Abstract
The regulatory region of Drosophila proliferating cell nuclear antigen (PCNA) gene consists of a promoter region (−168 to +24 with respect to the transcription initiation site) and an upstream region containing three homeodomain protein binding sites (HDB) (−357 to −165). The PCNA gene regulatory regions with HDB (−607 to +137) or without HDB (−168 to +137) were fused with the lacZ and transgenic flies were established by P-element-mediated transformation. Male transgenic flies were crossed with wild-type females, and zygotic expression of the lacZ was monitored by quantitative β-galactosidase assay, at various stages of development. Expression of the lacZ was high in embryos, first and second instar larvae, and adult females, and low at other stages of development. Only a marginal difference in expression was observed between flies carrying the homeodomain protein binding region and those not carrying it. Spatial pattern of the lacZ expression in the embryo visualized by immunostaining with the anti-lacZ antibody was similar to the distribution of the endogenous PCNA protein. Here, too, only a marginal difference was observed between transgenic flies carrying two different constructs of the PCNA lacZ. In genetic crossing experiments of transgenic flies with those carrying mutation in homeobox genes, no significant change in the lacZ expression pattern was observed. However, when male transgenic flies were crossed with female flies homozygous for a torso gain-of-function allele, repression of the lacZ expression was observed in the central region of the embryo. Because these local changes in the lacZ expression depend on the homeodomain protein binding region, unidentified homeodomain proteins are probably involved. Our results suggest that the promoter region is practically sufficient for expression of the PCNA gene and that the homeodomain protein binding region functions as a silencer when torso is activated ectopically.
Keywords: Homeodomain protein binding region, Drosophila proliferating cell nuclear antigen, Transgenic flies
THE proliferating cell nuclear antigen (PCNA), known as an accessary protein of DNA polymerase δ, is required for simian virus 40 DNA replication as well as for cellular DNA replication (Tsurimoto et al., 1990; Jaskulski et al., 1988; Liu et al., 1989). PCNA is also involved in DNA repair (Shivji et al., 1992). The amino acid sequence of the PCNA protein has been highly conserved in a wide range of species from yeast to human (reviewed by Moriuchi, 1990).
We earlier cloned and sequenced the Drosophila PCNA gene and its 5′-Hanking region (Yamaguchi et al., 1990). The presence of clusters of 10-bp sequences, similar to the binding consensus for Drosophila homeodomain proteins (Desplan et al., 1988; Hoey and Levine, 1988; Muller et al., 1988), was noted in the region from nucleotide positions −165 to −357 of the Drosophila PCNA gene. DNase I footprint analysis revealed that the homeodomain proteins encoded by a pair rule gene, even-skipped (eve), and a dorso-ventral gene, zerknullt (zen), can specifically bind to these sequences in vitro (Yamaguchi et al., 1990). Studies in vivo and in cultured cells revealed that zen could repress expression of the PCNA gene (Yamaguchi et al., 1991b). However, the repression appeared to be indirect because the target region of zen for the repression was mapped in the promoter region of the PCNA gene to which zen did not bind in vitro (Yamaguchi et al., 1991b; Hirose et al., 1994). Therefore, it remained to be determined whether or not the homeodomain protein binding region of the PCNA gene truly functions in vivo.
In the present study, we used transgenic flies carrying the PCNA gene regulatory regions fused with the lacZ (Yamaguchi et al., 1991b) to examine effects of the homeodomain protein binding region on the PCNA gene promoter function in vivo. We found that the region containing homeodomain protein binding sites exerts only a marginal effect on the promoter during normal developmental processes, but has the potential to modulate the promoter locally, because spatial change of the lacZ expression that depended on the homeodomain protein binding region was observed in torso mutant embryos.
MATERIALS AND METHODS
Establishment of Transgenic Flies
The plasmid p5′-607DPCNAlacZW8HS (Yamaguchi et al., 1991b) contains the PCNA gene fragment spanning from −607 to +137 fused with the lacZ in a P-element vector. The plasmid p5′-168DPCNAlacZW8HS (Yamaguchi et al., 1991b) contains the PCNA gene fragment spanning from −168 to +137 fused with the lacZ in a P-element vector. By P-element-mediated transformation as described by Robertson et al. (1988), 25 and 13 independent transformant lines were established for the p5′-607DPCNAlacZW8HS and the p5′-168DPCNAlacZW8HS, respectively. These transgenic fly strains and their chromosomal linkages are listed in Table 1.
TABLE 1.
THIRTY-EIGHT TRANSFORMANTS CARRYING THE lacZ GENE FUSED TO THE PCNA GENE 5′-FLANKING SEQUENCES
| P-Element Plasmids | Strains | Chromosome Linkage |
| p5′-607DPCNAlacZW8HS | 52 | II |
| 57 | X | |
| 60 | X | |
| 67 | III | |
| 88 | II | |
| 95A | III | |
| 95B | II | |
| 96 | II | |
| 109 | X | |
| 128 | II | |
| 133 | III | |
| 143A | III | |
| 143B | III | |
| 158 | III | |
| 164 | III | |
| 183 | II | |
| 186 | X | |
| 208 | III | |
| 218 | X | |
| 237 | II | |
| 244A | III | |
| 244B | III | |
| 248A | III | |
| 258 | II | |
| p5′-168DPCNAlacZW8HS | 5 | II |
| 10 | HI | |
| 19A | III | |
| 19B | II | |
| 21 | X | |
| 22 | III | |
| 24A | II | |
| 24B | X | |
| 70 | II | |
| 72 | II | |
| 84 | II | |
| 89 | II | |
| 98 | II |
Analysis of Expression Patterns for PCNA lacZ and Endogenous PCNA
Quantitative measurement of β-galactosidase activity in extracts (Friedell and Searles, 1992).
Male transgenic flies were crossed with female wild-type (Canton S) flies. Fifty to 100 individuals each of dechorionated embryos, larvae, pupae, and adult flies were homogenized in 500 μl of ice-cold assay buffer (50 mM potassium phosphate, pH 7.5/1 mM MgCl2). Homogenates were centrifuged at 10,000 × g at 4°C for 5 min. For each assay, 50–200 μl of the supernatant was added to give 1 ml of assay buffer containing 1 mM chlorophenol red-β-d-galactopyranoside substrate (CPRG; Boehringer Mannheim). Reactions were incubated at 37°C in the dark. The substrate conversion was measured at 574 nm using a spectrophotometer at 0.25, 0.5, 0.75, 1, and 1.5 h after addition of the extract; the rate of color development was linear. The β-galactosidase activity was defined as absorbance units per hour per milligram of protein. To correct for endogenous β-galactosidase activity, extracts from the wild-type strain were included in each experiment and this background reading was subtracted from readings obtained with each transformant line. The protein concentration of the extracts was determined by the Bio-Rad protein assay.
Immunocytochemical method
Male transgenic flies were crossed with female white (w) flies to observe zygotic expression of the PCNA lacZ in early embryos (0–6 h old). Female transgenic flies were crossed with male w flies to observe the maternal contribution to the PCNA lacZ expression. To test for expression of the PCNA lacZ fusion gene in embryos carrying a mutation in Antennapedia (Antp, mapped on the third chromosome), males heterozygous for the Antp mutation (Antp w10/TM3) with the transgene on the X chromosome were crossed with Antp w10 (Lindsley and Zimm, 1992) heterozygous females. Mutations in Deformed (Dfd), fushi tarazu (ftz), proboscipedia (pb), Sex combs reduced (Scr 7F28), and Abdominal B (AbdB M1, AbdB M5, and AbdB rx23-1) are all mapped on the third chromosome (Lindsley and Zimm, 1992). Accordingly, genetic crossing experiments were conducted in the same manner. To test for expression of the PCNA lacZ in homozygous mutants of tor, male transgenic flies were crossed with female flies homozygous for the tor PM (Casanova and Struhl, 1989) or the tor RL3 (Casanova and Struhl, 1989) mutation. Because tor RL3 was a ts-mutant, crossing with the tor zRL3 was conducted at 28°C.
Expression patterns of the PCNA lacZ and the endogenous PCNA were analyzed by an immunohistochemical method, as described previously (Yamaguchi et al., 1991b). Embryos 0–6 h old were collected, dechorionated, fixed, and devitellinized. Embryos were incubated with the primary antibody followed by treatment with the secondary antibody, as described previously (Yamaguchi et al., 1991b). A polyclonal rabbit anti-Drosophila PCNA (Ng et al., 1990) (used at a dilution of 1 : 2000) and an alkaline phosphatase-conjugated goat anti-rabbit IgG (Promega-Biotec, preabsorbed against 0–16-h-old fixed embryos and used at a dilution of 1 : 2000) served as the primary and secondary antibodies for detection of PCNA. A monoclonal mouse anti-lacZ (Promega-Biotec, preabsorbed against 0–16-h-old fixed embryos and used at a dilution of 1 : 1000) and an alkaline phosphatase-conjugated goat anti-mouse IgG (Promega-Biotec, preabsorbed against 0–16-h-old fixed embryos and used at a dilution of 1 : 2000) served as primary and secondary antibodies, respectively, for detection of PCNA lacZ. Histochemical color reaction for alkaline phosphatase was done as described previously (Yamaguchi et al., 1991b). Samples were examined under a microscope (Nikon FXA-A1) and photographed on TMAX 100 films (Kodak).
RESULTS
Distribution of PCNA Protein During Early Embryogenesis
Distribution of PCNA protein during embryonic development was analyzed by staining whole-mount embryos with a polyclonal anti-Drosophila PCNA antibody (Ng et al., 1990). Specificity of the antibody was verified by immunoblotting analysis (Ng et al., 1990; Yamaguchi et al., 1991a). This antibody specifically reacted with the PCNA protein in crude extracts of 0–3-h-old embryos and with that produced in E. coli carrying the expression plasmid for Drosophila PCNA (Yamaguchi et al., 1991a).
Strong staining of nuclei above the weak staining of ooplasm was detected throughout 13 nuclear division cycles (Fig. 1A) and at the cellular blastoderm stage (Fig. 1B). At the onset of gastrulation, strong staining was observed in apparently mesodermal cells within the ventral furrow and staining in the nuclei of most of the surface cells remained visible (Fig. 1C,D). During the early phase of germ band extension (stage 7), strong staining of the PCNA was seen in regions of the cephalic furrow, the amnioproctodeal imagination, the anterior midgut primordium, and the invaginated mesoderm (Fig. 1E,F), although weak staining in ectodermal cell nuclei was visible. No significant staining was detected in the dorsal region corresponding to the amnioserosa at this and later stages. During the germ band extension stage, anti-PCNA staining was observed throughout the germ band with stronger staining in the mesodermal layer, as well as within the anterior midgut primordium, the posterior midgut primordium, and the proctodeal primordium (Fig. 1G–J). No significant staining signal was observed with embryos treated with the normal rabbit IgG as a primary antibody (Fig. 1L). At the end of the germ band extension, strong signals were associated with the neuroblast cells (Fig. 1K).
FIG. 1.
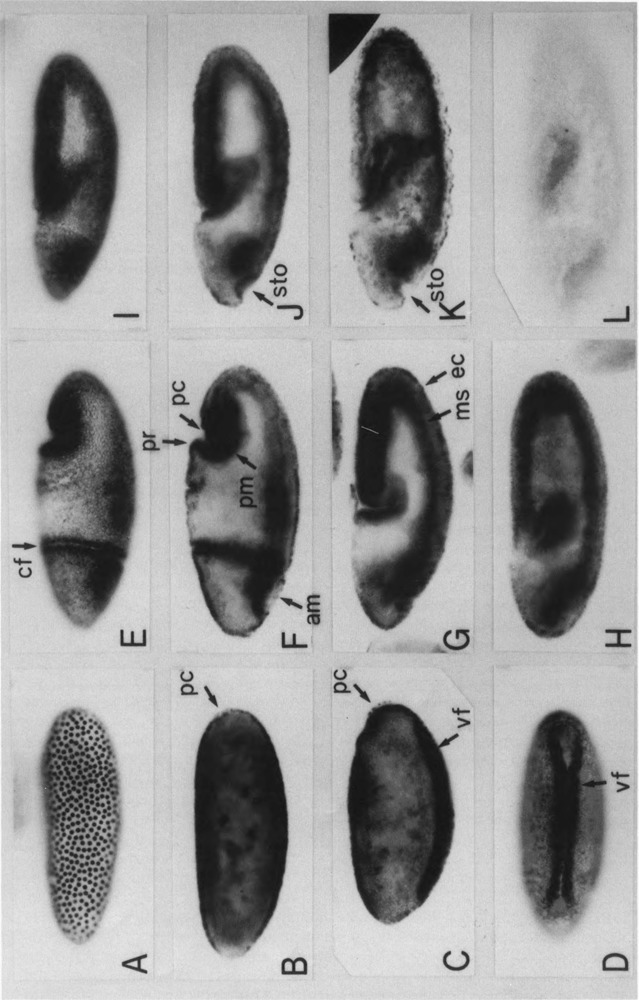
Distribution of PCNA protein in wild-type embryos. Whole-mount specimens of fixed embryos were immunostained with an anti-PCNA antibody. Embryos are oriented so that anteriors are to the left and ventrals are to the bottom [except in (D)]. Staging is according to Campos-Ortega and Hartenstein (1985). (A) Surface view of the syncytial blastoderm embryo (stage 4, 1320–1410 h). (B) Internal focal view of the blastoderm embryo (stage 5, 1410–1450 h), pole cells (pc). (C) Internal focal view of the early gastrulating embryo (stage 6, 1450–1500 h), ventral furrow (vf). (D) Ventral surface view of the early gastrulating embryo (stage 6). (E) Surface view of the embryo of the stage beginning of germ band extension (stage 7, 1500–1510 h), cephalic furrow (cf). (F) Internal focal view of the embryo shown in (E), anterior midgut primordium (am), proctodeum primordium (pr), posterior midgut primordium (pm). (G) Internal focal view of the stage 8 embryo (1510–1540 h) (germ band elongation), mesoderm (ms), ectoderm (ec). (H) Internal focal view of the stage 9 embryo (1540–1620 h). (I) Surface view of the stage 10 embryo (1620–1720 h). (J) Internal focal view of stage 10 embryo, stomodeum (sto). (K) Internal focal view of the early stage 11 embryo. (L) Stage 10 ebmbryo treated with normal rabbit IgG as a primary antibody.
Maternal and Zygotic Expression of the PCNA lacZ
The regulatory region of Drosophila PCNA gene is composed of a promoter region (−168 to + 24) and an upstream homeodomain protein binding region (−357 to −165) (Yamaguchi et al., 1991b). Transgenic flies carrying two different constructs of the plasmids p5′-607DPCNAlacZW8HS and p5′-168DPCNAlacZW8HS were established to examine the effect of a homeodomain protein binding region on the expression pattern of PCNA lacZ. The maternal and zygotic expressions of the PCNA lacZ were distinguished by comparing staining patterns in embryos from the cross of transgenic female × w male with that of w female × transgenic male.
In embryos from the cross of transgenic female × w male embryos, maternal storage of the PCNA lacZ in early embryos undergoing nuclear division cycle was detected (Fig. 2A,F). No significant difference was observed between flies carrying the p5′-607DPCNAlacZW8HS and those carrying the p5′-168DPCNAlacZW8HS. The PCNA lacZ protein appeared to be relatively stable, because the staining signal that appeared to come from maternal stores remained until the end of the germ band extension. Staining signals in Fig. 2 (panels E and J) are much stronger than those in Fig. 3 (panels J and T).
FIG. 2.
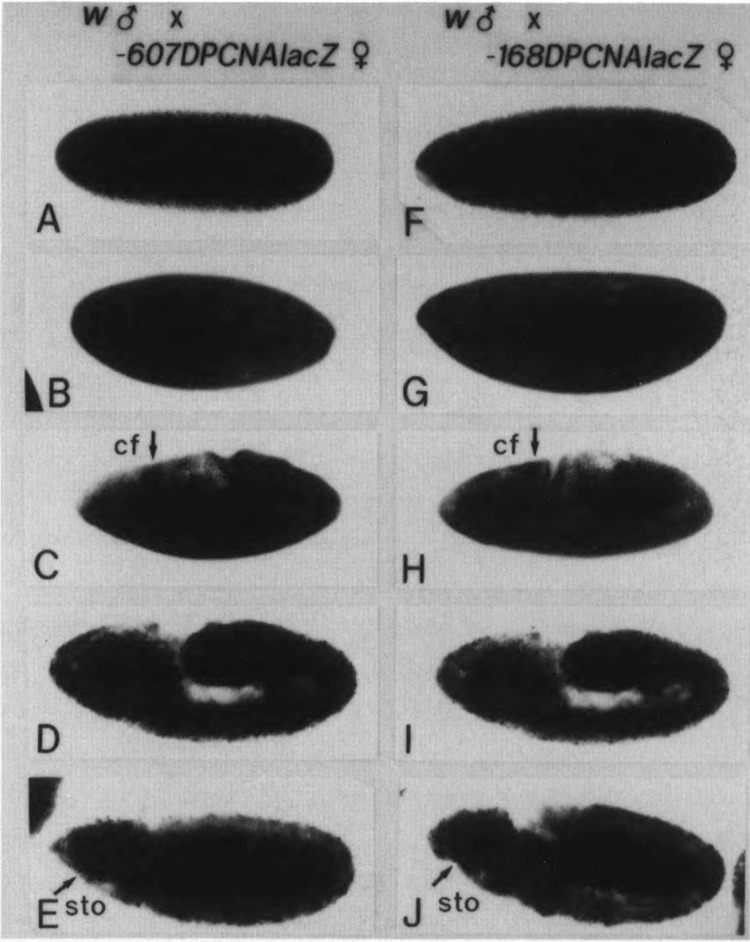
Maternal contribution to PCNA lacZ expression. Embryos from w males × p5′-607DPCNAlacZW8HS females (A–E) or w males × p5′-168DPCNAlacZW8HS females (F–J), as indicated at the top, were immunostained with anti-lacZ antibody. (A,F) Surface views of syncytial blastoderm embryos (stage 4, 1320–1410 h). (B,G) Surface views of cellular blastoderm embryos (stage 5, 1410–1450 h). (C,H) Surface views of stage 7 embryos (1300–1510 h), cephalic furrow (cf). (D,I) Internal focal views of stage 8 embryos (1510–1540 h). (E,J) Internal focal views of stage 10 embryos (1620–1720 h), stomodeum (sto).
FIG. 3.
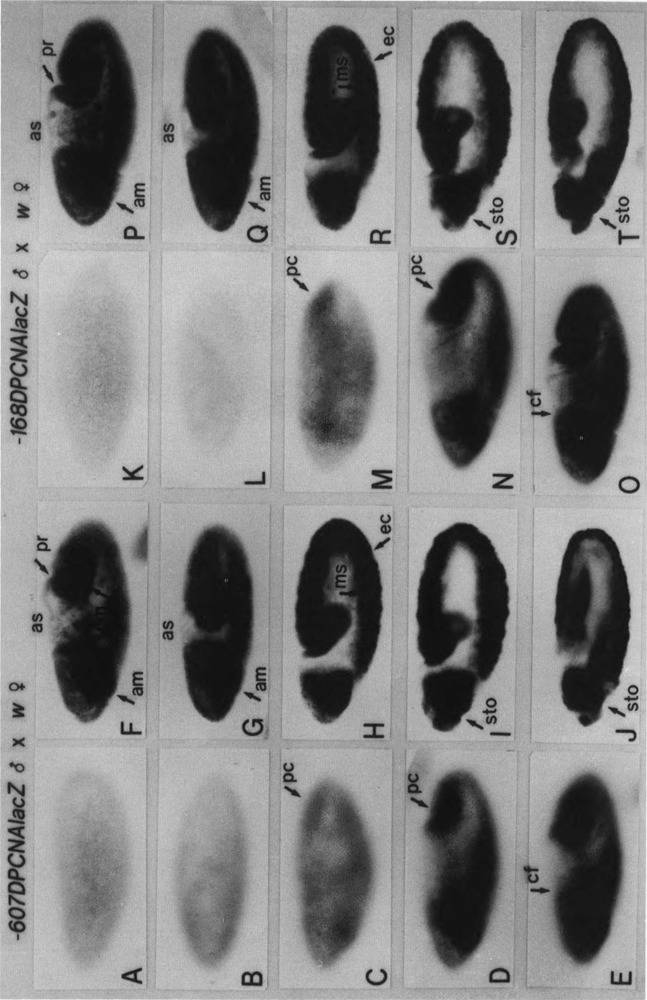
Zygotic expression of PCNA lacZ in embryos. Embryos from p5′-607DPCNAlacZW8HS males × w females (A–J) or p5′-168DPCNAlacZW8HS males × w females, as indicated at the top, were immunostained with an anti-lacZ antibody. Surface views of (A,K) syncytial blastoderm embryos (stage 4, 1320–1410 h), (B,L) cellular blastoderm embryos (stage 5, 1410–1450 h), (C,M) early gastrulating embryos (stage 6, 1450–1500 h), (D,N) embryos at the beginning of germ band elongation (early stage 7, 1500–1510 h), and (E,O) embryos at the early phase of germ band elongation (late stage 7) are shown; pole cells (pc) and cephalic furrow (cf). (F,P) Internal focal views of embryos shown in (E,O), anterior midgut primordium (am), proctodeum primordium (pr), posterior midgut primordium (pm), and aminoserosa (as). Internal focal views of (G,Q) stage 8 embryos (1510–1540 h), (H,R) stage 9 embryos (1540–1620 h), (I,S) stage 10 embryos (1620–1720 h), and (J,T) stage 11 embryos (1720–1920 h) are shown; mesoderm (ms), ectoderm (ec), stomodeum (sto).
In embryos from transgenic male × w female, no detectable expression of the PCNA lacZ was observed at syncytial blastoderm and cellular blastoderm stages (Fig. 3A,B,K,L). Zygotic expression of the PCNA lacZ became detectable at the gastrulating stage, although the signal was weak (Fig. 3C,M). In the early phase of germ band extension, strong staining of the PCNA was seen in regions corresponding to the cephalic furrow, the amnioproctodeal invagination, the anterior mid-gut primordium, and the invaginated mesoderm (Fig. 3D–F,N–P). This expression pattern is similar to the distribution of endogenous PCNA protein, hence the PCNA lacZ fusion constructs seem to reproduce expression of the endogenous PCNA gene. At later stages, anti-lacZ staining was observed throughout the germ band and also in the cephalic region, including the anterior midgut primordium, as well as in the posterior region, including the posterior midgut primordium and the proctodeal primordium (Fig. 3G–J,Q–T). No expression of PCNA lacZ was detected in the dorsal region, including the aminoserosa, throughout embryogenesis. No significant difference was observed between transgenic flies carrying the p5′-607DPCNAlacZW8HS and those carrying the p5′-168DPCNAlacZW8HS.
Because analyses by immunostaining are qualitative, quantitative measurements of β-galactosidase activity in the extracts of Drosophila bodies at various stages of development were made. Male flies from eight of 25 transformant lines carrying p5′-607DPCNAlacZW8HS and those from six out of 13 carrying p5′-168DPCNAlacZW8HS were crossed with wild-type females to observe the zygotic expression of the lacZ. Averaged values for β-galactosidase activities at various developmental stages are shown in Fig. 4. The expression of lacZ was high in embryos, first and second in-star larvae, and adult females and low in bodies of other stages of development. High expression of lacZ in the larva likely reflects the requirement of PCNA for the polytenization of larval tissues and cell proliferation in the imaginal discs. High expression of the lacZ in adult females represents the maternal contribution, as the expression was very low in adult males. Only a marginal difference was observed between transgenic flies carrying two different constructs of the transgene (Fig. 4). These results suggest that function of the homeodomain protein binding region located upstream of the PCNA gene is, if any, for the minor modulation of the promoter activity during normal developmental processes.
FIG. 4.
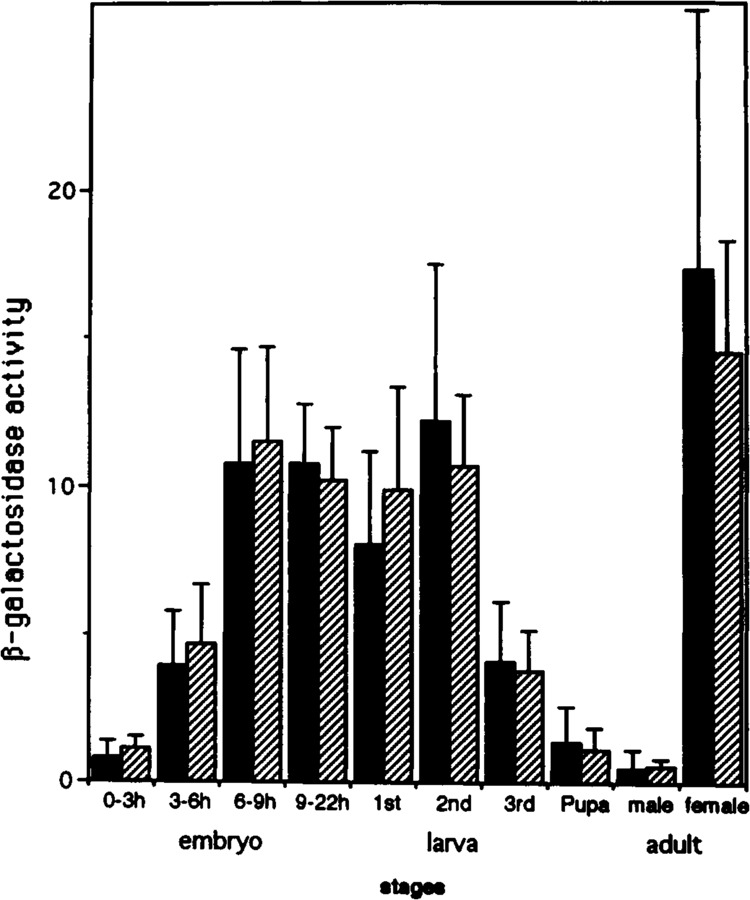
Expression of PCNA lacZ during development. Male transgenic flies were crossed with female wild-type flies and extracts were prepared from Drosophila bodies at various stages of development. The β-galactosidase activities in the extracts are expressed as absorbance units per h per mg protein. Black bars indicate the averaged value of eight independent transgenic strains carrying p5′-607DPCNAlacZW8HS. Hatched bars indicate the averaged value of six independent strains carrying the p5′-168DPCNAlacZW8HS. Deviation between independent strains is also indicated.
Expression Pattern of PCNA lacZ in Embryos Homozygousfor tor Mutation
In ongoing work, we observed the ectopic expression of PCNA lacZ in embryos homozygous for zen mutation. The zen, a homeobox gene, belongs to dorso-ventral genes. Therefore, we conducted genetic crossing experiments of transgenic flies with several other homeobox gene mutant flies, including Antp, Dfd, ftz, pb, Scr, and AbdB. No significant change in the PCNA lacZ expression pattern was observed with these crossings (data not shown).
We then extended the analysis with the tor mutant flies. The tor gene is a maternal effect gene and plays a key role in the formation of terminal structures of the embryo (Nüsslein-Volhard et al., 1987). The tor gene product acts within the embryo to induce the expression of zygotic genes required for the development of terminal structures. Loss-of-function tor mutations result in truncation of the anterior head skeleton, and structures posterior to the seventh abdominal segment do not form (Shupbach and Wieschaus, 1986). As no corresponding invagination is formed at the posterior end of the embryo, the pole cells are often left outside the embryo. The germ band does not extend along the dorsal side of the embryo; rather, it forms in abnormal folds at the ventral side of the embryo (Weigel et al., 1990). In contrast, embryos derived from the tor gain-of-function mutant mothers exhibit a pheno-type opposite that of embryos without tor activity (Klingler et al., 1988). The terminal parts of the anlagenplan are expanded at the expense of the central segmented region of the anlagenplan. The tor RL3 phenotype is caused by ectopic activation of the tor gene product at the central part of the embryos, which is normally restricted to the terminal regions (Casanova and Struhl, 1989). This is reflected by expression patterns of ftz, which is restricted to the center of the embryo (Klingler et al., 1988).
Male flies carrying the PCNA lacZ transgene were crossed with female flies homozygous for tor loss-of-function allele, tor PM, or tor gain-of-function allele, tor RL3. Although there was a slight repression of the lacZ expression at the posterior region of the tor PM embryos (data not shown), much more extensive change of the lacZ expression was observed with the tor RL3 embryos. As shown in Fig. 5, PCNA lacZ expression in the cephalic region, as well as in the posterior end region, was observed in tor RL3 embryos carrying either the p5′-607DPCNAlacZW8HS or the p5′-168DPCNAlacZW8HS. The PCNA lacZ expression in the region throughout the germ band was also detected with embryos carrying the p5′-168DPCNAlacZW8HS (Fig. 5E–H). In contrast, no expression in the central part of the presumptive mesodermal region was observed with embryos carrying the p5′-607DPCNAlacZW8HS (Fig. 5A–D). In light of these results, the upstream homeodomain protein binding region of the PCNA gene is probably responsible for repression of the PCNA lacZ expression in the central region of the tor RL3 embryos.
FIG. 5.
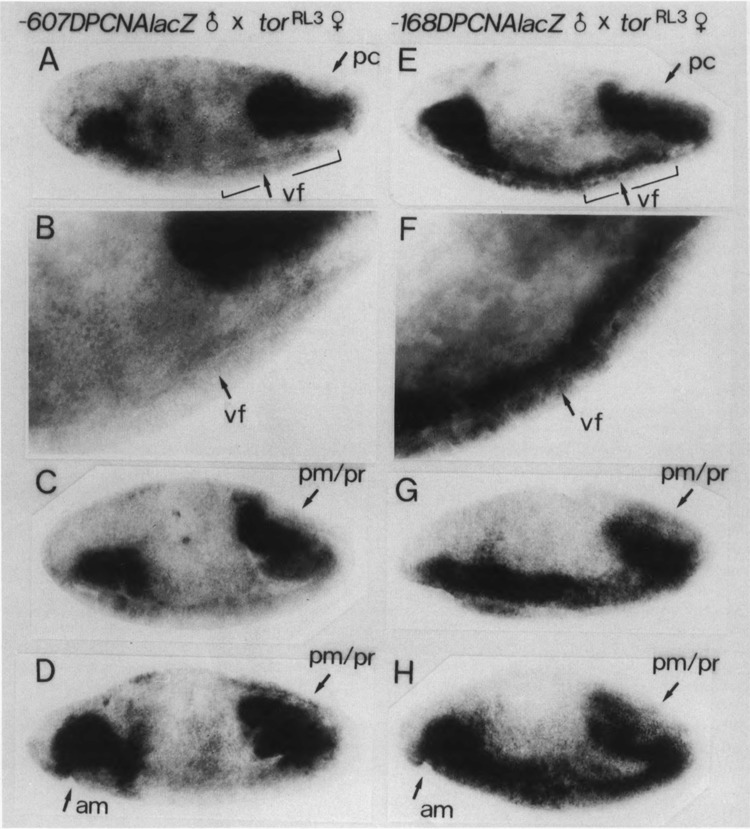
Expression of PCNA lacZ in tor RL3 mutant embryos. Embryos from p5′-607DPCNAlacZW8HS males × tor RL3 females (A–D) or p5′-168DPCNAlacZW8HS males × tor RL3 females (E–H), as indicated at the top of each panel, were immunostained with anti-lacZ antibody. Surface views of 3–6-h embryos are shown. Stages of the embryos shown in (D) and (H) are more advanced than those shown in (A–C) and (E–G). In (B) and (F), indicated parts of (A) and (E) are shown in higher magnifications; ventral furrow (vf), posterior midgut primordium (pm), proctodeum primordium (pr), anterior midgut primordium (am).
The expression pattern of endogenous PCNA gene in the tor RL3 embryos was also examined by immunostaining with the anti-PCNA antibody. As shown in Fig. 1 (panels C–F) and Fig. 6 (panels A–C), strong staining with the anti-PCNA antibody was observed in the region surrounding the ventral furrow of the wild-type embryo. In contrast, no or very little staining was observed in the central part of this region of the tor RL3 embryo (Fig. 6D–I). Therefore, torso is apparently involved in regulating both the transgene and the endogenous wild-type PCNA gene.
FIG. 6.
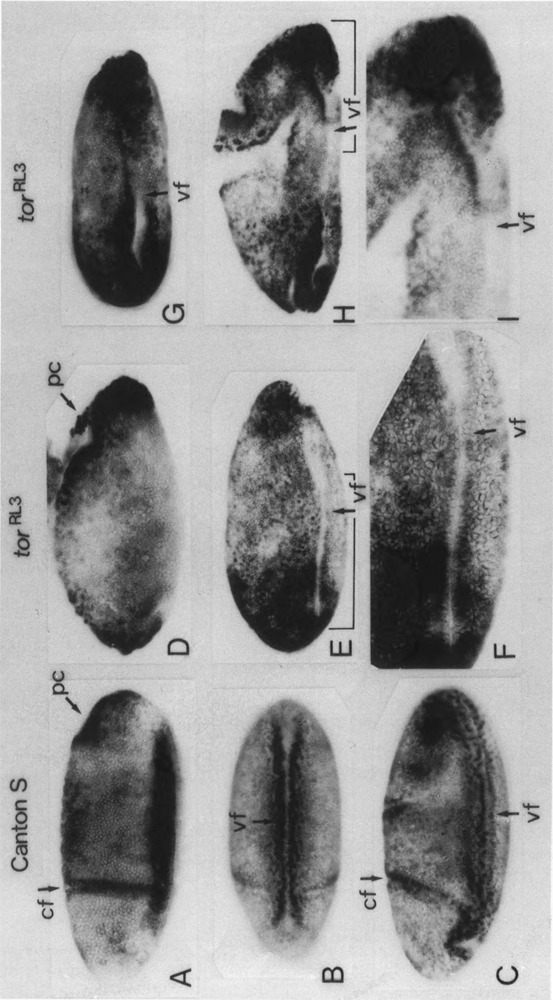
Distribution of PCNA protein in tor RL3 mutant embryos. Whole-mount specimens of fixed embryos were immunostained with an anti-PCNA antibody. Embryos are oriented so that anterior aspects are to the left and ventral areas are down [except (B) and (G)]. (A) Surface view of the early gastrulating wild-type (Canton S) embryo (stage 6, 1420–1500 h). (B) Ventral surface view of the early gastrulating wild-type embryo (stage 6). (C) Surface view of the wild-type embryo at initiation of germ band elongation (stage 7, 1500–1510 h). (D–I) Surface views of 2–6-h tor RL3 embryos. Ventral surface view is shown in (G). Stage of the embryo shown in (H) is more advanced than those in (D–G). In (F) and (I), indicated parts of (E) and (H) are shown in higher magnifications; cephalic fold (cf), pole cell (pc), ventral furrow (vf).
DISCUSSION
The binding consensus for homeodomain proteins that are located in the region from nucleotides −165 to −357 of the Drosophila PCNA gene is capable of binding homeodomain proteins in vitro (Yamaguchi et al., 1990). Interestingly, similar consensus sequences were found in the 5′-flanking regions of genes for the 180 kDa catalytic subunit (Hirose et al., 1991) and the 73 kDa sub-unit (Cotterill et al., 1992) of Drosophila DNA polymerase α. These observations suggest the possible involvement of homeodomain proteins in the regulation of Drosophila DNA replication-related genes. The present studies on transgenic flies revealed a minor role for the homeodomain protein binding region in the regulation of PCNA gene expression. Despite the minor role in the normal developmental process, the homeodomain protein binding region has the potential to regulate the PCNA gene expression extensively, because a significant difference in the lacZ expression pattern was observed between transgenic flies carrying the homeodomain protein binding region and those not carrying it, in the background of tor mutation.
Axton et al. (1994) have identified the plutonium (plu) gene that codes for the inhibitor of DNA replication; it locates in the 5′-flanking region of the PCNA gene. The plu gene is separated from the PCNA gene by less than 150 bp and the homeodomain protein binding region locates in the second intron and the 3′ exon of the plu. Thus, the homeodomain protein binding region may play a major role in regulating expression of the plu gene.
The tor gene is a maternal effect gene and its product is a key component for the formation of embryo terminal structures (Schupbach and Wieschaus, 1986; Klinger et al., 1988; Sprenger et al. 1989; Casanova and Struhl, 1989). Activation of the tor protein leads to phosphorylation of subordinate regulatory molecules, an event that controls the activation of multiple genes responsible for generating terminal structures. Several downstream genes such as Draf-1 (Nishida et al., 1988), tailless (Pignoni et al., 1990), huckebein (Weigel et al., 1990), and forkhead (Weigel et al., 1989) have been identified in this signal transduction system. We attempted genetic crossing experiments of transgenic flies with some of these downstream mutants, but no significant change of expression pattern of the PCNA lacZ was obtained. Therefore, global change of expressions of multiple genes as a result of mutation in a master gene like torso may be required to enhance the effect of the homeodomain protein binding region on PCNA promoter function. As none of the homeobox gene mutants so far examined affected the PCNA lacZ expression pattern, the involvement of multiple redundant genes in the regulation of the PCNA gene would need to be considered.
In the tor gain-of-function mutant, tor RL3 embryo, expression of PCNA lacZ in the central region of the embryo was lost, thereby suggesting that tor appears to negatively regulate PCNA gene expression. Unknown homeodomain proteins may be involved in the downstream of tor. The structure of the central region is not formed in this mutant; therefore, accurate regulation of DNA replication-related genes like PCNA by the tor-mediated signal transduction system is vital for formation of a normal body.
ACKNOWLEDGEMENTS
We are grateful to P. Fisher for providing the anti-Drosophila PCNA antibody; G. Struhl for the tor PM, tor RL3, AbdB M1, AbdB M5, and AbdB rx23-1 fly stocks; C. Nusslein-Volhard for the various homeobox mutant fly stocks; to C. Hama for the engrailed-lacZ fly stock; Y. Hiromi for the ftz-lacZ fly stock; and M. Ohara for assistance with the manuscript. This work was supported in part by grants-in-aid from the Ministry of Education, Science, and Culture, Japan.
REFERENCES
- Axton J. M., Shamanski F. L., Young L. M., Henderson D. S., Boyd J. B., and Orr-Weaver T. L. (1994), EMBO J 13, 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega J. A. and Hartenstein V. (1985), in The Embryonic Development of Drosophila melanogaster, Springer-Verlag, Berlin, pp. 210–217. [Google Scholar]
- Casanova J. and Struhl G. (1989), Genes Dev 3, 2025–2038. [DOI] [PubMed] [Google Scholar]
- Cotterill S., Lehman I. R., and McLachlan P. (1992), Nulceic Acids Res 20, 4325–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplan C., Theis J., and O’Farrell P. H. (1988), Cell 54, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell Y. C. and Searles L. L. (1992), Mol Cell Biol 12, 4571–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F., Yamaguchi M., and Matsukage A. (1994), J Biol Chem 269, 2937–2942. [PubMed] [Google Scholar]
- Hirose F., Yamaguchi M., Nishida Y., Masutani M., Miyazawa H., Hanaoka F., and Matsukage A. (1991), Nucleic Acids Res 19, 4991–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T. and Levine M. (1988), Nature 332, 858–861. [DOI] [PubMed] [Google Scholar]
- Jaskulski D., Deriel J. K., Mercer W. E., Calabretta B., and Baserga R. (1988), Science 240, 1544–1546. [DOI] [PubMed] [Google Scholar]
- Klinger M., Erdelyi M., Szabad J., and Nüsslein-Volhard C. (1988), Nature 335, 275–277. [DOI] [PubMed] [Google Scholar]
- Lindsley D. L. and Zimm G. G. (1992), in The Genome of Drosophila melanogaster, Academic Press, San Diego. [Google Scholar]
- Liu Y. C., Marraccino R. L., Keng P. C., Bambara R. A., Lord E. M., Chou W. G., and Zain S. B. (1989), Biochemistry 28, 2967–2974. [DOI] [PubMed] [Google Scholar]
- Moriuchi T. (1990), Med Sci Res 18, 911–915. [Google Scholar]
- Muller M., Affolter M., Leupin W., Otting G., Wuthrich K., and Gehring W. J. (1988), EMBO J 7, 4299–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L., Prelich G., Anderson C. W., Stillman B., and Fisher P. A. (1990), J Biol Chem 265, 11948–11954. [PubMed] [Google Scholar]
- Nishida Y., Hata M., Ayaki T., Ryo H., Yamagata M., Shimizu K., and Nishizuka Y. (1988), EMBO J 7, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Frohnhofer H. G., and Lehman R. (1987), Science 238, 1675–1681. [DOI] [PubMed] [Google Scholar]
- Pignoni F., Baldarelli R. M., Steingrimsson E., Diaz R. J., Patapoutain A., Merriam J. R., and Lengyel J. A. (1990), Cell 62, 151–163. [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., Phillis R. W., Johnson-Schlitz D. M., Benz W. K., and Engels W. R. (1988), Genetics 118, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbuch T. and Wieschaus E. (1986), Wilhelm Roux Arch Dev Biol 195, 302–317. [DOI] [PubMed] [Google Scholar]
- Shivji K. K., Kenny M. K., and Wood R. D. (1992), Cell 69, 367–374. [DOI] [PubMed] [Google Scholar]
- Sprenger F., Stevens L. M., and Nüsslein-Volhard C. (1989), Nature 338, 478–483. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., and Stillman B. (1990), Nature 346, 534–539. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jurgens G., Klingler M., and Jackle H. (1990), Science 248, 495–498. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jurgens G., Kuttner F., Seifert E., and Jackie H. (1989), Cell 57, 645–658. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Date T., and Matsukage A. (1991a), J. Cell Sci 100, 727–733. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Hirose F., Nishida Y., and Matsukage A. (1991b), Mol Cell Biol 11, 4909–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Nishida Y., Moriuchi T., Hirose F., Hui C.-c., Suzuki Y., and Matsukage A. (1990), Mol Cell Biol 10, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]


