Abstract
Background
Many transcription factors (TFs), such as those in the basic helix-loop-helix (bHLH) family, are important for regulating plant growth and plant responses to abiotic stress. The expression of OsbHLH035 is induced by drought and salinity. However, its functional role in rice growth, development, and the salt response is still unknown.
Results
The bHLH TF OsbHLH035 is a salt-induced gene that is primarily expressed in germinating seeds and seedlings. Stable expression of GFP-fused OsbHLH035 in rice transgenic plants revealed that this protein is predominantly localized to the nucleus. Osbhlh035 mutants show delayed seed germination, particularly under salt-stress conditions. In parallel, abscisic acid (ABA) contents are over-accumulated, and the expression of the ABA biosynthetic genes OsABA2 and OsAAO3 is upregulated; furthermore, compared with that in wild-type (WT) seedlings, the salt-induced expression of OsABA8ox1, an ABA catabolic gene, in germinating Osbhlh035 mutant seeds is downregulated. Moreover, Osbhlh035 mutant seedlings are unable to recover from salt-stress treatment. Consistently, sodium is over-accumulated in aerial tissues but slightly reduced in terrestrial tissues from Osbhlh035 seedlings after salt treatment. Additionally, the expression of the sodium transporters OsHKT1;3 and 1;5 is reduced in Osbhlh035 aerial and terrestrial tissues, respectively. Furthermore, genetic complementation can restore both the delayed seed germination and the impaired recovery of salt-treated Osbhlh035 seedlings to normal growth.
Conclusion
OsbHLH035 mediates seed germination and seedling recovery after salt stress relief through the ABA-dependent and ABA-independent activation of OsHKT pathways, respectively.
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0244-z) contains supplementary material, which is available to authorized users.
Keywords: ABA, bHLH, OsHKT, Salt stress, Transcription factor
Background
Salt stress severely affects plant growth and development. In fact, more than 20% of the land used for agriculture is of poor quality due to soil salinity (FAO, http://www.fao.org/home/en/). The adverse effects of salt stress on plant growth and development can be attributed to initial osmotic dehydration and subsequent ion toxicity. Osmotic stress signaling is known to be transduced via both ABA-dependent (e.g., mitogen-activated protein kinases [MAPKs]) and ABA-independent (e.g., via the OsDREB1 and OsDREB2 TFs) pathways (Kumar et al. 2013). Additionally, the maintenance of cellular ion homeostasis is important for plant survival under salt stress, especially in response to high cytosolic K+/Na+ ratios. In fact, many transporters, including HKTs (high-affinity potassium transporters), NHXs (Na+/H+ exchangers), and SOS (salt overly sensitive), act collectively to regulate intracellular sodium levels (Deinlein et al. 2014). The regulation of HKTs expression is critical for mediating salt stress tolerance in crops (Hamamoto et al. 2015). HKT transporters can be divided into two subfamilies. Subfamily I HKT genes (HKT1s) encode Na+-selective transporters, which are mostly located at the plasma membrane in xylem parenchyma cells. The functional role of HKT1s is responsible for retrieving Na+ from the xylem sap, which prevent aerial tissues from Na+ over-accumulation and toxicity. Subfamily II HKT transporter (HKT2) is involved in uptake of Na+ from the external medium (soil) when K+ is limiting. Therefore, when plants are exposed to salt, the phytohormone ABA, TFs, and transporters act as key components that integrate cellular signal transduction and stress-responsive gene expression to confer salt tolerance.
ABA plays a critical role in regulating seed dormancy, germination, and stress tolerance (Karssen et al. 1983; Koornneef et al. 1989; Groot and Karssen 1992; Qin and Zeevaart 2002; Finkelstein et al. 2008). To date, most genes related to ABA metabolism have been cloned and characterized (for review, see Taylor et al. 2000; Seo and Koshiba 2002; Nambara and Marion-Poll 2005; Ye et al. 2012). The first committed step of ABA biosynthesis involves the oxidative cleavage of a C40 9-cis-epoxycarotenoid, such as 9’-cis-neoxanthin or 9-cis-violaxanthin, to a C15 xanthoxin (the primary precursor) and a C25 by-product, which is catalyzed in plastids by 9-cis-epoxycarotenoid dioxygenase (NCED) (Schwartz et al. 2003; Xiong and Zhu 2003). At least 5 NCED genes have been documented in Arabidopsis, and 3 have been reported in rice (Tan et al. 2003; Welsch et al. 2008). Of the five AtNCEDs (AtNCED2, 3, 5, 6, and 9), only AtNCED3 is significantly induced by drought stress, and it controls the levels of endogenous ABA under drought-stress conditions (Iuchi et al. 2001). In rice, the expression of OsNCED3, OsNCED4, and OsNCED5 was induced after 1 h of salt stress and closely correlated with the endogenous ABA contents in roots (Welsch et al. 2008). ABA biosynthesis subsequently involves the conversion of xanthoxin to abscisic aldehyde (an intermediate), and the final steps of ABA production are catalyzed in the cytosol by AtABA2 and AtAAO3 in Arabidopsis (Seo et al. 2000; Rook et al. 2001; Cheng et al. 2002; Gonzalez-Guzman et al. 2002). For full catalytic activity, AtAAO3 requires a molybdenum cofactor, which is derived from AtABA3 function (Bittner et al. 2001; Xiong et al. 2001). Indeed, loss-of-function mutations in AtABA2, AtAAO3, and AtABA3 lead to ABA deficiency and early germination. Cellular ABA homeostasis is regulated by both the biosynthesis and degradation of ABA. ABA catabolism occurs via two main types of chemical reactions, hydroxylation and conjugation, with the hydroxylation of ABA at position C-8’ being the key regulatory step (Kushiro et al. 2004; Saito et al. 2004; Nambara and Marion-Poll 2005). Arabidopsis and rice possess 4 and 3 known ABA8ox genes, respectively (Kushiro et al. 2004; Saito et al. 2004; Saika et al. 2007). The four AtABA8ox genes, also known as CYP707As (CYP707A1, 2, 3, and 4), display complex spatiotemporal expression patterns and are important for many physiological processes. For example, CYP707A1 and CYP707A2 but not CYP707A3 or CYP707A4 are responsible for relieving the ABA-mediated inhibition of seed germination (Okamoto et al. 2006). These CYP707As are induced by osmotic stress presumably because the maintenance of endogenous ABA homeostasis under stress is necessary for plant survival (Saito et al. 2004). Although the biochemical and physiological roles of these ABA metabolic genes have been well characterized and documented, the mechanisms of their transcriptional regulation are still poorly understood.
Deciphering the roles of TFs is essential for revealing abiotic stress tolerance mechanisms in plants (Singh et al. 2002). In fact, several different types of TFs have been demonstrated to be involved in regulating abiotic stress responses (for review, see Nakashima et al. 2009; Todaka et al. 2012). For example, AtbHLH116/ICE1-CBF3/DREB1A [AtERF#031 (At4g25480)] acts in the cold-responsive signaling pathway via an ABA-independent pathway (Chinnusamy et al. 2003); ABI3, 4, and 5 (B3, AP2/ERF, and bZIP, respectively) act in the osmotic stress response pathway via an ABA-dependent pathway (Giraudat et al. 1992; Finkelstein et al. 1998; Finkelstein and Lynch 2000); and SUB1A (AP2/ERF) acts in submergence- and drought-responsive pathways (Fukao et al. 2011). Additionally, many TF types, including bHLH (Zhou et al. 2009; Li et al. 2010), WRKY (Tao et al. 2011), NAC (Hu et al. 2006), and AP2/ERF (Schmidt et al. 2013), are involved in the regulation of salt stress tolerance in rice. Approximately 7% of plant genes encode TFs. The Arabidopsis and rice (Oryza sativa subsp. japonica) genomes contain 2016 and 2424 TFs, respectively, that can be categorized into 58 and 56 families based on their DNA binding domains (Zhang et al. 2011). Basic helix-loop-helix (bHLH) TFs comprise the second largest TF family in plants and can affect many developmental and physiological processes (Feller et al. 2011). To date, at least 167 Arabidopsis and 177 rice bHLHs have been identified (Li et al. 2006; Carretero-Paulet et al. 2010). These genes are classified based on the proteins they encode, which usually contain a conserved bHLH domain (approximately 60 amino acids long), with the exception of a few atypical bHLH genes known as HLHs that lack the basic region (Li et al. 2006). A typical bHLH domain comprises two functionally distinct regions: a basic region for DNA binding and an HLH region for protein homodimerization or heterodimerization (Massari and Murre 2000; Toledo-Ortiz et al. 2003; Feller et al. 2011). Thus, HLH proteins can heterodimerize with bHLH proteins, thereby disrupting bHLH-bHLH interactions and preventing DNA binding (Sun et al. 1991; Toledo-Ortiz et al. 2003). bHLH proteins can recognize two types of target cis-acting elements: the E-box (5’-CANNTG-3’) and the G-box (5’-CACGTG-3’). Previous studies have demonstrated that several AtbHLHs are involved in regulating abiotic stress responses. AtbHLH116/ICE1 regulates the expression of CBF3/DREB1A, an AP2/ERF TF, that provides freezing tolerance through an ABA-independent pathway that is also known as the ICE-CBF regulon (Chinnusamy et al. 2003). OrbHLH1 and OrbHLH2, two ICE-like proteins in wild rice (Oryza rufipogon), positively regulate salt-stress responses in transgenic Arabidopsis through an ICE/CBF-independent and ABA-independent pathway, respectively (Zhou et al. 2009; Li et al. 2010). More recently, AtbHLH122 has been shown to confer tolerance to drought, salt, and osmotic stresses through an ABA-dependent pathway via the transcriptional repression of CYP707A3, thereby blocking ABA degradation (Liu et al. 2014). Notably, only approximately 10% of OsbHLHs have been characterized, in contrast to 38% of AtbHLHs (Heang and Sassa 2012).
To investigate which uncharacterized OsbHLHs may also be involved in the regulation of abiotic stress responses, we analyzed the gene expression profiles from publicly available microarray data (GSE6901) in the NCBI-GEO database, and several abiotic stress-responsive OsbHLHs were found, including OsbHLH035 (Additional file 1: Figure S1). In this study, we examined the roles of OsbHLH035 in rice during the germination and seedling stages. Using a reverse genetic approach with a Tos17-tagged mutant line, we found that OsbHLH035 mediates germination and confers recovery after salt stress relief through ABA-dependent and ABA-independent pathways, respectively.
Results
Characterization of the OsbHLH035 gene and its Tos17-tagged mutant line, NG7221
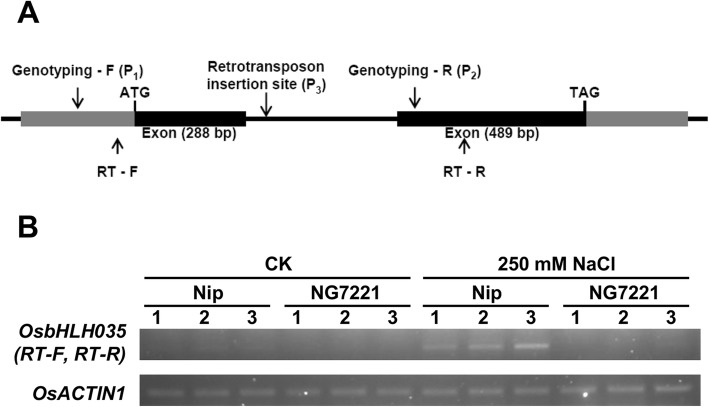
In silico analyses of gene expression profiles revealed that OsbHLH035 gene expression is upregulated by drought and salt treatments (Additional file 1: Figure S1). The OsbHLH035 gene contains two exons, and the encoded protein, which putatively binds G-boxes, contains a typical bHLH domain (residues 64 to 113) (Fig. 1a, Additional file 1: Figure S2A). To verify the microarray data, we compared the expression pattern of OsbHLH035 in aerial tissues from WT rice (Oryza sativa L. cv. Nipponbare, Nip) under normal and salt-treated conditions using RT-PCR. As shown in Fig. 1b, OsbHLH035 transcripts were barely detectable in aerial tissues under normal conditions, whereas they were present under salt-treated conditions. These data indicate that OsbHLH035 is reliably induced by salt stress in rice seedlings.
Fig. 1.
OsbHLH035 expression is induced by salt and is abolished in the NG7221 line. a The structure of the OsbHLH035 gene and the retrotransposon insertion site. b OsbHLH035 expression patterns in aerial tissues from third-leaf-stage WT and NG7221 seedlings. The seedlings were grown on basal medium for 13 days and then transferred to basal medium containing 0 (CK) or 250 mM NaCl for an additional day. The primer positions and sequences are shown in (a) and Additional file 2: Table S1. Arabic numerals in (b) represent three independent biological replicates within each genotype
To investigate the roles of OsbHLH035 in plant growth and salt-stress responses, we searched the rice Tos17 insertion mutant database and identified a retrotransposon insertional Osbhlh035 mutant line, NG7221. Based on its annotation, the retrotransposon is located in an OsbHLH035 gene intron (Fig. 1a, Additional file 1: Figure S2B). Homozygous mutants were identified from the segregating population by genomic DNA genotyping (Additional file 1: Figure S2C). RT-PCR analysis showed that OsbHLH035 transcripts were absent in aerial tissues from third-leaf-stage NG7221 seedlings under both normal and salt-treated conditions (Fig. 1b). Therefore, the NG7221 line is used as an Osbhlh035 mutant in this study.
Spatiotemporal OsbHLH035 expression and OsbHLH035 localization
Analyzing spatiotemporal gene expression patterns and protein localization can provide valuable insights into gene function. Thus, we monitored OsbHLH035 gene expression by detecting β-glucuronidase (GUS) signals in OsbHLH035::GUS transgenic plants from the germination stage until the third-leaf stage. In the germinating seeds, time-dependent expression of GUS was observed in the embryo, scutellum, aleurone layer, and endosperm after imbibition (Fig. 2a). In the aerial tissues, GUS was expressed in the hypocotyl and coleoptile at the postgermination stage but was difficult to detect in the third-leaf-stage seedlings (Fig. 2a, b). In terrestrial tissues, GUS signals were present in the root tip, vascular tissue, and lateral root initiation sites of both postgermination and third-leaf-stage seedlings (Fig. 2a, b).
Fig. 2.
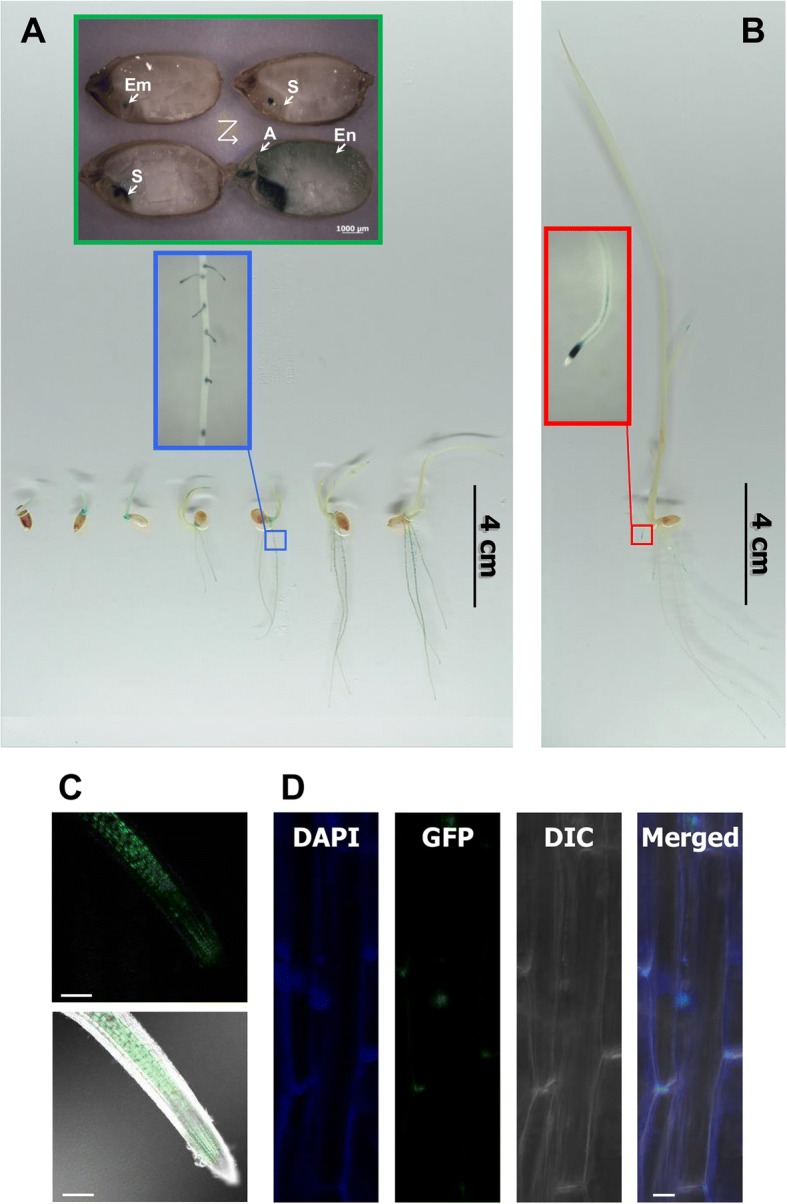
The spatiotemporal expression of OsbHLH035 and the subcellular localization of GFP-OsbHLH035 in stable rice transformants. a The germinating seeds and post-germination-stage seedlings. The Z-scheme represents the staining order of the water-imbibed transgenic seeds, which were collected within 2 days. Em, Embryo; S, Scutellum; A, Aleurone layer; and En, Endosperm. b A third-leaf-stage seedling. c The presence of GFP-OsbHLH035 in the root tip. The upper panel is fluorescence image; the lower panel is overlaid with transmitted light image. Scale bars, 200 μm. d GFP-fused OsbHLH035 protein is localized with the DAPI, a nuclear affinity dye, in rice cells. Scale bars, 5 μm
To validate the spatial expression of OsbHLH035 and the subcellular localization of OsbHLH035 protein in rice, an OsbHLH035::GFP-OsbHLH035 construct was transformed into the Osbhlh035 mutant via stable Agrobacterium-mediated transformation. Similar to its transcriptional expression pattern, the GFP-OsbHLH035 fusion protein was present in the root tip and vascular tissue (Fig. 2b vs. c) and was predominantly localized to the nucleus in rice cells and calli (Fig. 2d, Additional file 1: Figure S3).
Excess ABA leads to reduced germination rates in Osbhlh035 mutants
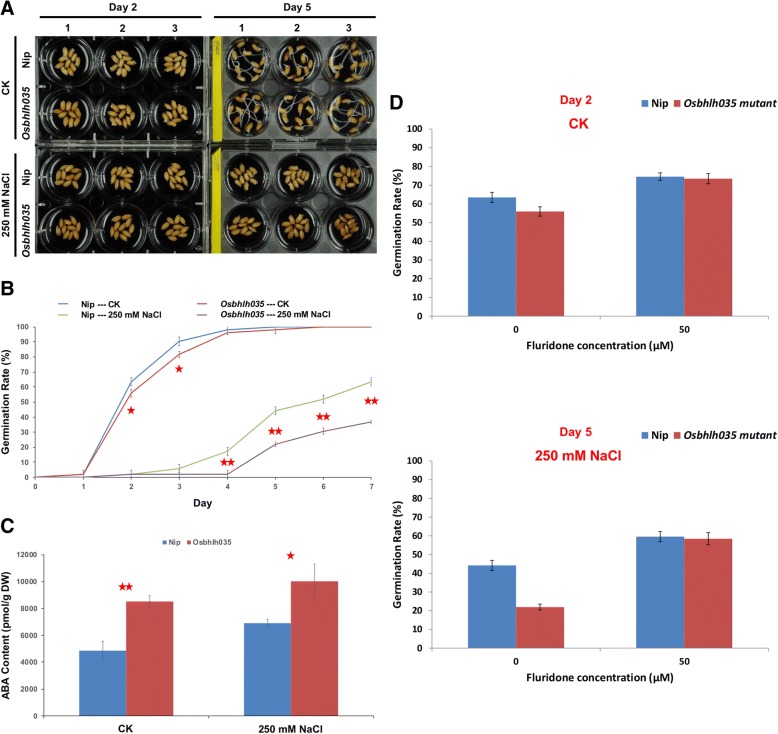
To investigate whether OsbHLH035 plays a role in regulating germination, we compared the germination rates of both WT and Osbhlh035 plants under normal and salt-treated conditions. Under normal conditions, the WT germination rates were 63.46% and 90.38% on days 2 and 3, respectively; however, the Osbhlh035 germination rates were reduced to 55.93% and 81.64%, respectively (Fig. 3a, b). Nevertheless, almost all of the WT and the Osbhlh035 seeds had germinated by day 4. Under salt-treated conditions, the WT germination rates were 17.31%, 44.23%, 51.92%, and 63.46% on days 4, 5, 6, and 7, respectively; however, they were 1.92%, 21.96%, 30.58%, and 36.75%, respectively, in the Osbhlh035 mutant. Notably, when normalized to the corresponding WT germination rates, the Osbhlh035 germination rates under normal conditions were reduced by 11.87% and 9.67% on days 2 and 3, respectively. However, the Osbhlh035 germination rates in the salt-treated conditions were reduced by 88.91%, 50.35%, 41.1%, and 42.09% on days 4, 5, 6, and 7, respectively. Taken together, these data indicate that OsbHLH035 may promote rice seed germination, especially under salt stress.
Fig. 3.
The delayed-germination phenotype of Osbhlh035 mutants is accompanied by the excess accumulation of endogenous ABA. a The germination of WT and Osbhlh035 seeds under normal and salt-treated conditions at 2 and 5 days after imbibition. Arabic numerals represent three independent biological replicates, and the seeds in each independent biological replicate were harvested from an individual rice plant. b The germination rates of WT and Osbhlh035 seeds under normal and salt-treated conditions. All germination rates represent the mean ± SD of two independent experiments performed with three independent biological replicates; 100 seeds were used in each independent biological replicate. c The endogenous ABA contents in both germinating WT and Osbhlh035 seeds grown under normal and salt-treated conditions on day 5. d The delayed-germination phenotype of Osbhlh035 mutants can be rescued by fluridone. The germination rates of WT and Osbhlh035 seeds under normal conditions at day 2 (upper panel) and salt-treated conditions at day 5 (lower panel). The seeds were grown on basal medium containing 0 (normal conditions, CK) or 250 mM NaCl (salt-treated conditions) supplemented with or without 50 μM fluridone. Samples for the ABA ELISA were harvested on day 5. Asterisks indicate significant differences in comparison with WT (*P < 0.05 and **P < 0.01) based on Student’s t-test
Because seed germination is inhibited by ABA, which antagonizes gibberellin signaling, we then measured the endogenous ABA contents in germinating WT and Osbhlh035 seeds. After 2 days under normal conditions, the ABA levels in germinating Osbhlh035 seeds were approximately 1.84-fold higher than those in WT seeds, and the corresponding germination rate in the Osbhlh035 mutant was lower than in WT (Fig. 3b, Additional file 1: Figure S4). Additionally, salt-induced ABA accumulation was observed in germinating WT and Osbhlh035 seeds after 5 days under salt-treated conditions (Fig. 3c). However, the ABA levels were approximately 1.45-fold higher in germinating Osbhlh035 seeds than in WT seeds, and the corresponding germination rate of the Osbhlh035 mutant was also lower than that of WT (Fig. 3b, c). To validate that the delayed-germination phenotype of Osbhlh035 mutants is due to excess ABA accumulation, the biosynthetic inhibitor fluridone was used in further tests. Indeed, the germination rates of WT and Osbhlh035 seeds were not statistically significantly different after 2 days under normal conditions or 5 days under salt-treated conditions in the presence of fluridone (Fig. 3d). Taken together, these data suggest that OsbHLH035 is involved in the repression of ABA-inhibited germination.
Expression of ABA metabolic genes in germinating WT and Osbhlh035 seeds
Because ABA metabolic genes have been well characterized in rice, we used q-PCR to investigate the expression profiles of these genes in germinating WT and Osbhlh035 seeds. In germinating WT seeds, the expression levels of the ABA biosynthetic genes OsABA2 and OsAAO3 under salt-treated conditions were approximately 2.01- and 2.06-fold, respectively, higher than those under normal conditions (Fig. 4a, b). However, the OsABA2 and OsAAO3 expression levels in germinating Osbhlh035 seeds were also higher than those in germinating WT seeds under either normal or salt-treated conditions. Moreover, the expression levels of the ABA catabolic gene OsABA8ox1 in germinating WT and Osbhlh035 seeds were comparable under normal conditions; however, the salt-induced expression of OsABA8ox1 was abolished in germinating Osbhlh035 seeds (Fig. 4c). These results agreed with the corresponding ABA levels detected under these conditions (Fig. 3c vs. Fig. 4). Taken together, these data reveal that the accumulation of excess ABA in germinating Osbhlh035 seeds may be caused not only by elevated ABA biosynthesis but also by reduced ABA catabolism under salt-treated conditions.
Fig. 4.
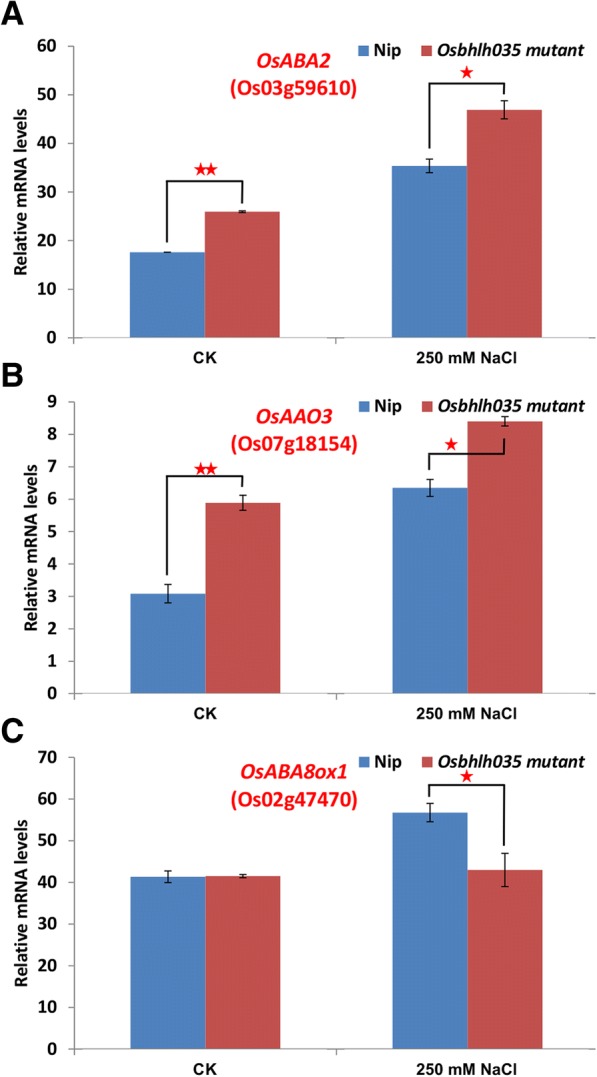
The expression of ABA metabolic genes in both germinating WT and Osbhlh035 seeds. ABA over-accumulation in germinating Osbhlh035 seeds is due to the upregulation of the biosynthetic genes OsABA2 (a) and OsAAO3 (b) and abolition of salt-induced OsABAox1 (c) expression. The experimental materials and methods are the same as those described in Fig. 3. Samples for the q-PCR assay were harvested on day 5. Asterisks indicate significant differences in comparison with WT (*P < 0.05 and **P < 0.01) based on Student’s t-test
OsbHLH035 is not involved in regulating plant growth during the seedling stage
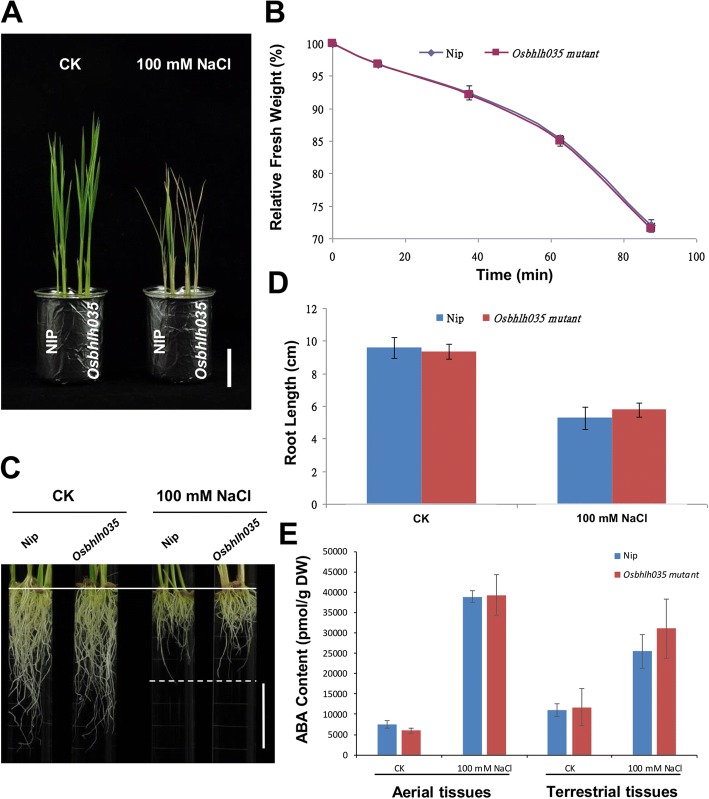
To understand the roles of salt-inducible OsbHLH035 during the seedling stage, we compared the plant growth and salt-stress responses in both WT and Osbhlh035 seedlings under normal and salt-treated conditions. Under normal conditions, there were no significant differences in the growth patterns of WT and Osbhlh035 aerial tissues on days 14 (Fig. 5a). Interestingly, there were also no obvious differences in the growth patterns and salt responses of the WT and Osbhlh035 aerial tissues under salt-treated conditions (Fig. 5a). Moreover, the water-loss rate and ABA contents in both WT and Osbhlh035 aerial tissues were comparable under either normal or salt-treated conditions (Fig. 5b and e). Furthermore, the primary root length and ABA contents of the Osbhlh035 terrestrial tissues were similar to those of the WT terrestrial tissues under either normal or salt-treated conditions (Fig. 5c-e). These data indicate that OsbHLH035 does not appear to be involved in regulating plant growth during the seedling stage.
Fig. 5.
A comparison of the growth patterns, salt responses, and ABA contents in both WT and Osbhlh035 seedlings under normal (CK) and salt-treated (100 mM NaCl) conditions. The seedlings were grown on basal medium for 7 days and then transferred to basal medium supplemented with 0 (CK) or 100 mM NaCl for an additional 7 days. a The growth patterns and salt responses of 14-day-old WT and Osbhlh035 aerial tissues. b The rates of water loss in 14-day-old WT and Osbhlh035 aerial tissues. c-d Characterization and quantification of root growth in both WT and Osbhlh035 seedlings. e The ABA contents in 14-day-old WT and Osbhlh035 seedlings. Scale bars in (a) and (c) represent 4 cm
Excess sodium accumulation in Osbhlh035 aerial tissues
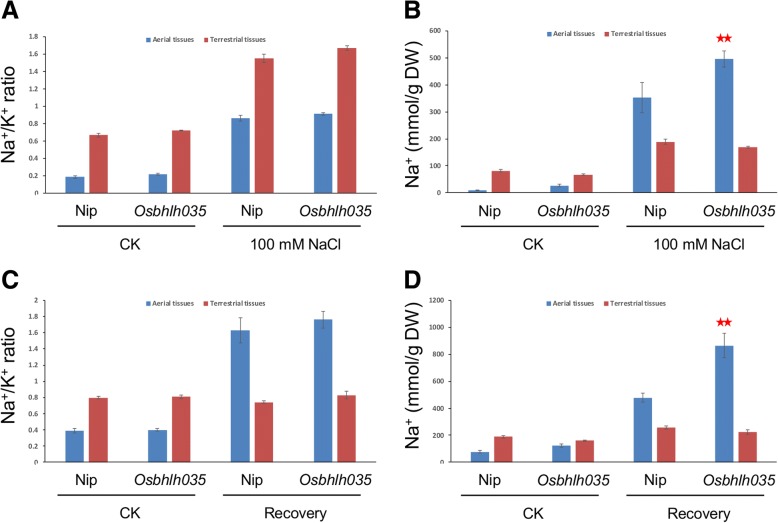
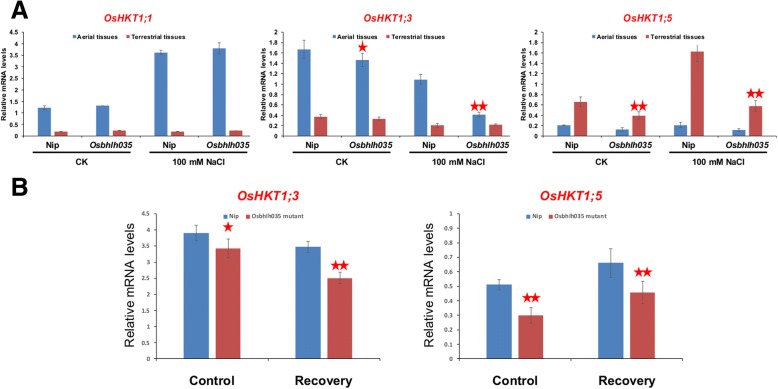
As mentioned above, OsbHLH035 is expressed in aerial and terrestrial tissues from the postgermination stage until the third-leaf stage (Fig. 2a), and its expression is enhanced by salt treatment (Fig. 1b). However, the roles of OsbHLH035 do not appear to include regulating seedling growth or salt tolerance at these stages and under these conditions (Fig. 5). To investigate the roles of OsbHLH035 in detail, we detected the sodium and potassium contents in both WT and Osbhlh035 seedlings by using inductively coupled plasma-optical emission spectrometry (ICP-OES). As shown in Fig. 6a, the sodium/potassium (Na+/K+) ratios in the aerial or terrestrial tissues from the WT and Osbhlh035 seedlings were comparable under either normal or salt-treated conditions; however, sodium was excessively accumulated in the Osbhlh035 aerial tissues, while it was slightly reduced in its terrestrial tissues (Fig. 6b). These data suggest that Na+ exclusion from the transpiration stream is impaired in the Osbhlh035 seedlings. Furthermore, q-PCR analysis revealed that OsHKT1;3 and OsHKT1;5 mRNA levels in Osbhlh035 aerial and terrestrial tissues, respectively, were lower than those in the corresponding WT tissues under either normal or salt-treated conditions (Fig. 7a).
Fig. 6.
Effects of salinity on the Na+/K+ ratio and Na+ content in both WT and Osbhlh035 seedlings. a-b Na+/K+ ratios and Na+ contents in aerial and terrestrial tissues from both salt-treated WT and Osbhlh035 seedlings. c-d Na+/K+ ratios and Na+ contents in aerial and terrestrial tissues from both salt-relieved WT and Osbhlh035 seedlings. The seedlings were grown on basal medium for 11 days and then transferred to basal medium containing 0 (CK) or 100 mM NaCl (salt-treated conditions) for an additional 3 days. After day 14, the seedlings were returned to basal medium for 7 days (Recovery). Asterisks indicate significant differences in comparison to WT (*P < 0.05 and **P < 0.01) based on Student’s t-test
Fig. 7.
Effects of salinity on the OsHKT1s expression profile in both WT and Osbhlh035 seedlings. a Quantification of OsHKT1;1, OsHKT1;3, and OsHKT1;5 mRNAs in aerial and terrestrial tissues from both salt-treated WT and Osbhlh035 seedlings. b Quantification of OsHKT1;3 and OsHKT1;5 mRNAs in aerial (left panel) and terrestrial (right panel) tissues, respectively, from both salt-relieved WT and Osbhlh035 seedlings. The seedlings were grown on basal medium for 11 days and then transferred to basal medium containing 0 (CK) or 100 mM NaCl (salt-treated conditions) for an additional 3 days. After day 14, the seedlings were returned to basal medium for 7 days (Recovery). Asterisks indicate significant differences in comparison to WT (*P < 0.05 and **P < 0.01) based on Student’s t-test
OsbHLH035 contributes to seedling recovery following the removal of salt stress
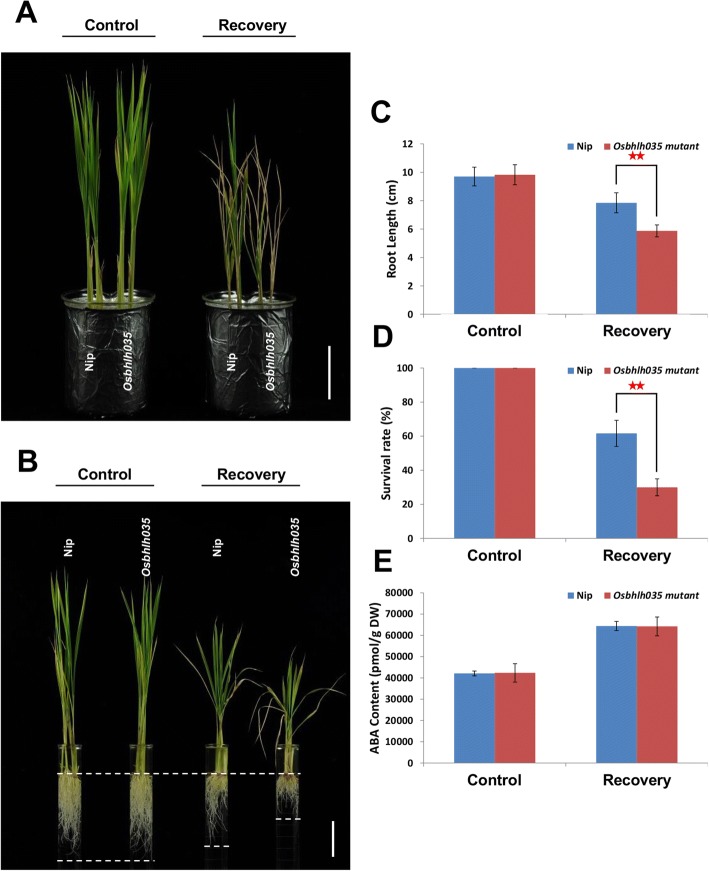
As mentioned previously, the ability to tolerate and recover from stress are both important for rice to manage abiotic stresses (Lenka et al. 2011; Zhang et al. 2012). Therefore, we further investigated the growth responses of WT and Osbhlh035 seedlings after the removal of salt stress. After a 7-day recovery period, new leaves sprouted in the WT seedlings but not in the Osbhlh035 seedlings (Fig. 8a). The primary roots of WT seedlings that had recovered from salt stress were longer than those of the corresponding Osbhlh035 seedlings (Fig. 8b, c). Moreover, the survival rate of WT seedlings after the removal of salt stress was approximately 61.67%, while the rate of Osbhlh035 seedlings was only approximately 30% (Fig. 8d). Although the endogenous ABA content was still high in both the WT and Osbhlh035 seedlings that had recovered from salt stress compared with controls, there were no significant difference between either the salt-stressed WT or Osbhlh035 seedlings postrecovery (Fig. 8e). Notably, sodium was continuously over-accumulated in salt-relieved Osbhlh035 aerial tissues (Fig. 6d). Moreover, the ratio of excessive Na+ in salt-relieved Osbhlh035 aerial tissues is higher than that in salt-treated Osbhlh035 aerial tissues (Fig. 6b vs. d). Besides, the expression of OsHKT1;3 and 1;5 was also reduced in salt-relieved Osbhlh035 aerial and terrestrial tissues, respectively (Fig. 7b). These data show that OsbHLH035 may play a positive, ABA-independent role in seedling recovery after salt stress.
Fig. 8.
Investigating growth responses, survival rate, and endogenous ABA contents in both WT and Osbhlh035 seedlings after recovery from salt stress. a-b The growth responses of aerial and terrestrial tissues in both WT and Osbhlh035 seedlings after relief from salt stress. c The primary root lengths of WT and Osbhlh035 seedlings after relief from salt stress. d The survival rate of WT and Osbhlh035 seedlings after relief from salt stress. All survival rates are shown as the mean ± SD of two independent experiments (n = 20 × 3 independent biological replicates within each genotype and/or experimental condition). e The endogenous ABA contents of WT and Osbhlh035 seedlings after recovery from salt stress. Scale bars in (a) and (b) represent 4 cm. The seedlings were grown on basal medium for 7 days and then transferred to basal medium containing 0 (control) or 100 mM NaCl (recovery) for an additional 7 days. After day 14, the seedlings were returned to basal medium for 7 days. Samples for the ABA ELISA were harvested on day 21. Asterisks indicate significant differences in comparison to WT (*P < 0.05 and **P < 0.01) based on Student’s t-test
Delayed germination and impaired salt-stress recovery of Osbhlh035 mutants can be rescued via genetic complementation
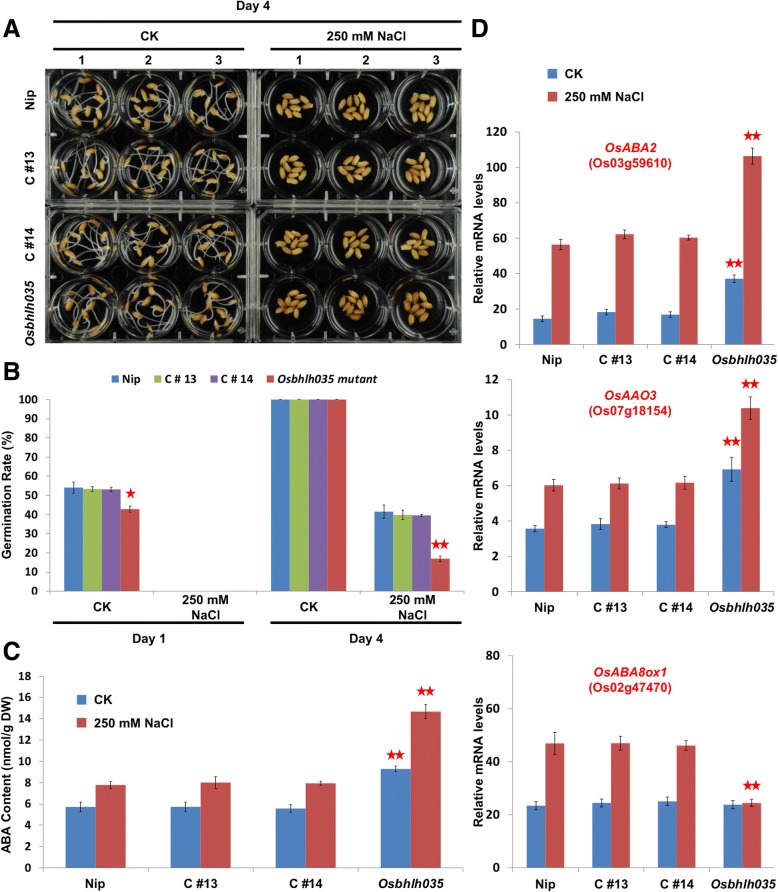
Genetic complementation was performed to validate the roles of OsbHLH035 in regulating seed germination and seedling recovery, and two representative complemented transformants (C#13 and C#14) were investigated. Compared with WT seeds germinated for 1 day under normal conditions or 4 days under salt-treated conditions, the corresponding Osbhlh035 seeds displayed a delayed-germination phenotype with excess ABA accumulation on day 4 after the beginning of imbibition. However, the germination rate and ABA contents of the germinating C#13 and C#14 seeds were comparable to those of WT seeds (Fig. 9a-c). Moreover, OsABA2, OsAAO3, and OsABA8ox1 expression in germinating C#13 and C#14 seeds was restored to levels similar to those of germinating WT seeds after 4 days under normal conditions or salt-treated conditions (Fig. 9d). These data demonstrate that OsbHLH035 plays a role in promoting germination by negatively regulating ABA-mediated seed germination.
Fig. 9.
Examination of seed germination, endogenous ABA contents, and ABA metabolic gene expression in WT, Osbhlh035, and genetically complemented transformant rice. a-b The germination characteristics and rates of seeds from WT, Osbhlh035, and genetically complemented lines under normal and salt-treated conditions. Arabic numerals represent three independent biological replicates, which were harvested from three different rice plants per genotype. All germination rates are shown as the mean ± SD of two independent experiments; n = 50 × 3 biological replicates within each genotype and/or experimental condition. c The endogenous ABA contents of WT, Osbhlh035, and the genetically complemented transformant lines under normal and salt-treated conditions on day 4. d OsABA2, OsAAO3, and OsABA8ox1 expression in WT, Osbhlh035, and the genetically complemented transformant lines under normal and salt-treated conditions on day 4. The seeds were grown on basal medium supplemented with 0 (CK) or 250 mM NaCl. Samples for the ABA ELISA and q-PCR assays were harvested on day 4. Asterisks indicate significant differences in comparison to WT (*P < 0.05 and **P < 0.01) based on Student’s t-test
Additionally, the excessive accumulation of sodium in Obhlh035 aerial tissues and the impaired ability of Osbhlh035 seedlings to recover from salt stress could also be rescued by genetic complementation. Under salt-treated conditions, the sodium levels in both the C#13 and C#14 aerial tissues were similar to those in corresponding WT tissues, with comparable levels of OsHKT1;3 and OsHKT1;5 expression (Fig. 10). After a 7-day recovery period, the primary root length and survival rates of both the C#13 and C#14 transformants were also comparable to those of WT seedlings (Fig. 11a-c). As expected, no obvious changes in endogenous ABA contents were observed between the WT, Osbhlh035, and two genetically complemented seedlings after 21 days under normal or post-salt-stressed conditions (Fig. 11d). These data suggest that OsbHLH035 might contribute to the ability of rice seedlings to recover from salt stress in an ABA-independent manner.
Fig. 10.
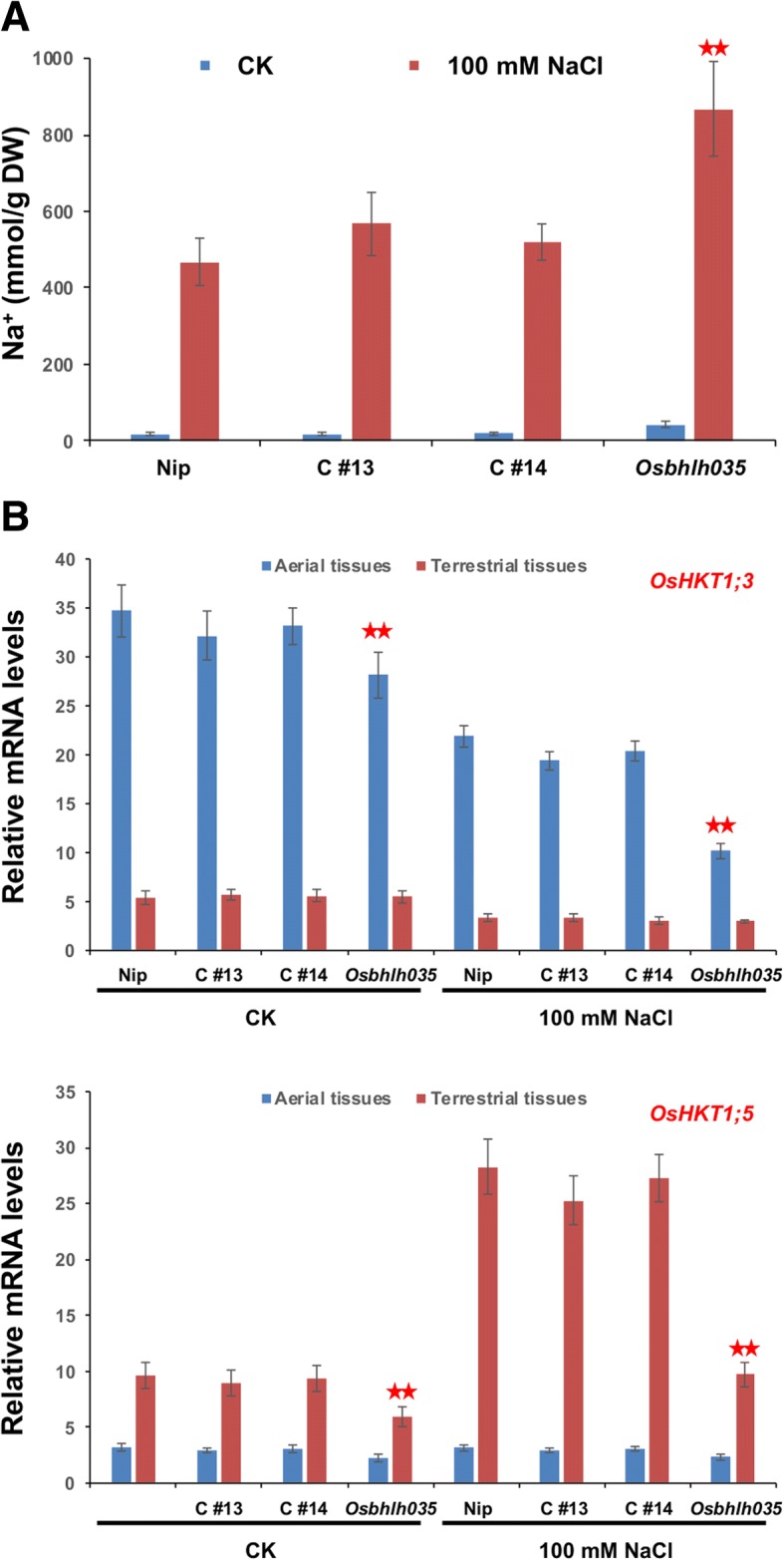
Na+ content a of the aerial tissues and the expression of OsHKT1;3 b and OsHKT1;5 c in WT, Osbhlh035, and genetically complemented transformant lines. The seedlings were grown on basal medium for 11 days and then transferred to basal medium supplemented with 0 (CK) or 100 mM NaCl for an additional 3 days. Asterisks indicate significant differences in comparison to WT (*P < 0.05 and **P < 0.01) based on Student’s t-test
Fig. 11.
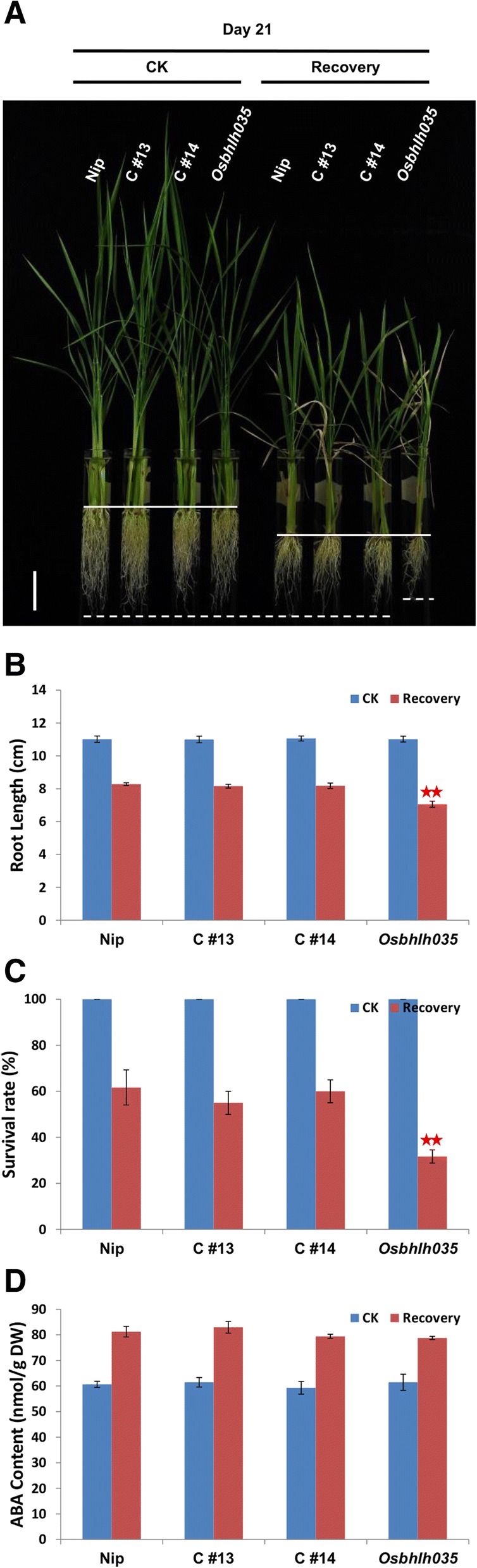
Phenotypic characterization and ABA contents after recovery from salt treatment. a-d Root elongation, survival rates, and endogenous ABA contents in WT, Osbhlh035, and genetically complemented transformant lines after salt stress removal. The seedlings were grown on basal medium for 7 days and then transferred to basal medium supplemented with 0 (CK) or 100 mM NaCl (recovery) for an additional 7 days, after which the seedlings were returned to basal medium for 7 days. Samples for the ABA ELISA and q-PCR assays were harvested on day 21. Asterisks indicate significant differences in comparison to WT (*P < 0.05 and **P < 0.01) based on Student’s t-test
Discussion
Plants are sessile and therefore must effectively cope with environmental changes to grow or survive. Thus, plants have evolved complicated gene regulatory networks, particularly diverse TFs, to respond to the surrounding environment. However, the functions of most TFs, as well as their involvement in plant growth, development and stress responses, are unknown. In this article, we conducted a functional study to address the roles of the TF OsbHLH035 on seed germination, salt tolerance, and the ability to recover from salt stress in rice.
OsbHLH035 mediates seed germination through an ABA-dependent pathway
ABA is important for coordinating the regulation of plant growth, development, and abiotic stress responses. However, the mechanisms that regulate the transcription of ABA metabolic genes in Arabidopsis and rice are poorly understood. In Arabidopsis, the ABA biosynthetic genes AtNCED6 and AtNCED9 contribute to ABA-mediated seed dormancy (Lefebvre et al. 2006). Notably, phylogenetic analysis reveals that plant NCEDs have undergone divergent evolution (Chen et al. 2011). In the rice genome, there are three putative NCEDs (see below for annotations) that are highly homologous to maize ZmVP14, the first plant NCED to be identified (Schwartz et al. 1997; Tan et al. 1997). To date, the rice orthologs of AtNCED6 and AtNCED9 have proven difficult to identify. In fact, both OsNCED1 (Os03g44380) and OsNCED3 (Os07g05940) expressions show no significant difference in germinating WT and Osbhlh035 seeds, and OsNCED2 (Os12g42280) transcripts are barely detectable under both normal and salt-treated conditions (data not shown). Apart from AtNCEDs, other biosynthetic genes such as AtABA2 and AtAAO3 as well as catabolic genes such as CYP707A1/AtABA8ox1 and CYP707A2/AtABAox2 are also involved in regulating seed germination. In germinating WT seeds, the expression levels of OsABA2, OsAAO3, and OsABA8ox1, which is a homolog of CYP707A1, 2, and 3 (Additional file 1: Figure S5), are upregulated under salt-treated conditions (Fig. 4). However, under either normal or salt-treated conditions, both OsABA2 and OsAAO3 are more highly expressed in germinating Osbhlh035 seeds than in corresponding WT seeds. No obvious differences in OsABA8ox1 transcription were detected between germinating WT and Osbhlh035 seeds under normal conditions, but the salt-induced expression of OsABA8ox1 is abolished in germinating Osbhlh035 seeds. Notably, the expression of OsbHLH035-GFP driven by a 2.0-kb OsbHLH035 native promoter can restore OsABA2, OsAAO3, and OsABA8ox1 expression and compensate for the germination deficiency in Osbhlh035 mutants (Fig. 9). These data show that the excess ABA accumulation in Osbhlh035 seeds germinating under salt stress is regulated at the transcriptional level due to not only the increased expression of ABA biosynthetic genes but also the inhibition of ABA catabolic gene expression. Given that OsbHLH035 is a G-box-binding TF, we searched for putative cis-acting elements in the upstream promoter regions of ABA metabolic genes. The 3807-bp and 3-kb regions upstream of the OsAAO3 and OsABA8ox1 initiation codons, respectively, were each found to contain one G-box element with a CACGTG core sequence. Because no G-box element was found in the promoter of OsABA2 despite its increased transcription in the germinating Osbhlh035 seeds, OsbHLH035 seems to suppress OsABA2 expression in an indirect manner. Interestingly, a functional G-box element was identified to be necessary for drought-induced AtNCED3 expression (Behnam et al. 2013). If this is the case, it cannot be excluded that OsbHLH035 suppresses OsAAO3 and OsABA8ox1 expression by interacting with G-box elements in their promoters. However, whether the transcriptional activity of OsAAO3 and OsABAox1 is directly regulated by OsbHLH035 needs to be confirmed.
OsbHLH035 provides the ability to recover from salt stress in an ABA-independent pathway
In this article, our data showed that no obvious differences were observed in plant growth, water-loss rate, or endogenous ABA contents in both WT and Osbhlh035 seedlings grown under normal or salt-treated conditions (Fig. 5). However, the Osbhlh035 mutant accumulates an excess of Na+ ions in the aerial tissues and is unable to recover after salt stress relief (Figs. 6 and 8). These data indicate that OsbHLH035 seems to play a fine-tuning role in regulating salt stress response. Thus, the salt-susceptibility phenotype of Osbhlh035 mutant becomes visible after salt stress relief. Besides, compared with that in aerial and terrestrial WT tissues, the expression of sodium transporter genes OsHKT1;3 and OsHKT1;5 is repressed in Osbhlh035 tissues, respectively (Fig. 7). Notably, genetic complementation can restore OsHKT1;3 and OsHKT1;5 expression and rescue the impaired salt-stress recovery of the Obhlh035 mutant (Figs. 10 and 11). These data suggest that OsbHLH035 confers the seedling recovery from salt stress through the ABA-independent activation of OsHKT1s. In Arabidopsis and rice, the HKT1s play a vital role in transferring Na+ from the xylem into xylem parenchyma cells (for review, see Su et al. 2015). This mechanism is used to prevent aerial tissues from Na+ over-accumulation and toxicity. Indeed, loss-of-function mutations in OsHKT1s, such as OsHKT1;1 and 1;5, lead to Na+ overaccumulation in the aerial tissues and display hypersensitivity to salt stress (Wang et al. 2015; Kobayashi et al. 2017). In fact, the NaCl-induced expression of OsHKT1;1 is directly regulated by a TF, OsMYBc (Wang et al. 2015). OsHKT1;3 and OsHKT1;5 are mainly expressed in the aerial and terrestrial tissues of rice seedlings, respectively (Garciadeblas et al. 2003; Kobayashi et al. 2017). Although the expression of OsHKT1;3 and 1;5 is downregulated in Osbhlh035 aerial and terrestrial tissues, respectively, no G-box element was found in their 3-kb promoter. These data indicate that OsbHLH035 mediates the expression of OsHKT1;3 and 1;5, most likely in an indirect manner.
Pleiotropic role of OsbHLH035 in regulating seed germination and salt-treated seedling recovery
In the plant bHLH TF gene family, the formation of homodimers or heterodimers is important for determining target gene expression. Dimeric molecules can specifically bind to certain cis-acting elements within the promoter regions of target genes, whose products can then mediate divergent physiological responses in plants. Therefore, plant bHLHs can play a dual or pleiotropic role in regulating growth and development based on their dimeric forms. For example, the POSITIVE REGULATOR OF GRAIN LENGTH 1-ANTAGONIST OF PGL1 (PGL1-APG) heterodimer, an OsHLH-OsbHLH complex, increases grain length and weight. However, the APG homodimer decreases grain length and weight (Heang and Sassa 2012). The function of OsbHLH068 not only is involved in regulating salt-stress responses but also may mediate flowering time in Arabidopsis (Chen et al. 2017). Thus, we assume that OsbHLH035 plays a pleiotropic role in regulating seed germination and seed recovery, presumably due to interactions with different TFs during the germination and seedling stages.
Conclusions
In the present study, we propose that OsbHLH035 plays a pleiotropic role in regulating seed germination and salt-treated seedling recovery in rice. During the germination stage, OsbHLH035 alters the expression of ABA metabolic genes, which reduces ABA levels and relieves the inhibitory effect of ABA on germination. However, OsbHLH035 also mediates the expression of OsHKT1 genes without altering ABA levels, which promotes seedling recovery from salt stress.
Methods
Plant materials, growth conditions, and experimental methods
Rice (Oryza sativa L.) cv. Nipponbare, an Osbhlh035 mutant (Tos17 line NG7221, obtained from the National Institute of Agrobiological Sciences [NIAS], Japan), and two independent transgenic complementation lines (C#13 and C#14, generated by Agrobacterium-mediated transformation) were analyzed in this study. Except in the germination test, the seeds used in all experiments were sterilized and imbibed at 37 °C for 2 days in the dark and then grown on a wire stand in a beaker at 28 °C under long-day conditions (16-h light/8-h dark cycle) with a light intensity of approximately 270 μE/s m2. For the germination test, seeds harvested from three independent plants were sterilized and grown directly in a Petri dish at 37 °C in the dark. The basal medium used in all of the experiments was Kimura B solution (Yoshida et al. 1976).
RNA extraction, RT-PCR, and q-PCR
Total RNA was extracted from various rice tissues using an RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. To avoid genomic DNA contamination, up to 3 μg of total RNA was treated after extraction with Turbo DNA-free™ DNase (Ambion) following the manufacturer’s instructions, and 1 μg of DNA-free total RNA was then subjected to first-strand cDNA synthesis (Invitrogen, Cat No: 18080–051). The q-PCRs were carried out in an ABI 7500 system using the SYBR® Green PCR Master Mix Kit (Applied Biosystems [ABI]). The initial amount of template cDNA in each RT-PCR and q-PCR was 50 and 10 μg, respectively. At least three independent biological replicates were performed for each experiment, and OsACTIN1 was used as an internal control for q-PCR normalization. The primer sequences used are provided in Additional file 2: Table S1.
Water loss test
The detached aerial tissues from 14-day-old rice seedlings were placed in plastic weigh boats under ambient conditions. The fresh weight of the tissue was measured after 12.5 min and every 25 min thereafter for a total of 87.5 min. At least three independent biological replicates were performed for each experiment.
ABA ELISA
Prior to the assay, the samples were vacuum dried at − 20 °C at least overnight for normalization to the final dry weight. ABA extraction, purification, and quantification were carried out as described previously (Lin et al. 2007), except that the samples were first ground using an SH-100 tissue homogenizer (Kurabo) before the addition of extraction buffer. At least three independent biological replicates containing two technical replicates each were performed for each experiment, and the data were presented as the average of two independent experiments.
Measurement of ion contents
Samples were harvested, washed twice with ultrapure water, and subsequently vacuum dried at least overnight to measure the dry weight. The dried samples were digested using nitric and hydrochloric acids in an approximately 4:1 ratio and boiled at 200 °C for 2 h. Ion contents were determined using an inductively coupled plasma optical emission spectrometer.
Transgene constructs and isolation of transgenic rice
The full-length coding sequences of OsbHLH035 and GFP were PCR-amplified either with or without stop codons and cloned into the pGEM-T Easy vector. As shown in parenthesis below, these fragments were subcloned into the binary vector pCAMBIA-1300 where their expression was driven either by a 2.0-kb native promoter (OsbHLH035::OsbHLH035-GFP and OsbHLH035::GFP-OsbHLH035) or a CaMV 35S promoter (35S::OsbHLH035-GFP and 35S::GFP-OsbHLH035). After the constructs were confirmed by sequencing, the OsbHLH035::GFP-OsbHLH035 construct was transformed into the Osbhlh035 mutant for subcellular OsbHLH035 localization and genetic complementation assays. Because the stable T0 rice transformants were chimeric and to avoid the undesirable side effects of multiple T-DNA insertions, T1 seeds were harvested from individual panicles of independent transformant lines that showed hygromycin resistance and sensitivity at a 3:1 ratio, normalized to seed viability, and used to further screen for homozygous transgenic rice. Additionally, the 2.0-kb OsbHLH035 promoter was cloned into pCAMBIA-1305.1 (OsbHLH035::GUS) and transformed into the rice cv. Tainung 67 background for further investigation of the spatiotemporal expression of the OsbHLH035 gene.
Additional files
Figure S1. Abiotic stress-responsive OsbHLHs. The fold change among each OsbHLH gene under different abiotic stresses is calculated as a ratio normalizing the data to it corresponding nontreated control. Red and green colors represent up- and downregulation, respectively. C, cold stress; D, drought stress; S, salt stress; and H, heat stress. Figure S2. Prediction of the conserved domains in OsbHLH035 and characterization of the NG7221 mutant line. (A) The ScanProsite tool in ExPASy (http://www.expasy.org/) predicts the presence of a typical bHLH domain (residues 64 to 113) in OsbHLH035. (B) Based on its annotation in the rice Tos17 insertion mutant database (https://tos.nias.affrc.go.jp/), NG7221 is a single retrotransposon insertional line. (C) The identification of homozygous NG7221 mutants via PCR genotyping of gDNA. The primer positions and sequences are shown in Fig. 1a and Additional file 2: Table S1, respectively. Arabic numbers in (C) represent three independent biological replicates within each genotype. Figure S3. GFP-fused OsbHLH035 protein is predominantly localized to the nucleus in rice calli. The husk-removed seeds harboring OsbHLH035::GFP-OsbHLH035 were placed on callus induction medium (Tran and Sanan-Mishra 2015) for 7 days and then the calli were subjected to GFP visualization by a confocal microscopy. Scale bars, 10 μm. Figure S4. The endogenous ABA contents in both germinating WT and Osbhlh035 seeds. The husk-removed seeds were grown on basal medium for 2 days and then subjected to ABA ELISAs. Figure S5. Phylogenetic analysis of AtCYP707As and OsABA8oxs using the neighbor-joining method. Numbers next to the descendant indicate confidence values based on the bootstrap method. (PDF 570 kb)
Table S1. Primers used in this study. (PDF 663 kb)
Acknowledgments
We thank the Live-Cell Imaging Core Lab at the Institute of Plant and Microbial Biology (IPMB), Academia Sinica, Taipei, Taiwan, for performing the confocal microscopy and Mr. Jiang-Tai Wu (IPMB, Academia Sinica) for assistance with the transient expression assays. We are also grateful to Miss Lin-Yun Kuang (Transgenic Plant Core Facility, Academia Sinica) for providing Agrobacterium-mediated stable transformation services.
Funding
This work was supported by the Ministry of Science and Technology, Taipei, Taiwan (Grant No. NSC-102-2811-B-002-146 to M.-C.C.).
Availability of data and materials
All data supporting the conclusions of this article are provided within the article and its additional files.
Abbreviations
- ABA
Abscisic acid
- bHLH
Basic helix-loop-helix
- GFP
Green fluorescent protein
- GUS
β-glucuronidase
- Nip
Nipponbare
- q-PCR
Quantitative PCR
- RT-PCR
Reverse transcription-PCR
- TFs
Transcription factors
Authors’ contributions
H-CC was responsible for designing and conducting the experiments, data assembly, and writing the manuscript. W-HC, C-YH, and Y-SC were responsible for providing the experimental facilities and mentoring the experiments. M-CC conceived the study and supervised the experiments. All authors have read and approved the submission of the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Behnam B, Iuchi S, Fujita M, Fujita Y, Takasaki H, Osakabe Y, Yamaguchi-Shinozaki K, Kobayashi M, Shinozaki K. Characterization of the promoter region of an gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res. 2013;20:315–324. doi: 10.1093/dnares/dst012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner F, Oreb M, Mendel RR. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem. 2001;276:40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Hsieh-Feng V, Liao PC, Cheng WH, Liu LY, Yang YW, Lai MH, Chang MC. The function of OsbHLH068 is partially redundant with its homolog, AtbHLH112, in the regulation of the salt stress response but has opposite functions to control flowering in Arabidopsis. Plant Mol Biol. 2017;94:531–548. doi: 10.1007/s11103-017-0624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Hwang SG, Chen SM, Shii CT, Cheng WH. ABA-mediated heterophylly is regulated by differential expression of 9-cis-epoxycarotenoid dioxygenase 3 in lilies. Plant Cell Physiol. 2011;52:1806–1821. doi: 10.1093/pcp/pcr117. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19:371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Yeung E, Bailey-Serres J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell. 2011;23:412–427. doi: 10.1105/tpc.110.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciadeblas B, Senn ME, Banuelos MA, Rodriguez-Navarro A. Sodium transport and HKT transporters: the rice model. Plant J. 2003;34:788–801. doi: 10.1046/j.1365-313X.2003.01764.x. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodriguez PL. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SP, Karssen CM. Dormancy and germination of abscisic acid-deficient tomato seeds : studies with the sitiens mutant. Plant Physiol. 1992;99:952–958. doi: 10.1104/pp.99.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N. HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol. 2015;32:113–120. doi: 10.1016/j.copbio.2014.11.025. [DOI] [PubMed] [Google Scholar]
- Heang D, Sassa H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One. 2012;7:e31325. doi: 10.1371/journal.pone.0031325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DL, Breekland AE, Koornneef M. Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta. 1983;157:158–165. doi: 10.1007/BF00393650. [DOI] [PubMed] [Google Scholar]
- Kobayashi NI, Yamaji N, Yamamoto H, Okubo K, Ueno H, Costa A, Tanoi K, Matsumura H, Fujii-Kashino M, Horiuchi T, Nayef MA, Shabala S, An G, Ma JF, Horie T. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017;91:657–670. doi: 10.1111/tpj.13595. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Hilhorst HW, Karssen CM. In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 1989;90:463–469. doi: 10.1104/pp.90.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K, Kumar M, Kim SR, Ryu H, Cho YG. Insights into genomics of salt stress response in rice. Rice. 2013;6:27. doi: 10.1186/1939-8433-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006;45:309–319. doi: 10.1111/j.1365-313X.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- Lenka SK, Katiyar A, Chinnusamy V, Bansal KC. Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J. 2011;9:315–327. doi: 10.1111/j.1467-7652.2010.00560.x. [DOI] [PubMed] [Google Scholar]
- Li F, Guo S, Zhao Y, Chen D, Chong K, Xu Y. Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang wild rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2010;29:977–986. doi: 10.1007/s00299-010-0883-z. [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol. 2007;143:745–758. doi: 10.1104/pp.106.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014;201:1192–1204. doi: 10.1111/nph.12607. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. CYP707A1 and CYP707A2, which encode abscisic acid 8’-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002;128:544–551. doi: 10.1104/pp.010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;26:421–433. doi: 10.1046/j.1365-313X.2001.2641043.x. [DOI] [PubMed] [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, Arimura S, Miyao A, Hirochika H, Kamiya Y, Tsutsumi N, Nambara E, Nakazono M. Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8’-hydroxylase in rice. Plant Cell Physiol. 2007;48:287–298. doi: 10.1093/pcp/pcm003. [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M. Arabidopsis CYP707As encode (+)-abscisic acid 8’-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004;134:1439–1449. doi: 10.1104/pp.103.037614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Mieulet D, Hubberten HM, Obata T, Hoefgen R, Fernie AR, Fisahn J, Segundo BS, Guiderdoni E, Schippers JH, Mueller-Roeber B. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell. 2013;25:2115–2131. doi: 10.1105/tpc.113.113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JA. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003;131:1591–1601. doi: 10.1104/pp.102.017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- Seo M, Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;7:41–48. doi: 10.1016/S1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci U S A. 2000;97:12908–12913. doi: 10.1073/pnas.220426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5:430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Su Y, Luo W, Lin W, Ma L, Kabir MH. Model of cation transportation mediated by high-affinity potassium transporters (HKTs) in higher plants. Biol Proced Online. 2015;17:1. doi: 10.1186/s12575-014-0013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/MCB.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35:44–56. doi: 10.1046/j.1365-313X.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR. Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci U S A. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Kou Y, Liu H, Li X, Xiao J, Wang S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot. 2011;62:4863–4874. doi: 10.1093/jxb/err144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor IB, Burbidge A, Thompson AJ. Control of abscisic acid synthesis. J Exp Bot. 2000;51:1563–1574. doi: 10.1093/jexbot/51.350.1563. [DOI] [PubMed] [Google Scholar]
- Todaka D, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K. Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice. 2012;5:6. doi: 10.1186/1939-8433-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TN, Sanan-Mishra N. Effect of antibiotics on callus regeneration during transformation of IR 64 rice. Biotechnol Rep (Amst) 2015;7:143–149. doi: 10.1016/j.btre.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W. The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 2015;168:1076–1090. doi: 10.1104/pp.15.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Wust F, Bar C, Al-Babili S, Beyer P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 2008;147:367–380. doi: 10.1104/pp.108.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/tpc.13.9.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003;133:29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye N, Jia L, Zhang J. ABA signal in rice under stress conditions. Rice. 2012;5:1. doi: 10.1186/1939-8433-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. Laboratory manual for physiological studies of rice. Manila: International Rice Research Institute; 1976. Routine procedure for growing rice plants in culture solution. [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39:D1114–D1117. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhao X, Wang W, Pan Y, Huang L, Liu X, Zong Y, Zhu L, Yang D, Fu B. Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One. 2012;7:e43274. doi: 10.1371/journal.pone.0043274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li F, Wang JL, Ma Y, Chong K, Xu YY. Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J Plant Physiol. 2009;166:1296–1306. doi: 10.1016/j.jplph.2009.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Abiotic stress-responsive OsbHLHs. The fold change among each OsbHLH gene under different abiotic stresses is calculated as a ratio normalizing the data to it corresponding nontreated control. Red and green colors represent up- and downregulation, respectively. C, cold stress; D, drought stress; S, salt stress; and H, heat stress. Figure S2. Prediction of the conserved domains in OsbHLH035 and characterization of the NG7221 mutant line. (A) The ScanProsite tool in ExPASy (http://www.expasy.org/) predicts the presence of a typical bHLH domain (residues 64 to 113) in OsbHLH035. (B) Based on its annotation in the rice Tos17 insertion mutant database (https://tos.nias.affrc.go.jp/), NG7221 is a single retrotransposon insertional line. (C) The identification of homozygous NG7221 mutants via PCR genotyping of gDNA. The primer positions and sequences are shown in Fig. 1a and Additional file 2: Table S1, respectively. Arabic numbers in (C) represent three independent biological replicates within each genotype. Figure S3. GFP-fused OsbHLH035 protein is predominantly localized to the nucleus in rice calli. The husk-removed seeds harboring OsbHLH035::GFP-OsbHLH035 were placed on callus induction medium (Tran and Sanan-Mishra 2015) for 7 days and then the calli were subjected to GFP visualization by a confocal microscopy. Scale bars, 10 μm. Figure S4. The endogenous ABA contents in both germinating WT and Osbhlh035 seeds. The husk-removed seeds were grown on basal medium for 2 days and then subjected to ABA ELISAs. Figure S5. Phylogenetic analysis of AtCYP707As and OsABA8oxs using the neighbor-joining method. Numbers next to the descendant indicate confidence values based on the bootstrap method. (PDF 570 kb)
Table S1. Primers used in this study. (PDF 663 kb)
Data Availability Statement
All data supporting the conclusions of this article are provided within the article and its additional files.