Abstract
Background
The gene encoding Phosphatidylinositol-4-Phosphate 5-Kinase (PIP5K1C) has been recently implicated in pain regulation. Interestingly, a recent cross-tissue and cross-phenotypic epigenetic analysis identified the same gene in alcohol use disorder (AUD). Given the high comorbidity between AUD and chronic pain, we hypothesized that genetic variation in PIP5K1C might contribute to susceptibility to AUD.
Methods
We conducted a case-control association study of genetic variants in PIP5K1C. Association analyses of 16 common PIP5K1C single nucleotide polymorphisms (SNPs) were conducted in cases and controls of African (427 cases and 137 controls) and European Ancestry (488 cases and 324 controls) using standard methods. In addition, given the prominent role of the opioid system in pain signaling, we investigated the effects of acute alcohol exposure on PIP5K1C expression in humanized transgenic mice for the mu-opioid receptor that included the OPRM1 A118G polymorphism, a widely used mouse model to study analgesic response to opioids in pain. PIP5K1C expression was measured in the thalamus and basolateral amygdala (BLA) in mice after short-term administration (single 2g/kg dose) of alcohol or saline using immunohistochemistry and analyzed by two-way ANOVA.
Results
In the case-control association study using a NIAAA discovery sample, eight SNPs in PIP5K1C were significantly associated with AUD in the African ancestry group (p < 0.05 after correction; rs4807493, rs10405681, rs2074957, rs10432303, rs8109485, rs1476592, rs10419980, and rs4432372). However, a replication analysis using an independent sample (N= 3801) found no significant associations after correction for multiple testing. In the humanized transgenic mouse model with the OPRM1 polymorphism, PIP5K1C expression was significantly different between alcohol and saline-treated mice, regardless of genotype, in both the thalamus (p < 0.05) and BLA (p < 0.01).
Conclusions
Our discovery sample shows that genetic variants in PIP5K1C are associated with AUD in the African ancestry group, and acute alcohol exposure leads to upregulation of PIP5K1C, potentially explaining one mechanism underlying the increased risk for chronic pain conditions in individuals with AUD.
Keywords: PIP5K1C, genetics, pain, alcohol, translational research
Introduction
Alcohol use disorder (AUD) is characterized by excessive and chronic use of alcohol which leads to negative physiological, psychological, and societal consequences and contributes to nearly 4% of deaths worldwide (Stahre et al., 2014, Association, 2013, Rehm et al., 2014, Rehm et al., 2009). While the pathophysiology of AUD is not yet fully understood, there is strong evidence suggesting a genetic component in addition to environmental factors (Tawa et al., 2016). Twin studies have established that AUD heritability ranges from 40% to 70% (Enoch and Goldman, 2001, Agrawal and Lynskey, 2008, Kendler et al., 2012). Adoption studies have also suggested that developing alcoholism in adoptees is more associated with their biological parents than with their adoptive parents (Heath, 1995, Sigvardsson et al., 1996). Despite the strong evidence supporting genetic factors underlying AUD etiology, identifying universal genetic risk variants has been challenging (Tawa et al., 2016). Several factors contribute to this difficulty, including diverse clinical phenotypes, complex genetic architecture, and involvement of multiple genes with small effect sizes. In addition, the presence of rare and de novo genetic mutations and epigenetic effects further complicate the identification of genetic risk variants for AUD (Tawa et al., 2016, Robison and Nestler, 2011).
A recent cross-tissue and cross-phenotypic epigenetic analysis of genome-wide methylomic variation in AUD identified the Phosphatidylinositol-4-Phosphate 5-Kinase gene (PIP5K1C) as a top target associated with AUD (Lohoff et al., 2017). PIP5K1C is highly expressed in the brain (Wright et al., 2014) and regulates receptor-mediated calcium signaling, actin cytoskeleton dynamics, and synaptic vesicle trafficking (Di Paolo et al., 2004, Legate et al., 2012, Ling et al., 2002, Unoki et al., 2012, van den Bout and Divecha, 2009, Vasudevan et al., 2009, Volpicelli-Daley et al., 2010, Wang et al., 2004, Xu et al., 2010, Yu et al., 2011, Wang et al., 2007). Above all, PIP5K1C plays a vital role in production of the protein signaling substrate, Phosphatidylinositol 4,5-biphosphate (PIP2) (White et al., 2013, Di Paolo et al., 2004, Wright et al., 2014). PIP2 is found in dorsal root ganglia (DRG) neurons of mice, and, following hydrolysis, may initiate multiple pain signaling pathways. Due to its role in PIP2 production, PIP5K1C has been implicated as a critical regulator of nociceptive signaling and sensitization and may be a therapeutic target for chronic pain (Wright et al., 2014).
The potential role of epigenetic regulation of PIP5K1C by alcohol is intriguing given the high comorbidity between AUD and chronic pain (Katon et al., 1985, Egli et al., 2012, Zale et al., 2015, Apkarian et al., 2013). Previous research estimates that 32.6 million American adults currently meet criteria for an AUD and that 25.3 million American adults experience daily chronic pain (Grant et al., 2015, Nahin, 2015). Therefore, the overlap between individuals suffering from AUD and chronic pain is significant. In a study of treatment-seeking AUD patients, 73% individuals who identified alcohol as their drug of choice reported experiencing moderate-to-severe pain in the previous 30 days (Larson et al., 2007). Another study found that problem drinkers reported a higher prevalence of pain in the past 30 days compared to non-problem drinkers (Brennan et al., 2005). Additionally, preclinical research indicates that alcohol preference and pain covary in rodents genetically selected for alcohol consumption (Chester et al., 2002, Kampov-Polevoy et al., 1996, Kimpel et al., 2003). Despite the high comorbidity of AUD and chronic pain, the underlying neurobiology is poorly understood.
Several candidate genes have been suggested to affect systems involved in alcohol dependence and pain (Zubieta et al., 2003, Mobascher et al., 2010, Tiihonen et al., 1999, Wang et al., 2001, Bardin et al., 2009, Grande et al., 2004, Nilsen et al., 2007, Scarinci et al., 1994, Van Houdenhove and Luyten, 2006). One of the most studied genetic targets is the µ-opioid receptor encoded by the µ-opioid receptor 1 gene (OPRM1). The µ-opioid receptor regulates analgesic response to pain and also mitigates the rewarding effects of many drugs (Crist and Berrettini, 2014). Furthermore, naltrexone, a FDA approved drug antagonizig µ-opioid receptor, reduced alcohol self-administration in animal models and alcohol craving and relapse in clinical populations experiencing AUD (Altshuler et al., 1980, Myers et al., 1986, Volpicelli et al., 1986, Volpicelli et al., 1992, O’Malley et al., 1992). Naltrexsone prescribed at a low dose has been used for chronic pain treatment (Hota et al., 2016). Specifically, OPRM1 A118G is the most common and studied non-synonymous single nucleotide polymorphism (SNP) with a global minor allele, G, frequency of 19% (Crist and Berrettini, 2014). In preclinical studies, humanized transgenic mouse models for the 118A>G SNP in OPRM1 gene have been widely used to investigate effects of the genetic variation on sensitivity to pain, analgesic effect of opioids, and alcohol addiction (Mura et al., 2013). The A118G genetic variant in human affects pain sensitivity and is a major determinant of striatal dopamine response to alcohol (Fillingim et al., 2005, Lotsch et al., 2006, Vossen et al., 2010, Crist and Berrettini, 2014, Bonenberger et al., 2015, Ramchandani et al., 2011). Therefore, using the mouse models, it may be possible to explain a complicated mechanism by which alcohol may work as an analgesic although chronic heavy drinking is often linked to chronic pain.
To investigate the role of PIP5K1C in alcohol dependence, we conducted a case-control association study, including discovery and replication cohorts. In addition, we used a humanized OPRM1 A118G transgenic mouse model to explore the interaction of alcohol, OPRM1 genetic variation, and PIP5K1C expression.
Materials/Subjects and Methods:
Genetic Association Analysis of PIP5K1C in a Clinical Population
915 individuals with AUD and 461 healthy controls were recruited to the National Institute on Alcohol Abuse and Alcoholism (NIAAA) at the National Institutes of Health in Bethesda, Maryland, USA. A total of 812 individuals were of European ancestry (EA) (488 AUD cases, 324 controls) and a total of 564 individuals were of African ancestry (AA) (427 AUD cases, 137 controls). Alcohol dependence was diagnosed using the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID) (First, 2007). Subjects were diagnosed with DSM-IV alcohol dependence (AD), but we used AUD throughout the manuscript for clarity.
16 common PIP5K1C single nucleotide polymorphisms (SNPs) were extracted from genotype data generated using the Illumina Human OmniExpress-12 and Illumina Human OmniExpressExome v1.2 arrays (Illumina Inc., San Diego, CA). After confirming Hardy-Weinberg Equilibrium, exploratory χ2 tests were performed using PLINK version 1.07 (Purcell et al., 2007). The Benjamini-Hochberg procedure for false discovery rate was used to correct for multiple comparisons.
In addition, linear regression models were used to explore a variety of alcohol-related phenotypes. These phenotypes were severity of alcohol dependence (as measured by the Alcohol Dependence Severity (ADS) Score) (Skinner and Allen, 1982), heavy drinking days in the previous 90 days (as measured by the Timeline Follow Back (TLFB)) (Sobell, 1992), average drinks per day in the previous 90 days (as measured by the TLFB), Clinical Institute Withdrawal Assessment-Alcohol revised (CIWA-AR) score (Sullivan et al., 1989), alcohol craving (as measured by the Penn Alcohol Craving Scale) (Flannery et al., 1999), severity of nicotine dependence (as measured by the Fagerstrӧm Test of Nicotine Dependence (FTND)) (Heatherton et al., 1991), depressive symptoms (as measured by Comprehensive Psychopathological Rating Scale (CPRS)) (Mattila-Evenden et al., 1996), anxiety (as measured by the CPRS) (Svanborg and Asberg, 1994), neuroticism (as measured by the Neuroticism-Extraversion-Openness Personality Inventory) (Costa and McCrae, 1992), and trait anxiety (as measured by the State-Trait Anxiety Inventory) (Spielberger, 2010). Age, gender, and ancestry informative markers (AIM) for African and European Ancestry were used as covariates.
Haplotype analyses were conducted using both PLINK and HaploView software (Barrett et al., 2005) with haplotype blocks defined using the default D’/LOD method in Haploview. Haplotype association tests for alcohol dependence were conducted in PLINK and were corrected for multiple comparisons using permutation tests (10,000 permutations).
Exploratory χ2 tests, using the same methods as described above, were replicated using data from the Study of Addiction: Genetics and Environment (SAGE) database. SAGE is a component of the Gene Environment Association Studies initiative (GENEVA), a genome-wide association study of common genetic variants that may be related to addiction (dbGaP accession phs000092.v1.p1). Of the 16 SNPs used in the NIAAA sample, 15 (all but rs12984273) were available in the SAGE database. Whole genotype genotyping was completed using the Illumina ILMN-Human-1 platform (Illumina Inc., San Diego, CA). A total of 2,635 individuals were of EA (1244 AUD cases, 1391 controls) and a total of 1,166 individuals were of AA (671 AUD cases, 495 controls).
DNA methylation levels were generated on Illumina Human Methylation 450 microarrays in a subset of individuals as published previously (Lohoff et al., 2017). PIP5K1C promoter probe cg17845617, previously associated with AUD in brain and blood in our previous work (Lohoff et al., 2017) was assessed for association to genotype data with general linear models.
PIP5K1C Expression in OPRM1 Mouse Model
We used a humanized mouse line in which the μ-opioid receptor (OPRM1) was replaced with either the major 118A allele or 118G SNP from human exon 1 of OPRM1. These mice were bred at the NIAAA animal facility. Mice heterozygous for the respective human allele were bred to homozygosity to establish the h/mOPRM1–118AA and h/mOPRM1–118GG lines. Methods used to create this humanized mouse lines were previously described elsewhere (Ramchandani et al., 2011). Of the twelve knock-in mice and 12 wild-type mice, half were treated with a single dose of 2g/kg ethanol and half were treated with a single dose of saline.
Briefly, 30 minutes after ethanol or saline injection, the animals were deeply anesthetized with pentobarbital sodium (50 mg/kg, i.p.) before being transcardially perfused with 50 ml of ice-cold saline (0.9% NaCl) and 150 ml of 4% paraformaldehyde (PFA). Following perfusion, brains were removed, post-fixed in 4% PFA for 2 hours, dehydrated in a 30% sucrose solution for 48 hours, snap frozen in powdered dry ice, and stored at −80°C until further use. Coronal sections (40 µm) of the regions of interest were taken by a cryostat (Leica CM3050), collected in cryoprotectant solution and stored at −20°C.
The PIP5K1C staining protocol was adapted from the studies by Wright et al. (Wright et al., 2014). Briefly, free-floating sections were washed with 0.1 M PBS and then incubated for 15 min with 3% H2O2 in 0.1 M PBS. Tissue was then pre-blocked in 0.1 M PBS containing 0.2% Triton X-100 and 5% normal goat serum for 30 min. Sections were incubated in the primary antibody rabbit anti-PIP5K1C (VECTASTAIN Elite ABC HRP Kit (Peroxidase, Rabbit IgG), PK-6101, Vector Laboratories, Burlingame, CA) for two days at 4°C on a shaker. Negative controls were performed using goat serum 5% without primary antibody. Following incubation, sections were rinsed with 1× PBS (3 times, 5 minutes) and incubated with biotinylated secondary antibody (1:100, goat anti-rabbit vector ABC Elite Kit, PK-6101, Vector Laboratories, Burlingame, CA) for 30 minutes.
After rinsing, the color reaction for light microscopy was performed using first the avidin-biotin peroxidase solution (Vector ABC Elite Kit, PK-6101, Vector Laboratories, Burlingame, CA) and then adding the 3 3’-diaminobenzidine tetrahydrochloride (DAB) (D5905, Sigma, St. Louis, MO) according to the manufacturer’s instructions, resulting in a brown color staining. Sections were then rinsed with PBS (2 times, 15 min), mounted on gelatin-coated slides, and processed the next day through alcohol-xylene for light microscopy examination.
Pictures were taken on a Leica light microscope DM6000CS (Leica Microsystem Inc., Bannockbern, IL, USA) at 20X or 40X magnification and BioQuant imaging software (R&M Biometrics, Nashville, TN) was used for cell density analysis. Two coronal sections (two levels), containing the thalamus and basolateral amygdala (BLA), both left and right side, were analyzed for PIP5K1C positive cells stained as dark brown color for two regions (coordinates: BLA – Bregma: −1.06 to −1.94 mm and thalamus – Bregma: −1.22 to −1.94 mm._ENREF_41(Paxinos et al., 2001) Results are depicted as the positively stained cells per square mm. Blue spots were also observed due to unwanted damage to tissues, which we excluded for the analysis.
Data for PIP5K1C positive cells are reported as mean values ± standard error as number per square millimeter. The data from each experiment was analyzed by a student’s t-test. For all statistical analyses, differences between control and experimental groups were considered significant if p < 0.05.
Results
Genetic Association Analysis of PIP5K1C in a Clinical Population
PIP5K1C Genetic Analyses in Discovery Data
Demographic characteristics of the whole NIAAA clinical sample of 915 alcohol-dependent individuals and 461 healthy controls are shown in Supplementary Table 1 (Top). The sample was separated based on ancestry; demographics for a total of 812 individuals of European ancestry (488 AUD cases, 324 controls) and 564 individuals of African ancestry (AA; 427 AUD cases, 137 controls) are displayed in Supplementary Table 1 (Top). No alcohol-related phenotypes, tested using linear regression, were significantly associated with a diagnosis of AUD in either the EA or AA groups. Years of education, drinking days in the last 90 days, heavy drinking days in the last 90 days, ADS score, and FTND score differed significantly (p< 0.0001) between cases and controls in both the EA group and the AA group (Supplementary Table 1, Top).
The genotype information for the 16 PIP5K1C SNPs is shown in Table 1. Given large differences in AIM Europe and AIM Africa scores, we conducted analyses separately and used age, gender, and AIM for African and European Ancestry as covariates. Hardy–Weinberg equilibrium (HWE) was not significantly deviated in the poplution (data not shown). The single marker association analyses revealed that 1 SNP (rs12460780) was significantly associated with a diagnosis of AUD in the EA group (Table 1, Top). However, this association did not hold for multiple corrections. In the AA sample, 9 SNPs (rs4807493, rs10405681, rs2074957, rs10432303, rs757454, rs8109485, rs1476592, rs10419980, and rs4432372) were indicated as significantly associated with a diagnosis of AUD (Table 1, Bottom). Of these 9 SNPs, 8 remained significantly associated with AUD after correction for multiple comparisons (rs4807493, rs10405681, rs2074957, rs10432303, rs8109485, rs1476592, rs10419980, and rs4432372) (Table 1, Bottom).
Table 1.
Results from single marker association analyses in NIAAA discovery sample (Cases n= 915, Controls n= 461).
| European Ancestry | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||
| SNP | BP | Minor Allele | Minor Allele Freq. (Case) | Minor Allele Freq. (Control) | χ2 | p-value | Odds Ratio | p-value (after correction) | |||||||||||
| rs4807493 | 3631036 | A | 0.2023 | 0.1687 | 2.85 | 0.09139 | 1.249 | 0.2041 | |||||||||||
| rs3746124 | 3632595 | A | 0.05943 | 0.07562 | 1.659 | 0.1978 | 0.7724 | 0.2877 | |||||||||||
| rs10405681 | 3634019 | G | 0.2111 | 0.1775 | 2.772 | 0.09594 | 1.24 | 0.2041 | |||||||||||
| rs1004323 | 3643093 | G | 0.3924 | 0.3457 | 3.635 | 0.05659 | 1.223 | 0.2041 | |||||||||||
| rs12460780 | 3645947 | A | 0.4775 | 0.5356 | 5.257 | 0.02186* | 0.7922 | 0.2041 | |||||||||||
| rs2270083 | 3646642 | G | 0.06865 | 0.08824 | 2.112 | 0.1461 | 0.7616 | 0.2598 | |||||||||||
| rs12984273 | 3647901 | G | 0.4447 | 0.4032 | 2.674 | 0.102 | 1.185 | 0.2041 | |||||||||||
| rs2074957 | 3653525 | G | 0.4652 | 0.4164 | 3.74 | 0.05312 | 1.219 | 0.2041 | |||||||||||
| rs10432303 | 3656652 | A | 0.4641 | 0.4969 | 1.679 | 0.1951 | 0.8767 | 0.2877 | |||||||||||
| rs757454 | 3660619 | A | 0.4631 | 0.418 | 3.209 | 0.07322 | 1.201 | 0.2041 | |||||||||||
| rs8109485 | 3664094 | G | 0.2033 | 0.1867 | 0.6752 | 0.4113 | 1.111 | 0.5062 | |||||||||||
| rs11672559 | 3676507 | A | 0.07526 | 0.07944 | 0.09513 | 0.7578 | 0.9431 | 0.8083 | |||||||||||
| rs1476592 | 3679740 | G | 0.1951 | 0.1776 | 0.7761 | 0.3784 | 1.122 | 0.5045 | |||||||||||
| rs10419980 | 3685116 | A | 0.2582 | 0.2477 | 0.2281 | 0.633 | 1.057 | 0.7234 | |||||||||||
| rs740873 | 3698333 | A | 0.05236 | 0.0743 | 3.259 | 0.07103 | 0.6884 | 0.2041 | |||||||||||
| rs4432372 | 3698842 | G | 0.3432 | 0.3467 | 0.02122 | 0.8842 | 0.9846 | 0.8842 | |||||||||||
| African Ancestry | |||||||||||||||||||
|
| |||||||||||||||||||
| SNP | BP | Minor Allele | Minor Allele Freq. (Case) | Minor Allele Freq. (Control) | χ2 | p-value | Odds Ratio | p-value (after correction) | |||||||||||
| rs4807493 | 3631036 | A | 0.06206 | 0.1058 | 5.898 | 0.01516* | 0.559 | 0.03455* | |||||||||||
| rs3746124 | 3632595 | A | 0.2082 | 0.2226 | 0.2572 | 0.6121 | 0.9183 | 0.6121 | |||||||||||
| rs10405681 | 3634019 | G | 0.07629 | 0.1241 | 5.906 | 0.01509* | 0.583 | 0.03455* | |||||||||||
| rs1004323 | 3643093 | G | 0.216 | 0.2701 | 3.437 | 0.06376 | 0.7445 | 0.102 | |||||||||||
| rs12460780 | 3645947 | A | 0.5035 | 0.4489 | 2.475 | 0.1157 | 1.245 | 0.1683 | |||||||||||
| rs2270083 | 3646642 | G | 0.1185 | 0.146 | 1.425 | 0.2326 | 0.7868 | 0.2862 | |||||||||||
| rs12984273 | 3647901 | G | 0.4798 | 0.5296 | 2.03 | 0.1542 | 0.8192 | 0.2056 | |||||||||||
| rs2074957 | 3653525 | A | 0.4894 | 0.4051 | 5.913 | 0.01503* | 1.408 | 0.03455* | |||||||||||
| rs10432303 | 3656652 | A | 0.3392 | 0.2482 | 7.928 | 0.004867** | 1.555 | 0.02596* | |||||||||||
| rs757454 | 3660619 | A | 0.4778 | 0.5474 | 4.031 | 0.04468* | 0.7562 | 0.07942 | |||||||||||
| rs8109485 | 3664094 | G | 0.2676 | 0.365 | 9.519 | 0.002033** | 0.6358 | 0.01627* | |||||||||||
| rs11672559 | 3676507 | A | 0.1851 | 0.1642 | 0.6132 | 0.4336 | 1.156 | 0.4955 | |||||||||||
| rs1476592 | 3679740 | G | 0.3016 | 0.4088 | 10.8 | 0.001016** | 0.6248 | 0.01626* | |||||||||||
| rs10419980 | 3685116 | A | 0.408 | 0.4963 | 6.554 | 0.01047* | 0.6995 | 0.03455* | |||||||||||
| rs740873 | 3698333 | A | 0.1635 | 0.1825 | 0.5321 | 0.4657 | 0.8758 | 0.4968 | |||||||||||
| rs4432372 | 3698842 | A | 0.3047 | 0.2299 | 5.668 | 0.01727* | 1.468 | 0.03455* | |||||||||||
p < 0.05,
p < 0.01,
p < 0.001
Analysis controls for age, gender, and Africa and Europe AIM scores
We previously identified higher DNA methylation levels in AUD subjects at a PIP5K1C promoter CpG, cg17845617 (Lohoff et al., 2017). Using a subset of N=70 controls, we identified significant associations after Bonferroni correction for multiple comparisons of rs4807493, rs8109485, rs1476592, and rs10419980 as methylation quantitative trait loci (mQTLs) from among the above significant SNPs (Table 2).
Table 2.
Association analysis of significantly associated PIP5K1C variants to DNA methylation at cg17845617 in PIP5K1C.
| SNP | R squared | F stat | DF |
A/G
β value |
error | p value |
Bonferroni P value |
G/G β value |
error | p value |
Bonferroni
P value |
| rs8109485 | 0.26 | 11.42 | 3_66 | -0.027 | 0.008 | 8E-04 | 0.007 | 0.049 | 0.018 | 0.009 | 0.069 |
| rs1476592 | 0.25 | 10.89 | 3_66 | -0.025 | 0.008 | 0.001 | 0.01 | 0.047 | 0.018 | 0.011 | 0.089 |
| rs10419980 | 0.19 | 7.61 | 3_66 | -0.054 | 0.017 | 0.002 | 0.014 | -0.03 | 0.017 | 0.059 | 0.476 |
| rs4807493 | 0.14 | 5.24 | 3_66 | -0.076 | 0.024 | 0.002 | 0.017 | -0.07 | 0.023 | 0.006 | 0.052 |
| rs2074957 | 0.089 | 3.20 | 3_66 | -0.023 | 0.009 | 0.016 | 0.126 | -0.02 | 0.012 | 0.094 | 0.754 |
| rs10405681 | 0.13 | 5.08 | 3_66 | ##### | 0.008 | 0.269 | 1 | 0.066 | 0.023 | 0.006 | 0.05 |
| rs10432303 | 0.021 | 0.69 | 3_66 | -0.014 | 0.013 | 0.273 | 1 | -0.01 | 0.013 | 0.281 | 1 |
| rs4432372 | 0.020 | 0.68 | 3_66 | -0.010 | 0.01 | 0.315 | 1 | -0 | 0.012 | 0.944 | 1 |
Linkage Disequilibrium Structure in Discovery Data
To confirm our single marker association finding, haplotype analyses were run separately for EA and AA samples. Four main haplotype blocks were found based on the linkage disequilibrium (LD) plots for the 16 SNPs that span the PIP5K1C gene. Haplotype structure analyses revealed that the LD plots for the EA and AA groups differed, indicating different haplotype structures in AA and EA populations (Supplementary Figure 1). Haplotype structures of the two subgroups are shown with D’ (Supplementary Figure 1 a,c) and R2 (Supplementary Figure 1 b,d) values.
Haplotype-Based Association in Discovery Data
Haplotype-based association tests were also run separated by ancestry to look for associations between haplotypes and AUD (Supplementary Table 2). In the EA sample, the block 1 haplotype GGAG (frequency= 0.372, p= 0.0606) and the block 4 haplotype AG (frequency=0.0609, p= 0.0593) were nearly significantly associated with AUD. The haplotype AG contains alleles from 1 SNP (rs12460780) found to be significant in the single SNP association prior to accounting for multiple comparisons. However, similar to the single SNP association test, the significance of the AG haplotype did not withstand correction for multiple comparisons.
In the AA sample, the block 2 haplotype AAG (frequency= 0.319) was significantly associated with AUD (p< 0.01) (Supplementary Table 3). This result is consistent with the single marker association test. That is, the haplotype AAG contains alleles from 3 SNPs (rs2074957, rs10432303, rs757454) that were found to be significantly associated with AUD. Furthermore, the block 3 haplotype GG (frequency= 0.291) was significantly associated with AUD (p< 0.01) in the AA sample. This result is again consistent with results found in the single marker analyses as the haplotype GG contains alleles from 2 SNPs (rs8109485, rs11672559) that were found to be significantly associated with AUD.
PIP5K1C Genetic Analyses in Replication Data
Demographic characteristics for the SAGE sample, in which 2,635 individuals were of EA (1244 AUD cases, 1391 controls) and 1,166 individuals of AA ancestry (671 AUD cases, 495 controls) are shown in Supplementary Table 1 (Bottom). No alcohol-related phenotypes, tested with linear regression, were significantly associated with a diagnosis of AUD in either the EA or AA samples (Supplementary Table 1, Bottom).
The genotype information for the 15 PIP5K1C SNPs are shown in Table 3. Given the results in our discovery analysis, we conducted analyses separated by ancestry. The single marker association analyses did not reveal significant associations between the 15 SNPs and AUD diagnosis in either the EA or AA samples (Table 3).
Table 3.
Results from single marker association analyses in SAGE replication sample.
| European Ancestry | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||||||||||||||
| SNP | BP | Minor Allele | Minor Allele Freq. (Case) | Minor Allele Freq. (Control) | χ2 | p-value | Odds Ratio | p-value (after correction) | ||||||||||||||||||||||
| rs4807493 | 3582036 | A | 0.1969 | 0.2129 | 2.061 | 0.1512 | 0.9064 | 0.6727 | ||||||||||||||||||||||
| rs3746124 | 3583595 | A | 0.05189 | 0.05288 | 0.02575 | 0.8725 | 0.9803 | 0.9464 | ||||||||||||||||||||||
| rs10405681 | 3585019 | C (G) | 0.2022 | 0.2173 | 1.803 | 0.1794 | 0.9129 | 0.6727 | ||||||||||||||||||||||
| rs1004323 | 3594093 | G | 0.3797 | 0.3986 | 1.953 | 0.1622 | 0.9238 | 0.6727 | ||||||||||||||||||||||
| rs12460780 | 3596947 | A | 0.5 | 0.4802 | 2.058 | 0.1514 | 1.083 | 0.6727 | ||||||||||||||||||||||
| rs2270083 | 3597642 | G | 0.08011 | 0.07458 | 0.5612 | 0.4538 | 1.081 | 0.8508 | ||||||||||||||||||||||
| rs2074957 | 3604525 | C (G) | 0.4451 | 0.456 | 0.6255 | 0.429 | 0.957 | 0.8508 | ||||||||||||||||||||||
| rs10432303 | 3607652 | A | 0.4731 | 0.4665 | 0.2275 | 0.6334 | 1.027 | 0.9464 | ||||||||||||||||||||||
| rs757454 | 3611619 | A | 0.4433 | 0.4543 | 0.6367 | 0.4249 | 0.9567 | 0.8508 | ||||||||||||||||||||||
| rs8109485 | 3615094 | G | 0.201 | 0.1979 | 0.07672 | 0.7818 | 1.019 | 0.9464 | ||||||||||||||||||||||
| rs11672559 | 3627507 | T (A) | 0.06436 | 0.06619 | 0.07172 | 0.7889 | 0.9705 | 0.9464 | ||||||||||||||||||||||
| rs1476592 | 3630740 | G | 0.1841 | 0.1824 | 0.02564 | 0.8728 | 1.011 | 0.9464 | ||||||||||||||||||||||
| rs10419980 | 3636116 | T (A) | 0.2468 | 0.2468 | 9.50E-07 | 0.9992 | 1.000 | 0.9992 | ||||||||||||||||||||||
| rs740873 | 3649333 | T (A) | 0.06356 | 0.05804 | 0.7005 | 0.4026 | 1.102 | 0.8508 | ||||||||||||||||||||||
| rs4432372 | 3649842 | G | 0.3368 | 0.3387 | 0.02154 | 0.8833 | 0.9915 | 0.9464 | ||||||||||||||||||||||
| African Ancestry | ||||||||||||||||||||||||||||||
|
| ||||||||||||||||||||||||||||||
| SNP | BP | Minor Allele | Minor Allele Freq. (Case) | Minor Allele Freq. (Control) | χ2 | p-value | Odds Ratio | p-value (after correction) | ||||||||||||||||||||||
| rs4807493 | 3582036 | A | 0.06408 | 0.07692 | 1.45 | 0.2285 | 0.8217 | 0.927 | ||||||||||||||||||||||
| rs3746124 | 3583595 | A | 0.2228 | 0.2247 | 0.01177 | 0.9136 | 0.9892 | 0.927 | ||||||||||||||||||||||
| rs10405681 | 3585019 | C (G) | 0.07836 | 0.09919 | 3.101 | 0.07826 | 0.7721 | 0.927 | ||||||||||||||||||||||
| rs1004323 | 3594093 | G | 0.2187 | 0.2237 | 0.08355 | 0.7725 | 0.9712 | 0.927 | ||||||||||||||||||||||
| rs12460780 | 3596947 | G | 0.494 | 0.5 | 0.08108 | 0.7758 | 0.9764 | 0.927 | ||||||||||||||||||||||
| rs2270083 | 3597642 | G | 0.1442 | 0.1557 | 0.5875 | 0.4434 | 0.9138 | 0.927 | ||||||||||||||||||||||
| rs2074957 | 3604525 | T (A) | 0.4671 | 0.4615 | 0.07297 | 0.7871 | 1.023 | 0.927 | ||||||||||||||||||||||
| rs10432303 | 3607652 | A | 0.3048 | 0.2864 | 0.9159 | 0.3386 | 1.092 | 0.927 | ||||||||||||||||||||||
| rs757454 | 3611619 | A | 0.4888 | 0.4949 | 0.08519 | 0.7704 | 0.9758 | 0.927 | ||||||||||||||||||||||
| rs8109485 | 3615094 | G | 0.2914 | 0.2931 | 0.008392 | 0.927 | 0.9916 | 0.927 | ||||||||||||||||||||||
| rs11672559 | 3627507 | T (A) | 0.1677 | 0.1791 | 0.5262 | 0.4682 | 0.9229 | 0.927 | ||||||||||||||||||||||
| rs1476592 | 3630740 | G | 0.3321 | 0.3067 | 1.684 | 0.1944 | 1.124 | 0.927 | ||||||||||||||||||||||
| rs10419980 | 3636116 | T (A) | 0.4222 | 0.4249 | 0.01814 | 0.8929 | 0.9886 | 0.927 | ||||||||||||||||||||||
| rs740873 | 3649333 | T (A) | 0.1729 | 0.1846 | 5.33E-01 | 0.4654 | 0.9233 | 0.927 | ||||||||||||||||||||||
| rs4432372 | 3649842 | A | 0.2761 | 0.2682 | 0.179 | 0.6723 | 1.041 | 0.927 | ||||||||||||||||||||||
PIP5K1C Expression in OPRM1 Mouse Model
To confirm whether alcohol stimulates PIP5K1C expression and genetic variants of μ-opioid receptor (OPRM1) differentiate the expression, we injected a single dose of 2g/kg of ethanol to OPRM1 humanized mouse lines with either the major 118A or minor 118G allele SNP, h/mOPRM1–118AA and h/mOPRM1–118GG mice, and performed immunohistochemistry to examine PIP5K1C expression. The 118A>G SNP in OPRM1 gene leads to altered pain threshold and analgesic effect because of changes in the opioid receptor expression and signaling cascades, which may affect PIP5K1C expression by alcohol (Mura et al., 2013, Walter et al., 2013).
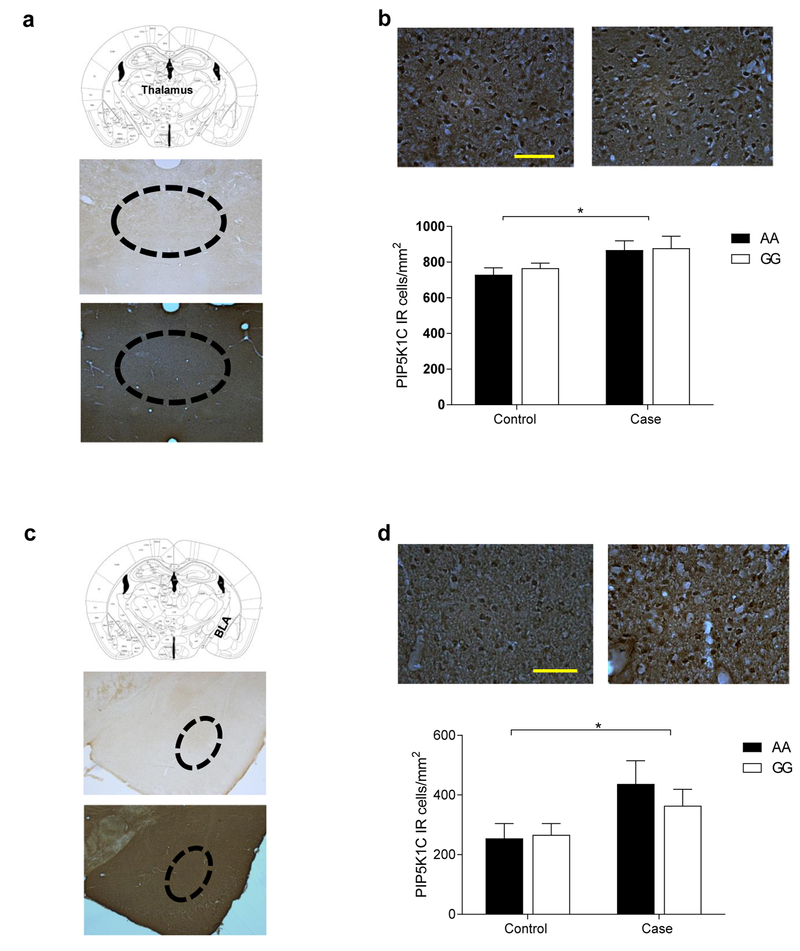
In mice acutely treated with ethanol, there was significantly higher PIP5K1C expression in both the thalamus and basolateral amygdala (BLA) (p< 0.05) than in saline-treated mice (Figure 1). However, interaction was not observed between genotype and group in neither the thalamus nor the BLA. Thus, there were no significant differences in PIP5K1C expression between AA and GG mice in either the alcohol or saline-treated mice in both brain regions (Figure 1 b,d).
Figure 1. Treatment with ethanol induced a significant increase in PIP5K1C expression in thalamus and basolateral amygdala (BLA).
Treatment with ethanol induced a significant increase in PIP5K1C expression. Mice were homozygous for either the h/mOPRM1–118AA (n= 12) or h/mOPRM1–118GG (n= 12). Half of each group were either treated with a single dose of 2g/kg of ethanol or a single dose of saline. See details in the methods under PIP5K1C Expression in OPRM1 Mouse Model. (a) Schematic picture and location of thalamus with negative control and positive control for immunohistochemistry (IHC) experiment. (b) Alcohol-induced PIP5K1C was evaluated in mice using IHC DAB staining. Treatment with one dose of ethanol (2g/kg) induced significantly higher PIP5K1C expression. Data represent mean number of PIP5K1C immunoreactive cells per square millimeter from two sections per brain, left and right parts of the Thalamus. The cells were counted under ×20 magnification and the scale bar represents 100 μm, and the results are presented as mean ± S.E.M.*p< 0.05. (c) Schematic picture and location of the BLA with negative and positive controls on lower magnification (5x, the scale bar represents 500 μm). (d) Data represent the mean number of PIP5K1C immunoreactive cells per square millimeter from two sections per brain. Left and right side of the BLA were counted. Cells were counted under ×40 magnification and the scale bar represents 100 μm. Results are presented as mean ± S.E.M Treatment with a single dose of ethanol (2g/kg) had significantly higher PIP5K1C expression. *p< 0.05.
Discussion
In this study, we explored, for the first time, the association between PIP5K1C variants and AUD. We found in our discovery clinical data an association between common PIP5K1C variants and AUD in individuals of African American descent. The haplotype analysis confirmed the results of our initial single SNP association and indicated that SNPs within the PIP5K1C gene were associated with a diagnosis of AUD in the AA group. The two most significant (p< 0.01) SNPs that withstood correction for multiple comparisons and were confirmed by the haplotype analysis were rs10432303 and rs8109485. Both SNPs are intronic variants that are near a repeating element. However, no association between AUD and PIP5K1C variants was observed in the EA sample. It is possible that other ethnicity-specific genetic background differences could have an effect on PIP5K1C variants, and thus an association between AUD and PIP5K1C expression was only observable in the AA sample.
Our findings in our discovery clinical dataset were not observed in our larger replication dataset. Thus, we did not conduct a haplotype analysis in the replication sample. One potential confounding factor in our study might have been a spurious association due to population stratification. However, we carefully controlled for this potential effect using AIM scores. Furthermore, clinical heterogeneity may have confounded the results, as some cases had other substance dependence, such as cocaine, cannabis or opioid, as well as other psychiatric comorbidities: Different individuals had different types of drug use or psychiatric disorders, so comorbidities were not consistent across participants. Another limitation was that in both our discovery and replication samples, the case and control groups were not matched exactly regarding age and gender. In the NIAAA discovery sample, the majority of both cases and controls in the AA and EA groups were male. However, in the SAGE replication sample, while the majority of cases in the AA and EA groups were male, the majority of the controls of both subgroups were females. Consequently, we statistically controlled for gender, in addition to age, in our analyses. Further research with case and control groups matched for age and gender is recommended.
Given the recent report of possible epigenetic regulation of PIP5K1C in AUD (Lohoff et al., 2017), we performed a cursory analysis in a small subset of control individuals to assess the potential for the identified genotypes to act as mQTLs. Not surprisingly, four out of the eight SNPs associated with AUD after correction for multiple testing were significantly associated with PIP5K1C promoter DNA methylation, suggesting a portion of alcohol-related epigenetic associations identified previously may be affected by genetic variation. Interestingly, according to the GTEx database (GTExConsortium, 2015), all of the SNPs associated with PIP5K1C methylation were also markedly associated with expression in different types of tissues (data not shown here but available on the GTEx Porter, http://gtexportal.org). Only rs10419980 was, however, associated with brain expression, supporting the potential biological relevance of these variants. In all cases, homozygotes and heterozygotes were associated with lower PIP5K1C DNA methylation. In light of our previous observations of AUD-associated DNA methylation increases, these findings are consistent with a protective effect of the ancestral allele. Furthermore, these findings are consistent with the observations in the European American sample where the ORs for the minor allele are greater than 1 as opposed to those in the African American sample. This data further suggests that controlling for underlying genetic architecture in epigenetic studies is critical to interpreting the results independent of confounds potentially associated with different ancestry. In the future, additional studies in larger cohorts are needed to evaluate to what degree epigenetic variation in combination with genetic variation could contribute to susceptibility to AUD and pain.
Although the comorbidity between AUD and chronic pain has been well documented, a strong genetic risk variant has not yet been identified (Katon et al., 1985, Egli et al., 2012, Zale et al., 2015, Apkarian et al., 2013). Research indicates that the OPRM1 A118G polymorphism is implicated in both pain and AUD, but evidence supporting its role as a genetic link underlying chronic pain and AUD comorbidity is not strong. Given the role of PIP5K1C in pain signaling, we used the transgenic humanized mouse line in which the OPRM1 gene was replaced with the major 118A human allele of OPRM1. The OPRM1 A118G is understood to play a role in modulating pain and alcohol, but the underlying neurobiology is not yet fully understood. In order to create sample homogeneity and prevent possible confounding factors, we used mice that were bred to homozygosity to establish the h/mOPRM1–118AA line. The results from our mouse model provide evidence of a relationship between ethanol treatment and PIP5K1C expression in both the BLA and thalamus. No interaction was observed between genotype and treatment group, indicating that the OPRM1 A118G polymorphism is not involved in mediating PIP5K1C expression. Inhibition of respective receptors mediating the pain response has not produced enough analgesic effect on chronic pain. It is likely that other convergent signaling pathways beyond the receptor level play an important role in explaining the underlying pain mechanism in AUD (Gold and Gebhart, 2010). Alternatively, the A118G genotype (rs1799971) in OPRM1 gene by itself may not be closely associated with addictions. When cis-expression QTL SNPs were analyzed for association with heroin addiction, four SNPs (rs9478495, rs3778150, rs9384169, and rs562859) in OPRM1 intron 1 were significantly associated. Notably, haplotype analyses revealed that A allele of the functional getetic variant (rs1799971) was significantly correlated with the addiction only when rs3778150-C was present (Hancock et al., 2015). In a genome-wide association study, rs73568641, located at upstream of OPRM1 gene, was highly associated with a therapeutic methadone dose, a selective μ-opioid receptor agonist for treatment of opiod dependence (OD), in OD subjects (Smith et al., 2017). Therefore, other unknown genetic variants in OPRM1 gene or a neighboring gene may participate in PIP5K1C expression by alcohol.
Mice shortly treated with ethanol exhibited significantly higher PIP5K1C expression than control mice treated with saline. Our results are particularly interesting because our case group was treated with a single injection of 2g/kg of ethanol exhibited significantly higher PIP5K1C expression than control mice treated with saline. This may indicate that acute alcohol exposure affects PIP5K1C expression. Our study employed an acute injection of ethanol as an exploratory measure to determine whether an interaction between alcohol, PIP5K1C, and OPRM1 expression was possible. This was based on results from the original study piloting the use of the h/mOPRM1–118AA line which indicated that a single injection of alcohol was associated with substantial dopamine release (Ramchandani et al., 2011). Future research should investigate the effect of chronic ethanol exposure on targeted PIP5K1C expression. Additionally, our analysis was limited to thalamus and BLA given the well-established role of the thalamus in nociception signaling and that of the BLA in cue-induced alcohol seeking (Head and Holmes, 1911, Yen and Lu, 2013, Apkarian, 2013, Jones, 2007). Future research could expand this analysis to other brain regions relative to pain signaling such as cingulate cortex. Other limitations of our study were that we did not perform behavior tests in our mouse model and that we could not analyze a pain index in clinical populations based on PIP5K1C SNPs due to lack of pain history data. Including a behavioral task measuring pain sensitivity by PIP5K1C and alcohol or histories of pain by the genotypes in AUD would lend to a more complete understanding of the interaction of alcohol, pain and PIP5K1C expression.
To the best of our knowledge, using a translational research approach, our data is the first to reveal the relationship between AUD and PIP5K1C genetic variants specifically in the AA population from the discovery sample.Thus, our study implicates PIP5K1C as an interesting new target for the investigation of the genetic link underlying comorbid AUD and chronic pain.
Supplementary Material
Acknowledgements
This research was supported by the National Institutes of Health (NIH) intramural funding ZIA-AA000242 (Section on Clinical Genomics and Experimental Therapeutics; to FWL), Division of Intramural Clinical and Biological Research of the National Institute on Alcohol Abuse and Alcoholism (NIAAA).
We thank Erick Singley (Office of the Clinical Director, NIAAA) and Monte J. Phillips (Section on Clinical Genomics and Experimental Therapeutics, NIAAA) for technical support.
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
References
- Agrawal A, Lynskey MT (2008) Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction 103:1069–1081. [DOI] [PubMed] [Google Scholar]
- Altshuler HL, Phillips PE, Feinhandler DA (1980) Alteration of ethanol self-administration by naltrexone. Life Sciences 26:679–688. [DOI] [PubMed] [Google Scholar]
- Apkarian AV, Bushnell, M C., & Schweinhardt P. (2013) Representation of pain in the brain, in in Wall and Melzack’s Textbook of Pain (6th ed.), in Wall and Melzack’s Textbook of Pain (6th ed.), pp 111–128, Elsevier, Toronto [Google Scholar]
- Apkarian AV, Neugebauer V, Koob G, Edwards S, Levine JD, Ferrari L, Egli M, Regunathan S (2013) Neural mechanisms of pain and alcohol dependence. Pharmacol Biochem Behav 112:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association A (2013) Diagnotic and Statistical Manual of Mental Disorders, American Psychiatric Association, Washington, DC. [Google Scholar]
- Bardin L, Malfetes N, Newman-Tancredi A, Depoortere R (2009) Chronic restraint stress induces mechanical and cold allodynia, and enhances inflammatory pain in rat: Relevance to human stress-associated painful pathologies. Behav. Brain Res. 205:360–366. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- Bonenberger M, Plener PL, Groschwitz RC, Gron G, Abler B (2015) Polymorphism in the micro-opioid receptor gene (OPRM1) modulates neural processing of physical pain, social rejection and error processing. Exp. Brain Res. 233:2517–2526. [DOI] [PubMed] [Google Scholar]
- Brennan PL, Schutte KK, Moos RH (2005) Pain and use of alcohol to manage pain: prevalence and 3-year outcomes among older problem and non-problem drinkers. Addiction 100:777–786. [DOI] [PubMed] [Google Scholar]
- Chester JA, Price CS, Froehlich JC (2002) Inverse genetic association between alcohol preference and severity of alcohol withdrawal in two sets of rat lines selected for the same phenotype. Alcohol Clin Exp Res 26:19–27. [PubMed] [Google Scholar]
- Costa PT, McCrae RR (1992) Neo Personality Inventory-Revised (NEO PI-R), Psychological Assessment Resources; Odessa, FL. [Google Scholar]
- Crist RC, Berrettini WH (2014) Pharmacogenetics of OPRM1. Pharmacol Biochem Behav 123:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, Moskowitz HS, Gipson K, Wenk MR, Voronov S, Obayashi M, Flavell R, Fitzsimonds RM, Ryan TA, De Camilli P (2004) Impaired PtdIns(4,5)P2 synthesis in nerve terminals produces defects in synaptic vesicle trafficking. Nature 431:415–422. [DOI] [PubMed] [Google Scholar]
- Egli M, Koob GF, Edwards S (2012) Alcohol dependence as a chronic pain disorder. Neurosci Biobehav Rev 36:2179–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Goldman D (2001) The genetics of alcoholism and alcohol abuse. Curr Psychiatry Rep 3:144–151. [DOI] [PubMed] [Google Scholar]
- Fillingim RB, Kaplan L, Staud R, Ness TJ, Glover TL, Campbell CM, Mogil JS, Wallace MR (2005) The A118G single nucleotide polymorphism of the mu-opioid receptor gene (OPRM1) is associated with pressure pain sensitivity in humans. J. Pain 6:159–167. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer Robert L., Gibbon Miriam, and Williams Janet B.W. (2007) Structured Clinical Interview for DSM-IV-TR Axis I Disorders - Non-patient Edition (SCID-I/NP, 1/2007 revision), in Series Structured Clinical Interview for DSM-IV-TR Axis I Disorders - Non-patient Edition (SCID-I/NP, 1 /2007 revision), Biometrics Research Department New York State Psychiatric Institute 1051 Riverside Drive - Unit 60 New York, New York: 10032 [Google Scholar]
- Flannery BA, Volpicelli JR, Pettinati HM (1999) Psychometric properties of the Penn Alcohol Craving Scale. Alcohol. Clin. Exp. Res. 23:1289–1295. [PubMed] [Google Scholar]
- Gold MS, Gebhart GF (2010) Nociceptor sensitization in pain pathogenesis. Nature medicine 16:1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande LA, Loeser JD, Ozuna J, Ashleigh A, Samii A (2004) Complex regional pain syndrome as a stress response. Pain 110:495–498. [DOI] [PubMed] [Google Scholar]
- Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, Pickering RP, Ruan WJ, Smith SM, Huang B, Hasin DS (2015) Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III . JAMA psychiatry 72:757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTExConsortium (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Levy JL, Gaddis NC, Glasheen C, Saccone NL, Page GP, Hulse GK, Wildenauer D, Kelty EA, Schwab SG, Degenhardt L, Martin NG, Montgomery GW, Attia J, Holliday EG, McEvoy M, Scott RJ, Bierut LJ, Nelson EC, Kral AH, Johnson EO (2015) Cis-Expression Quantitative Trait Loci Mapping Reveals Replicable Associations with Heroin Addiction in OPRM1. Biological psychiatry 78:474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head H, Holmes G (1911) SENSORY DISTURBANCES FROM CEREBRAL LESIONS1. Brain 34:102–254. [Google Scholar]
- Heath AC (1995) Genetic influences on alcoholism risk: a review of adoption and twin studies. Alcohol Research and Health 19:166. [PMC free article] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO (1991) The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br. J. Addict. 86:1119–1127. [DOI] [PubMed] [Google Scholar]
- Hota D, *Department of Pharmacology AIIoMS, Bhubaneswar, India, Srinivasan A, Pharmacology Do, Dutta P, Department of Endocrinology PIoMEaR, Chandigarh, India, Bhansali A, Department of Endocrinology PIoMEaR, Chandigarh, India, Chakrabart A, Pharmacology Do (2016) Off-Label, Low-Dose Naltrexone for Refractory Painful Diabetic Neuropathy. Pain Medicine 17:790–791. [Google Scholar]
- Jones EG (2007) The thalamus (2nd ed.), Cambridge University Press, Cambridge, UK [Google Scholar]
- Kampov-Polevoy AB, Kasheffskaya OP, Overstreet DH, Rezvani AH, Viglinskaya IV, Badistov BA, Seredenin SB, Halikas JA, Sinclair JD (1996) Pain sensitivity and saccharin intake in alcohol-preferring and -nonpreferring rat strains. Physiol Behav 59:683–688. [DOI] [PubMed] [Google Scholar]
- Katon W, Egan K, Miller D (1985) Chronic pain: lifetime psychiatric diagnoses and family history. Am J Psychiatry 142:1156–1160. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Chen X, Dick D, Maes H, Gillespie N, Neale MC, Riley B (2012) Recent advances in the genetic epidemiology and molecular genetics of substance use disorders. Nat Neurosci 15:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Brown MM, Froehlich JC (2003) Pain thresholds in alcohol preferring and non-preferring rats: diurnal and repeated trial line differences. Alcohol Clin Exp Res 27:1921–1928. [DOI] [PubMed] [Google Scholar]
- Larson MJ, Paasche-Orlow M, Cheng DM, Lloyd-Travaglini C, Saitz R, Samet JH (2007) Persistent pain is associated with substance use after detoxification: a prospective cohort analysis. Addiction 102:752–760. [DOI] [PubMed] [Google Scholar]
- Legate KR, Montag D, Bottcher RT, Takahashi S, Fassler R (2012) Comparative phenotypic analysis of the two major splice isoforms of phosphatidylinositol phosphate kinase type Igamma in vivo. J Cell Sci 125:5636–5646. [DOI] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA (2002) Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420:89–93. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Sorcher JL, Rosen AD, Mauro KL, Fanelli RR, Momenan R, Hodgkinson CA, Vendruscolo LF, Koob GF, Schwandt M, George DT, Jones IS, Holmes A, Zhou Z, Xu M-J, Gao B, Sun H, Phillips MJ, Muench C, Kaminsky ZA (2017) Methylomic profiling and replication implicates deregulation of PCSK9 in alcohol use disorder. Molecular psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotsch J, Stuck B, Hummel T (2006) The human mu-opioid receptor gene polymorphism 118A > G decreases cortical activation in response to specific nociceptive stimulation. Behav. Neurosci. 120:1218–1224. [DOI] [PubMed] [Google Scholar]
- Mattila-Evenden M, Svanborg P, Gustavsson P, Asberg M (1996) Determinants of self-rating and expert rating concordance in psychiatric out-patients, using the affective subscales of the CPRS. Acta Psychiatr. Scand. 94:386–396. [DOI] [PubMed] [Google Scholar]
- Mobascher A, Brinkmeyer J, Thiele H, Toliat MR, Steffens M, Warbrick T, Musso F, Wittsack HJ, Saleh A, Schnitzler A, Winterer G (2010) The val158met polymorphism of human catechol-O-methyltransferase (COMT) affects anterior cingulate cortex activation in response to painful laser stimulation. Mol. Pain 6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura E, Govoni S, Racchi M, Carossa V, Ranzani GN, Allegri M, van Schaik RH (2013) Consequences of the 118A>G polymorphism in the OPRM1 gene: translation from bench to bedside?, in J Pain Res, Vol. 6, J Pain Res, pp 331–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RD, Borg S, Mossberg R (1986) Antagonism by naltrexone of voluntary alcohol selection in the chronically drinking macaque monkey. Alcohol 3:383–388. [DOI] [PubMed] [Google Scholar]
- Nahin RL (2015) Estimates of pain prevalence and severity in adults: United States, 2012. The journal of pain : official journal of the American Pain Society 16:769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen KB, Sand T, Westgaard RH, Stovner LJ, White LR, Bang Leistad R, Helde G, Ro M (2007) Autonomic activation and pain in response to low-grade mental stress in fibromyalgia and shoulder/neck pain patients. Eur. J. Pain 11:743–755. [DOI] [PubMed] [Google Scholar]
- O’Malley SS, Jaffe AJ, Chang G, Schottenfeld RS, Meyer RE, Rounsaville B (1992) Naltrexone and Coping Skills Therapy for Alcohol Dependence: A Controlled Study. Archives of general psychiatry 49:881–887. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ, Franklin KBJ (2001) The mouse brain in stereotaxic coordinates. 2nd ed., Academic Press, San Diego. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandani VA, Umhau J, Pavon FJ, Ruiz-Velasco V, Margas W, Sun H, Damadzic R, Eskay R, Schoor M, Thorsell A, Schwandt ML, Sommer WH, George DT, Parsons LH, Herscovitch P, Hommer D, Heilig M (2011) A genetic determinant of the striatal dopamine response to alcohol in men. Molecular psychiatry 16:809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm J, Dawson D, Frick U, Gmel G, Roerecke M, Shield KD, Grant B (2014) Burden of disease associated with alcohol use disorders in the United States. Alcohol Clin Exp Res 38:1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J (2009) Alcohol and Global Health 1 Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet 373:2223–2233. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ (2011) Transcriptional and Epigenetic Mechanisms of Addiction. Nat Rev Neurosci 12:623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarinci IC, McDonald-Haile J, Bradley LA, Richter JE (1994) Altered pain perception and psychosocial features among women with gastrointestinal disorders and history of abuse: a preliminary model. Am. J. Med. 97:108–118. [DOI] [PubMed] [Google Scholar]
- Sigvardsson S, Bohman M, Cloninger CR (1996) Replication of the Stockholm Adoption Study of alcoholism. Confirmatory cross-fostering analysis. Archives of general psychiatry 53:681–687. [DOI] [PubMed] [Google Scholar]
- Skinner HA, Allen BA (1982) Alcohol dependence syndrome: measurement and validation. J. Abnorm. Psychol. 91:199–209. [DOI] [PubMed] [Google Scholar]
- Smith AH, Jensen KP, Li J, Nunez Y, Farrer LA, Hakonarson H, Cook-Sather SD, Kranzler HR, Gelernter J (2017) Genome-wide association study of therapeutic opioid dosing identifies a novel locus upstream of OPRM1. Molecular psychiatry 22:346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell LC, Sobell MB (1992) Timeline Follow-back: A technique for assessing self-reported ethanol consumption, in Measuring Alcohol Consumption: Psychosocial and Biochemical Methods, Measuring Alcohol Consumption: Psychosocial and Biochemical Methods (RAYE Z. LITTEN JPA ed, Humana Press, Totowa, NJ. [Google Scholar]
- Spielberger CD (2010) State‐Trait anxiety inventory, Wiley Online Library. [Google Scholar]
- Stahre M, Roeber J, Kanny D, Brewer RD, Zhang X (2014) Contribution of excessive alcohol consumption to deaths and years of potential life lost in the United States. Preventing chronic disease 11:E109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Sykora K, Schneiderman J, Naranjo CA, Sellers EM (1989) Assessment of alcohol withdrawal: the revised clinical institute withdrawal assessment for alcohol scale (CIWA-Ar). Br. J. Addict. 84:1353–1357. [DOI] [PubMed] [Google Scholar]
- Svanborg P, Asberg M (1994) A new self-rating scale for depression and anxiety states based on the Comprehensive Psychopathological Rating Scale. Acta psychiatrica Scandinavica 89:21–28. [DOI] [PubMed] [Google Scholar]
- Tawa EA, Hall SD, Lohoff FW (2016) Overview of the Genetics of Alcohol Use Disorder. Alcohol Alcohol 51:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiihonen J, Hallikainen T, Lachman H, Saito T, Volavka J, Kauhanen J, Salonen JT, Ryynanen OP, Koulu M, Karvonen MK, Pohjalainen T, Syvalahti E, Hietala J (1999) Association between the functional variant of the catechol-O-methyltransferase (COMT) gene and type 1 alcoholism. Mol. Psychiatry 4:286–289. [DOI] [PubMed] [Google Scholar]
- Unoki T, Matsuda S, Kakegawa W, Van NT, Kohda K, Suzuki A, Funakoshi Y, Hasegawa H, Yuzaki M, Kanaho Y (2012) NMDA receptor-mediated PIP5K activation to produce PI(4,5)P(2) is essential for AMPA receptor endocytosis during LTD. Neuron 73:135–148. [DOI] [PubMed] [Google Scholar]
- van den Bout I, Divecha N (2009) PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci 122:3837–3850. [DOI] [PubMed] [Google Scholar]
- Van Houdenhove B, Luyten P (2006) Stress, depression and fibromyalgia. Acta Neurol. Belg. 106:149–156. [PubMed] [Google Scholar]
- Vasudevan L, Jeromin A, Volpicelli-Daley L, De Camilli P, Holowka D, Baird B (2009) The beta- and gamma-isoforms of type I PIP5K regulate distinct stages of Ca2+ signaling in mast cells. J Cell Sci 122:2567–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley LA, Lucast L, Gong LW, Liu L, Sasaki J, Sasaki T, Abrams CS, Kanaho Y, De Camilli P (2010) Phosphatidylinositol-4-phosphate 5-kinases and phosphatidylinositol 4,5-bisphosphate synthesis in the brain. J Biol Chem 285:28708–28714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli JR, Alterman AI, Hayashida M, O’Brien CP (1992) Naltrexone in the Treatment of Alcohol Dependence. Archives of general psychiatry 49:876–880. [DOI] [PubMed] [Google Scholar]
- Volpicelli JR, Davis MA, Olgin JE (1986) Naltrexone blocks the post-shock increase of ethanol consumption. Life Sciences 38:841–847. [DOI] [PubMed] [Google Scholar]
- Vossen H, Kenis G, Rutten B, van Os J, Hermens H, Lousberg R (2010) The genetic influence on the cortical processing of experimental pain and the moderating effect of pain status. PLoS One 5:e13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter C, Doehring A, Oertel BG, Lotsch J (2013) micro-opioid receptor gene variant OPRM1 118 A>G: a summary of its molecular and clinical consequences for pain. Pharmacogenomics 14:1915–1925. [DOI] [PubMed] [Google Scholar]
- Wang T, Franke P, Neidt H, Cichon S, Knapp M, Lichtermann D, Maier W, Propping P, Nothen MM (2001) Association study of the low-activity allele of catechol-O-methyltransferase and alcoholism using a family-based approach. Mol. Psychiatry 6:109–111. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lian L, Golden JA, Morrisey EE, Abrams CS (2007) PIP5KI gamma is required for cardiovascular and neuronal development. Proc Natl Acad Sci U S A 104:11748–11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Li WH, Wang J, Xu K, Dong P, Luo X, Yin HL (2004) Critical role of PIP5KI{gamma}87 in InsP3-mediated Ca(2+) signaling. J Cell Biol 167:1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC, Sanger Institute Mouse Genetics P, Tannahill D, Logan DW, Macarthur DG, Flint J, Mahajan VB, Tsang SH, Smyth I, Watt FM, Skarnes WC, Dougan G, Adams DJ, Ramirez-Solis R, Bradley A, Steel KP (2013) Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BD, Loo L, Street SE, Ma A, Taylor-Blake B, Stashko MA, Jin J, Janzen WP, Frye SV, Zylka MJ (2014) The lipid kinase PIP5K1C regulates pain signaling and sensitization. Neuron 82:836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Wang P, Petri B, Zhang Y, Tang W, Sun L, Kress H, Mann T, Shi Y, Kubes P, Wu D (2010) Integrin-induced PIP5K1C kinase polarization regulates neutrophil polarization, directionality, and in vivo infiltration. Immunity 33:340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CT, Lu PL (2013) Thalamus and pain . Acta anaesthesiologica Taiwanica : official journal of the Taiwan Society of Anesthesiologists 51:73–80. [DOI] [PubMed] [Google Scholar]
- Yu YL, Chou RH, Chen LT, Shyu WC, Hsieh SC, Wu CS, Zeng HJ, Yeh SP, Yang DM, Hung SC, Hung MC (2011) EZH2 regulates neuronal differentiation of mesenchymal stem cells through PIP5K1C-dependent calcium signaling. J Biol Chem 286:9657–9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zale EL, Maisto SA, Ditre JW (2015) Interrelations between pain and alcohol: An integrative review. Clin Psychol Rev 37:57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta JK, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y, Koeppe RA, Stohler CS, Goldman D (2003) COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science 299:1240–1243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.