderson et al., 1988; Gunderson et al., 1990; Sadowski et al., 1993), although this factor has not yet been definitively identified. All of the nematode spliceosomal RNAs also have a common PSE sequence (Thomas et al., 1990), although the role of this sequence in expression has not been demonstrated. A common PSE sequence has also been proposed for the Drosophila snRNA genes (Saba et al., 1986; Beck et al., 1984). Because the sea urchin probably contains two temporally regulated sets of genes for all the spliceosomal snRNAs (our unpublished results), one of which is tandemly repeated and the other present in low copy number, an attractive possibility is that the tandemly repeated gene sets share a common PSE element and the low copy number set share a second PSE element. Temporal regulation of the tandemly repeated genes could then be accomplished simply by regulating the expression of the factors that interact with the tandemly repeated PSE element. Despite the lack of sequence similarity among the sea urchin U1E, U2E, and U6E tandemly repeated genes, we have recently obtained evidence that these three PSE sequences interact with the same factor (Li, J.-M. and W.F.M., unpublished results). Temporal regulation of this factor could account for the coordinate regulation of the tandemly repeated snRNA genes. There must be a distinct PSE factor that is required for expression of the U2L gene, and it is possible that the same factor could be involved in expression of the other spliceosomol snRNA genes expressed late in embryogenesis and in adults.
ACKNOWLEDGEMENTS
This work was supported by grant GM27789 from the NIH. We thank Drs. Weinzierl and Tjian for a generous gift of monoclonal antibodies to Drosophila TBP. We thank Brian Weldelburg and Jian-Ming Li for unpublished results and for many helpful discussions.
REFERENCES
- Beck E., Jorcano J. L., and Alonso A. (1984), J Mol Biol 173, 539–542. [DOI] [PubMed] [Google Scholar]
- Colin A. M., Catlin T. L., Kidson S. H., and Maxson R. (1988), Proc Natl Acad Sci USA 85, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg J. E. and Lund E. (1989), in Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles (Birnstiel M. L., ed.), Springer-Verlag, Berlin, pp. 38–70. [Google Scholar]
- DiLiberto M., Lai Z., Fei H., and Childs G. (1989), Genes Dev 3, 973–985. [DOI] [PubMed] [Google Scholar]
- Forbes D. J., Kirschner M. W., Caput D., Dahlberg J. E., and Lund E. (1984), Cell 38, 681–689. [DOI] [PubMed] [Google Scholar]
- Gruber A., Soldati D., Burri M., and Schümperli D. (1991), Biochim Biophys Acta Gene Struc Exp 1088, 151–154. [DOI] [PubMed] [Google Scholar]
- Gunderson S. I., Knuth M. W., and Burgess R. R. (1990), Genes Dev 4, 2048–2060. [DOI] [PubMed] [Google Scholar]
- Gunderson S. I., Murphy J. T., Knuth M. W., Steinberg T. H., Dahlberg J. H., and Burgess R. R. (1988), J Biol Chem 263, 17603–17610. [PubMed] [Google Scholar]
- Hoey T., Weinzierl R. O. J., Gill G., Chen J.-L., Dynlacht B. D., and Tjian R. (1993), Cell 72, 247–260. [DOI] [PubMed] [Google Scholar]
- Korf G. M., Botros I. W., and Stumph W. E. (1988), Mol Cell Biol 8, 5566–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf G. M. and Stumph W. E. (1986), Biochemistry 25, 2041–2047. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. (1985), Proc Natl Acad Sci USA 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo S. M., Marzluff W. F., Seufert A. C., Dean W. L., Schultz G. A., Simerly C., and Schatten G. (1988), Dev Biol 127, 349–361. [DOI] [PubMed] [Google Scholar]
- Lund E. and Dahlberg J. E. (1987), Genes Dev 1, 39–46. [DOI] [PubMed] [Google Scholar]
- Lund E., Kahan B., and Dahlberg J. E. (1985), Science 229, 1271–1274. [DOI] [PubMed] [Google Scholar]
- Lührmann R., Kastner B., and Bach M. (1990), Biochim Biophys Acta Gene Struct Exp 1087, 265–292. [DOI] [PubMed] [Google Scholar]
- Maniatis T. and Reed R. (1987), Nature 325, 673–678. [DOI] [PubMed] [Google Scholar]
- Maxson R., Mohun T. J., Cohn R., and Kedes L. (1983), Annu Rev Genet 17, 239–277. [DOI] [PubMed] [Google Scholar]
- McMahon A. P., Flytzanis C. N., Hough-Evans B. R., Katula K. S., Britten R. J., and Davidson E. H. (1985), Dev Biol 108, 420–430. [DOI] [PubMed] [Google Scholar]
- Morris G. F. and Marzluff W. F. (1985), Mol Cell Biol 5, 1143–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. F., Price D. H., and Marzluff W. F. (1986), Proc Natl Acad Sci USA 83, 3674–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash M. A., Kozak S. E., Angerer L. M., Angerer R. C., Schatten H., Schatten G., and Marzluff W. F. (1987), J Cell Biol 104, 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash M. A., Sakallah S., Santiago C., Yu J.-C., and Marzluff W. F. (1989), Dev Biol 134, 289–296. [DOI] [PubMed] [Google Scholar]
- Nijhawan P. and Marzluff W. F. (1979), Biochemistry 8, 1353–1360. [DOI] [PubMed] [Google Scholar]
- Parry H. D., Scherly D., and Mattaj I. W. (1989), Trends Biochem Sci 14, 15–19. [Google Scholar]
- Phillips S. C. and Turner P. C. (1991), Nucleic Acids Res 19, 1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba J. A., Busch H., Wright D., and Reddy R. (1986), J Biol Chem 261, 8750–8753. [PubMed] [Google Scholar]
- Sadowski C. L., Henry R. W., Lobo S. M., and Hernandez N. (1993), Genes Dev 7, 1535–1548. [DOI] [PubMed] [Google Scholar]
- Santiago C. and Marzluff W. F. (1989), Proc Nad Acad Sci USA 86, 2572–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southgate C. and Busslinger M. (1989), EMBO J 8, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B., Li J.-M., Sakallah S., and Marzluff W. F. (1991), Dev Biol 148, 284–294. [DOI] [PubMed] [Google Scholar]
- Stefanovic B. and Marzluff W. F. (1992), Mol Cell Biol 12, 650–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson K. A., Yu J.-C., and Marzluff W. F. (1992), Nucleic Acids Res 20, 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K. and Birnstiel M. L. (1986), EMBO J 5, 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter-Crazzolara C. and Keller W. (1991), Gene Exp 1, 91–102. [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Grossniklaus U., Herr W., and Hernandez N. (1988), Genes Dev 2, 1764–1778. [DOI] [PubMed] [Google Scholar]
- Thomas J., Lea K., Zucker-Aprison E., and Blumenthal T. (1990), Nucleic Acids Res 18, 2633–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendelburg B. W. and Marzluff W. F. (1992), Nucleic Acids Res 20, 3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]


