Abstract
Background
Cytoplasmic male sterility (CMS) is a natural phenomenon of pollen abortion caused by the interaction between cytoplasmic genes and nuclear genes. CMS is a simple and effective pollination control system, and plays an important role in crop heterosis utilization. Circular RNAs (circRNAs) are a vital type of non-coding RNAs, which play crucial roles in microRNAs (miRNAs) function and post-transcription control. To explore the expression profile and possible functions of circRNAs in the soybean CMS line NJCMS1A and its maintainer NJCMS1B, high-throughput deep sequencing coupled with RNase R enrichment strategy was conducted.
Results
CircRNA libraries were constructed from flower buds of NJCMS1A and its maintainer NJCMS1B with three biological replicates. A total of 2867 circRNAs were identified, with 1009 circRNAs differentially expressed between NJCMS1A and NJCMS1B based on analysis of high-throughput sequencing. Of the 12 randomly selected circRNAs with different expression levels, 10 showed consistent expression patterns based on high-throughput sequencing and quantitative real-time PCR analyses. Tissue specific expression patterns were also verified with two circRNAs by quantitative real-time PCR. Most parental genes of differentially expressed circRNAs were mainly involved in biological processes such as metabolic process, biological regulation, and reproductive process. Moreover, 83 miRNAs were predicted from the differentially expressed circRNAs, some of which were strongly related to pollen development and male fertility; The functions of miRNA targets were analyzed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG), and the target mRNAs were significantly enriched in signal transduction and programmed cell death. Furthermore, a total of 165 soybean circRNAs were predicted to contain at least one internal ribosome entry site (IRES) element and an open reading frame, indicating their potential to encode polypeptides or proteins.
Conclusions
Our study indicated that the circRNAs might participate in the regulation of flower and pollen development, which could provide a new insight into the molecular mechanisms of CMS in soybean.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5054-6) contains supplementary material, which is available to authorized users.
Keywords: Soybean (Glycine max (L.) Merr.), Cytoplasmic male sterility, CircRNAs, High-throughput sequencing, Parental genes, Binding miRNAs
Background
Soybean (Glycine max (L.) Merr.) is an important oil and protein crop, but soybean yield is low. Utilization of heterosis has been proved to be one of the effective methods to increase crop yield, and cytoplasmic male sterility (CMS) plays an important role in heterosis utilization [1]. CMS is a maternally inherited phenotype caused by the interaction of cytoplasmic genes and nuclear genes, which lead to pollen abortion but normal pistil development [2, 3]. To date, CMS has been observed in over 200 plant species [4] and is often obtained in wild germplasm or by inter- or intra-subspecies backcrossing [5]. In soybean, CMS was first reported by Davis [6], after which many studies on CMS were conducted [7, 8]. In recent years, transcriptomics [9], proteomics [10], microRNA [11], and DNA methylation of CMS have been explored [12], but the function of circular RNAs (circRNAs) in soybean CMS has not been reported.
CircRNAs are a class of endogenous noncoding RNAs, which do not have 5′ caps and 3′ tails and form a ring structure with covalent bonds [13]. CircRNAs were first reported based on deep sequencing of RNA by Salzman et al. [14], which were neglected for decades because they were considered as transcriptional noise or reverse transcription PCR artifacts by-product [15]. With the development of high-throughput sequencing technology and bioinformatics, circRNAs have been identified in all domains of life, including eukaryotes [15, 16], archaea [17], bacteria [18], and viruses [19]. Previous studies showed that circRNAs were more resistant to degradation by RNase R than their linear counterparts [20], and had tissue, cell-type, and developmental-stage specific expression patterns [15, 21, 22]. Li et al. [23] demonstrated that the circRNAs located in the nucleus could promote transcription of their parental genes via specific RNA-RNA interactions, but another recent study revealed that circRNAs derived from organelle genome could also regulate gene expression [24]. Furthermore, circRNAs act as miRNA sponges and prevent them from inhibiting their target mRNAs [25, 26]. Most recent studies have demonstrated that some circRNAs can be translated into polypeptides or proteins by translation initiation element internal ribosome entry site (IRES) or N6-methyladenosine (m6A) [27, 28].
Although not as comprehensive as in animals, the exploration of circRNAs in plants is increasing. Differential expression of circRNAs has been reported in plants, for example, 27 rice exonic circRNAs were associated with phosphate starvation responsive expression [16], 163 tomato circRNAs with chilling responsive expression [29], and 62 wheat circRNAs with dehydration stress specific expression [30]. Moreover, some circRNAs showed time, tissue, species, or developmental-stage specific expression patterns in plants [16, 24, 31, 32]. Unlike the positive regulation of circRNAs in animals [23], over-expression of circRNAs in rice and tomato reduced expression level of their parental genes [33, 34]. In soybean, a total of 5372 circRNAs were identified, of which approximately 80% were generated from paralogous genes [35]. Meanwhile, Zhao et al. [35] found that up-regulation of circRNAs might decrease the activity of target miRNAs and increase expression of the related mRNAs.
To explore the expression profile and possible functions of circRNAs in the soybean CMS line NJCMS1A vs. its maintainer line NJCMS1B, high-throughput deep sequencing coupled with RNase R enrichment strategy was conducted. Target miRNAs of differentially expressed circRNAs and the correlated mRNAs were predicted using bioinformatics methods, and their potential functions were further analyzed. Our study investigated the possible role of circRNAs in CMS for the first time, and the results showed that the circRNAs might participate in the regulation of flower and pollen development in soybean.
Methods
Plant materials and sample collection
The soybean cytoplasmic male-sterile line NJCMS1A was developed through consecutive backcross. The cultivar N8855 was the donor and cultivar N2899 (designated as NJCMS1B afterwards) was a recurrent parent [8, 36, 37]. NJCMS1A and NJCMS1B are near-isogenic lines of isonuclear alloplasmic type with similar nucleus but different cytoplasm. NJCMS1A and NJCMS1B were grown in the summer of 2016 at Dangtu Experimental Station, National Center for Soybean Improvement, Nanjing Agricultural University, Maanshan, Anhui, China. Male sterile plants were identified by observing dehiscence of anthers, germination rate of pollen, and morphological traits of plants at maturity. Because it is difficult to determine development stage of pollen based on flower bud size in soybean, during the flowering period, the flower buds of different sizes were collected from NJCMS1A or NJCMS1B plants and mixed, with three biological replicates per line. Meanwhile, roots, stems and leaves of soybean were also collected at the flowering period with three biological replicates. The tissues were immediately frozen in liquid nitrogen and stored at − 80 °C for further use.
Total RNA extraction and RNase R treatment
Total RNA from flower buds of NJCMS1A and NJCMS1B lines was extracted using the RNAprep pure Plant Kit (Qiagen, DEU) according to the manufacturer’s protocol, DNA contamination was removed using the DNase I contained in the Kit. For RNase R-treated total RNA samples, the purified DNaseI-treated total RNA was incubated for 15 min at 37 °C with 3 units RNase R per μg of total RNA (Epicentre, Shanghai, CN). RNA was subsequently purified using an RNase MinElute Cleaning Kit (Qiagen, DEU). To obtain accurate and sufficient transcriptome data, RNA from three biological replicates of NJCMS1A and NJCMS1B lines were sequenced.
CircRNA library construction and sequencing
rRNA-depleted RNAs in 5 μg RNA per sample was obtained using the Epicentre Ribo-zero™ rRNA Removal Kit (Epicentre, USA) and were further treated with RNase R (Epicentre, USA) for Trizol extraction. Sequencing libraries were prepared using NEBNext Ultra Directional RNA LibraryPrep Kit (NEB, USA) following manufacturer’s instructions. The libraries were preliminarily quantified by Qubit and diluted to 1 ng/ul. The insert size of the libraries was detected by Agilent 2100/Caliper, which was expected to be distributed around 250~ 300 bp. The effective concentration of the libraries was accurately quantified by qRT-PCR, and the effective concentration was greater than 2 nM to ensure the libraries quality. Briefly, fragmentation was carried out using divalent cations under elevated temperature in NEBNext First-strand Synthesis Reaction Buffer. First-strand cDNA was synthesized using random hexamer primer and M-MuLV reverse transcriptase (RNaseH). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In the reaction buffer, dTTP in dNTPs was replaced by dUTP. Remaining overhangs were blunted via exonuclease/polymerase. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure was ligated for hybridization. In order to obtain cDNA fragments of 150–200 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA). Subsequently, 3 μl USER Enzyme (NEB, USA) was added to adaptor-ligated cDNA of 150–200 bp and kept at 37 °C for 15 min followed by 5 min at 95 °C before PCR. PCR was performed with Phusion high-fidelity DNA polymerase, universal PCR primer, and index (X) primer. Finally, the library was purified (AMPure XP system) and then qualified by the Agilent Bioanalyzer 2100 system. Clustering of the index-coded samples was performed on a cBot Cluster Generation System using HiSeq PE Cluster Kit v4 cBot (Illumina) according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina Hiseq 2500 platform and 125 bp paired-end reads were generated. Library construction and sequencing were carried out by Novogene (Novogene, Beijing, China).
Identification of circRNAs and differential expression analysis
Raw data (raw reads in fastq format) was first processed by a custom perl script. Sequence reads with adapter, ploy-N, and low-quality were eliminated. The remaining reads were used for read quality and GC content estimation, and downstream calculation. Soybean reference genome and gene annotation were downloaded from the website at https://phytozome.jgi.doe.gov/pz/portal.html#. The index of the reference genome was built using Bowtie v2.0.6, paired-end reads were aligned to the reference genome (soybean genome version 2.0 in Phytozome) using TopHat v2.0.9. 20-mers from 5′ and 3′ end of the unmapped reads were extracted and aligned independently to reference sequences using Bowtie v2.0.6. Anchor sequences were extended by find_circ [15], such that the complete read was aligned and the breakpoints were flanked by GU/AG splice sites. Subsequently, the back-spliced reads with at least two supporting reads were annotated as circRNAs.
Expression level of circRNAs was normalized by transcript per million (TPM) through the following criteria [38]: Normalized expression = (mapped reads)/(total reads) * 1000000. Differential expression between two groups was performed using DESeq2 (version 1.6.3) [39]. Differentially expressed circRNAs were identified with the cutoff threshold of |log2 (fold-changes)| ≥ 2 based on the method used by Wang et al. [30] and Liu et al. [31].
Prediction of miRNA targets of circRNAs, mRNA targets of miRNA, and annotation of functions
miRNA binding sites of differentially expressed circRNAs were predicted by psRobot_tar in psRobot [40]. Meanwhile, the circRNA-miRNA interaction network was delineated by Cytoscape [41]. The obtained miRNAs were used to predict the target mRNAs by psRobot with default parameters [40]. The parental genes of differentially expressed circRNAs and the predicted target mRNAs were classified into different functional processes based on Gene Ontology (GO) term enrichment using the Web Gene Ontology Annotation Plot (WEGO) [42] and agriGO [43, 44], respectively. The KOBAS [45] was used for KEGG pathways enrichment analysis. The gene annotation of A. thaliana at http://www.arabidopsis.org/ was used to define functions of homologous genes in soybean.
PCR amplification and sanger sequencing
To validate the soybean circRNAs identified in this study, a set of 12 differentially expressed circRNAs were amplified with divergent primer. As a control, a pair of convergent primers were designed for gma-circRNA0002 (Additional file 1). All primers were synthesized by Invitrogen (Shanghai, China). The total and RNase R-treated RNA of each sample were used as templates. PCR products were separated using agarose gel, and each band was excised and purified using the AxyPrep DNA Gel Extraction Kit (Axygen, Suzhou, China) for Sanger-sequencing.
Quantitative real-time PCR validation
Quantitative real-time PCR (qRT-PCR) was carried out to validate differential expressional levels of circRNAs. Divergent primers were designed in order to obtain amplicon from circle template (Additional file 1). According to the instruction of the iScript Select cDNA Synthesis Kit (BIO-RAD, USA), 1 μg of the total RNA untreated with RNase R was reverse-transcribed with random primers. Expression of circRNAs was quantified using the iTaq Universal SYBR Green Supermix (BIO-RAD, USA) on the Bio-Rad CFX96 machine (CFX96 Touch, BIO-RAD, USA). All real-time PCR assays were performed with three biological replicates, and the expression of the housekeeping gene GADPH was used as a reference for data normalization. The qRT-PCR aliquot contained 2 μL cDNA, 0.6 μL of each upstream and downstream primers (10 μM), 10 μL Takara SYBR Premix Ex Taq, and 6.8 μL RNase-free ddH2O and performed with an initial denaturation at 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s, 60 °C for 30 s. The amplification curve and melting curve were examined to evaluate specific amplification. The circRNAs relative expression levels (log2 values) were calculated using the 2–ΔΔCt method. Student’s t-test was performed to compare differences of circRNAs expression between NJCMS1A and NJCMS1B. The probability threshold of significance was P < 0.05.
Protein-coding potential prediction of circRNAs
The IRES element is required to initiate translation of a mRNA sequence without a 5′-cap structure [46]. If a circRNA has at least one IRES element, it may be able to encode a protein. To predict the IRES elements in soybean circRNAs, we blasted sequences of the circRNAs to all the IRES sequences in the website (http://iresite.org/) at an E-value < 0.05 [47]. To predict the ORFs of predictive circRNAs, we used the prediction algorithm at the website (https://github.com/kadenerlab/cORF_pipeline). Briefly, the longest ORF spanning the circRNA junction was selected for further analysis. The possible coding products of the circRNAs with protein-coding potential were used to predict the conserved domains using the Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Results and discussion
Identification of circRNAs in soybean
Total amount of sequence for each RNA libraries was ≥9 Gb with Q20 ≥ 93%, Q30 ≥ 85%, and an error rate ≤ 0.05 (Table 1), the depth and accuracy of high quality sequence was sufficient for subsequent analysis.
Table 1.
Summary of circRNA sequencing data
| Sample | Replicates | Number of raw reads | Number of clean reads | Clean bases (Gb) | Error rate (100%) | Q20 (100%) | Q30 (100%) | GC content (100%) |
|---|---|---|---|---|---|---|---|---|
| NJCMS1A-1 | 84,474,228 | 80,699,898 | 12.1 | 0.05 | 93.38 | 85.41 | 54.16 | |
| NJCMS1A | NJCMS1A-2 | 67,528,014 | 63,913,764 | 9.58 | 0.02 | 97.85 | 94.07 | 53.75 |
| NJCMS1A-3 | 98,550,080 | 92,713,788 | 13.9 | 0.03 | 97.29 | 93.12 | 54.55 | |
| NJCMS1B-1 | 74,471,198 | 68,964,198 | 10.34 | 0.03 | 97.39 | 93.08 | 53.11 | |
| NJCMS1B | NJCMS1B-2 | 77,162,554 | 70,527,006 | 10.58 | 0.03 | 97.04 | 93.03 | 53.94 |
| NJCMS1B-3 | 88,067,914 | 84,489,922 | 12.68 | 0.05 | 93.53 | 85.55 | 54.67 |
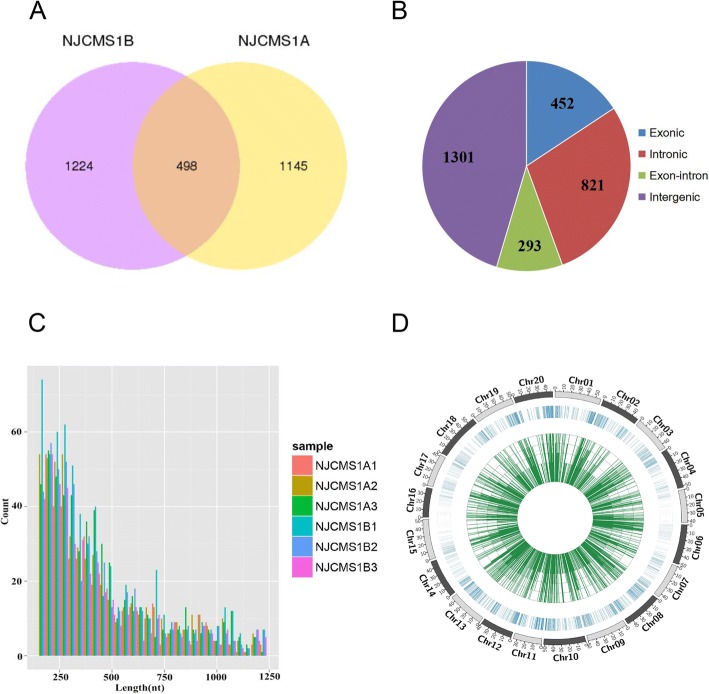
A total of 2867 circRNAs were identified, which included 1722 from NJCMS1B and 1643 from NJCMS1A (Fig. 1a, Additional file 2). Among them, 452 (15.8%) were derived from exons of a single protein-coding gene (exonic circRNAs), 821 (28.6%) from introns (intronic circRNAs), 293 (10.2%) from exons and introns of one or more genes (exon-intron circRNAs), and 1301 (45.4%) from intergenic regions (intergenic circRNAs) (Fig. 1b). CircRNAs primarily derived from exons in several plants have been reported, e.g. 5152 (85.7%) circRNAs in A. thaliana [16], 615 (72.0%) in tomato [29], and 1453 (93.7%) in maize [48]. However, only 2494 (46.4%) exonic circRNAs were identified in soybean by Zhao et al. [35], which was similar to our result. The length of circRNAs in this study ranged from 150 to 44,756 bp, but most (92.8%) were < 2000 bp (Fig. 1c). The length distribution of soybean circRNAs in this study was similar to that reported by Zhao et al. [35]. Additionally, the sequence alignment between the circRNAs inform this study and previous study (Zhao et al. [35]) using blastn indicated that only 78 circRNAs were homologous (Additional file 3) at e-value <1e− 5 and identity > 85%. The small proportion of homologous circRNAs in these two studies might be associated with tissue specific expression pattern of circRNA (Additional file 4) and use of different circRNAs prediction software. Besides, the number of circRNAs in different chromosomes and the densities of circRNAs in different chromosomal regions were also different (Fig. 1d).
Fig. 1.
CircRNA sequencing data. (a) Venn diagram shows the number of the identified circRNAs in NJCMS1A and NJCMS1B. (b) Source statistics of the circRNAs. (c) Sequence length distribution of the circRNAs in different samples. (d) Circle plot shows the distribution of the identified circRNAs in soybean chromosomes and their expression levels. The outermost layer represents all soybean chromosomes (Chr01-Chr20). The middle blue lines show the distribution of the circRNAs in soybean chromosomes, while the denser lines indicate more circRNA distribution. The innermost green lines show expression levels of the circRNAs, and the height of the lines indicates the level of expression
Expression profiling of circRNAs and qRT-PCR validation
A total of 1009 circRNAs with at least two-fold (log2) change of expression level between NJCMS1A and NJCMS1B were identified by high-throughput sequencing. Of these, a total of 360 circRNAs were up-regulated, 649 circRNAs were down-regulated in NJCMS1A vs. NJCMS1B (Additional file 5).
The back splicing sites and expression profiles of the 12 randomly selected from 1009 circRNAs were further experimentally validated. We treated total RNA samples with RNase R to verify the special stability (resistance to exonuclease-mediated degradation) of circRNAs and eliminated interference from linear RNA. Divergent primers were designed to guarantee that amplifications were from circular templates and a pair of convergent primers for gma-circRNA0002 were designed as a control. Total RNA samples and RNase R-treated samples were used as reaction templates to amplify circRNAs. As a result, all 12 pairs of divergent primers yielded amplification products with the expected length from cDNAs that with and without RNase R treatment. To demonstrate that the amplification products from the divergent primers were derived from the corresponding circRNAs and spanning the junction sites, we collected the amplification products for further detection by Sanger sequencing (Fig. 2 and Additional file 6). As shown in Fig. 3, among the 12 random-selected circRNAs, 10 showed consistent expression patterns with RNA-seq results. The coincidence rate between qRT-PCR and RNA-seq was 83.3%. Two inconsistent expression of circRNAs candidates (gma-circ2468 and gma-circ2848) may be caused by the low expression levels of the two circRNAs.
Fig. 2.
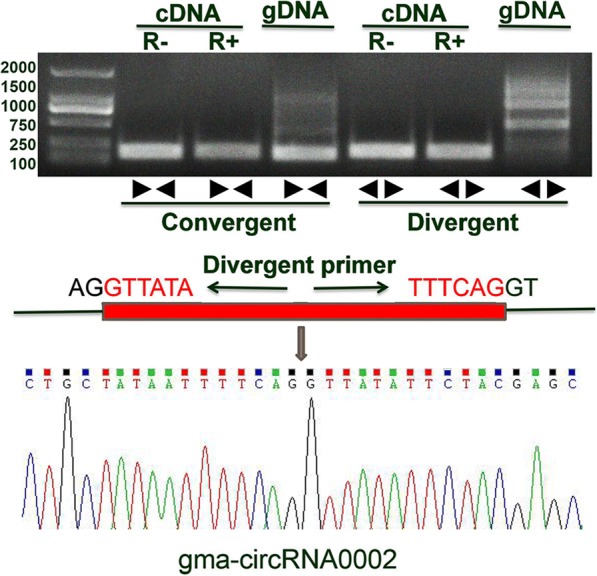
An example of circRNA (gma-circRNA0002) that was validated via amplification and sequencing. R+ and R- represent samples with and without RNase R treatment, respectively
Fig. 3.
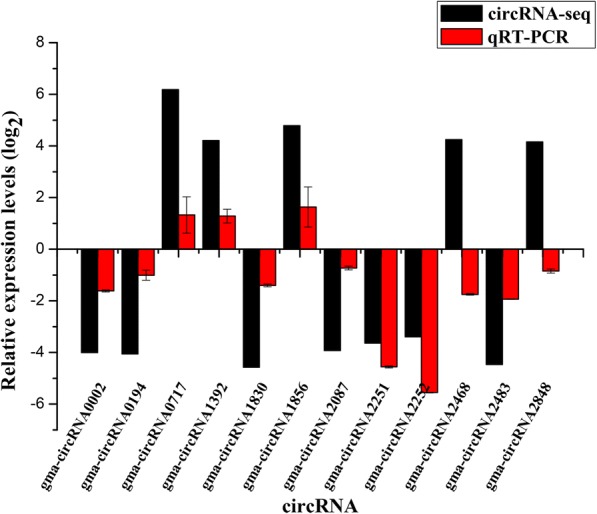
Validation of differentially expressed circRNAs in NJCMS1A and NJCMS1B by qRT-PCR. The x-axis represents the names of circRNAs, while the y-axis represents the circRNA relative expression level generated from qRT-PCR analysis and high-throughput sequencing. Expression of the GADPH gene was used as the internal reference. All qRT-PCR reactions were performed with three biological replicates, and the error bars indicate the standard errors of the means of 2–ΔΔCt, with NJCMS1B as a control
Tissue specific expression patterns validation by qRT-PCR
Previous studies showed that circRNAs were tissue-preferentially expressed [15, 24]. To verify this, two highly expressed circRNAs (gma-circRNA0002 and gma-circRNA2483) based on the high throughput sequencing were selected for qRT-PCR analysis, these two circRNAs were expressed only in leaves and flower buds of NJCMS1A and NJCMS1B, but not in roots and stems (Fig. 4). The expression level of gma-circRNA0002 was only significantly different in the flower buds, while the expression levels of gma-circRNA2483 were significantly different in the leaves and flower buds between NJCMS1A and NJCMS1B.
Fig. 4.
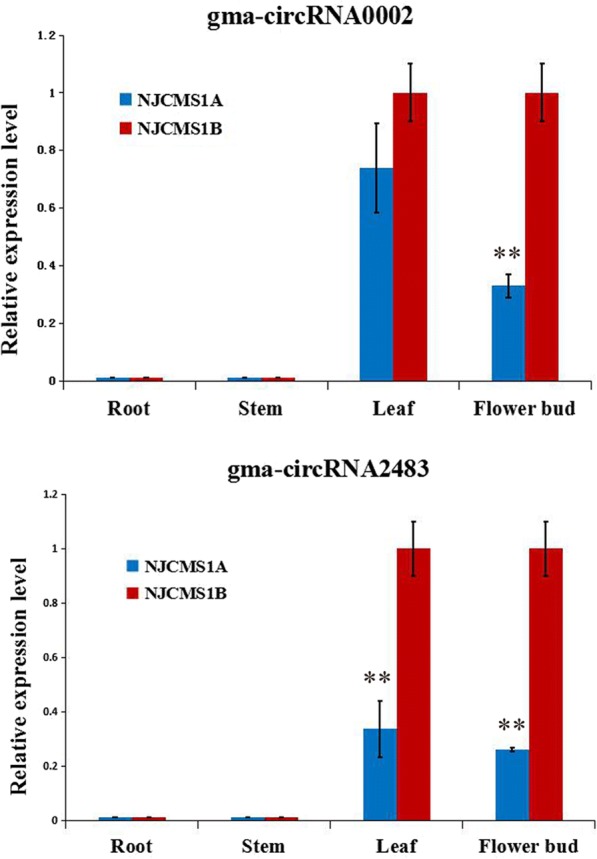
Tissue specific expression patterns validation by qRT-PCR. The relative expression levels of gma-circRNA0002 and gma-circRNA2483 were obtained from roots, stems, leaves and flower buds with three biological replicates by qRT-PCR,, and the error bars indicated the standard error of the mean of 2–ΔΔCt, with NJCMS1B as a control. The sign “**” represents P < 0.01 according to student t-test, which indicated extremely significantly differences between NJCMS1A and NJCMS1B
Functional categorization of parental genes of differentially expressed circRNAs
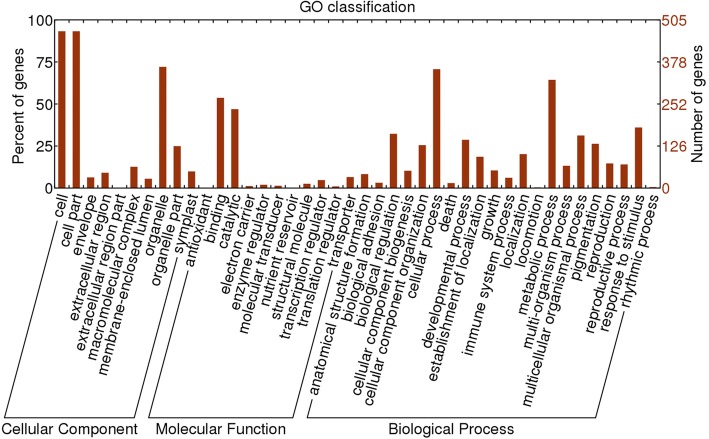
It has been reported that circRNAs could regulate expression of their parental genes [33, 34]. To understand the possible functions of circRNAs in CMS of soybean, the parental genes of the differentially expressed circRNAs were predicted. A total of 545 parental genes were obtained from the 1009 differentially expressed circRNAs. GO classification showed that these genes are involved in a wide range of biological processes, such as in metabolic process (GO:0008152), biological regulation (GO:0065007), and cellular process (GO:0009987) (Fig. 5). Interestingly, a fraction of parental genes was classified into the categories of reproduction (GO:0000003) and reproductive process (GO:0022414). Among the cellular components, cell (GO:0005623), cell part (GO:0044464), and organelle (GO:0043226) accounted for a large proportion. In the molecular functions, the two main categories were binding (GO:0005488) and catalytic (GO:0003824). GO analysis of the parental genes showed that the differentially expressed circRNAs from NJCNS1A and NJCMS1B were associated with various functions in different biological processes, cellular components, and molecular function, indicating that circRNAs may play an important role in the fertility of soybean. KEGG pathway analysis identified 7 pathways including valine, leucine and isoleucine degradation, selenocompound metabolism, RNA transport, synthesis and degradation of ketone bodies, ascorbate and aldarate metabolism, ubiquitin mediated proteolysis, and porphyrin and chlorophyll metabolism (Additional file 7). These pathways are mainly related to amino acid degradation and material metabolism.
Fig. 5.
Gene Ontology (GO) annotation of parental genes of the differentially expressed circRNAs between NJCMS1A and NJCMS1B
Putative functions of parental genes of differentially expressed circRNAs in flower development and male fertility
Based on the gene annotation of A. thaliana, the function of many parental genes were related to flower development and male fertility. We randomly enumerated 20 parental genes (Table 2) and selected four of them to describe their functions in detail. Glyma.06G173500, the parental gene of gma-circRNA0717, was a homolog of the Glucan Synthase-Like 8 (GSL8) gene in A. thaliana. GSL8 and Glucan Synthase-Like 10 (GSL10) were two members of the A. thaliana GSL gene family, which are independently required for male gametophyte development and plant growth. Experiments showed that GSL8 and GSL10 T-DNA insertions led to pollen sterility [49]. GLS8 was believed to be involved in the synthesis of cell wall component callose [50]. It was hypothesized that gma-circRNA0717 may play a vital role in male gametophyte development. Glyma.13G230500, the parental gene of gma-circRNA1856, was an ortholog of the maternal effect embryo arrest 18 (MEE18) gene in A. thaliana. Mutation of MEE18 may affect pollen gametogenesis, pollen germination, pollen tube growth, polarity or guidance, and pollen tube-embryo sac interactions or fertilization [51]. X-ray induced 1 gene (XRI1) was a novel DNA repair factor and was essential for male and female meiosis, and homozygous XRI1 mutants caused complete sterility in A. thaliana [52]. In this study, Glyma.18G164900, the parental gene of gma-circRNA2481, was a homolog of the XRI1 gene. Glyma.18G118100, the parental gene of gma-circRNA2454, was a homolog of squalene epoxidase 1 (SQE1) gene in A. thaliana. Previous studies showed that SQE1 mutants displayed severe growth defects in A. thaliana, including short stature, short roots, and complete infertility [53]. We speculated that differences in circRNAs expression levels may influence the functions of pollen and male gametophytes, and result in CMS in NJCMS1A by interacting with their parental genes.
Table 2.
Twenty fertility-related parental genes of differentially expressed circRNAs
| circRNAs ID | Parental genes | Homologue Genes (Arabidopsis thaliana) | Function description |
|---|---|---|---|
| gma-circRNA0457 | Glyma.04G155600 | AT2G24120 | male gametophyte development |
| gma-circRNA0717 | Glyma.06G173500 | AT2G36850 | male gametophyte development; callose component |
| gma-circRNA1324 | Glyma.09G266600 | AT3G61050 | male gametophyte development; pollen tube growth |
| gma-circRNA2270 | Glyma.16G218700 | AT5G22110 | male gametophyte development; embryo development |
| gma-circRNA2801 | Glyma.20G205100 | AT1G06750 | male gametophyte development; pollen tube growth |
| gma-circRNA0057 | Glyma.01G140600 | AT1G71820 | pollen germination; pollen tube growth |
| gma-circRNA0685 | Glyma.06G135600 | AT3G03810 | pollen tube development; chloroplast component |
| gma-circRNA0914 | Glyma.07G002300 | AT1G78900 | pollen development |
| gma-circRNA1244 | Glyma.09G103400 | AT2G37270 | pollen tube development |
| gma-circRNA1301 | Glyma.09G194900 | AT5G65930 | pollen germination; plasma membrane component |
| gma-circRNA1432 | Glyma.10G217200 | AT1G76850 | pollen germination; pollen tube growth |
| gma-circRNA1856 | Glyma.13G230500 | AT2G34090 | pollen gametogenesis; pollen germination; pollen tube-embryo sac interactions |
| gma-circRNA2481 | Glyma.18G164900 | AT5G48720 | pollen development; DNA repair; male meiosis |
| gma-circRNA2791 | Glyma.20G182800 | AT3G10380 | pollen germination; pollen tube growth |
| gma-circRNA1934 | Glyma.14G119800 | AT1G43850 | flower development; embryo development |
| gma-circRNA2102 | Glyma.15G000300 | AT1G16710 | flower development; protein amino acid acetylation |
| gma-circRNA2454 | Glyma.18G118100 | AT1G58440 | flower development; endomembrane system component |
| gma-circRNA2637 | Glyma.19G212600 | AT4G02300 | flower development; cell wall component |
| gma-circRNA0464 | Glyma.04G163400 | AT5G61150 | negative regulation of flower development |
| gma-circRNA2163 | Glyma.15G265300 | AT1G79280 | negative regulation of flower development; stamen development |
Characterization of binding miRNAs of differentially expressed circRNAs
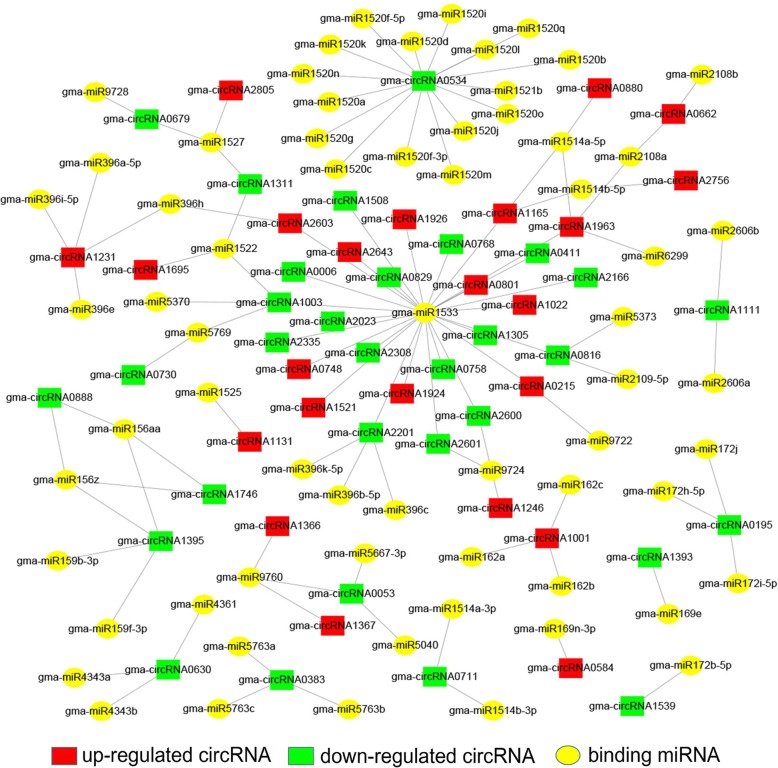
CircRNAs have been reported to act as miRNA sponges regulating gene expression [25]. To verify if circRNAs have a similar function in soybean, we predicted potential binding sites of miRNAs of the differentially expressed circRNAs. We observed that 72 differentially expressed circRNAs, of which 28 were up-regulated and 44 were down-regulated, contained 83 predicted circRNA-binding miRNAs. Of these circRNAs, only 24 had two to four miRNA binding sites (Additional file 8). Based on the predicted results, a circRNA-miRNA interaction network was delineated by Cytoscape (Fig. 6). The results showed that a single circRNA could target different miRNAs; for example, gma-circRNA0534 targeted to 16 miRNAs. Meanwhile, a single miRNA could be targeted by diverse circRNAs. For instance, gma-miR1533 was targeted by 27 circRNAs.
Fig. 6.
CircRNA-miRNA interaction network for differentially expressed circRNAs in NJCMS1A and NJCMS1B. Red squares represent up-regulated circRNAs, green squares represent down-regulated circRNAs, and yellow circles represent binding miRNAs. The figure comprises 72 differentially expressed circRNAs and their binding miRNAs. Of these circRNAs, 28 are up-regulated and 44 are down-regulated
Among the predicted miRNAs, three miRNAs (gma-miR169e, gma-miR4349, and gma-miR4993) were differentially expressed in our previous study [11]. In addition, we found that some miRNAs, such as miR156, miR162, miR169, and miR172, were important for pollen development and male fertility in soybean. miR156 is one of the highly conserved miRNA families, which was first reported in A. thaliana [54]. Squamosa promoter binding like (SPL) family proteins are involved in almost all physiological and biochemical processes such as morphogenesis, development stage transition, sporulation, flower and fruit development, external stress response, and hormone signal transduction [55]. Previous studies showed that miR156 regulated the development stage transition [56], flowering process [57], fertility maintenance [58], and fruit ripening [59] in plants by regulating members of the SPL family. Gma-circRNA0888, gma-circRNA1395, and gma-circRNA1746 could all target gma-miR156aa and gma-miR156z, which are members of miR156 family and probably involved in flower development. Furthermore, gma-circRNA1001 was predicted to target gma-miR162a, gma-miR162b, and gma-miR162c. NADP-dependent isocitrate dehydrogenase (NADP-ICDH) was targeted by gma-miR162a, gma-miR162b, and gma-miR162c according to the degradome analysis performed prior by NJCMS1A and NJCMS1B [11]. NADPH is a key cofactor in the cellular redox homeostasis, and is essential in the metabolism of reactive oxygen species (ROS) [60]. In A. thaliana, NADP-ICDH activity is regulated by molecules involved in ROS, including hydrogen peroxide (H2O2) [61]. Our previous study showed that these three miRNAs had no significant differences in expression levels between the NJCMS1A and NJCMS1B, but the target gene NADP-ICDH was down-regulated in NJCMS1A as revealed by qRT-PCR analysis [11]. The decreased NADPH level may result in significant ROS accumulation and male sterility in soybean. Moreover, gma-miR169e was targeted by gma-circRNA1393, and gma-miR169e could target dihydrolipoyl dehydrogenase (E3), an important part of the pyruvate dehydrogenase complex. Pyruvate dehydrogenase complex is a critical pathway that supports energy generation for pollen and pollen tube growth [62]. In our previous study, qRT-PCR analysis showed that the target gene E3 was down-regulated in NJCMS1A [11]. In addition, gma-circRNA0195 was predicted to target gam-miR172j, which could target Glyma.01 g188400, the homolog of the APETALA2 (AP2) of A. thaliana. In A. thaliana, miR172 could control flowering time and floral organ formation by regulating expression of the AP2-like transcription factor. A previous study indicated that over-expression of miR172 caused flower development abnormalities, and its phenotype was similar to ap2 mutants, leading to abnormal gametophyte development [63].
Functional annotation of predicted target mRNAs
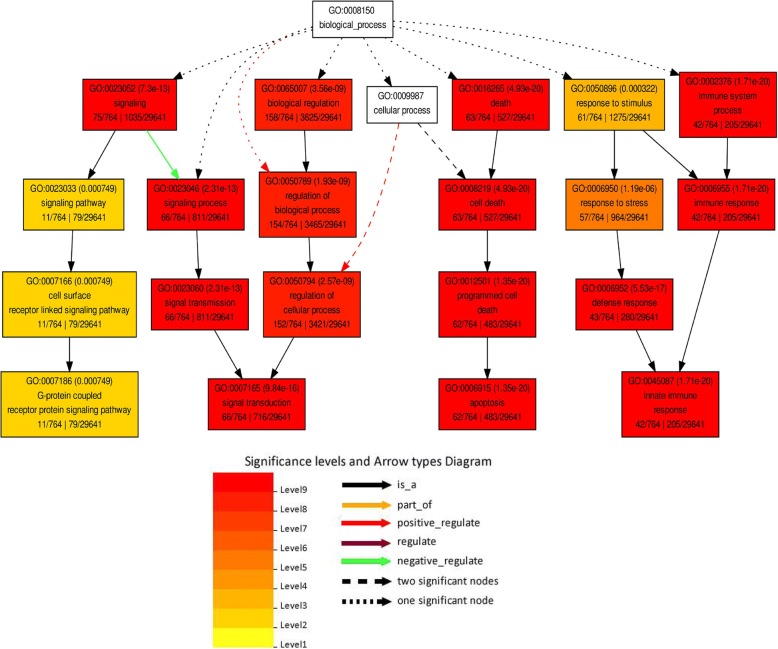
To further explore the role of circRNAs in CMS in soybean, the 83 binding miRNAs of differentially expressed circRNAs were used to predict their possible target mRNAs by psRobot with the default parameters. In total, 1166 target mRNAs were predicted and further used for functional analysis (Additional file 8). A direct acyclic graph (DAG) of biological processes was obtained using the agriGO online server (Fig. 7). The DAG can indicate submitted terms and the inter-relationships between terms. From this figure, we could identify several pathways with significant enrichment, especially those involved in signal transduction and programmed cell death (PCD). A previous study showed that pollen development depended on the interaction of multiple signaling pathways, in which calmodulin was a key element [64]. Disruption of signaling pathways might cause abnormal pollen development, resulting in male sterility in soybean. In addition, PCD in plants is a cellular process similar to apoptosis, which contains fragmentation of nuclear DNA and is controlled by mitochondrion-driven signals [3]. Plant PCD plays a role in development processes such as senescence, seed germination, organ development, root tip elongation, xylem and aerenchyma formation, and disease resistance [65]. The development of plant male gametophytes in anthers requires cooperative interactions between sporophytic (anther wall) and gametophytic (microspore) cells as well as proper PCD-controlled cellular degeneration of the tapetum and the anther wall tissue [66]. Therefore, premature or delayed PCD leads to abnormality of pollen development and tapetal function, and even male sterility [67–69]. For example, the PET1-CMS cytoplasm in sunflower causes premature PCD of the tapetal cells, which then leads to abnormal anther development [70]. The KEGG pathway analysis was also used to further explore the function of predicted target mRNAs of differentially expressed circRNAs. The target mRNAs were significantly enriched in six pathways: beta-Alanine metabolism, fatty acid degradation, lysine degradation, ascorbate and aldarate metabolism, limonene and pinene degradation, and mRNA surveillance pathway (Table 3). Several studies have shown that in most plant species, the contents of amino acids, proteins, and soluble sugars in male sterile lines were lower than those in their maintainer lines [71, 72]. In this study, the pathways of amino acid degradation and material metabolism may be important causes of CMS in soybean. GO categories and KEGG pathway analyses showed that a large number of target mRNAs were involved in pollen development and male fertility, which implied that the differentially expressed circRNAs might play an important role in CMS of soybean.
Fig. 7.
A direct acyclic graph (DAG) illustrating the biological process category generated from the Gene Ontology (GO) annotation of the predicted target miRNAs. The nodes in the image are classified into ten levels, which are associated with corresponding specific colors. The smaller of the term’s adjusted p-value, the more significant statistically, and the node’s color is darker and redder. Inside the box of the significant terms, the information include: GO term, adjusted p-value, GO description, item number mapping the GO in the query list and background, and total number of the query list and background. Different arrow types are also shown in the annotation diagram
Table 3.
KEGG pathway enrichment of target mRNAs of differentially expressed circRNAs in soybean
| Pathway ID | Description | Gene count | P-value |
|---|---|---|---|
| gmx00410 | beta-Alanine metabolism | 9 | 0.004 |
| gmx00071 | Fatty acid degradation | 7 | 0.027 |
| gmx00310 | Lysine degradation | 5 | 0.030 |
| gmx00053 | Ascorbate and aldarate metabolism | 6 | 0.032 |
| gmx00903 | Limonene and pinene degradation | 3 | 0.034 |
| gmx03015 | mRNA surveillance pathway | 12 | 0.045 |
Pathway with the threshold of P-value < 0.05 was listed
Prediction of protein-coding potential of soybean circRNAs
Recent studies have demonstrated that some circRNAs can be translated into polypeptides or proteins by translation initiation element internal ribosome entry site (IRES) [27]. To verify whether soybean circRNAs have a similar function, we predicted the protein-coding potential by blasting sequences of the circRNAs to all the IRES sequences at the website. A total of 165 soybean circRNAs contained at least one IRES element and an open reading frame (Additional file 9), which might have protein-coding potential. Furthermore, conserved domains of the possible protein-coding products were predicted by Conserved Domain Database (Additional file 10) [73], which might have important functions. Recent studies have shown that the protein-coding products of circRNAs can influence the function of their parental genes by interacting directly or indirectly with them [74, 75]. In this study, some parental genes of circRNAs with protein-coding potential were associated with flower and pollen development. For example, gma-circRNA0736 was predicted to encode an 81- amino acid protein, and its parental gene was a homolog of VERNALIZATION INDEPENDENCE 4 (VIP4) in A. thaliana. The VIP4 mutants could cause slightly early flowering and variable fertility under standard growth conditions [76]. Gma-circRNA1793 was predicted to encode a 154-amino acid protein, and its parental gene was a homolog of no pollen germination related 2 (NPGR2) in A. thaliana. The NPGR2 encodes a calmodulin-binding protein that is essential for pollen germination [77]. The protein-coding products of these circRNAs may affect flower and pollen development in soybean by affecting the function of the parental genes.
Conclusion
In this study, a total 2867 circRNAs, of which 1009 were differentially expressed between the soybean CMS line NJCMS1A and its maintainer NJCMS1B, were identified by high-throughput deep sequencing. Tissue specific expression patterns were verified by quantitative real-time PCR. The parental genes of differentially expressed circRNAs were mainly enriched in biological processes such as metabolic process, biological regulation, and reproductive process. A large number of parental genes were related to flower development and male fertility. A total of 83 binding miRNAs were predicted among the differentially expressed circRNAs, which included well-known flower and pollen development-related miRNAs. The target mRNAs predicted for the 83 binding miRNAs were significantly enriched in signal transduction and programmed cell death. A total of 165 soybean circRNAs contained at least one IRES element and an open reading frame (ORF), indicating their potential to encode polypeptides or proteins. Our study indicated that circRNAs might participate in the regulation of flower and pollen development, which could provide a new insight into the molecular mechanisms of CMS in soybean.
Additional files
Table S1. Divergent primers for validation of 12 randomly-selected differentially expressed circRNAs and qRT-PCR. (XLSX 10 kb)
Table S2. The circRNAs identified in NJCMS1A and NJCMS1B. (XLSX 254 kb)
Table S3. The homologous circRNAs identified in this study vs. previous study of Zhao et al. (XLSX 3455 kb)
Figure S1. Venn diagram shows the number of tissue preferentially expressed circRNAs in different tissues of soybean. (PDF 93 kb)
Table S4–1. The up-expressed circRNAs identified in NJCMS1A and NJCMS1B; Table S4–2. The down-expressed circRNAs identified in NJCMS1A and NJCMS1B. (XLSX 106 kb)
Figure S2. Junction sites were confirmed by Sanger sequencing. (PDF 2526 kb)
Table S5. KEGG pathway enrichment of parental genes of differentially expression circRNAs in soybean. (XLSX 9 kb)
Table S6. Predicted circRNA-miRNA-mRNA connection for differentially expressed circRNAs in NJCMS1A and NJCMS1B. (XLSX 48 kb)
Table S7. CircRNAs with protein-coding potential. (XLSX 42 kb)
Table S8. Conserved domains of the predicted protein-coding products. (XLSX 72 kb)
Acknowledgements
We would thank Novogene Bioinformatics Technology Co.Ltd. (Beijing, China) for conducting the whole-genome circRNA sequencing.
Funding
This work was financially supported by grants from the National Key R&D Program of China (2016YFD0101500, 2016YFD0101504), the National Hightech R&D Program of China (2011AA10A105), and the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT_17R55, PCSIRT13073).
Availability of data and materials
The sequencing data have been deposited in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) under accession number SRP160000.
Abbreviations
- AP2
APETALA2
- circRNA
circular RNA
- CMS
Cytoplasmic male sterility
- DAG
Direct acyclic graph
- E3
dihydrolipoyl dehydrogenase
- GO
Gene Ontology
- GSL10
Glucan Synthase-Like 10
- GSL8
Glucan Synthase-Like 8
- IRES
Internal ribosome entry site
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- m6A
N6-methyladenosine
- MEE18
Maternal effect embryo arrest 18
- miRNA
microRNA
- NADP-ICDH
NADP-dependent isocitrate dehydrogenase
- NPGR2
No pollen germination related 2
- PCD
Programmed cell death
- qRT-PCR
Quantitative real time PCR
- ROS
Reactive oxygen species
- SPL
Squamosa promoter-binding protein-like
- SQE1
Squalene epoxidase 1
- VIP4
VERNALIZATION INDEPENDENCE 4
- WEGO
Web Gene Ontology Annotation Plot
- XRI1
X-ray induced 1
Authors’ contributions
JG, SY, and LC conceived and designed the experiments. LC performed the experiments and analyzed the data. LC, XD, HZ, TH, YL, TW, XL, and LJ contributed reagents, materials, and analysis tools. LC and SY wrote the paper. JG, SY, QS, and LC revised the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The plant materials were collected from germplasms bank of National Center for Soybean Improvement. The collection and usage of samples followed the ethics of the People’s Republic of China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Linfeng Chen, Email: linfeng_chen2015@163.com.
Xianlong Ding, Email: xlding2012@163.com.
Hao Zhang, Email: haozhang1223@163.com.
Tingting He, Email: ht142857@163.com.
Yanwei Li, Email: wwyixiao2013@163.com.
Tanliu Wang, Email: tanliuwtl@126.com.
Xiaoqiang Li, Email: 15261866251@163.com.
Ling Jin, Email: msjinling@163.com.
Qijian Song, Email: qijian.song@ars.usda.gov.
Shouping Yang, Email: spyung@126.com.
Junyi Gai, Email: sri@njau.edu.cn.
References
- 1.Havey MJ. The use of cytoplasmic male sterility for hybrid seed production. Springer Netherlands. 2004;2004:623–634.
- 2.Hanson MR. Plant mitochondrial mutations and male sterility. Annu Rev Genet. 1991;25:461–486. doi: 10.1146/annurev.ge.25.120191.002333. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, Liu YG. Male sterility and fertility restoration in crops. Annu Rev Plant Biol. 2014;65:579–606. [DOI] [PubMed]
- 4.Hu J, Wang K, Huang W, Liu G, Gao Y, Wang J, et al. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell. 2012;24:109–122. doi: 10.1105/tpc.111.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujii S, Yamada M, Fujita M, Itabashi E, Hamada K, Yano K, et al. Cytoplasmic-nuclear genomic barriers in rice pollen development revealed by comparison of global gene expression profiles among five independent cytoplasmic male sterile lines. Plant Cell Physiol. 2010;51:610–620. doi: 10.1093/pcp/pcq026. [DOI] [PubMed] [Google Scholar]
- 6.Davis WH. Route to hybrid soybean production. 1985; US Patent No, 4545146.
- 7.Sun H, Zhao LM, Huang M. Studies on cytoplasmic-nuclear male sterile soybean. Chin Sci Bull. 1994;39:175–176. [Google Scholar]
- 8.Gai JY, Cui ZL, Ji DF, Ren ZJ, Ding DR. A report on the nuclear cytoplasmic male sterility from a cross between two soybean cultivars. Soy Genet Newsl. 1995;22:55–58. [Google Scholar]
- 9.Li JJ, Han SH, Ding XL, He TT, Dai JY, Yang SP, et al. Comparative transcriptome analysis between the cytoplasmic male sterile line NJCMS1A and its maintainer NJCMS1B in soybean (Glycine max (L.) Merr.) PLoS One. 2015;10:e0126771. doi: 10.1371/journal.pone.0126771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li JJ, Ding XL, Han SH, He TT, Zhang H, Yang LW, et al. Differential proteomics analysis to identify proteins and pathways associated with male sterility of soybean using iTRAQ-based strategy. J Proteomics. 2016;138:72–82. doi: 10.1016/j.jprot.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Ding XL, Li JJ, Zhang H, He TT, Han SH, Li YW, et al. Identification of miRNAs and their targets by high-throughput sequencing and degradome analysis in cytoplasmic male-sterile line NJCMS1A and its maintainer NJCMS1B of soybean. BMC Genomics. 2016;17:24. doi: 10.1186/s12864-015-2352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YW, Ding XL, Wang X, He TT, Zhang H, Yang LS, et al. Genome-wide comparative analysis of DNA methylation between soybean cytoplasmic male-sterile line NJCMS5A and its maintainer NJCMS5B. BMC Genomics. 2017;18:596. doi: 10.1186/s12864-017-3962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 17.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Innocenti N, Nguyen HS, D'hérouël AF, Aurell E. An observation of circular RNAs in bacterial RNA-seq data. arXiv preprint. 2016;arXiv(1606):04576. [Google Scholar]
- 19.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a herpesvirus saimiri noncoding RNA. Science. 2010;328(5985):1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 24.Darbani B, Noeparvar S, Borg S. Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in barley. Front Plant Sci. 2016;7:776. doi: 10.3389/fpls.2016.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 26.Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–1741. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legnini I, Timoteo GD, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun Y, Fan X, Mao M, Song X, Ping W, Yang Z, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo J, Wang Q, Zhu B, Luo Y, Gao L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Biophys Res Commun. 2016;479:132–138. doi: 10.1016/j.bbrc.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Yang M, Wei S, Qin F, Zhao H, Suo B. Identification of circular RNAs and their targets in leaves of triticum aestivum L. under dehydration stress. Front Plant Sci. 2017;7:2024. doi: 10.3389/fpls.2016.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu T, Zhang L, Chen G, Shi T. Identifying and characterizing the circular RNAs during the lifespan of Arabidopsis leaves. Front Plant Sci. 2017;8:1278. doi: 10.3389/fpls.2017.01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Liu Y, Li D, Li L, Zhang Q, Wang S, et al. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front Plant Sci. 2017;8:413. doi: 10.3389/fpls.2017.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan JJ, Zhou ZJ, Niu YJ, Sun XY, Deng ZP. Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Sci Rep. 2017;7:8594. doi: 10.1038/s41598-017-08806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao W, Cheng YH, Zhang C, You QB, Shen XJ, Guo W, et al. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci Rep. 2017;7:5636. doi: 10.1038/s41598-017-05922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding DR, Gai JY, Cui ZL, Yang SP, Qiu JX. Development and verification of the cytoplasmic-nuclear male sterile soybean line NJCMS1A and its maintainer NJCMS1B. Chin Sci Bull. 1999;44:191–192. doi: 10.1007/BF02884752. [DOI] [Google Scholar]
- 37.Ding DR, Gai JY, Cui ZL, Qiu JX. Development of a cytoplasmic-nuclear male-sterile line of soybean. Euphytica. 2002;124:85–91. doi: 10.1023/A:1015683526982. [DOI] [Google Scholar]
- 38.Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One. 2010;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu HJ, Ma YK, Chen T, Wang M, Wang XJ. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012;40:W22–W28. doi: 10.1093/nar/gks554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W67. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017; 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed]
- 45.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 46.Annecatherine P, Herve P. Translational control of gene expression: role of iress and consequences for cell transformation and angiogenesis. Prog Nucleic Acid Res Mol Biol. 2002;72:367–413. doi: 10.1016/S0079-6603(02)72075-8. [DOI] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L, Zhang P, Fan Y, Huang J, Lu Q, Li Q, et al. Transposons modulate transcriptomic and phenotypic variation via the formation of circular RNAs in maize. BioRxiv. 2017; 10.1101/100578.
- 49.Töller A, Brownfield L, Neu C, Twell D, Schulze-Lefert P. Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant J. 2008;54:911–923. doi: 10.1111/j.1365-313X.2008.03462.x. [DOI] [PubMed] [Google Scholar]
- 50.Chen XY, Liu L, Lee E, Han X, Rim Y, Chu H, et al. The Arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol. 2009;150:105–113. doi: 10.1104/pp.108.133918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boavida LC, Shuai B, Yu HJ, Pagnussat GC, Sundaresan V, Mccormick S. A collection of ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics. 2009;181:1369–1385. doi: 10.1534/genetics.108.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dean PJ, Siwiec T, Waterworth WM, Schlögelhofer P, Armstrong SJ, West CE. A novel ATM-dependent X-ray-inducible gene is essential for both plant meiosis and gametogenesis. Plant J. 2009;58:791–802. doi: 10.1111/j.1365-313X.2009.03814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lalanne E, Michaelidis C, Moore JM, Gagliano W, Johnson A, Patel R, et al. Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics. 2004;167:1975–1986. doi: 10.1534/genetics.104.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 55.Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu JH, Zhao YX, Qin YT, Xiao HL. Discovery of MicroRNAs associated with the S type cytoplasmic male sterility in maize. J Integr Agr. 2013;12:229–238. doi: 10.1016/S2095-3119(13)60222-1. [DOI] [Google Scholar]
- 57.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 58.Xing S, Salinas M, Höhmann S, Berndtgen R, Huijser P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010;22:3935–3950. doi: 10.1105/tpc.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Usami T, Horiguchi G, Yano S, Tsukaya H. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development. 2009;136:955–964. doi: 10.1242/dev.028613. [DOI] [PubMed] [Google Scholar]
- 60.Leterrier M, Barroso JB, Valderrama R, Corpas FJ. NADP-dependent isocitrate dehydrogenase from Arabidopsis roots contributes in the mechanism of defence against the nitro-oxidative stress induced by salinity. ScientificWorldJournal. 2012;2012:694740. [DOI] [PMC free article] [PubMed]
- 61.Leterrier M, Barroso JB, Palma JM, Corpas FJ. Cytosolic NADP-isocitrate dehydrogenase in Arabidopsis leaves and roots. Biol Plantarum. 2012;56:1–6.
- 62.Selinski J, Scheibe R. Pollen tube growth: where does the energy come from? Plant Signal Behav. 2014;9:e977200. doi: 10.4161/15592324.2014.977200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rato C, Monteiro D, Hepler PK, Malhó R. Calmodulin activity and cAMP signalling modulate growth and apical secretion in pollen tubes. Plant J. 2004;38:887–897. doi: 10.1111/j.1365-313X.2004.02091.x. [DOI] [PubMed] [Google Scholar]
- 65.Reape TJ, Mccabe PF. Apoptotic-like regulation of programmed cell death in plants. Apoptosis. 2010;15:249–256. doi: 10.1007/s10495-009-0447-2. [DOI] [PubMed] [Google Scholar]
- 66.Hong M. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- 67.Papini A, Mosti S, Brighigna L. Programmed-cell-death events during tapetum development of angiosperms. Protoplasma. 1999;207:213–221. doi: 10.1007/BF01283002. [DOI] [Google Scholar]
- 68.Kawanabe T, Ariizumi T, Kawai-Yamada M, Uchimiya H, Toriyama K. Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 2006;47:784–787. doi: 10.1093/pcp/pcj039. [DOI] [PubMed] [Google Scholar]
- 69.Ji C, Li H, Chen L, Xie M, Wang F, Chen Y, et al. A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol Plant. 2013;6:1715–1718. doi: 10.1093/mp/sst046. [DOI] [PubMed] [Google Scholar]
- 70.Balk J, Leaver CJ. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell. 2001;13:1803–1818. doi: 10.1105/tpc.13.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazzafera P. Biochemical characterization of anthers from male-sterile coffee plants. Revista Brasileira De Genetica. 1991;2:413–423. [Google Scholar]
- 72.Ma ZH, Sun CQ, Wang JH, Pan YP. Study on biochemical characteristics of hot pepper (Capsicum annuum) cytoplasmic male sterile line with yellow-green seedling marker. Acta Agriculturae Jiangxi. 2012;2012:26–30. [Google Scholar]
- 73.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang YB, Gao XY, Zhang ML, Yan S, Sun CJ, Xiao FZ, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2017;110(3):1. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang ML, Huang N, Yang XS, Luo JY, Yan S, Xiao FZ, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 76.Zhang H, Van Nocker S. The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J. 2002;31:663–673. doi: 10.1046/j.1365-313X.2002.01380.x. [DOI] [PubMed] [Google Scholar]
- 77.Golovkin M, Reddy AS. A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc Natl Acad Sci U S A. 2003;100:10558–10563. doi: 10.1073/pnas.1734110100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Divergent primers for validation of 12 randomly-selected differentially expressed circRNAs and qRT-PCR. (XLSX 10 kb)
Table S2. The circRNAs identified in NJCMS1A and NJCMS1B. (XLSX 254 kb)
Table S3. The homologous circRNAs identified in this study vs. previous study of Zhao et al. (XLSX 3455 kb)
Figure S1. Venn diagram shows the number of tissue preferentially expressed circRNAs in different tissues of soybean. (PDF 93 kb)
Table S4–1. The up-expressed circRNAs identified in NJCMS1A and NJCMS1B; Table S4–2. The down-expressed circRNAs identified in NJCMS1A and NJCMS1B. (XLSX 106 kb)
Figure S2. Junction sites were confirmed by Sanger sequencing. (PDF 2526 kb)
Table S5. KEGG pathway enrichment of parental genes of differentially expression circRNAs in soybean. (XLSX 9 kb)
Table S6. Predicted circRNA-miRNA-mRNA connection for differentially expressed circRNAs in NJCMS1A and NJCMS1B. (XLSX 48 kb)
Table S7. CircRNAs with protein-coding potential. (XLSX 42 kb)
Table S8. Conserved domains of the predicted protein-coding products. (XLSX 72 kb)
Data Availability Statement
The sequencing data have been deposited in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) under accession number SRP160000.