Abstract
MicroRNAs (miRNAs) being proved to be involved in the carcinogenesis of numerous tumors. MicroRNA-124 (miR-124), identified as a tumor suppressor, has been demonstrated to exert pivotal roles in multiple processes of tumorigenesis. The present study demonstrated that miR-124 was low-expressed in human hepatocellular carcinoma (HCC) tissues and cell lines. In addition, overexpression of miR-124 through infected with miR-124 lentivirus inhibited the proliferation and migration of HCC in vitro and tumorigenesis in vivo, whereas inhibition of miR-124 expression can reverse the process. Moreover, Baculoviral IAP Repeat Containing 3 (BIRC3) was identified as a target gene of miR-124. The BIRC3 mRNA expression was increased in HCC tissues and negatively correlated with miR-124 expression. Knockdown of BIRC3 recovered the miR-124-induced inhibiting effect on HCC progression. Furthermore, we found that up-regulation of miR-124 significantly inhibited p-P65, p-IκBα and c-Myc proteins expression. However, the effect of miR-124 up-regulation on HCC development was partly reversed by BIRC3 restoration. In conclusion, our data proved that miR-124 inhibits the proliferation and migration of HCC at least partly through targeting BIRC3 and regulating NF-κB signaling pathway, and it may be a therapeutic target for HCC prognosis.
Keywords: miR-124, BIRC3, Hepatocellular carcinoma, migration, NF-κB signal pathway
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and aggressive malignance in the world 1. Due to the occurrence of liver cancer metastasis, the prognosis and survival of HCC patients remain undesirability despite there are a large number of advanced facilities and medical technology 2. Therefore, the underlying molecular mechanism under investigation is urgently needed for the diagnosis and treatment of HCC patients.
microRNAs (miRNAs) are a class of short endogenously and noncoding RNAs, which have functional of regulating the expression of genes through binding directly to the 3'untranslated region (3'-UTR) of the target messenger RNAs (mRNAs). miRNAs affect the pathological development of HCC through its own alteration and dysfunction 3-5. Recent research found that miRNAs were involved in various biological processes of tumor, such as cellular differentiation, apoptosis, and mutation 6, 7. miRNA-124, a novel tumor-related miRNA, inhibited tumor development by regulating target genes expression. For example, miR-124 inhibits cell proliferation, metastasis and invasion in CRC by targeting ROCK1 8. miR-124-1 suppresses tumorigenesis in hepatocellular carcinoma by targeting CASC3 9. miR-124 reverses EMT and inhibits invasion in endometrial cancer cells via the direct repression of IQGAP1 expression 10. Although a large number of studies have demonstrated the importance of miR-124 in tumor progression, the potential molecular mechanism and regulatory signaling pathway involved in this process remain unclear.
Baculoviral IAP Repeat Containing 3 (BIRC3) is the main regulator of apoptosis that belongs to the inhibitor of apoptosis proteins (IAP) family. BIRC3 exerts a functional role in many malignancies, including oral squamous cell carcinoma, glioblastoma, pancreatic cancer. In addition, it plays an important role in regulating nuclear factor-κB (NF-κB) signaling pathway, which is involved in the development of cancer 11, 12.
In this study, we found that miR-124 inhibited the proliferation and migration of HCC cells through targeting BIRC3, and the miR-124-BIRC3 axis participated in the HCC process by regulating the NF-κB signaling pathway. Our results suggest that miR-124 functions as a tumor suppressor and it may be a therapeutic target for HCC prognosis.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of Human Research at Qingdao University, Department of Hepatobiliary Surgery. The written information were provided from all patients enrolled in the study for the publication of the data.
Human tissue specimens and cell lines
A total of 18 paired HCC tissues and adjacent non-cancerous tissues were obtained immediately following hepatic carcinectomy at the Affiliated Hospital of Qingdao University. The fresh samples were snap-frozen in liquid nitrogen after resection to extract RNA. Human HCC cell lines SMMC7721, HepG2, and HCCLM3 were purchased from the American Type Culture Collection (ATCC, Mannasas, VA, USA), HCC cell line SK-HEP1 and normal hepatic cell line HL7702 were purchased from Shanghai Genechem Co.Ltd (Shanghai, China). The cells were routinely maintained in Dulbecco's modified Eagle's medium (DMEM, HyClone, Logan, UT, USA), including 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA) and 1% penicillin-Streptomycin Solution at 37 °C under 5% CO2.
Real time quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was extracted using TRIzol Reagent (Invitrogen, USA) as per manufacturer's instructions. cDNA was synthesized from reverse transcription with RNA through miRNA reverse transcription kit (TaKaRa, ShangHai, China). The PCR amplification for the quantification was performed with specific primers and carried out using the SYBR-Green PCR system (Takara Bio, Inc.). PCR reaction procedure was 40 cycles of 95 °C 5 s, 60 °C 30 s, 94 °C 90 s. The primer sequences for the miR-124 and U6, BIRC3 and β-actin were listed in Table 2. The data were normalized using the U6 and β-actin transcripts for miRNA and genes, respectively. The relative expression of each gene was quantified by the 2-ΔΔCt method. Each reaction was performed in triplicate.
Table 2.
The sequences of primers used for qRT-PCR
| Primer | Sequence (5'→3') | |
|---|---|---|
| miR-124 | ||
| forward | GGGCGTGTTCACAGCGGAC | |
| reverse | CAGTGCGTGTCGTGGAGT | |
| U6 | ||
| forward | ATTGGAACGATACAGAGAAGATT | |
| reverse | GGAACGCTTCACGAATTTG | |
| BIRC3 | ||
| forward | CCAAGTGGTTTCCAAGGTGT | |
| reverse | TGGGCTGTCTGATGTGGATA | |
| β-actin | ||
| forward | TGACGTGGACATCCGCAAAG | |
| reverse | CTGGAAGGTGGACAGCGAGG |
Western blotting
Cells were harvested in the RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) and the protein concentration of each sample was determined using BCA protein assay reagent kit (Thermofisher Scientific, Inc) after transfection of 48 h. The supernatants containing total protein were mixed with corresponding volume of 5×SDS loading buffer (Beyotime Biotechnology, Shanghai, China) and heated at 100 °C for 10 min. The lysates of 20 μg were electrophoresed on 10% SDS-PAGE and transferred to PVDF membranes. The proteins were blocked containing 5% defatted milk for 1 h at room temperature. Subsequently, primary antibodies were incubated at 4 °C overnight, which consisted of the following: BIRC3 (Abcam, cat: ab32059, 1:1000); p-P65 (Abcam, cat: ab6503, 1:1000); P65 (Abcam, cat: ab16502, 1:1000); IκBα (CST, cat: #4812, 1:1000); p-IκBα (CST, cat: #2859, 1:1000); c-myc (CST, cat: 5605, 1:1000); GAPDH (Novus Biologicals, cat: 2D4A7, 1:5,000). Followed by incubation with the horseradish peroxidase conjugated secondary antibodies (anti-rabbit, 1:10,000; cat: #7074; or anti-mouse, 1:10,000; cat: #7076; both from CST, Inc.) for 1.5 h at room temperature. To ensure that equal amounts of sample protein were applied for electrophoresis, GAPDH was used as an internal control.
Luciferase reporter assay
All stable transfection were completed using Effectene transfection reagent (Invitrogen). Human embryonic kidney 293T (HEK293T) cells were seeded in 12-well plates and were transiently added LV-miR-124 and reporter gene plasmids. The cells were harvested for 48 h and luciferase activity was measured using a dual reporter assay kit (Promega, WI, USA) according to the manufacturer's instructions. The Renilla luciferase activity was used as normalization.
MTT and colony formation assays
Cell proliferation was performed using MTT assay kit (Solarbio, Shanghai, China) according to the manufacturer's instructions. The cells were seeded in 96-well plates and were cultured for 24 h, 48 h, 72 h and 96 h after infected with LV-miR-124 or LV-miR-CON, respectively. For colony formation assay, 8×103 cells were added into 6-well plate and suspended in DMEM medium for 10-15 day with refreshment of medium every 3 days. Then, these dishes were fixed with 100% methanol for 15 min and stained with crystal violet for 10 min. The number of colonies were observed and calculated from random area using Photoshop software. All experiments were performed in triplicate.
Wound-healing assay
Cells were seeded in twelve-well plate with DMEM medium and infected with lentiviral vectors (LV-miR-124 or LV-miR-CON) for 48 h until cell density reached 90%. The cell monolayer was scratched using 200 yellow tip and then exfoliated cells were washed with 1×PBS. Subsequently, the cells were cultured for additional 24h. The speed of wound closure was observed and photographed using Inverted microscopyy (Nikon, Tokyo, Japan). Cell mobility was assessed by measuring three randomly perpendicular wound width.
The “Transwell” cell migration assay
Corning Incorporateed Transwell Chambers (Corning, 8 μm, NY, USA) were used to detect the migration capacity in this research. Placed the chambers into the 24-well plate that contained culture medium supplemented with 20% FBS as a chemo-attractant. For HCCLM3, 2×104 cells were suspended in serum-free medium and loaded onto the upper chamber. Cells were incubated at 37 °C and allowed to invade through the membrane pores. Subsequently, the non-invading cells were removed using swab after 15 h and the lower side of the membrane were fixed with 100% methanol and stained with crystal violet. Cells that migrated onto the lower surface were counted from representative areas using ImageJ software (NIH, Bethesda, MD).
Immunofluorescence staining
The cells were planted and adhered to glass slides (NEST, USA) into 24-well plate and then infected with viral vector for addiontial 48 h. The cells were collected and washed with cold PBS, and then fixed with 4% paraformaldehyde on ice for 15 min. Subsequently, they were permeabilized with 0.3% Triton X-100 and blocked with 5% normal goat serum for 30 min at room temperature. The fixed cells were incubated overnight at 4 ˚C with antibody against anti-P65 (1:100, Abcam, USA), and followed by incubation with secondary antibody: Alexa Fluor 488-conjugated goat anti-rabbit IgG, Cy5/FITC-labeled anti-rabbit IgG (1:1000, CWBIO, China). The cover slips were visualized with DAPI staining (dilution: 1:1000, Sigma, USA) for 6 min in the dark. The fluorescence images were finally observed and analyzed by Fluorescence microscope (Nikon, Tokyo, Japan).
Immunohistochemistry (IHC)
The paraffin-embedded tissues were deparaffinized using xylene and rehydrated through graded concentrations of alcohol solution. Subsequently, the slides were incubated with 0.3% hydrogen peroxide for 30 min to inactivate endogenous peroxidase. Followed by tissue sections with citrate buffer to repair the antigen in a microwave. The slides were rinsed with 1×PBS for 15 min and followed by blocked containing goat serum for 1 h at room temperature. The primary antibodies were incubated at 4 °C overnight: a rabbit BIRC3-antibody (1:100, Abcam, cat: ab32059, USA) and a rabbit P65-antibody (1:100, Abcam, cat: ab16502, USA). After washing for 15 min with PBS, the sections were covered with secondary antibody for 1 h at room temperature. Then the sections were visualized with 3, 3'-diaminobenzidine (DAB) chromogenic fluid and counterstained with hematoxylin (CWBIO, Beijing, China).
In vivo tumorigenicity assay
Female BalB/C nude mice were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China) aged 4-6 weeks. Animals were subcutaneously injected HCCLM3-LV-miR-CON or HCCLM3-LV-miR-124 cells containing 1×106 into the dorsal flanks of mouse to detect the vivo tumorigenic capacity. The tumor growth was assessed with electronic calipers by recording tumor length and width for 4 weeks. Subsequently, the animals were sacrificed and tumor mass was collected to weigh and photograph. Animal experiments were done in accordance with protocols approved by the Animal Care and Use Committee of Qingdao University.
Statistical analysis
Statistical testing was conducted with the assistance of SPSS 17.0 software and GraphPad Prism 6. All assays were conducted three times and data were presented as mean ± SD. Statistical differences were determined by t test. Results were considered significant when P values < 0.05.
Results
Expression of miR-124 is significantly decreased in HCC tissues and cell lines
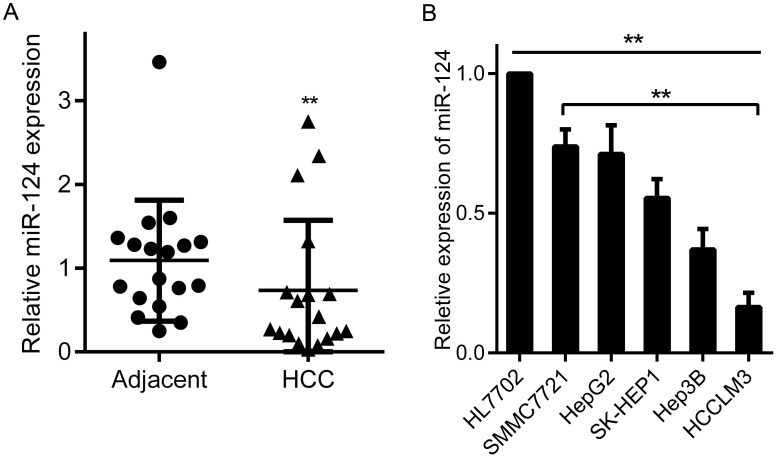
Studies have shown that miR-124 is abnormally expressed in a variety of tumors 13. Here, we firstly evaluated the expression of miR-124 in 18 pairs of HCC tumor tissues and matched adjacent non-cancerous tissues by qRT-PCR, and U6 RNA as an endogenous control. The results manifested that miR-124 expression was significantly decreased in tumor tissues than that in adjacent non-tumor tissues (Figure 1A and Table 1). Moreover, the expression of miR-124 in HCC cell lines (SMMC7721, HepG2, SK-HEP1, Hep3B and HCCLM3) were strikingly lower than normal hepatic cell line HL7702. Particularly, miR-124 has relatively highest expression in SMMC7721 cell line and the lowest expression in HCCLM3, which is cell line with strong migration ability (p < 0.01) (Figure 1B).
Figure 1.
Aberrant expression of miR-124 in HCC tissues and cell lines. (A) miR-124 expression levels were analyzed by qRT-PCR in 18 pairs of HCC tumor tissues and adjacent non-tumor tissues. (B) The expression of miR-124 was analyzed by qRT-PCR in HCC cell lines (SMMC7721, HepG2, SK-HEP1, Hep3B and HCCLM3) and normal hepatic cell line (HL7702) (**p < 0.01).
Table 1.
Relative miR-124 expression in tumor tissues compared to the matched adjacent non-tumor tissues
| Subjects | Gender | Ages | Degrees of differentiation | TNM Stages | Lymphonode metastasis | Relative miR-124 expression |
|---|---|---|---|---|---|---|
| HCC01 | M | 65 | Low | I | No | 2.11 |
| HCC02 | M | 54 | Low | II | No | 0.69 |
| HCC03 | M | 56 | Low | II | No | 0.61 |
| HCC04 | F | 64 | Low | II | No | 0.42 |
| HCC05 | F | 65 | Low | III | Yes | 0.68 |
| HCC06 | M | 40 | Low | II | No | 0.71 |
| HCC07 | M | 50 | High | II | No | 2.75 |
| HCC08 | F | 57 | High | II | No | 0.20 |
| HCC09 | M | 49 | Low | II | No | 0.27 |
| HCC10 | M | 56 | Medium | III | Yes | 0.16 |
| HCC11 | M | 56 | Medium | I | No | 0.25 |
| HCC12 | F | 21 | High | II | No | 0.23 |
| HCC13 | M | 59 | High | III | Yes | 0.08 |
| HCC14 | M | 62 | Low | III | Yes | 0.10 |
| HCC15 | M | 59 | Medium | II | No | 1.22 |
| HCC16 | M | 59 | Medium | I | No | 1.32 |
| HCC17 | M | 47 | Low | II | No | 0.34 |
| HCC18 | M | 42 | Low | II | No | 0.03 |
miR-124 inhibits the capacity of HCC cell proliferation and migration in vitro
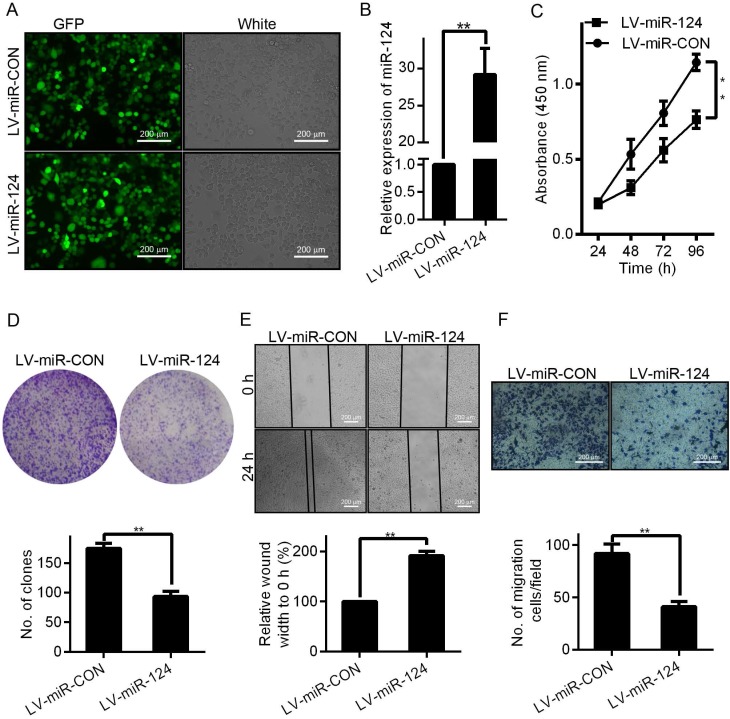
To investigate whether miR-124 affects HCC cell morphology and biological properties, we evaluated the proliferation and migration of HCC cells through a series of functional experiments. First, we successfully constructed and infected the miR-124 overexpression lentivirus vector (LV-miR-124) and corresponding control vector (LV-miR-CON) into HCCLM3 cells (Figure 2A). Compared with control group (LV-miR-CON), the expression of miR-124 was significantly increased when infected with LV-miR-124 (Figure 2B). Second, the effect of miR-124 on proliferation ability of HCC was evaluated by MTT and clone formation assays. The results demonstrated that the proliferation ability of HCCLM3 cells was reduced when infected with LV-miR-124 compared with control group (Figure 2C and 2D). Then, wound healing and Transwell assays were performed to detect the effect of miR-124 on migration ability of HCC cells. Consistently, the migration ability was also dramatically decreased when overexpression of miR-124 (Figure 2E and 2F). Taken together, these results supported that miR-124 plays an important role in regulating the biological characteristics of HCC.
Figure 2.
miR-124 up-regulation inhibits the proliferation and migration of HCC cells in vitro. (A) Observed green fluorescence under the fluorescence microscope and verified the infection efficiency of lentivirus in HCCLM3 cells. (B) Validated the expression of miR-124 when infected with LV-miR-124 or corresponding negative control (LV-miR-CON) by qRT-PCR. The cell proliferative ability was detected by MTT (C) and clone formation assays (D). The cell migration ability was validated by wound-healing (E) and transwell assays (F) (**p < 0.01).
BIRC3 is a functional target of miR-124 in HCC
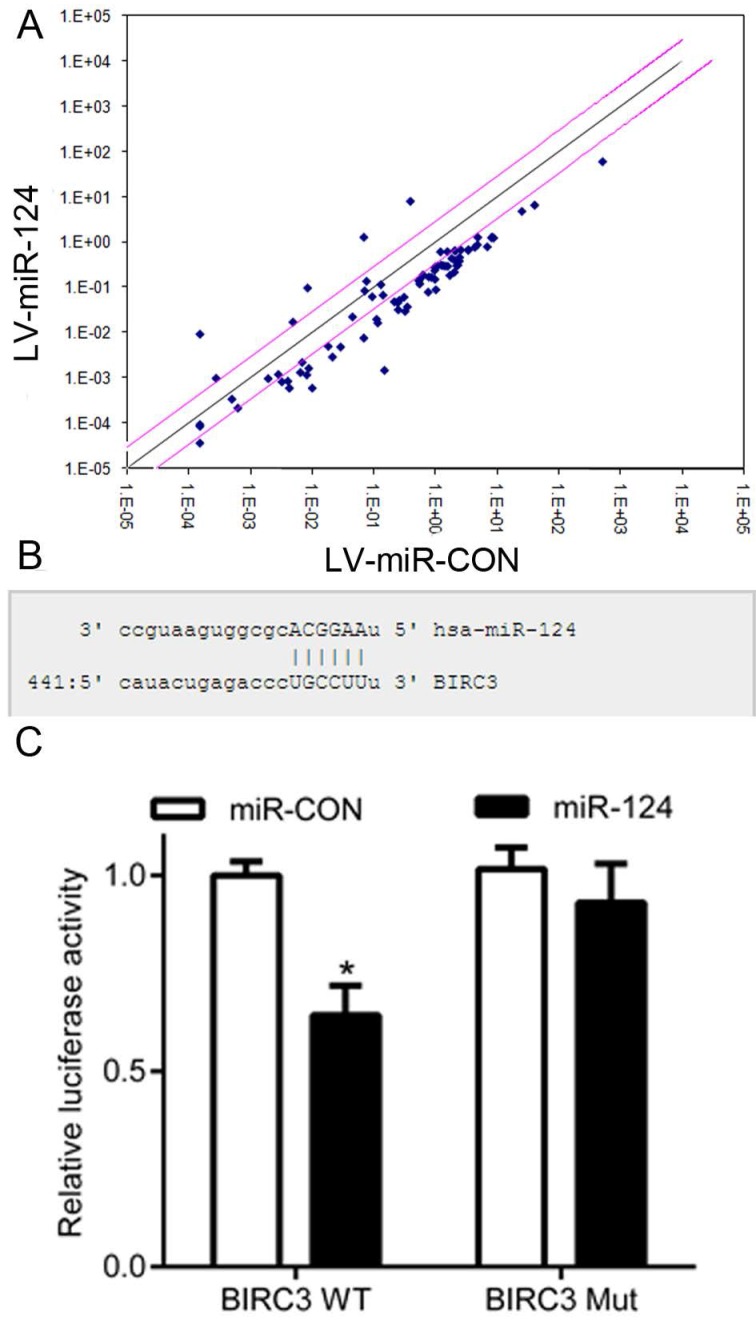
It has been reported that miRNA plays an important role in mainly regulating the expression of target genes 14, 15. In the present study, we preferred use of NF-κB Signaling Pathway PCR Array (QIAGEN, German, Cat No APH-025) to detect the expression of genes, which are involved in the NF-κB signaling pathway (Figure 3A). Subsequently, the potential targets of miR-124 were predicted using publicly available databases, such as MiRanda, PicTar and TargetScan. Then, we used the luciferase reporter assay to detect whether BIRC3 is a downstream target of miR-124 (Figure 3B). The HEK293T cells were co-transfected with pcDNAmiR-124 and BIRC3-WT-3'UTR or BIRC3-Mut-3'UTR. The result showed that miR-124 reduced the luciferase activity of wild type BIRC3 (BIRC3-WT) (p < 0.05), while no effect on the mutant type BIRC3 (BIRC3-Mut) (p > 0.05) (Figure 3C). These data indicate that BIRC3 was a potential target of miR-124 through directly targeting 3′-UTR.
Figure 3.
BIRC3 is identified as a functional target of miR-124. (A) Screening the potential targets of miR-124 using NF-κB Signaling Pathway PCR Array. (B) Binding sites of miR-124 in the BIRC3 3'UTR region. (C) Relative luciferase activity was analyzed after infected with LV-miR-124 and co-transfected reporter gene plasmids into HEK293T cells (*p < 0.05).
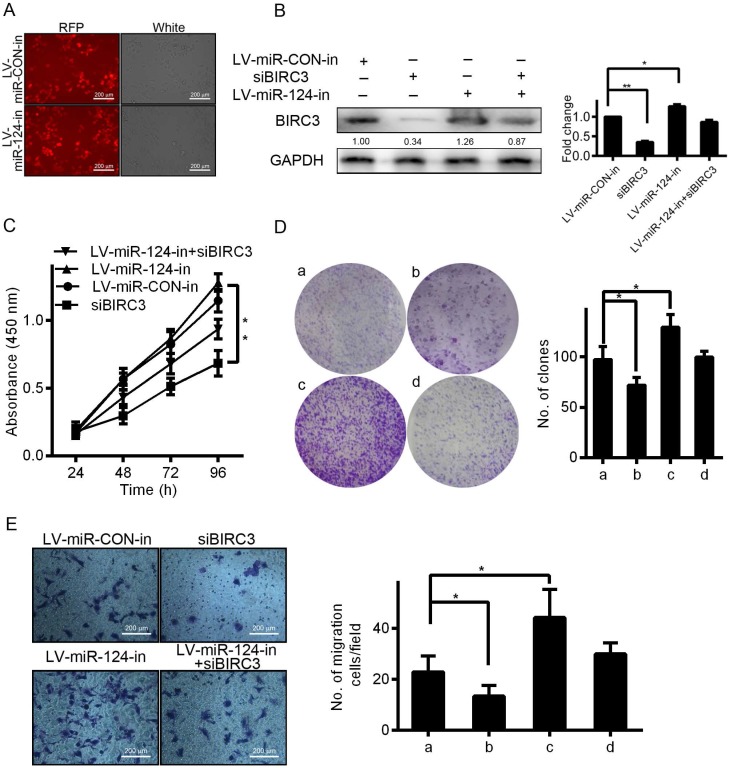
Knockdown of BIRC3 partially reverses the effect of miR-124 on HCC cell proliferation and migration
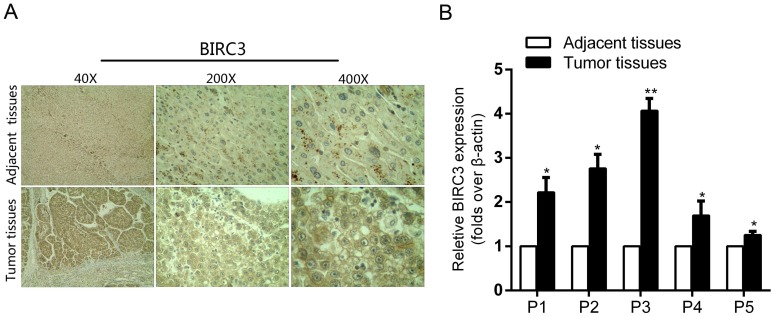
In order to explore the effect of BIRC3 on miR-124-regulated proliferation and migration, we first examined the BIRC3 expression by IHC staining and qRT-PCR assays. Our results showed that the expression of BIRC3 in tumor tissues was significantly higher than in adjacent non-tumor tissues (Figure 4A and 4B). And then we transfected miR-124 interference lentivirus (LV-miR-124-in) into SMMC7721 cells to detect infection efficiency (Figure 5A). Furthermore, a series of functional experiments have been completed to elucidate that the regulation of miR-124 on HCC progression by inhibiting BIRC3. As shown in Figure 5B, BIRC3 protein levels were restored when co-transfected with BIRC3 and miR-124 interference vectors compared with single transfected with siBIRC3. In addition, the reduction of BIRC3 can restore the promotion of cell proliferation and migration caused by miR-124 down-regulation (Figure 5C—5E). Collectively, these data indicated that the inhibitory effect of miR-124 on HCC cells can be achieved by targeting BIRC3.
Figure 4.
The expression of BIRC3 in HCC tumor tissues and matched adjacent tissues. (A) The expression of BIRC3 in HCC tumor tissues was determined by the IHC staining. (B) The relative expression of BIRC3 mRNA in the 5 paired tumor and adjacent tissues was analyzed by qRT-PCR assay (*p < 0.05, **p < 0.01).
Figure 5.
BIRC3 is involved in miR-124 suppresses proliferation and migration of HCC cells. (A) The expression of RFP fluorescence was observed under the fluorescence microscope by infected with LV-miR-124-in. (B) BIRC3 protein expression level was confirmed by western blotting in SMMC7721 cells. The cell growth curves (C), cell proliferation (D) and cell migration (E) were performed by MTT, colony formation and Transwell assays, respectively. The experiments were independently repeated three times. a: LV-miR-CON-in, b: siBIRC3, c: LV-miR-124-in, d: LV-miR-124-in+siBIRC3 (*p < 0.05, **p < 0.01).
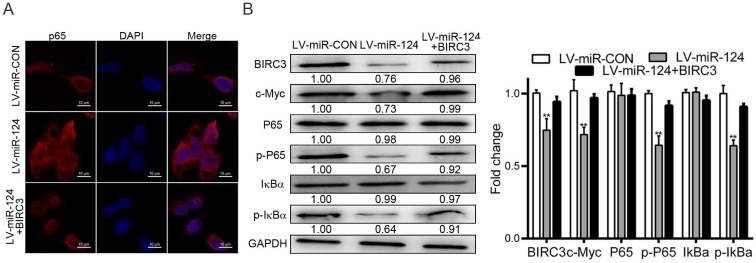
miR-124 affects the development of HCC by targeting BIRC3 and regulating NF-κB signaling pathway
NF-κB signaling pathway has drawn widely concerned in tumor formation and tumorigenesis 16, which is best known for its important role in regulating the expression of most gene families 17. To investigate the molecular mechanism of miR-124 and BIRC3 involved in the development of HCC, we preferred to detect the expression of P65 by immunofluorescence assay. The result showed that P65 was activated when BIRC3 participates in the regulation of miR-124 (Figure 6A). Previous research reported that BIRC3 is a specific transcriptional target of P65 18, which is consistent with our research. Next, the member protein levels of the NF-κB signaling pathway, including P65, p-P65, IκBα, p-IκBα and c-Myc, were detected by western blotting. The results found that overexpression of miR-124 significantly inhibited p-P65, p-IκBα and c-Myc protein expression. However, up-regulation of BIRC3 reversed the inhibition of miR-124 on the above mentioned protein expression (Figure 6B). Taken together, the results indicated that miR-124 inhibits the development of HCC through targeting BIRC3 and regulating NF-κB pathway.
Figure 6.
miR-124 affects the development of HCC by targeting BIRC3 and regulating the NF-κB pathway. (A) The expression of P65 was assessed using immunofluorescence assay when BIRC3 was involved. (B) Protein expression level of NF-κB signaling pathway, including BIRC3, p-P65, p-IκBα and c-Myc, were detected by western blotting. GAPDH was used as a negative control (**p < 0.01).
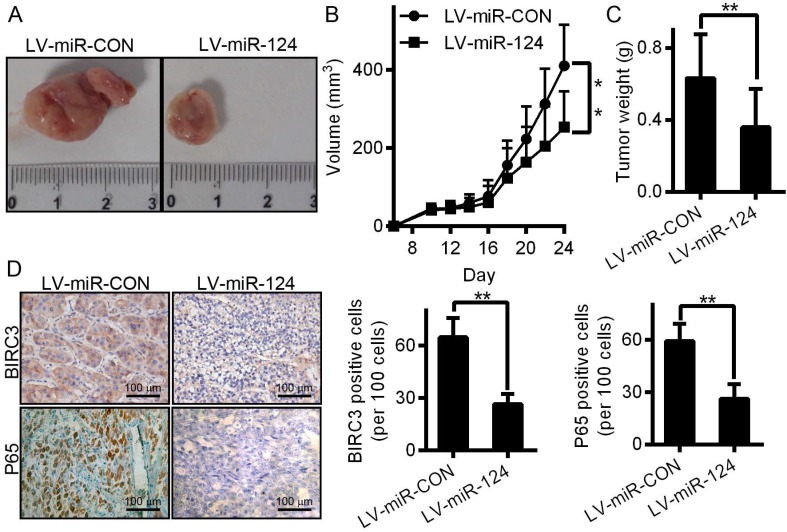
miR-124 inhibits the tumorigenesis of HCC in vivo
To better illustrate the function of miR-124 in the progression of HCC, xenograft tumor were constructed and implanted into BalB/C nude mice to examine miR-124-mediated inhibition of tumor growth in vivo (Figure 7A). Consistent with the results observed in vitro, tumor growth, including tumor volume and tumor weight, were also significantly decreased compared with the control group (Figure 7B and 7C). Meanwhile, we examined the expression of BIRC3 and P65 derived from mouse tissue samples using immunohistochemistry assay. The results showed that the expression of BIRC3 and P65 after infected with LV-miR-124 were lower than that of infected with LV-miR-CON (Figure 7D). Taken together, these data illustrated that miR-124 can effectively suppress the HCC tumorigenesis and growth in vivo.
Figure 7.
miR-124 inhibits the tumorigenesis of HCC in vivo. Mouse models were constructed through infected with LV-miR-124 or LV-miR-CON. The tumor size (A) growth curve (B) and tumor weight (C) were measured and calculated in two treatment groups. (D) The expression of BIRC3 and P65 were detected by immunohistochemistry (**p < 0.01).
Discussion
An increasing number of evidence indicates that miRNAs play a crucial role in both formation and development of cancer 19. Among the numerous miRNAs, miR-124 has been reported to be lowly expressed in many malignant tumors. Meanwhile, the downregulation of miR-124 will increase the cell proliferation and cycle progression in different cancer 20-23. Previous studies have reported that miR-124 mostly acts as a tumor suppressor gene in HCC 24, 25. In the present study, we firstly detected the expression of miR-124 in HCC. We found that miR-124 was significantly decreased in HCC tissues and cell lines. Furthermore, with many functional experiments, including MTT, clone formation, Transwell, wound-healing and tumorigenicity assays, we proved that overexpression of miR-124 inhibited HCC cell proliferation and migration in vitro and tumor growth in vivo. In contrast, knockdown of miR-124 can reverse the process. Together, these findings imply that miR-124 plays an important role in regulating the development of HCC.
BIRC3, also known as cIAP2, a member of the inhibitor of apoptotic proteins (IAP) families that inhibits apoptosis by directly inhibiting caspases cascade 26. It has been reported that BIRC3 was involved in multiple biological processes, including cell proliferation, migration and apoptosis 27, and high expression of BIRC3 was found in variety of human malignant tumors 28. For example, Gressot 29 et al reported that BIRC3 accelerated malignant transformation of low-grade gliomas (LGG) to high-grade gliomas (HGG) as a facilitator of malignant progression in glioma. Wang 30 et al found that up-regulation of BIRC3 resulted in apoptosis evasion and therapeutic resistance in glioblastoma both in vivo and in vitro. In our study, we found that BIRC3 was frequently observed to be highly expressed in HCC tumor tissues, which is negatively correlated with miR-124 expression. Further investigation showed that BIRC3 was a functional target of miR-124 in HCC. On the one hand, aberrant expression of miR-124 suppressed the activity of the luciferase reporter vector containing the BIRC3 3'UTR. On the other hand, down-regulation of BIRC3 partially reversed the effect of miR-124 reduction on HCC cell proliferation and migration. These results suggested that the regulation of miR-124 on HCC progression is partly achieved through targeting of BICR3.
Recent studies have shown that BIRC3 plays a pivotal role in regulation of NF-κB signaling pathway, which control the development of tumor promotion and tumor suppression 31-34. Sequence analysis of the BIRC3 promoter revealed two critical NF-κB and two potential activator protein-1 (AP-1)-binding sites 35. It is well known that NF-κB signaling pathway has drawn widely concerned in tumor formation and tumorigenesis. In the present study, our results manifested that the protein expression of BIRC3, p-P65, p-IκBα and c-Myc were apparently decreased in correlation with miR-124 overexpression. Whereas BIRC3 restoration eliminated the inhibition of miR-124 up-regulation on the above proteins. Thus, miR-124 may affect the development of HCC mainly through targeting BIRC3 and regulating NF-κB signaling pathway.
In conclusion, our data indicate that miR-124 was apparently down-regulated and greatly suppressed HCC cell proliferation and metastasis in vitro and in vivo. We identified BIRC3, as a novel target of miR-124, participated in the progression of HCC by regulating NF-κB signaling pathway. These findings give new insights to investigate the molecular mechanism underlying HCC development, and miR-124 can be used as a candidate diagnostic and treatment biomarker for HCC.
Acknowledgments
This work was supported by grants from the National Natural Science Foundations of China (Grant No. 81372632, 81560474 and 81402579), the Special Funds of Post-doctoral innovation projects of Shandong Province of China (Grant No. 201303063), the Application of post-doctoral funds of Qingdao city, Shandong Province of China (Grant No. 20130118), and the talent development program of The Chinese academy of science (Grant No.2014-91).
References
- 1.Guo X, Yanna, Ma X, An J, Shang Y, Huang Q. et al. A meta-analysis of array-CGH studies implicates antiviral immunity pathways in the development of hepatocellular carcinoma. PloS one. 2011;6:e28404. doi: 10.1371/journal.pone.0028404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang AC, Massague J. Molecular basis of metastasis. The New England journal of medicine. 2008;359:2814–23. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatti I, Lee A, Lund J, Larvin M. Small RNA: a large contributor to carcinogenesis? Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2009;13:1379–88. doi: 10.1007/s11605-009-0887-6. [DOI] [PubMed] [Google Scholar]
- 4.Schetter AJ, Harris CC. Alterations of microRNAs contribute to colon carcinogenesis. Seminars in oncology. 2011;38:734–42. doi: 10.1053/j.seminoncol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu WK, Law PT, Lee CW, Cho CH, Fan D, Wu K. et al. MicroRNA in colorectal cancer: from benchtop to bedside. Carcinogenesis. 2011;32:247–53. doi: 10.1093/carcin/bgq243. [DOI] [PubMed] [Google Scholar]
- 6.Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K. et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer research. 2008;68:10307–14. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Y, Ji B, Song R, Wang S, Li T, Zhang X. et al. Accurate detection for a wide range of mutation and editing sites of microRNAs from small RNA high-throughput sequencing profiles. Nucleic acids research. 2016;44:e123. doi: 10.1093/nar/gkw471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L, Xu Z, Ren X, Chen K, Xin S. MicroRNA-124 (MiR-124) Inhibits Cell Proliferation, Metastasis and Invasion in Colorectal Cancer by Downregulating Rho-Associated Protein Kinase 1(ROCK1) Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2016;38:1785–95. doi: 10.1159/000443117. [DOI] [PubMed] [Google Scholar]
- 9.Xu L, Dai W, Li J, He L, Wang F, Xia Y. et al. Methylation-regulated miR-124-1 suppresses tumorigenesis in hepatocellular carcinoma by targeting CASC3. Oncotarget. 2016;7:26027–41. doi: 10.18632/oncotarget.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong P, Ihira K, Xiong Y, Watari H, Hanley SJ, Yamada T. et al. Reactivation of epigenetically silenced miR-124 reverses the epithelial-to-mesenchymal transition and inhibits invasion in endometrial cancer cells via the direct repression of IQGAP1 expression. Oncotarget. 2016;7:20260–70. doi: 10.18632/oncotarget.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alhourani E, Othman MA, Melo JB, Carreira IM, Grygalewicz B, Vujic D. et al. BIRC3 alterations in chronic and B-cell acute lymphocytic leukemia patients. Oncology letters. 2016;11:3240–6. doi: 10.3892/ol.2016.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortese D, Sutton LA, Cahill N, Smedby KE, Geisler C, Gunnarsson R. et al. On the way towards a 'CLL prognostic index': focus on TP53, BIRC3, SF3B1, NOTCH1 and MYD88 in a population-based cohort. Leukemia. 2014;28:710–3. doi: 10.1038/leu.2013.333. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A. et al. Reprogramming of miRNA networks in cancer and leukemia. Genome research. 2010;20:589–99. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY. et al. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology (Baltimore, Md) 2014;60:598–609. doi: 10.1002/hep.27118. [DOI] [PubMed] [Google Scholar]
- 15.Bai R, Weng C, Dong H, Li S, Chen G, Xu Z. MicroRNA-409-3p suppresses colorectal cancer invasion and metastasis partly by targeting GAB1 expression. International journal of cancer Journal international du cancer. 2015;137:2310–22. doi: 10.1002/ijc.29607. [DOI] [PubMed] [Google Scholar]
- 16.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 17.Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nature reviews Immunology. 2009;9:778–88. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, Laver T, Hong SW, Twitty GB Jr, Devos A, Devos M. et al. An NF-kappaB p65-cIAP2 link is necessary for mediating resistance to TNF-alpha induced cell death in gliomas. Journal of neuro-oncology. 2011;102:367–81. doi: 10.1007/s11060-010-0346-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q. et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer cell. 2011;19:232–43. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Cui Z, Hu Y. MicroRNA-124 suppresses Slug-mediated lung cancer metastasis. European review for medical and pharmacological sciences. 2016;20:3802–11. [PubMed] [Google Scholar]
- 21.Du S, Li H, Sun X, Li D, Yang Y, Tao Z. et al. MicroRNA-124 inhibits cell proliferation and migration by regulating SNAI2 in breast cancer. Oncology reports. 2016;36:3259–66. doi: 10.3892/or.2016.5163. [DOI] [PubMed] [Google Scholar]
- 22.Fu Y, Xiong J. MicroRNA-124 enhances response to radiotherapy in human epidermal growth factor receptor 2-positive breast cancer cells by targeting signal transducer and activator of transcription 3. Croatian medical journal. 2016;57:457–64. doi: 10.3325/cmj.2016.57.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada M, Jinnin M, Wang Z, Hirano A, Tomizawa Y, Kira T. et al. The expression of miR-124 increases in aged skin to cause cell senescence and it decreases in squamous cell carcinoma. Bioscience trends. 2016;10:454–9. doi: 10.5582/bst.2016.01102. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Yue X, Cui Y, Zhang J, Wang K. MicroRNA-124 suppresses growth of human hepatocellular carcinoma by targeting STAT3. Biochemical and biophysical research communications. 2013;441:873–9. doi: 10.1016/j.bbrc.2013.10.157. [DOI] [PubMed] [Google Scholar]
- 25.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP. et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–89. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 26.Gan H, Liu H, Zhang H, Li Y, Xu X, Xu X. et al. SHh-Gli1 signaling pathway promotes cell survival by mediating baculoviral IAP repeat-containing 3 (BIRC3) gene in pancreatic cancer cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:9943–50. doi: 10.1007/s13277-016-4898-0. [DOI] [PubMed] [Google Scholar]
- 27.Smolewski P, Robak T. Inhibitors of apoptosis proteins (IAPs) as potential molecular targets for therapy of hematological malignancies. Current molecular medicine. 2011;11:633–49. doi: 10.2174/156652411797536723. [DOI] [PubMed] [Google Scholar]
- 28.Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M. et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122–30. doi: 10.1182/blood-2015-07-659144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gressot LV, Doucette T, Yang Y, Fuller GN, Manyam G, Rao A. et al. Analysis of the inhibitors of apoptosis identifies BIRC3 as a facilitator of malignant progression in glioma. Oncotarget. 2016;8:12695–04. doi: 10.18632/oncotarget.8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D, Berglund A, Kenchappa RS, Forsyth PA, Mule JJ, Etame AB. BIRC3 is a novel driver of therapeutic resistance in Glioblastoma. Scientific reports. 2016;6:21710. doi: 10.1038/srep21710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim C, Pasparakis M. Epidermal p65/NF-kappaB signalling is essential for skin carcinogenesis. EMBO molecular medicine. 2014;6:970–83. doi: 10.15252/emmm.201303541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Zhou W, Zhong Y, Huo Y, Fan P, Zhan S. et al. Overexpression of G protein-coupled receptor GPR87 promotes pancreatic cancer aggressiveness and activates NF-kappaB signaling pathway. Molecular cancer. 2017;16:61. doi: 10.1186/s12943-017-0627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamato A, Soda M, Ueno T, Kojima S, Sonehara K, Kawazu M. et al. Oncogenic activity of BIRC2 and BIRC3 mutants independent of nuclear factor-kappaB-activating potential. Cancer science. 2015;106:1137–42. doi: 10.1111/cas.12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi D, Deaglio S, Dominguez-Sola D, Rasi S, Vaisitti T, Agostinelli C. et al. Alteration of BIRC3 and multiple other NF-kappaB pathway genes in splenic marginal zone lymphoma. Blood. 2011;118:4930–4. doi: 10.1182/blood-2011-06-359166. [DOI] [PubMed] [Google Scholar]
- 35.Piro G, Giacopuzzi S, Bencivenga M, Carbone C, Verlato G, Frizziero M. et al. TAK1-regulated expression of BIRC3 predicts resistance to preoperative chemoradiotherapy in oesophageal adenocarcinoma patients. British journal of cancer. 2015;113:878–85. doi: 10.1038/bjc.2015.283. [DOI] [PMC free article] [PubMed] [Google Scholar]