Abstract
By using a combination of Northern blot hybridization with strand-specific DNA probes, S1 nuclease protection, and sequencing of oligo-dT-primed cDNA clones, we have identified a 0.8 kb poly(A)-containing RNA encoded by the H-strand of the mouse mitochondrial D-loop region. The 5′ end of the RNA maps to nucleotide 15417, a region complementary to the start of tRNAPro gene and the 3′ polyadenylated end maps to nucleotide 16295 of the genome, immediately upstream of tRNAPhe gene. The H-strand D-loop region encoded transcripts of similar size are also detected in other vertebrate systems. In the mouse, rat, and human systems, the 3′ ends of the D-loop encoded RNA are preceded by conserved sequences AAUAAA, AAUUAA, or AACUAA, that resemble the polyadenylation signal. The steady-state level of the RNA is generally low in dividing or in vitro cultured cells, and markedly higher in differentiated tissues like liver, kidney, heart, and brain. Furthermore, an over 10-fold increase in the level of this RNA is observed during the induced differentiation of C2C12 mouse myoblast cells into myotubes. These results suggest that the D-loop H-strand encoded RNA may have yet unknown biological functions. A 20 base pair DNA sequence from the 3′ terminal region containing the conserved sequence motif binds to a protein from the mitochondrial extracts in a sequence-specific manner. The binding specificity of this protein is distinctly different from the previously characterized H-strand DNA termination sequence in the D-loop or the H-strand transcription terminator immediately downstream of the 16S rRNA gene. Thus, we have characterized a novel poly(A)-containing RNA encoded by the H-strand of the mitochondrial D-loop region and also identified the putative ultimate termination site for the H-strand transcription.
Keywords: Mouse mitochondria, D-loop encoded RNA, H-Strand transcription
MITOCHONDRIA from all vertebrate species investigated contain closed circular DNA genomes of about 16 kb. The primary function of vertebrate mitochondrial DNA is to code for 13 polypeptides that are associated with the mitochondrial energy-transducing complexes, and also 22 tRNAs and two rRNAs that are components of the mitochondrial translation system (Attardi and Schatz, 1988; Clayton, 1984, 1991). Analysis of steady-state mitochondrial RNA (Aloni and Attardi, 1971; Battey and Clayton, 1978; Rastl and Dawid, 1979) and RNA pulse labeled in isolated mitochondria (Bhat et al., 1985; Gelfand and Attardi, 1981), coupled with the DNA sequence data (Anderson et al., 1981, 1982; Bibb et al., 1981), demonstrated that both strands of the mitochondrial DNA are transcribed; the L-strand encodes eight tRNAs and one protein, whereas the H-strand encodes 14 tRNAs, two rRNAs, and 12 proteins.
The displacement-loop structure, termed the D-loop, is a ubiquitous feature of the vertebrate mitochondrial genomes (Clayton, 1984) that consists of about 0.8–1 kb triple-stranded structure located between genes encoding tRNAPro on the L-strand and tRNAPhe on the H-strand. This structure is created by the displacement of the parental H-strand by a 0.6–0.8 kb DNA complementary to the L-strand (Clayton, 1984; Doda et al., 1981). In vitro transcriptional analysis and RNA mapping in different vertebrate systems have shown that the D-loop houses both the H- and L-strand transcription promoters (Bogenhagen et al., 1994, 1986; Bogenhagen and Yoza, 1986; Bogenhagen and Romaneli, 1988; Chang and Clayton, 1984, 1986b, 1986c; Christianson and Clayton, 1986) and also the binding sites for the mitochondrial transcription factor mtTF1 (Fisher and Clayton, 1988; Fisher et al., 1991), which appears to have an important role in transcription activation and/or promoter selection (Fisher et al., 1991). Additionally, the H-strand replication and the L-strand transcription in a number of different vertebrates are initiated at the same point immediately downstream of the L-strand transcription promoter (Bhat et al., 1989; Cairns and Bogenhagen, 1986; Chang and Clayton, 1986a; Chang et al., 1985). In the mouse system it has been demonstrated that the transcripts with identical 5′ ends map to this site, but with heterogeneous 3′ ends mapping to the three highly conserved sequence regions, CSB I, CSB II, and CSB III. At least some of these transcripts act as primers for the synthesis of D-loop H-strand DNA (Bhat et al., 1989; Chang et al., 1985). There is compelling evidence that a major part, if not all, of the L-strand of the D-loop DNA in the vertebrates is transcribed. Recent studies also suggest that a significant portion of H-strand within the D-loop region of the rat mitochondrial genome may be transcribed (Sbisa et al., 1990).
Although initial studies suggested that H-strand transcription is initiated at multiple sites (Cairns and Bogenhagen, 1986; Manam and Van Tule, 1979, reviewed by Clayton, 1984), more recent results from various laboratories provide evidence for a single (or a major) initiation site 10–30 nucleotides upstream of the tRNAPhe gene (Bhat et al., 1989; Bogenhagen et al., 1986; Cairns and Bogenhagen, 1986; Chang and Clayton, 1986a; see Clayton, 1991). It has also been demonstrated that a majority of the H-strand transcripts terminate at a tridecamer sequence region, located within the tRNALeu gene immediately downstream of the 16S rRNA gene (Christianson and Clayton, 1988). A significant portion of the transcription complexes escape termination at this site, allowing the transcription of downstream tRNA and mRNA genes. A consensus sequence from this region has indeed been shown to have termination function under both in vivo and in vitro conditions (Christianson and Clayton, 1988; Hess et al., 1991), and 34–36 kDa mitochondrial protein factors binding to this motif have been characterized (Daga et al., 1993; Hess et al., 1991; Kruse et al., 1989). Patients suffering from a complex neuromuscular disease MELAS carry mitochondrial DNA mutations that affect the transcription termination at this site (Goto et al., 1990; Hess et al., 1991; Kobayashi et al., 1990). Thus, although the mechanism of H-strand transcription termination, yielding the rRNA species that represents the most abundant of the mitochondrial steady-state transcripts, is becoming increasingly clear, currently little is known about the site as well as the nature of termination of the relatively less abundant H-strand transcripts downstream of the tRNAThr gene. In this article we present evidence that the H-strand of the mouse mitochondrial D-loop DNA is transcribed as a relatively stable poly(A)-containing RNA. Our results also show that this transcript terminates at a position immediately upstream of the tRNAPhe gene and downstream of a conserved sequence motif that binds to a specific mitochondrial factor.
EXPERIMENTAL PROCEDURES
Mitochondrial DNA Clones and Synthetic DNAs
The details of the two mouse mitochondrial DNA clones p644 and p407 used in this study have been described previously (Bhat et al., 1989). Briefly, p644 consists of a BglII (nucleotide 15329) and XbaI (nucleotide 15973) fragment cloned in pGEM 7z and p407 consists of a XbaI (nucleotide 15974) and HaeIII (nucleotide 85) fragment cloned in pGEM 7z. In addition, the following single- and double-stranded synthetic DNAs were used.
S1 primer
ACTGTATGGTGTATGTCAGA. This single-stranded DNA is complementary to the L-strand sequence 15610–15629 of the mouse mitochondrial genome.
Double-stranded DNA UPHSS
CCGTGAA-CCAAAACTCTAATCATACTCTA. This 29 base pair sequence (nucleotides 16246–16275) is located immediately upstream of the H-strand transcription start site of the mouse mitochondrial genome.
Double-stranded DNA HSS
ATTACGCAATAAACATTAACAA. This 22 base pair sequence (nucleotides 16274–16295) is from the H-strand transcription start site of the mouse mitochondrial genome.
Double-stranded DNA TAS
ACTCAACTA-TAAATTCACAAC. The conserved sequence motif TAS (termination associated sequence) located close to the 3′ end of the D-loop H-strand DNA in almost all vertebrates is believed to be involved in the termination of H-strand D-loop DNA (Christianson and Clayton, 1986; Clayton, 1991; Doda et al., 1981; Mackay et al., 1986).
Double-stranded DNA TERM
CCAATAACCTTCTCTAGGTTA. The sequence motif from the tRNALeu gene (H-strand sequence 2681–2660), immediately downstream of the mouse mitochondrial 16S rRNA gene. The human homolog of this conserved sequence motif binds to 34–36 kDa protein(s) (Daga et al., 1993) and functions as a selective terminator (Fisher and Clayton, 1988), allowing a relatively small population of transcripts to proceed further as pre-mRNAs.
Conserved region oligonucleotide
CCATCGTGATGTCTTATTTAAGGGGAA. This single-stranded DNA (nucleotides 15830–15857 of the mouse genome) showing 26 out of 27 nucleotide conservation in the D-loop regions of the human and bovine mitochondrial DNA was used for cDNA screening.
Isolation and Analysis of RNA
Mitochondria were isolated from freshly harvested cells and tissues by the differential centrifugation method and treated with digitonin (75 μg/mg protein) as described previously (Bhat et al., 1982). In some experiments, crude mitochondria were banded through a discontinuous sucrose gradient (Vijayasarathy et al., 1989) before treatment with digitonin. Total RNA from liquid nitrogen-frozen tissues and cultured cells, as well as freshly prepared mitoplasts, was isolated by using the guanidinium thiocyanate solubilization, and extraction with phenol-chloroform as described previously (Bhat et al., 1985). RNA was resolved by electrophoresis on formaldehyde-containing gels, transblotted to Nytran membranes and hybridized with the 32P-labeled probes under moderate or high stringency conditions as described previously (Carter and Avadhani, 1991). The hybridization probes were labeled by the Klenow extension of primers using [32P]dCTP. The double-stranded DNA probes were labeled to a specific activity of about 109 cpm/μg by extension of random primers using a kit supplied by Pharmacia. Strand-specific DNA probes were prepared by the extension of M13 probe primer using M13 single-stranded DNA templates such that only the complementary strand around the phage arm is labeled, leaving the insert DNA in a single-stranded form for hybridization (Hu and Messing, 1982). Hybridization with the M13 single strand-specific probes was carried out under moderate stringency without denaturing the probes (Bhat et al., 1985, 1989).
D-Loop DNA Analysis
DNA was isolated from fresh mitochondrial preparations using the proteinase K digestion followed by phenol-chloroform extraction and iso-propanol precipitation essentially as described by Williams (1986). DNA was resolved on 1.2% agarose gels and blotted onto Nytran membranes for Southern hybridization using standard conditions, except that the alkaline denaturation step was omitted. In some experiments, DNA samples were denatured by heating at 90°C for 5 min in TE buffer containing 60% formamide before loading on the gel. The blots were hybridized with the L-and H-strand-specific D-loop DNA probes as described in the Northern blot analysis.
Screening of cDNA Libraries
Libraries of oligo-dT-primed cDNAs for the mouse liver and rat liver poly(A) RNAs were constructed in the lambda ZapII plasmid vector (Stratagene Corp.). A library for human liver RNA in UniZap vector was a gift from Dr. Dinesh Tewari. The mouse and rat liver cDNA libraries were screened with the 32P-labeled p-407 DNA, and the human liver library was screened with 5′ end-labeled conserved region primer. About 5 × 105 plaques from each of the libraries were screened by plaque hybridization using 32P-labeled DNA probes. Positive clones were isolated by repeated screening; the DNA inserts were subcloned in pGEM 5z, and were sequenced by the dideoxy chain termination method of Sanger et al. (1982).
S1 Nuclease Protection
S1 nuclease protection was carried out with the H-strand-specific probe generated by the extension of the S1 primer essentially as described previously (Berk and Sharp, 1977). About 100 ng of the 5′ end-labeled primer was annealed to about 16 μg of denatured p-644 template DNA and extended using 18 units of Klenow enzyme (Bhat et al., 1985). The DNA was digested with BglII and the resultant 301-nucleotide-long probe was gel purified. About 2 × 105 cpm of the probe was annealed with 60 μg of total cell RNA or 5 μg of poly(A) RNA subjected to S1 analysis as described (Bhat et al., 1985).
DNA-Protein Binding Assays by Gel Mobility Shift
Dignam et al. (1983). Briefly, mitoplasts (1 g) were lysed by homogenization in a buffer containing 20 mM Tris-Cl (pH 7.8), 10 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol, 20% glycerol, 0.2% NP-40, 1 mM PMSF, 0.5 mg/l leupeptin, and 0.75 mg/l of pepstatin, and the soluble fraction was separated by centrifugation by 105,000 × g for 30 min at 0–4°C. The supernatant fraction was dialyzed against the same buffer for 3–4 h, clarified by centrifugation in an Eppendorf table top centrifuge for 3 min at 0–4 °C, and used for binding to heparin-agarose resin. About 100 mg of heparin-agarose preequilibrated with the homogenization buffer was added to the extract and mixed with gentle agitation for 30 min at 0–4°C. The resin was pelleted by centrifugation in an Eppendorf centrifuge for 1 min, washed three times with the homogenization buffer, and suspended in 250 μl of homogenization buffer containing 400 mM KC1. The contents were mixed for 5–10 min by inversion to elute the bound proteins and then centrifuged in an Eppendorf centrifuge for 3 min. The supernatant fraction was dialyzed for 1 h against the homogenization buffer using Millipore dialysis membrane and was frozen in aliquots at −80°C. Nuclear factors were prepared from rat liver nuclei banded through 2 M sucrose essentially as described by Parker and Topol (1984). DNA-protein binding was assayed by gel mobility shift analysis (Carter et al., 1992) using 4% acrylamide gels and Tris-acetate-EDTA buffer system. The synthetic double-stranded DNA were labeled at the 5′ end using [γ32P]ATP and polynucleotide kinase. Protein-DNA binding reactions were carried out in 20 μl final volumes by incubating 0.1−0.2 ng of probe DNA (10,000 cpm) with 5 μl of mitochondrial or nuclear protein extract (20–50 μg protein), and 1 μg of dI : dC as described previously (Carter et al., 1992). In some experiments, about 20 ng of bacterially expressed, purified human mtTF1 factor, a kind gift from Dr. D. A. Clayton, was used in DNA binding assays. Competing DNAs were preincubated with the mitochondrial protein extract or purified factor for 5 min before addition of the labeled probe. Binding was carried out for 25 min at room temperature and the DNA-bound protein complexes were resolved by electrophoresis at 4% polyacrylamide-TAE (Tris-acetate-EDTA) gels.
RESULTS
RNA Species Encoded by the Mouse Mitochondrial D-Loop Region
Figure 1 shows the structure of the mouse mitochondrial D-loop region. The positions of the tRNAThr, tRNAPro, and tRNAPhe genes and the 12S rRNA gene flanking both sides of the D-loop region are based on the DNA sequence reported by Bibb et al. (1981). The position of the 0.6–0.8 kb D-loop (H-strand) DNA, which displaces the parental H-strand creating the D-loop structure, is based on DNA and RNA mapping reported by Doda et al. (1981) and Chang et al. (1985). The dotted lines represent the major species of cappable RNAs encoded by the L-strand with identical 5′ ends mapping to the L-strand transcription start site (LSS) (Bhat et al., 1989). The identification of the major H-strand transcription start site (HSS) was based on in vitro transcription (Clayton 1984; Chang and Clayton 1984) and physical mapping with in vitro capped RNA (Bhat et al., 1989). The 0.8 kb RNA and its larger precursor (indicated by solid lines) represent the D-loop region H-strand encoded RNAs identified in the present study.
FIG. 1.
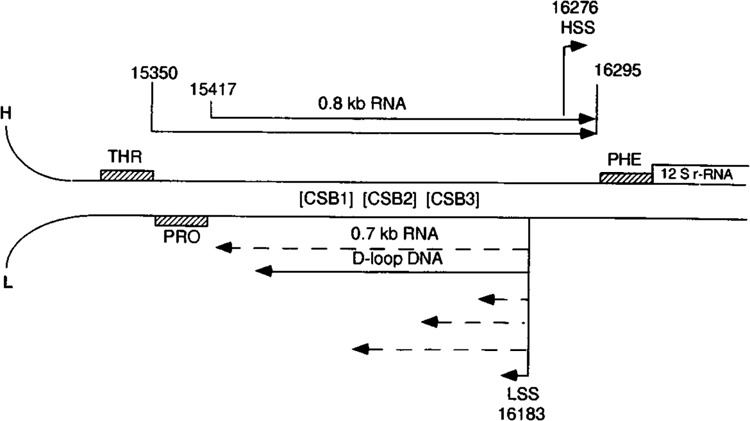
The genetic and transcriptional map of the mouse mitochondrial D-loop region. The precise map positions of the 0.8 kb H-strand coded RNA, heterogeneous size class of the L-strand coded RNAs, including the 0.7 kb RNAs (discontinuous lines) and the D-loop H-strand DNA of the mouse mitochondrial genome are presented. The map positions of the tRNAs and rRNAs are according to Bibb et al. (1981).
Northern blot analysis of total RNA from mouse liver, kidney, heart, and Ehrlich ascites tumor cells is presented in Fig. 2. The p-407 DNA contains the 5′ end of the D-loop region including the H- and L-strand transcription promoters, origin of H-strand replication, the gene for tRNAPhe, and the 5′ terminal 15 nucleotide region of the 12S rRNA gene. As seen from Fig. 2A, this probe hybridizes with two closely migrating species of about 0.8 and 0.7 kb RNAs and a more abundant RNA of <100 nucleotides. Further, the L-strand-specific probe hybridizes with the 0.7 kb and a <0.1 kb species (Fig. 2B), whereas the H-strand-specific probe hybridized with the larger 0.8 kb RNA in addition to a shorter (<0.1 kb) RNA species (Fig. 2C). In keeping with our previous observation (Bhat et al., 1989), the 0.7 kb RNA hybridizing with the L-strand appears to be the cappable RNA of similar size mapping between the L-strand transcription start site and the tRNAPro gene. The nature of the smaller RNA hybridizing with the L-strand probe remains unknown, although, as indicated in our previous study (Bhat et al., 1989), it might represent the free RNA primers shown to be present as part of the steady-state pool. The H-strand-specific probe hybridizes to the 0.8 and a <0.1 kb species, both of which are sensitive to RNAse (Fig. 2D, lane 3) and insensitive to DNAse (Fig. 2D, lane 2). Detection of a stable RNA hybridizing to the H-strand probe was surprising in view of the current notion that this region of the H-strand is the only nontranscribed part of the mitochondrial genome (Annex and Williams, 1990, reviewed by Clayton, 1991). Based on the size and relative abundance, the <0.1 kb RNA probably represents tRNAPhe.
FIG. 2.
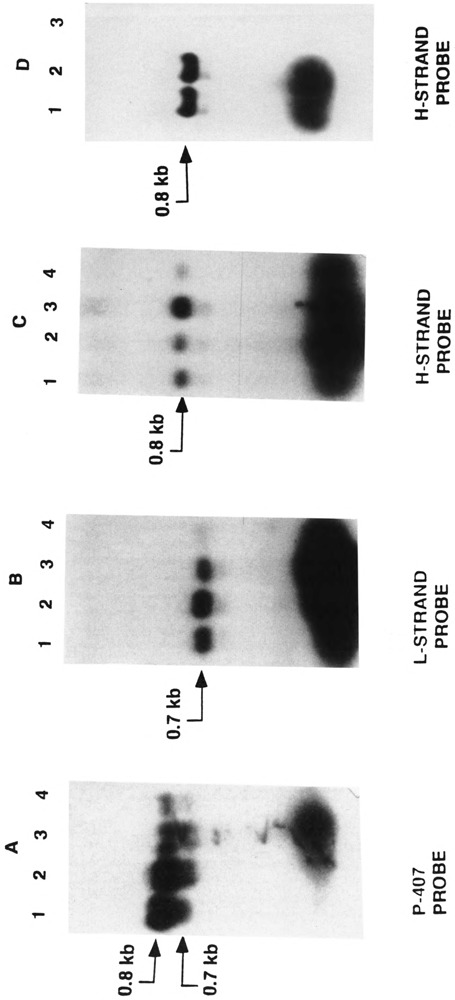
Northern blot hybridization of RNA with mouse mitochondrial D-loop DNA probes. The p-407 double-stranded DNA from the mouse mitochondrial D-loop region, and the strand-specific MIS DNA probes were labeled with 32P and used for hybridization with RNA from mouse Ehrlich ascites cells and different mouse tissues as described under Experimental Procedures. (A) Lane 1: RNA from mouse heart lane 2: mouse kidney, lane 3: mouse liver, and lane 4: Ehrlich ascites cells. (B,C) Lane 1: liver RNA, lane 2: kidney RNA, lane 3: heart RNA, and lane 4: Ehrlich ascites cell RNA. (D) The effects of DNAse and RNAse on the hybridization pattern were determined. Lane 1: untreated mouse liver RNA, lane 2: mouse liver RNA treated with two units of RQ1 DNAse, and lane 3: mouse liver RNA treated with one unit of pancreatic RNAse. Both DNAse and RNAse treatments were carried out on ice for 15 min prior to loading on the gel. RNA (30 μg) was loaded in each case. The amount of RNA in individual lanes did not vary more than 15% as judged by the ethidium bromide staining of the 18S and 28S rRNA bands. The RNA size was determined based on therelative migration of known molecular weight markers and was viewed by ethidium bromide staining. Lanes in (A) were hybridized with 32P-labeled p-407, double-stranded DNA probe, lanes in (B) were hybridized with the L-strand-specific M13 single-stranded DNA probe, and lanes in (C) and (D) were hybridized with the H-strand-specific M13 single-stranded DNA probe.
The cellular distribution of the 0.8 kb RNA was investigated by the Northern blot hybridization of RNA isolated from different subcellular fractions. As shown in Fig. 3, the D-loop H-strand-specific DNA probe hybridizes with two major RNA species of 0.8 kb and <0.1 kb from both total hepatocellular RNA and also RNA from a purified mitochondrial fraction (see lanes 1 and 2). A very minor band immediately above the 0.8 kb RNA band may represent the 0.88 kb precursor RNA detected in the S1 protection experiment described in Fig. 4. The relative intensity of the 0.8 kb band with the mitochondrial RNA is nearly 10-20-fold higher than that with the total cellular RNA, suggesting its true mitochondrial location. It is also seen that RNA from purified nuclei or the cytosolic fractions (lanes 3 and 4) contains no detectable level of this species, further confirming its mitochondrial origin as well as intramitochondrial location.
FIG. 3.
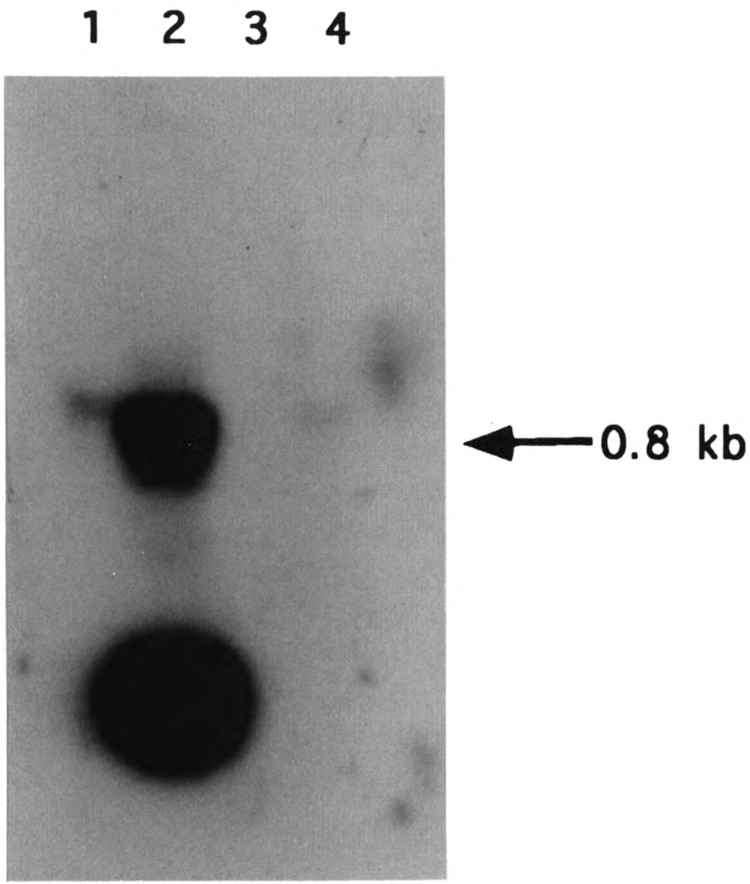
Subcellular location of the RNA hybridizing with the D-loop region H-strand probe. Total mouse liver RNA (25 μg) (lane 1) and RNA (25 μg) from mouse liver mitochondria (lane 2), cytosol (lane 3), and RNA (10 μg) from mouse liver nuclei (lane 4) were hybridized with the H-strand-specific M13 DNA probe as described in Fig. 2. Mitochondria were prepared by sucrose density banding as described under Experimental Procedures. The 100,000 × g supernatant fraction of the liver homogenate was used as the cytosolic fraction, and nuclei were prepared by sedimentation through 2.2 M sucrose in 20 mM Tris-Cl (pH 8.0), 0.5 mM CaCl2, and were used for RNA isolation.
FIG. 4.
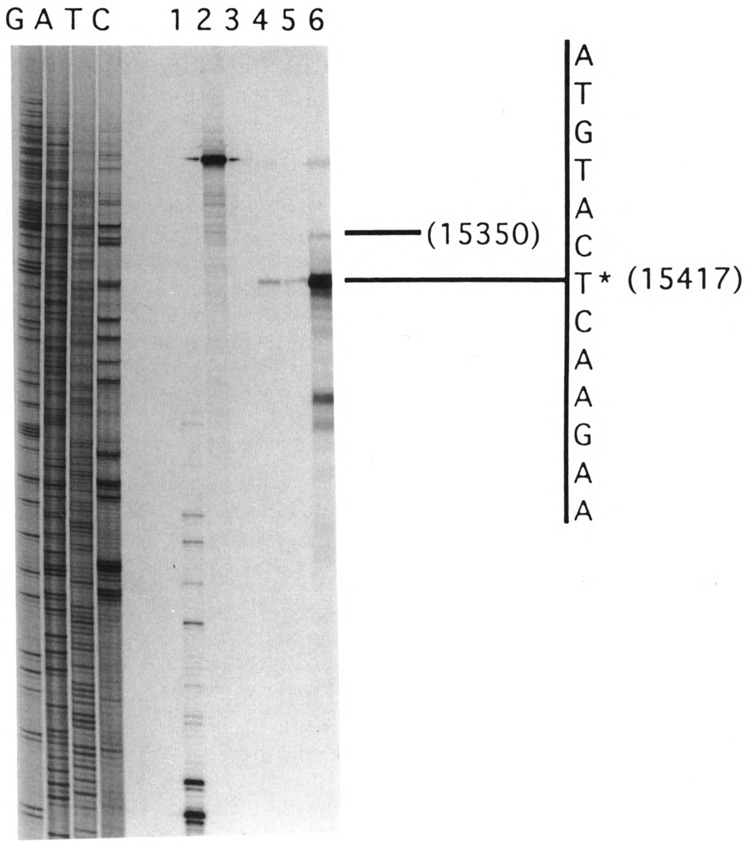
The 5′ terminus of the D-loop region H-strand coded RNA. The 5′ terminus was mapped by S1 nuclease protection. The 301-nucleotide-long probe was prepared by the Klenow extension of the 5′ end-labeled S1 primer using p-644 template DNA as described under Experimental Procedures. About 60 μg of total cell RNA or 5 μg of poly(A) RNA was used for the S1 analysis. Lane 1: 105 cpm probe with no RNA and 50 units of S1, lane 2: 104 cpm of the probe and no S1, lanes 3–6: 2 × 105 cpm of the probe and 150 units of S1 and 60 μg yeast tRNA (lane 3), 60 μg mouse liver RNA (lane 4), 60 μg mouse kidney RNA (lane 5), and 5 μg of mouse liver poly(A) RNA (lane 6). The sequence ladder marked GATC was created using the S1 primer and p-644 template DNA.
The 5′ and 3′ Map Positions of the 0.8 kb RNA
The 5′ end of the putative H-strand D-loop region encoded RNA was mapped by S1 nuclease protection. As shown in Fig. 4, S1 protection analysis using a 301-nucleotide-long 5′ end-labeled H-strand-specific probe yields a major protected fragment of 213 nucleotides with both total cellular RNA and poly(A) RNA mapping to nucleotide position 15417. This position corresponds to the nucleotide immediately upstream of the start of tRNAPro transcribed from the opposite strand (see Fig. 1 for details). Additionally, the poly(A) RNA yields a minor protected species of 278 nucleotides mapping to nucleotide 15350 of the genome, immediately downstream of the H-strand encoded tRNAThr gene. As expected, the intensities of protected fragments with poly(A) RNA are several fold higher than with total hepatocellular RNA.
Results of S1 analysis and also Northern blot hybridization with oligo-dT cellulose-bound RNA (latter results not shown) suggested that the 0.8 kb RNA is not only polyadenylated, but it is also detected at a relatively high abundance in the total RNA from liver and other tissues. Further, the Northern blot presented in Fig. 5 shows that RNAs of comparable size hybridizing to the H-strand D-loop region probe are also present in other species (human, bovine, and rat). For these reasons we have used a relatively new and a straightforward strategy of isolating oligo-dT-primed cDNAs, followed by DNA sequencing, to map the 3′ ends of the RNA from different species. A partial nucleotide sequence of the clone isolated from the mouse cDNA library is presented in Fig. 6. It is seen that the H-strand coded RNA contains a 3′ poly(A) tail, and the sequence data suggest that the 3′ terminus of this RNA corresponds to nucleotide positions 16293–16295 of the template strand. Because the last two nucleotides of the template strand (i.e., H-strand) are dTs, it is unknown if the first two dA residues of the 3′ poly(A) tail are part of the template-dependent transcription or posttranscriptionally added residues. Figure 6 also shows partial sequences of the rat and human cDNAs encoded by this region. The 3′ end of the rat RNA corresponds to nucleotide 16273 of the rat genome and that of the human RNA encoded by the D-loop region corresponds to nucleotide position 227 of the genome. These results also demonstrate that, as observed for the mouse system, the rat and human RNAs are polyadenylated. The precise map positions of the mouse mitochondrial D-loop H-strand encoded RNAs, and also the H- and L-strand transcription start sites, are presented in Fig. 1.
FIG. 5.
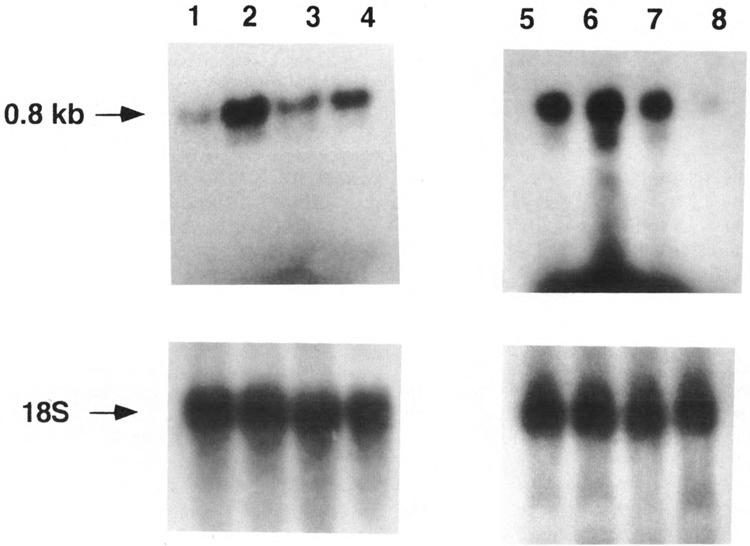
Steady-state levels of the D-loop H-strand coded RNA in different species and different tissues. Preparation of the H-strand-specific M13 probe and details of Northern blot hybridization were as described in Fig. 2. Lane 1: Hela cell RNA, lane 2: rat liver RNA, lane 3: rat hepatoma RNA, lane 4: bovine liver RNA, lane 5: mouse liver RNA, lane 6: mouse heart RNA, lane 7: mouse brain RNA, and lane 8: RNA from mouse neuroblastoma cells. About 25 μg of RNA was loaded in each case and the loading level was verified by stripping the probes off the blots and rehybridizing with 32P-labeled 18S rRNA probe.
FIG. 6.

Identification of the 3′ end and polyadenylation sites of the H-strand encoded RNA by cDNA sequencing. cDNA libraries for the mouse, rat, and human liver RNA were screened using D-loop DNA probes and sequenced as described under Experimental Procedures. Partial sequence of all the three cDNAs are shown. The consensus polyadenylation sites and also the 3′ poly(A) tails are shown in bold letters.
Protein Binding by Sequence Upstream of the Putative Termination Site
DNA-protein binding, DNAse I footprinting, and in vitro transcription studies have demonstrated that the mouse mitochondrial DNA sequence (nucleotides 16255–16267) close to the H-strand transcription promoter binds to protein factor mtTF1, which has a positive role in mitochondrial transcription activation (Fisher and Clayton, 1988; Fisher et al., 1987, 1991). To understand the functional significance of the putative H-strand transcription termination site, located immediately downstream of the mtTF1 binding region, we have carried out DNA-protein binding by gel mobility shift using synthetic double-stranded DNA probes from these two regions. As shown in Fig. 7, the UPHSS probe (upstream of HSS, sequence 16244–16273) forms a fast migrating complex with mitochondrial protein extract (lane 2). This band probably represents a complex with bound mtTF1 based on the known affinity of this sequence region for binding to this transcription factor (Fisher et al., 1991) and also similarly to gel migration of the complex with bacterially expressed, purified mtTF1 (see lane 3). The HSS DNA (H-strand transcription start site, sequence 16274–16295), on the other hand, forms a distinctly different slow migrating complex with mitochondrial extract (lane 5), suggesting that it has a different specificity. The HSS-bound complex consistently resolves on the gel as a diffused band, probably due to its relative instability during electrophoresis. The protein binding specificity of the HSS DNA was further compared using synthetic DNA with known protein binding sequence motifs, such as UPHSS, TAS, and the 16S rRNA terminator sequence (TERM) for competition. As shown in Fig. 8A, the mitochondrial protein complex with HSS DNA is effectively competed by 25–50 molar excess of unlabeled HSS DNA (lanes 3 and 4), but not by UPHSS DNA (lanes 5 and 6), TAS DNA (lanes 7 and 8), or TERM DNA (lanes 9 and 10), further implying the specificity of protein binding. The nature of protein binding to HSS DNA was further studied by comparing the gel mobility shift patterns with mitochondrial and nuclear protein factors. Results presented in Fig. 8B show that proteins from liver nuclei (lanes 4 and 5) form a number of undefined slow migrating complexes that are not competed even with 100 molar excess of unlabeled HSS DNA. These results suggest that a specific mitochondrial protein is involved in complex formation with the conserved sequence region immediately upstream of the 0.8 kb D-loop H-strand coded RNA termination site.
FIG. 7.
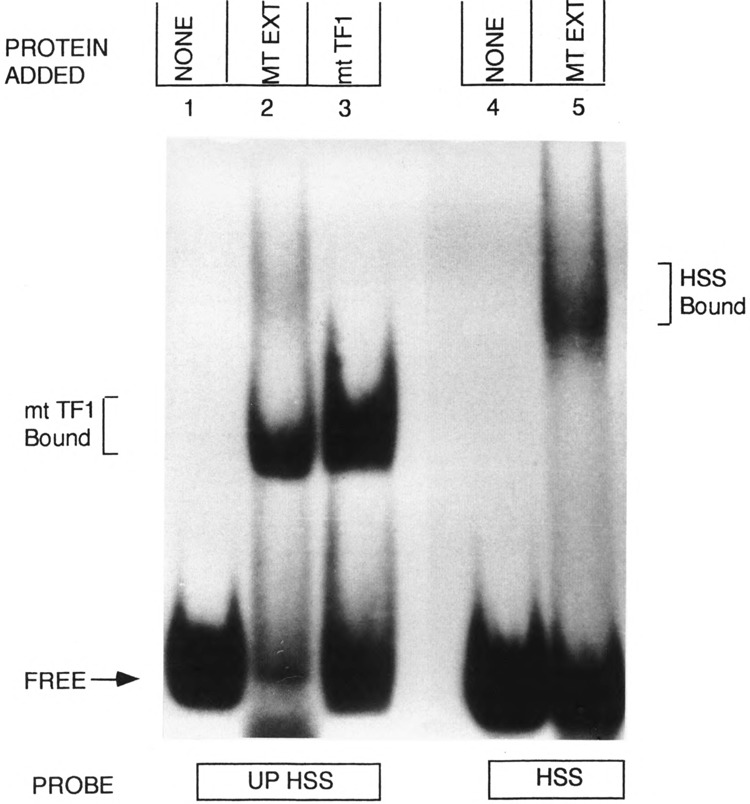
Protein binding to the H-strand transcription termination region DNA. DNA-protein binding by gel mobility shift analysis was carried out using 32P-labeled UPHSS (lanes 1–3) and HSS DNA (lanes 4 and 5) as described under Experimental Procedures. Lanes 1 and 4 represent controls with no added protein. In lanes 2 and 5: μg of mouse liver mitochondrial protein, and in lane 3: 20 ng of bacterially expressed, purified human mtTF1 were used. Both the protein binding and the electrophoresis were carried out at 4°C.
FIG. 8.
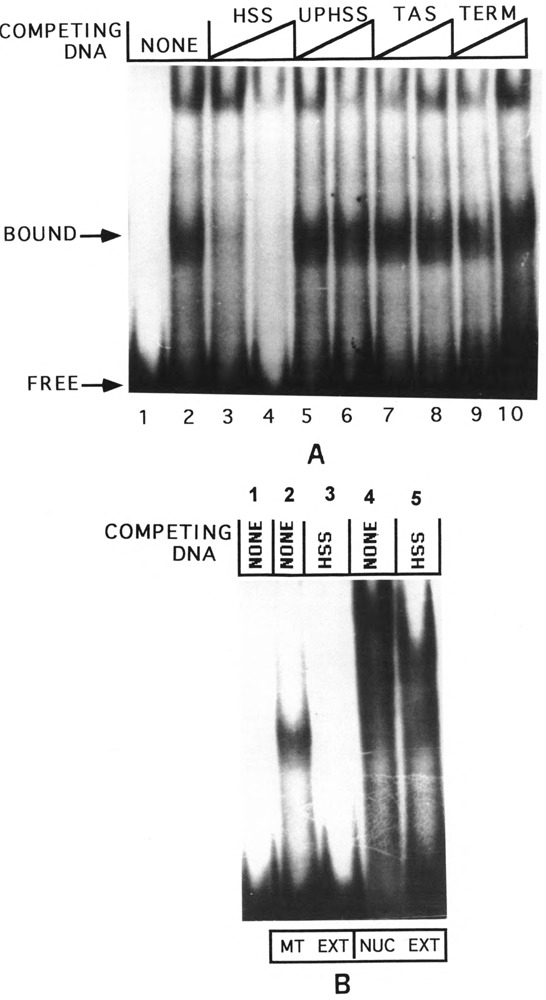
Specificity of protein binding to the termination region DNA probe HSS. Details of gel mobility shift analysis using 32P-labeled HSS DNA probe were as described in Fig. 7. (A) Abilities of known protein binding motifs, UPHSS, TAS, and TERM, to compete for protein binding to HSS were compared. Lane 1 represents a control with no protein, and in lanes 2–10, 25 μg of liver mitochondrial protein were added. In lanes 3–10, 25- or 50-fold molar excess of unlabeled DNAs was added as follows: lanes 3 and 4: HSS DNA, lanes 5 and 6: UPHSS DNA, lanes 7 and 8: TAS DNA, and lanes 9 and 10: TERM DNA. (B) Protein binding patterns of HSS DNA probe with 25 μg of liver mitochondrial and nuclear protein extracts were compared. Lane 1: no protein added, lanes 2 and 3: mitochondrial extract, and lanes 4 and 5: nuclear extract. In lanes 3 and 5: 100 molar excess of unlabeled HSS DNA was added.
Existence of the 0.8 kb RNA as a Free DNA-Nonassociated Form
The H-strand DNA encoded by the L-strand of the mitochondrial D-loop region from various animal species exists in association with the template strand as a triple-stranded structure (Annex and Williams, 1990; Doda et al., 1981). We have therefore carried out experiments to determine if the H-strand coded 0.8 kb RNA also exists in association with D-loop DNA. DNA isolated from the sucrose density gradient purified mouse liver and kidney mitochondria was resolved on an agarose gel before and after denaturation and hybridized with strand-specific D-loop DNA probes. As shown in Fig. 9A and C, the L- and H-strand-specific probes, respectively, hybridize to a minor slow migrating DNA at the top of the gel, probably representing the nearly entire mitochondrial genome. Generally poor hybridization in these lanes is probably due to poor transfer of native double-stranded DNA. It is also seen that the L-strand probe hybridizes with the nearly entire mitochondrial DNA in addition to a faster migrating DNA of about 0.6 kb from the denatured DNA samples (Fig. 9B). The faster migrating band in Fig. 9B (lanes 1 and 2) is known to be the D-loop H-strand DNA creating the triple-stranded structure (Clayton, 1984; Doda et al., 1981; Annex and Williams, 1990). As shown in lane 3 (Fig. 9B), the L-strand probe also hybridizes with a minor 0.7 kb RNA species corresponding to the L-strand transcript shown in Fig. 2B. In contrast, the H-strand probe exclusively hybridizes with the nearly intact mitochondrial DNA and fails to hybridize with any faster migrating species (Fig. 9D, lanes 1 and 2). As expected, however, this probe hybridizes with a 0.8 kb RNA from total cell RNA (lane 3). These results together suggest that a major fraction of the 0.8 kb H-strand encoded RNA exists as free RNA.
FIG. 9.
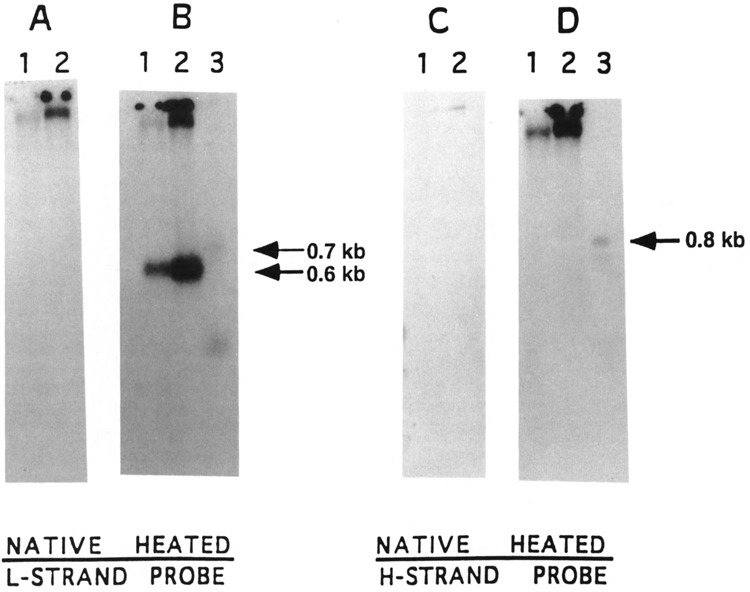
Mouse liver and kidney mitochondrial D-loop DNA analysis. DNA isolated from purified mouse liver (lane 1) and kidney (lane 2) mitochondria was analyzed for D-loop DNA and possible H-strand RNA by hybridization with the L- and H-strand-specific probes, respectively. (A,C) Native DNA samples (500 ng) and (B,D) heat-denatured DNA samples were used for electrophoresis on a 1.2% agarose gel. In both (B) and (D), lane 3 contained 30 μg of mouse liver RNA. Lanes in (A) and (B) were hybridized with the L-strand probe and those in (C) and (D) were hybridized with the H-strand-specific probe. The positions of D-loop DNA and the 0.7 kb RNA bands in (B) and the 0.8 kb RNA band in (D) are indicated by arrows.
Variations in Different Tissues and During Myogenesis
The relative abundance of the D-loop H-strand encoded RNA in different tissues was compared by the Northern blot analysis of total RNA. As shown in Fig. 2C (lane 4) and Fig. 5 (lanes 1, 3, and 8), cultured cells like Ehrlich ascites, rat hepatoma, Hela cells, and mouse neuroblastoma cells contain relatively low levels of the RNA, whereas tissues like liver, heart, brain, and kidney (Fig. 2C, lanes 1–3, Fig. 5, lanes 2, 4–7) contain several fold higher abundance of this species. It was also observed that the level of this RNA was markedly increased during myogenesis. As seen from Fig. 10A, mouse C2C12 myoblasts contain relatively low levels of the 0.8 kb RNA in both subconfluent and confluent stages (lanes 2 and 3), whereas fully differentiated myotubes contain > 10-fold higher levels (lane 1). The relative levels of COX I and COX II mRNAs, which are also H-strand encoded species, are increased by only twofold during the differentiation of myotubes from precursor myoblasts (Fig. 10C). It is also interesting that the levels the 0.7 kb L-strand transcript in cells from the three stages of differentiation do not reflect the relative abundance of the 0.8 kb RNA (Fig. 10A,B). Thus, although the physiological role of this RNA currently remains unclear, our results demonstrate a selective accumulation or increased transcription of the 0.8 kb H-strand encoded RNA in tissues and cells that are differentiated to form multicellular aggregates, such as the myotube.
FIG. 10.
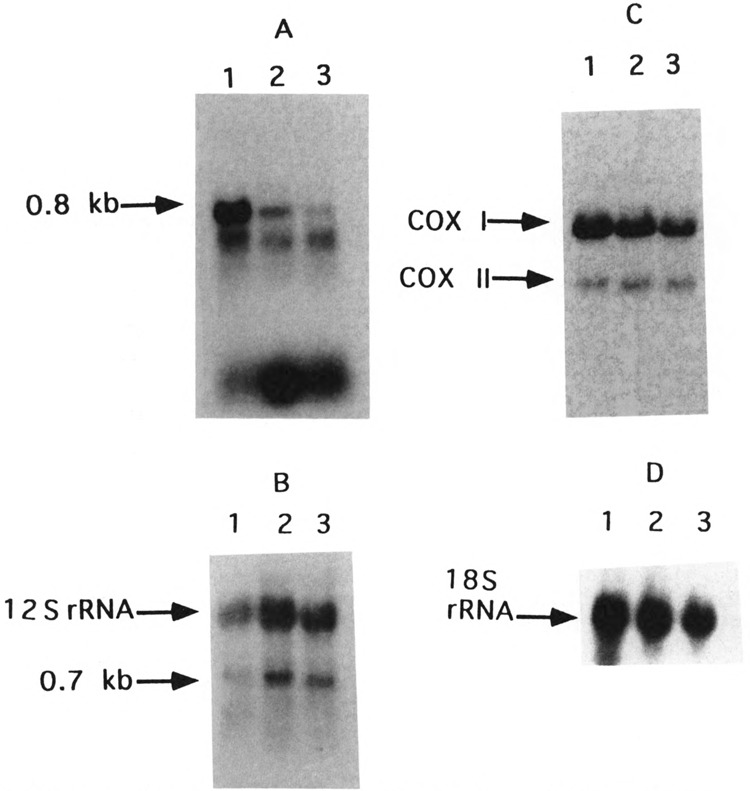
Accumulation of the D-loop H-strand encoded RNA during myogenesis. C2C12 myoblasts were grown to confluency in DMEM high glucose medium containing 10% fetal calf serum and induced to form myotubes by supplementing the medium with 10% heat-inactivated horse serum according to the ATCC manual. RNA (30 μg each) from subconfluent (lane 3) and confluent (lane 2) myoblasts and induced myotubes (lane 1) was electrophoretically resolved in duplicates and blotted as described under Experimental Procedures. The blot in (A) was hybridized with 32P-labeled H-strand-specific probe. (B) The blot from (A) was stripped off the probe and rehybridized with a mixture of L-strand-specific probe and the mitochondrial 12S rRNA probe. (C) A representative blot as in (A) was hybridized with a mitochondrial DNA clone containing the COX I and COX II genes. (D) The blot from (C) was stripped off the probe and rehybridized with the cytoplasmic 18S rRNA probe. RNA contents between individual lanes and replicates varied by less than 10% as judged by ethidium bromide staining of the 18S and 28S rRNAs.
DISCUSSION
Results presented in this article demonstrate that the entire H-strand of murine mitochondrial DNA is transcribed. The 0.8 kb transcript encoded by the D-loop region contains a 3′ poly(A) tail and maps between the region complementary to the start of tRNAPro gene (encoded by the L-strand) and the 5′ end of the tRNAPhe gene encoded by the H-strand (Fig. 1). Identification of this transcript provides a new insight regarding the mechanism of mitochondrial transcription termination, particularly that of H-strand. It also poses new questions on the H-strand transcription initiation and the possible role of the mitochondrial D-loop region on the regulation of mitochondrial biogenesis.
It is well established that nearly the entire length of the L-strand from the D-loop region, except perhaps the 100–250 nucleotide stretch upstream of the LSP, is transcribed. However, it is generally believed that much of the H-strand within the D-loop region is not transcribed. Our results demonstrate that the H-strand transcription proceeds beyond the tRNAPro gene and terminates immediately upstream of the tRNAPhe gene in the mouse and rat systems and about 350 nucleotides upstream in the human mitochondrial system. The results of SI nuclease protection have identified a minor species of transcript mapping to the 3′ end of tRNAThr, and a prominent species mapping to the site on the H-strand corresponding to the start of tRNAPro, encoded by the opposite strand. A majority of the mitochondrial mRNA coding genes on the animal mitochondrial genomes are interrupted by tRNA genes (Anderson et al., 1981, 1982; Bibb et al., 1981; Montoya et al., 1981; Ojala et al., 1981). It was hypothesized that the secondary structure created by the folding of the tRNAs or antisense tRNA sequences provides the necessary signal for processing of precursor RNA yielding various mitochondrial mRNA species (Manam and Van Tule, 1979; Montoya et al., 1981; Ojala et al., 1981). In support of this hypothesis, our results show a major processing at a site antisense to the start of tRNAThr. A comparable 5′ map position was recently described for the rat mitochondrial D-loop region coded RNA (Sbisa et al., 1990).
The 3′ end formation for nuclear poly(A)-containing mRNA is known to involve two distinct signals: a poly(A) addition signal AAUAAA located close to the 3′ end of mRNA, and a signal for transcription termination, located more than 300 bases downstream (Manley et al., 1989; Proudfoot, 1989). It is interesting that some features of the eukaryotic transcription termination are conserved in the vertebrate mitochondrial system, except that the transcription termination may occur immediately past the polyadenylation signal. Transcripts of similar size hybridizing to the H-strand of the D-loop region are detected in at least three other vertebrate systems investigated (bovine, rat, human), and in all cases, canonical polyadenylation signals AAUAAA, AAUUAAA, or AACUAAA are detected 4–12 nucleotides upstream of the poly(A) start site. Further, a sequence search indicated that the consensus polyadenylation signals are conserved at analogous positions in almost all vertebrate mitochondrial genomes for which DNA sequence data are available (Anderson et al., 1981, 1982; Bibb et al., 1981; Cairns and Bogenhagen, 1986; Cantatore et al., 1989; Desjardins and Morais, 1990; Roe et al., 1985). Recently, it was shown that a nucleotide sequence AAUAA(C/U)AUUCUU is involved in the 3′ end formation of Yeast mitochondrial mRNAs (Min and Zassenhaus, 1993). Interestingly, this sequence exhibits near perfect to very high sequence homology to the 3′ terminal regions of the vertebrate D-loop H-strand coded RNAs described here (Fig. 11). Based on these, we postulate that some aspects of mitochondrial RNA 3′ end formation are conserved in both unicellular and multicellular organisms. Two distinct mechanisms are likely for the 3′ end formation of mammalian mitochondrial D-loop encoded 0.8 kb RNA: 1) a unique termination-linked polyadenylation or 2) a 3′ processing-linked polyadenylation of a precursor RNA similar to those responsible for the generation of other mitochondrial mRNAs. In view of current belief that the H-strand transcription complex terminates within the triple-stranded D-loop region, the termination-linked polyadenylation is suggested to be the more likely mechanism. It remains to be seen if the conserved sequence motifs from the vertebrate mitochondrial DNA contain signals for transcription termination as well as 3′ polyadenylation.
FIG. 11.

Nucleotide sequence similarities between the 3′ ends of the Yeast mitochondrial mRNA (Min and Zassenhaus, 1993) and the D-loop H-strand coded RNAs from different vertebrates.
The functional significance of the region immediately upstream of the putative transcription termination site was investigated by DNA-protein binding studies using gel mobility shift analysis. We tested the 22 nucleotide sequence from the putative termination region because it contains the conserved AT-rich sequence resembling the polyadenylation signal. Additionally, this sequence is immediately downstream of the nucleotide sequence that binds to the mitochondrial transcription factor mtTF1 (Fisher et al., 1991). As shown in Fig. 7, the 22 bp DNA probe binds to a protein factor from mouse liver mitochondrial extract. Further, use of the upstream mtTF1 binding sequence both as a probe and competitor (Figs. 7 and 8) indicates that the putative terminator region binding protein is different from the purified human mtTF1. Our results also show that protein binding to this region is not competed by synthetic DNA corresponding to the TAS sequence, or the 16S rRNA terminator sequence (TERM), suggesting a distinctly different sequence specificity for protein binding by the H-strand transcription termination region conserved sequence motif identified in this study. Thus, although we have not yet functionally analyzed the sequence immediately upstream of the termination site, its ability to bind a specific protein(s) indirectly suggests a possible role in the termination process.
The H-strand transcription initiation sites in human (Bogenhagen et al., 1984; Chang and Clayton, 1986c; Montoya et al., 1982), mouse (Bhat et al., 1989; Chang and Clayton, 1986a), bovine (King and Low, 1987a, 1987b), and Xenopus (Bogenhagen et al., 1986) systems have been mapped using various approaches, and in all cases the major transcription starts at about 10–30 nucleotides upstream of the tRNAPhe gene. Although the D-loop regions of all of these and also other vertebrate systems as described above contain AT-rich sequence resembling the polyadenylation signal, at present the precise termination sites of only the mouse, rat, and human mitochondrial DNA are available. In the mouse system, where the D-loop region is relatively small (875 nucleotides), the H-strand transcription initiation site and the polyadenylation signal, suggested to be important in transcription termination, are virtually juxtaposed, or perhaps even overlapping. Although the mtTF1 binding sequence motif is 8–16 nucleotides upstream of the transcription start site, one can envision a complex set of mechanisms separating the events for transcription initiation from those of termination. In the case of human mitochondrial DNA with a relatively larger D-loop region (over 1 kb), the picture is less complex; the putative termination signal occurs about 363 nucleotides upstream (nucleotide 227) of the H-strand transcription initiation at nucleotide 590. Additional information on the H-strand transcription start sites of the rat, and termination sites of bovine, Xenopus, and other cell systems will be necessary to develop a comprehensive picture of the spatial organization of transcription start and stop sites in these vertebrate systems.
Several studies have ascribed wide-ranging functional significance to the vertebrate mitochondrial D-loop region. Because this region of the mitochondrial DNA from higher vertebrates houses both the H- and L-strand transcription promoters, and sites for binding transcription factor mtTF1, it is believed to play an important role in transcription regulation. The D-loop region also contains the origin of H-strand replication, and conserved sequence motifs termed TAS near the 5′ end of the D-loop structure (Doda et al., 1981; Mackay et al., 1986), which bind to specific proteins (Madsen et al., 1993). It is believed that a regulated balance between the rate of H-strand DNA initiation and the rate of termination at or near the TAS motifs may regulate the rate of H-strand replication, and thus the entire mitochondrial DNA replication process and copy number. A number of studies have shown a close correlation between the mitochondrial content of triple-stranded D-loop structure and cellular growth rates, and/or, the oxidative capacity of the tissues (Annex and Williams, 1990; King and Low, 1987b; Williams, 1986). Tissues like heart and muscle with very high oxidative capacity contain up to 80% of their mitochondrial DNA with D-loop structures (Annex and Williams, 1990). The precise underlying mechanisms relating the DNA triple-stranded structure with higher metabolic activities of the mitochondrion, however, remain unclear. Results of the present study show that mitochondria from relatively fast dividing or cultured cells contain a very low steady-state level of the H-strand coded RNA. The generally low cellular level of this RNA is probably the reason why it was not readily detected in a number of previous studies from various laboratories that involved the use of tissue culture cells. Mitochondria from differentiated tissues, on the other hand, contain 5–15-fold higher levels of this RNA (Fig. 5). Furthermore, a 10–20-fold accumulation of this RNA species is observed during in vitro-induced myogenesis (Fig. 10). These results collectively suggest that the D-loop region H-strand encoded RNA has some functional significance particularly associated with the differentiated state of the cells. It should be noted that the level of this RNA does not reflect the frequency of the D-loop structure; hepatic cells containing relatively low contents of D-loops contain very high levels of the RNA, and C2C12 myoblasts containing a high level of D-loop DNA contain a low level of the RNA. Furthermore, cardiac and skeletal muscle tissues contain high levels of both D-loop DNA and this H-strand encoded transcript. Thus, although the precise function of the D-loop encoded RNA remains unknown, it is likely to have some functional significance associated with transcription regulation or nuclear-mitochondrial interaction.
ACKNOWLEDGEMENTS
We are thankful to Dr. David Clayton for a generous gift of the human mtTF1 used in this study, and for critically reading the manuscript. We also thank Dr. Robert S. Carter for helping with the computer analysis and his comments on the manuscript. Finally, we are thankful to Ms. Vasantha Reddy for excellent technical assistance. This research was supported by National Institutes of Health Grant CA-22762 and a grant from the Council for Tobacco Research-U.S.A. Inc.
REFERENCES
- Aloni Y. and Attardi G. (1971), J Mol Biol 28, 251–267. [DOI] [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de-Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., and Young I. G. (1981), Nature 290, 457–465. [DOI] [PubMed] [Google Scholar]
- Anderson S., de-Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., and Young I. G. (1982), J Mol Biol 156, 683–717. [DOI] [PubMed] [Google Scholar]
- Annex H. B. and Williams R. S. (1990), Mol Cell Biol 10, 5671–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi G. and Schatz G. (1988), Annu Rev Cell Biol 4, 289–333. [DOI] [PubMed] [Google Scholar]
- Battey J. and Clayton D. A. (1978), Cell 14, 143–156. [DOI] [PubMed] [Google Scholar]
- Berk A. J. and Sharp P. A. (1977), Cell 12, 721–732. [DOI] [PubMed] [Google Scholar]
- Bhat K. S., Avdalovic N., and Avadhani N. G. (1989), Biochemistry 28, 763–769. [DOI] [PubMed] [Google Scholar]
- Bhat K. S., Bhat N. K., Kulkarni G. R., Iyengar A., and Avadhani N. G. (1985), Biochemistry 24, 5818–5825. [DOI] [PubMed] [Google Scholar]
- Bhat N. K., Niranjan B. G., and Avadhani N. G. (1982), Biochemistry 21, 2452–2460. [DOI] [PubMed] [Google Scholar]
- Bibb M. G., Van Etten R. A., Wright C. T., Walberg M. W., and Clayton D. A. (1981), Cell 26, 167–180. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Applegate E. F., and Yoza B. K. (1984), Cell 36, 1105–1113. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F. and Romaneli M. F. (1988), Mol Cell Biol 8, 2917–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F. and Yoza B. K. (1986), Mol Cell Biol 6, 2543–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Yoza B. K., and Cairns S. S. (1986), J Biol Chem 26, 8488–8494. [PubMed] [Google Scholar]
- Cairns S. S. and Bogenhagen D. F. (1986), J Biol Chem 261, 8481–8487. [PubMed] [Google Scholar]
- Cantatore P., Roberti M., Rainaldi G., Gadaleta M. N., and Saccone C. (1989), J Biol Chem 264, 10965–10975. [PubMed] [Google Scholar]
- Carter R. S. and Avadhani N. G. (1991), Arch Biochem Biophys 288, 97–106. [DOI] [PubMed] [Google Scholar]
- Carter R. S., Bhat N. K., Basu A., and Avadhani N. G. (1992), J Biol Chem 267, 23418–23426. [PubMed] [Google Scholar]
- Chang D. D. and Clayton D. A. (1984), Cell 36, 635–643. [DOI] [PubMed] [Google Scholar]
- Chang D. D. and Clayton D. A. (1986a), Mol Cell Biol 6, 1446–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D. and Clayton D. A. (1986b), Mol Cell Biol 6, 3253–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D. and Clayton D. A. (1986c), Mol Cell Biol 6, 3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Hauswirth W. W., and Clayton D. A. (1985), EMBO J 4, 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W. and Clayton D. A. (1986), Proc Natl Acad Sci USA 83, 6277–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W. and Clayton D. A. (1988), Mol Cell Biol 8, 4502–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. (1984), Annu Rev Biochem 53, 573–594. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. (1991), Annu Rev Cell Biol 7, 453–478. [DOI] [PubMed] [Google Scholar]
- Daga A., Micol V., Hess D., Aebersold R., and Attardi G. (1993), J Biol Chem 268, 8123–8130. [PubMed] [Google Scholar]
- Desjardins P. and Morais R. (1990), J Mol Biol 212, 599–634. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., and Roeder R. G. (1983), Nucleic Acids Res 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doda J. N., Wright C. T., and Clayton D. A. (1981), Proc Natl Acad Sci USA 78, 6116–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. P. and Clayton D. A. (1988), Mol Cell Biol 8, 3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. P., Liskowsky T. G., Breen A. M., and Clayton D. A. (1991), J Biol Chem 266, 9153–9160. [PubMed] [Google Scholar]
- Fisher R. P., Topper J. N., and Clayton D. A. (1987), Cell 50, 247–258. [DOI] [PubMed] [Google Scholar]
- Gelfand R. and Attardi G. (1981), Mol Cell Biol 1, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Nonaka I., and Horai S. (1990), Nature 348, 651–653. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Parisi M. A., Bennett J. L., and Clayton D. A. (1991), Nature 351, 236–239. [DOI] [PubMed] [Google Scholar]
- Hu N. -t. and Messing J. (1982), Gene 17, 271–277. [DOI] [PubMed] [Google Scholar]
- King T. C. and Low R. L. (1987a), J Biol Chem 262, 6204–6213. [PubMed] [Google Scholar]
- King T. C. and Low R. L. (1987b), J Biol Chem 262, 6214–6220. [PubMed] [Google Scholar]
- Kobayashi Y., Momoi M. Y., Tominaga K., Momoi T., Nihei K., Yanagisawa M., Kagawa Y., and Ohta S. (1990), Biochem Biophys Res Commun 173, 816–822. [DOI] [PubMed] [Google Scholar]
- Kruse B., Narasimhan N., and Attardi G. (1989), Cell 58, 391–397. [DOI] [PubMed] [Google Scholar]
- Manam S. and Van Tule G. C. (1979), J Biol Chem 262, 10272–10279. [PubMed] [Google Scholar]
- Min J. and Zassenhaus H. P. (1993), Mol Cell Biol 13, 4167–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya J., Christianson T., Levens D., Rabinowitz M., and Attardi G. (1982), Proc Natl Acad Sci USA 79, 7195–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay S. L., Olivo P. D., Laipis P. J., and Hauswirth W. W. (1986), Mol Cell Biol 6, 1261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen C. S., Ghivizziani S., and Hauswirth W. W. (1993), Mol Cell Biol 13, 2162–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Proudfoot N. J., and Platt T. (1989), Genes Dev 3, 2218–2244. [DOI] [PubMed] [Google Scholar]
- Montoya J. D., Ojala D., and Attardi G. (1981), Nature 290, 465–470. [DOI] [PubMed] [Google Scholar]
- Ojala D., Montoya J., and Attardi G. (1981), Nature 290, 470–474. [DOI] [PubMed] [Google Scholar]
- Parker C. S. and Topol J. (1984), Cell 36, 357–369. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. (1989), Trends Biochem Sci 14, 105–110. [DOI] [PubMed] [Google Scholar]
- Rastl E. and Dawid I. B. (1979), Cell 18, 501–510. [DOI] [PubMed] [Google Scholar]
- Roe B. A., Ma D.-P., Wilson R. K., and Wong J. H.-H. (1985), J Biol Chem 260, 9759–9774. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., and Peterson G. B. (1982), Proc Natl Acad Sci USA 162, 729–773. [DOI] [PubMed] [Google Scholar]
- Sbisa E., Nardelli M., Tanzariello F., Tullo A., and Saccone C. (1990), Curr Genet 17, 247–253. [DOI] [PubMed] [Google Scholar]
- Vijayasarathy C., Bhat N. R., and Avadhani N. G. (1989), J Biol Chem 264, 7772–7775. [PubMed] [Google Scholar]
- Williams R. S. (1986), J Biol Chem 261, 12390–12394. [PubMed] [Google Scholar]


