Abstract
Osteosarcoma is the most common primary malignant bone tumor. However, our understanding of the molecular mechanism underlying its pathogenesis is incomplete. Studies have shown that aberrant expression of NK homeobox (NKX) genes may be involved in the oncogenesis of various cancers. Here, through migration screening assay, we found that a series of NKX genes inhibit the migration of osteosarcoma cells. Among these genes, NKX2-2 is a bona fide tumor suppressor for osteosarcoma. Overexpression of NKX2-2 decreases the migration, invasion, proliferation and colony formation of osteosarcoma cells in vitro and suppresses tumor growth and metastasis in vivo. Moreover, based on the results from both in vitro and in vivo studies, the transcriptional activation domain of NKX2-2 is important for its tumor suppressor function. Mechanistically, we revealed that NKX2-2 acts as a tumor suppressor partially by mediating the transcriptional downregulation of COL5A2, PLAU, SEMA7A and S1PR1 genes. In summary, our studies of NKX2-2 revealed new molecular mechanisms underlying osteosarcoma proliferation and metastasis and may provide a series of potential therapeutic targets for osteosarcoma.
Keywords: osteosarcoma, tumor suppressor, metastasis, NKX2-2, transcription factor
Introduction
Osteosarcoma (OS) is the most common primary tumor of bone and typically occurs during the adolescent growth spurt 1. It has a global incidence of approximately one to three cases annually per million. OS primarily arises in children and adolescents, with a second peak in incidence in those over the age of 50 2. Current therapies incorporate surgical resection and combination chemotherapy (doxorubicin and cisplatin with or without methotrexate), resulting in a 5-year overall survival rate of ~60%-70% in patients with localized disease 3. However, survival for patients with metastatic (most commonly lung metastasis) or relapsed osteosarcoma has remained unchanged over the past 30 years, with an overall 5-year survival rate of approximately 20% 4, 5. A comprehensive understanding of the molecular mechanisms underlying osteosarcoma pathogenesis is not yet available.
Proper regulation of gene expression is essential for normal cellular development. Aberrant gene expression results in improper cell growth and differentiation, which may induce different diseases, including cancers. The gain- and loss-of-function of transcription factors have been shown to be involved in oncogenesis 6. In addition to the well-established role in primary tumor proliferation, multiple oncogenic transcription factors (TFs) are now known to regulate metastasis. For example, c-Myc is an oncogene functions in both primary tumor formation and metastasis by regulating genes involved in cell cycle control, stem cell maintenance, apoptosis and epithelial-mesenchymal transition 7. p53, a prototypical tumor suppressor, is mutated in approximately half of all human cancers, leading to unrestrained proliferation and resistance to apoptosis 8. p53 knockout mice develop tumors that are not typically metastatic, while gain-of-function mutant isoforms of p53 have been shown to give rise to tumors with higher metastatic capabilities 9. Although many genes have been proposed as drivers of osteosarcoma, only TP53, RB1, CDKN2A and MYC have been verified with certainty 10.
The NK homeobox (NKX) family consists of a large group of genes that act as transcription factors with tissue-specific distributions 11. Abnormalities in NKX genes have been associated with various cancers 12. NKX2-1 behaves as both tumor suppressor and oncogene depending on the lung cancer subtype 13. NKX3-1 is a major tumor suppressor gene in prostate cancer 14, while in primary human TAL1-expressing leukemic cells, NKX3-1 expression does not depend on the Notch pathway, and its inactivation impairs cell proliferation 15. NKX2-2 contributes to oncogenic transformation in Ewing's sarcoma, while downregulation of NKX2-2 correlates with increased tumor malignancy in glioblastoma 16, 17. However, the function of NKX genes in osteosarcoma is still largely unknown.
This study aims to investigate the role of NKX genes in osteosarcoma and explore the underlying mechanism. A series of NKX genes inhibited the migration of 143B osteosarcoma cells. We chose NKX2-2 for further study and examined its tumor suppressor function, which depends on its transcriptional activation domain, both in vitro and in vivo. Mechanistically, we discovered a series of NKX2-2 downstream effectors responsible for its tumor suppressor function, including COL5A2, PLAU, SEMA7A and S1PR1. Our studies of NKX2-2 revealed new molecular mechanisms underlying osteosarcoma proliferation and metastasis and may provide potentially new therapeutic targets for osteosarcoma.
Material and Methods
Cell culture and Reagents
Human osteosarcoma cell lines (143B, U2OS and HOS) and HEK293T cells were purchased from American Type Culture Collection (ATCC). Cells were cultured according to the manufacturer's instructions. ZOS and ZOS-M, syngeneic human osteosarcoma cell lines derived from a primary tumor and metastasis, respectively, from the same patient, were described previously 18. The primary cell cultures were grown in Dulbecco's modified Eagle medium (Invitrogen) with 10% fetal bovine serum (GIBCO), penicillin (100U/mL) and streptomycin (100U/mL) in a humidified 5% CO2 at 37°C.
Plasmids
The full-length NKX2-1, NKX2-2, NKX2-3, NKX2-5, NKX2-8, NKX3-1, NKX6-1, NKX6-2 and NKX6-3 cDNAs were cloned into the lentiviral vector pLenti-CMV-EGFP-3FLAG-PGK-zeo (Obio Technology, Shanghai, China). The FLAG tag was introduced at the amino-terminus of NKX2-2, and its mutants, and the cDNAs were cloned into the pSIN lentivirus vector using standard techniques. Oligonucleotides were inserted into the pLKO.1-puro vector to construct the specific shRNA. Oligonucleotide sequences are listed in table 1.
Table 1.
shRNA sequence
| Gene name | Oligonucleotide sequence |
|---|---|
| COL5A2 | |
| shA-F | CCGGGGGAGTCAAGTAGGACTAATGCTCGAGCATTAGTCCTACTTGACTCCCTTTTTG |
| shA-R | AATTCAAAAAGGGAGTCAAGTAGGACTAATGCTCGAGCATTAGTCCTACTTGACTCCC |
| shB-F | CCGGGGAGTCAAGTAGGACTAATGCCTCGAGGCATTAGTCCTACTTGACTCCTTTTTG |
| shB-R | AATTCAAAAAGGAGTCAAGTAGGACTAATGCCTCGAGGCATTAGTCCTACTTGACTCC |
| PLAU | |
| shA-F | CCGGGCTGAAGATCCGTTCCAAGGACTCGAGTCCTTGGAACGGATCTTCAGCTTTTTG |
| shA-R | AATTCAAAAAGCTGAAGATCCGTTCCAAGGACTCGAGTCCTTGGAACGGATCTTCAGC |
| shB-F | CCGGGCATGACTTTGACTGGAATTGCTCGAGCAATTCCAGTCAAAGTCATGCTTTTTG |
| shB-R | AATTCAAAAAGCATGACTTTGACTGGAATTGCTCGAGCAATTCCAGTCAAAGTCATGC |
| SEMA7A | |
| shA-F | CCGGGCCATTGTTCCACTCTAAATACTCGAGTATTTAGAGTGGAACAATGGCTTTTTG |
| shA-R | AATTCAAAAAGCCATTGTTCCACTCTAAATACTCGAGTATTTAGAGTGGAACAATGGC |
| shB-F | CCGGGGTCAGTGCTGCAATCCATTACTCGAGTAATGGATTGCAGCACTGACCTTTTTG |
| shB-R | AATTCAAAAAGGTCAGTGCTGCAATCCATTACTCGAGTAATGGATTGCAGCACTGACC |
| S1PR1 | |
| shA-F | CCGGGCCGCAGCAAATCGGACAATTCTCGAGAATTGTCCGATTTGCTGCGGCTTTTTG |
| shA-R | AATTCAAAAAGCCGCAGCAAATCGGACAATTCTCGAGAATTGTCCGATTTGCTGCGGC |
| shB-F | CCGGCCACAAGCACTATATCCTCTTCTCGAGAAGAGGATATAGTGCTTGTGGTTTTTG |
| shB-R | AATTCAAAAACCACAAGCACTATATCCTCTTCTCGAGAAGAGGATATAGTGCTTGTGG |
Virus Production
HEK293T cells were seeded in 6-well cell culture plate and incubated overnight (16-20 h) at 37°C in a 5% CO2 atmosphere. After seeding, HEK293T were co-transfected with lentiviral expression constructs (3 μg), viral packing plasmid (psPAX2, 2 μg), and viral envelope plasmid (pMD2.G, 1 μg) using Lipofectamine 2000 (Invitrogen) according to manufacturer's recommendations. Viral supernatant was collected and filtered through a 0.45 μm filter at 48 h post-transfection and then stored at 4°C for short-term use or -80°C for long-term storage.
Viral Transduction
2.5×105 target cells were seeded in 6-well cell culture plate. Two milliliters of the corresponding culture medium and 200 μL viral supernatant supplemented with 8 μg/mL polybrene were added to each well and incubated for 24 h, followed by replacement with growth medium. Cells were selected using zeocin (5 μg/mL) or puromycin (0.5 μg/mL) prior to use in experiments.
Boyden chamber assays
The migration and invasion of osteosarcoma cells were examined using 24-well Boyden chambers with 8 μm inserts coated without (migration) or with Matrigel (invasion) as previously described 19. A total of 105 cells per well were plated on the inserts and cultured at 37°C in the upper chambers in serum-free medium. At the indicated time points, cells inside the chamber were removed with cotton swabs. Cells that remained on the lower surface of the filter were fixed, stained with crystal violet (0.005%, Sigma) and the number of cells per field of view was counted using phase-contrast microscope. The average cell counts from three inserts per condition were used to plot results.
Western blotting
Cells were lysed in RIPA buffer supplemented with protease inhibitor and phosphatase inhibitor cocktails (Thermo Scientific). Equal amounts of protein were resolved on 4-12% Bis-Tris/PAGE, transferred onto polyvinylidene difluoride membranes and blocked with buffer [5% (wt/vol) milk, TBST (TBS, pH 7.4, 0.05% Tween-20)] for 1 h at room temperature. Primary antibodies were diluted in Primary Antibody Dilution Buffer (Beyotime) as follows: Anti-Flag (1:2000; CST, 8146S) and β-tubulin (1:2000; Proteintech, 10094-1-AP). Secondary antibodies were anti-mouse or anti-rabbit IgG (1:10000, Promega). Blots were developed using Tanon™ High-sig ECL Western Blotting Substrate and imaged with a MiniChemi (SAGECREATION) imaging system.
RNA Extraction and Real-Time qPCR
Total RNA was extracted from samples (RNAprep pure Cell/Bacteria Kit; TIANGEN) and quantified using a Nanodrop 2000 spectrophotometer prior to storage at -80°C. One microgram of RNA was reverse transcribed using a HiScript II 1st Strand cDNA Synthesis Kit (Vazyme), according to the manufacturer's recommendations. Transcripts were quantified by real-time qPCR using a LightCycler 480 instrument (Roche Diagnostics) and ChamQ SYBR qPCR Master Mix (Vazyme) according to the manufacturer's instructions. qPCR primers are listed in table 2.
Table 2.
Primers used in this study
| Primer name | Primer sequence |
|---|---|
| GAPDH F | ATCAATGGAAATCCCATCACCA |
| GAPDH R | GACTCCACGACGTACTCAGCG |
| COL5A2 F | TAGGAACTGATGGTACTCCTGG |
| COL5A2-R | GGCCTATCGGACCCTGAATAC |
| PLAU-F | GGGAATGGTCACTTTTACCGAG |
| PLAU-R | GGGCATGGTACGTTTGCTG |
| SEMA7A-F | CACCAAGACCAGGCTTACGAT |
| SEMA7A-R | ACACGGGACACATTGAGAGGA |
| S1PR1-F | GCCTCTTCCTGCTAATCAGCG |
| S1PR1-R | GCAGTACAGAATGACGATGGAG |
In Vitro Proliferation assay
Cell proliferation was determined using the MTT assay. Briefly, cells were plated at a density of 2,000 (U2OS) or 1,200 (143B) cells per well in a 96-well plate. After 1, 2, 3, 4 or 5 days of incubation, MTT was added to each well under sterile conditions (at a final concentration of 5 mg/mL), and the plates were incubated for 4 h at 37°C. Untransformed MTT was removed by aspiration, and formazan crystals were dissolved in dimethyl sulfoxide (100 μL/ well). The plate was shaken for 10 min until the crystals dissolved. The absorbance [optical density (OD)] was measured at 490 nm with an Infinite F50 microplate reader (TECAN).
Colony formation assay
Briefly, osteosarcoma cells were plated in triplicate in 6-well plates at a density of 500 (U2OS) or 300 (143B) cells per well and cultured for 12 (U2OS) or 6 (143B) days. Then cell clones were washed three times with PBS, fixed with methanol for 10 minutes, and dyed with crystal violet for 10 minutes at room temperature. Afterwards, the dye was removed by washing and colonies containing more than 50 cells were counted.
Mouse xenograft model
Animal experiments were approved by the Animal Research Committee of Sun Yat-sen University Cancer Center and were performed in accordance with established guidelines. To model osteosarcoma in vivo, we used human osteosarcoma 143B cells expressing luciferase (143B-luc) 20. Cells were grown to confluency and resuspended in sterile phosphate buffer saline (PBS). 8x105 cells were orthotopically injected into the medullary cavity of the right tibiae of 4 week old immunocompromised nude BALB/C mice purchased from Charles River Laboratories (Beijing, China). The injection was performed using a BD Ultra-Fine™ Ⅱ 8mm x 30G needle in the tibial tuberosity. The resulting tumors were measured with a caliper every 3 days. The 143B-luc tumor volume was calculated using the formula V = 4/3 π [1/4 (D1+D2)] 2. The lungs of mice bearing the osteosarcoma tumor xenografts that stably expressed luciferase were analyzed using an IVIS Lumina Imaging System (Xenogen) weekly. When the majority of mice in the control group formed lung metastases, the animals were sacrificed via cervical dislocation, and the tumor weights were measured after careful resection. In addition, the lungs were harvested, fixed with 4% paraformaldehyde, and embedded in paraffin. To quantify the number of pulmonary metastatic lesions, sequential 3-mm thick sections of whole lungs were obtained. Sections were stained with H&E and examined under a light microscopy to identify the metastases, as previously described 21.
Total RNA isolation and sequencing
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. DNA was removed by digestion with RNase-free DNase (Sigma, St. Louis, MO). RNA quality was evaluated by gel electrophoresis and a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). The cDNA sequencing libraries were prepared using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) according to the manufacturer's recommenddations and index codes were added to attribute sequences to each sample. Single-end raw sequence reads (100bp/50bp) were generated using an Illumina HiSeq2000/2500 platform according to a standard protocol. All reads were directly aligned to human transcript reference sequences from the genome website. The DEGs (differentially expression greater than 1 fold with p values less than 0.005) were identified using the DESeq R package (1.12.0). The statistical analysis was performed using R language (www.R-project.org).
Data Resources
The following osteosarcoma GEO datasets were used for gene expression analyses: GSE39055 and GSE49003. Data preprocessing and differential expression analyses were described previously 22, 23.
Statistical analysis
Statistical analyses were performed with GraphPad Prism software (GraphPad Software, Inc.). Data are presented as means ± SD. Significance was determined using an unpaired/paired t test. A detailed description of the statistical analysis performed for each experiment is also presented in the figure legends. In mouse experiments, n represents number of mice utilized in each treatment group.
Results
Multiple NKX genes, including NKX2-2, suppress the migration of osteosarcoma cells
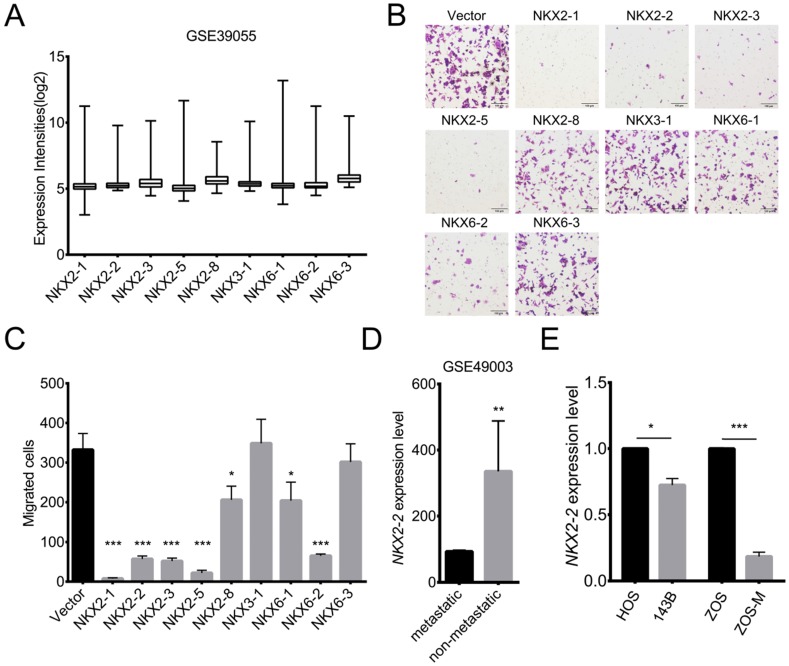
Although many studies have been performed, a comprehensive understanding of the molecular mechanisms underlying osteosarcoma pathogenesis is not available. Survival rates are poor and have remained stagnant due to the high propensity to form lung metastasis 24. Transcription factors have been shown to play important roles in tumorigenesis and metastasis 25. Abnormalities in NK homeobox genes have been associated with various cancers. However, the association of NKX genes with osteosarcoma is largely unknown. To determine which NKX genes are expressed in osteosarcoma, the GSE39055 dataset was analyzed. Fig. 1A shows 9 NKX genes that are expressed in osteosarcoma. To explore their functions, we overexpressed NKX genes in 143B cells, a highly metastatic osteosarcoma cell line. As shown in Fig. 1B and C, overexpression of NKX2-1, 2-2, 2-3, 2-5 and 6-2 dramatically suppressed the migration of 143B cells, while modest effects were observed for NKX2-8 and 6-1. Additionally, bioinformatics analyses of the GSE49003 dataset showed higher expression of NKX2-2 in non-metastatic osteosarcoma than in metastatic osteosarcoma (Fig. 1D), indicating its potential role in metastasis. To validate the relationship between NKX2-2 expression and osteosarcoma metastasis, we chose two pairs of well-established metastasis cell lines, 143B and HOS, ZOS-M and ZOS, and detected NKX2-2 expression. 143B and HOS cells originated from the same host, and the 143B cells, but not HOS cells, show high metastatic ability in vivo. ZOS and ZOS-M cells were established from the primary tumor and the skip metastasis of a patient with osteosarcoma, respectively. As shown in Fig. 1E, significantly lower NKX2-2 expression was observed in ZOS-M and 143B cells than in ZOS and HOS cells, consistent with its inhibitory role in the migration assay. Therefore, we chose NKX2-2 for further investigation.
Figure 1.
The results of the migration assay reveal multiple NXK genes, including NKX2-2, that inhibit migration. (A) Expression levels of NKX genes in osteosarcoma biopsy specimens (GSE39055: n=37). Values are presented as medians ± relative deviations. (B, C) The migration of 143B cells transduced with the indicated NKX genes was determined as described in the “Methods”. Representative images (100×) of the indicated stably transfected cell lines are shown (B). n = 5 fields per replicate; 3 replicates per condition, data are representative of three independent experiments (C). (D) Expression level of NKX2-2 in metastatic and non-metastatic osteosarcoma cell lines (GSE49003). (E) Real-time qPCR of NKX2-2 expression in osteosarcoma cells. The graph of the qPCR data shows average relative expression normalized to GAPDH. Data are representative of two independent experiments. Data are presented as the means ± SDs. Two-tailed unpaired t test: *P < 0.05, **P < 0.01 and ***P < 0.001.
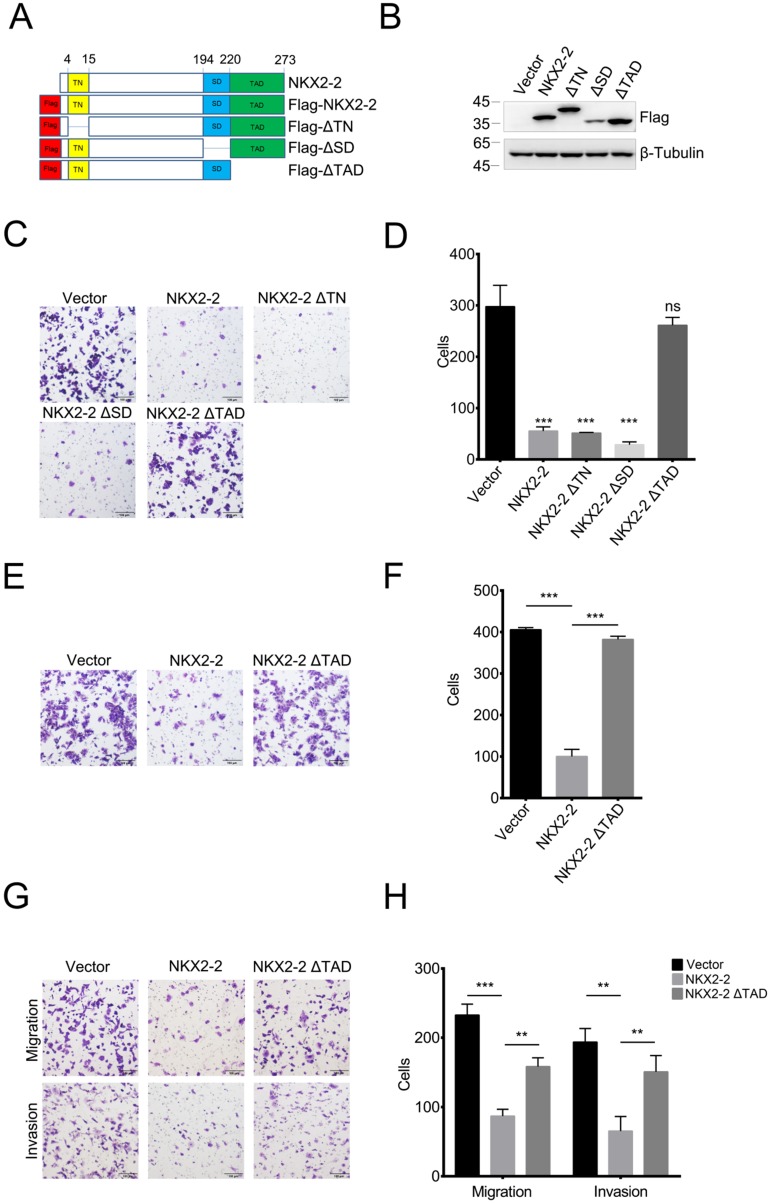
The transcriptional activation domain of NKX2-2 is responsible for its inhibitory effects on the migration and invasion of osteosarcoma cells
NKX2-2 contains three important domains: the transcriptional repression domain (TN), the specific domain (SD) and the transcriptional activation domain (TAD) 26. To determine which domain was responsible for the inhibitory effects on migration, we constructed FLAG-tagged NKX2-2 and a series of truncations and confirmed their expression levels (Fig. 2A and B). As shown in Fig. 2C and D, the truncations of the transcriptional repression domain and the specific domain did not affect the ability of NKX2-2 to inhibit the migration of 143B cells, while depletion of the transcriptional activation domain abolished its inhibitory effect on migration. In addition, NKX2-2 overexpression attenuated the ability of 143B cells to invade through Matrigel in a transwell invasion assay and depletion of the transcriptional activation domain (NKX2-2 ΔTAD) abolished its inhibitory function (Fig. 2E and F). The same results were obtained from U2OS cells in both migration and invasion assays (Fig. 2G and H).
Figure 2.
The TAD domain of NKX2-2 is essential for its inhibitory effects on migration and invasion. (A) Schematic showing the FLAG-tagged NKX2-2 constructs. The positions of the transcriptional repressor domain (TN), the NK2-specific domain (SD) and the transcriptional activation domain (TAD) are shown. (B) Western blot analysis of 143B cells infected with the indicated cDNA constructs (C, D) The migration of 143B cells transduced with indicated cDNA constructs was determined as described in the “Methods”. Representative images (100×) of the indicated stably transfected cell lines are shown (C). n = 5 fields per replicate; 3 replicates per condition, data are representative of three independent experiments (D). (E, F) The invasion of 143B cells transduced with the vector, NKX2-2 or NKX2-2 ΔTAD was determined as described in (C, D). (G, H) The migration and invasion of U2OS cells transduced with the vector, NKX2-2 or NKX2-2 ΔTAD were determined as described in (C, D). Data are presented as the means ± SDs. Two-tailed unpaired t test: **P< 0.01 and ***P < 0.001, ns: not significant.
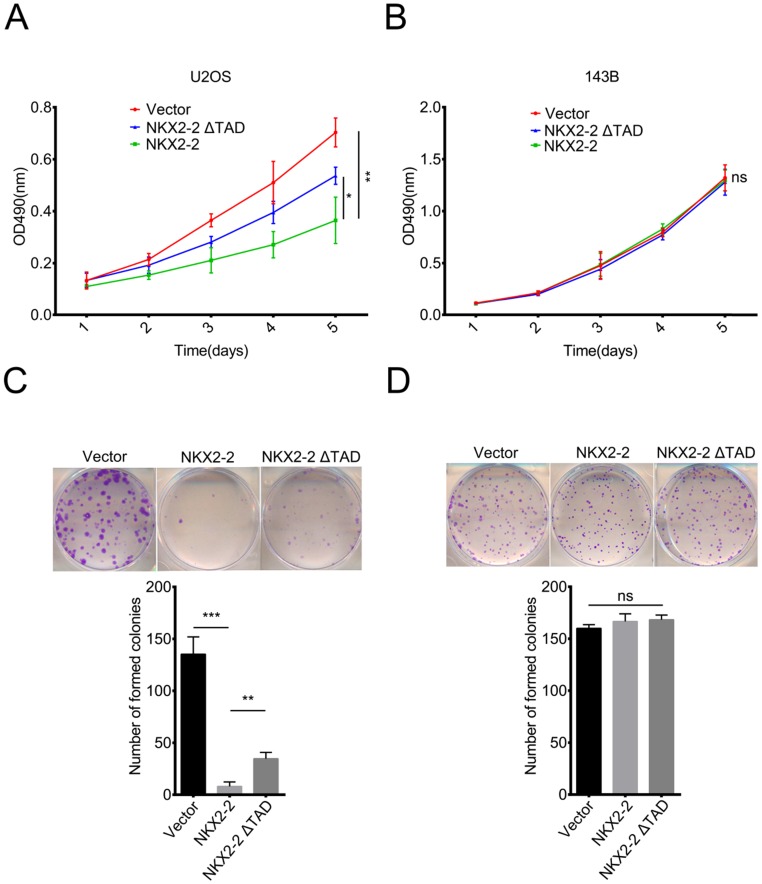
NKX2-2 overexpression impairs the proliferation of U2OS cells in vitro
In addition to its function in metastasis, we were curious about the roles of NKX2-2 in osteosarcoma proliferation and colony formation. According to the results of the MTT assay, NKX2-2 overexpression dramatically decreased the proliferation rate of U2OS cells, while NKX2-2 ΔTAD partially rescued its inhibitory effect on proliferation (Fig. 3A). Surprisingly, NKX2-2 overexpression did not affect the proliferation rate of 143B cells, indicating that NKX2-2 may not be a key regulator of 143B cell proliferation (Fig. 3B). Then we examined the effects of NKX2-2 on colony formation. As shown in Fig. 3C, NKX2-2 overexpression nearly completely abolished the formation of U2OS colonies, and the NKX2-2 ΔTAD mutant showed a moderate restoration. Although NKX2-2 overexpression did not affect the number of 143B colonies, the colony size was relatively smaller in the NKX2-2-overexpressing group than in the vector and NKX2-2 ΔTAD groups (Fig. 3D).
Figure 3.
NKX2-2 inhibits the proliferation of U2OS cells in vitro. (A and B) The proliferation of the vector control, NKX2-2 or NKX2-2 ΔTAD transduced U2OS (A) and 143B (B) was measured using MTT assays. The OD490 value was measured on the indicated day. (C and D) Osteosarcoma cells transduced with vector control, NKX2-2 or NKX2-2 ΔTAD were seeded in 6-well plates, allowed to form colonies for 12 (U2OS cells, C) or 6 (143B cells, D) days, stained with crystal violet, and then counted. (Error bars, SD; two-tailed unpaired t test: *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant).
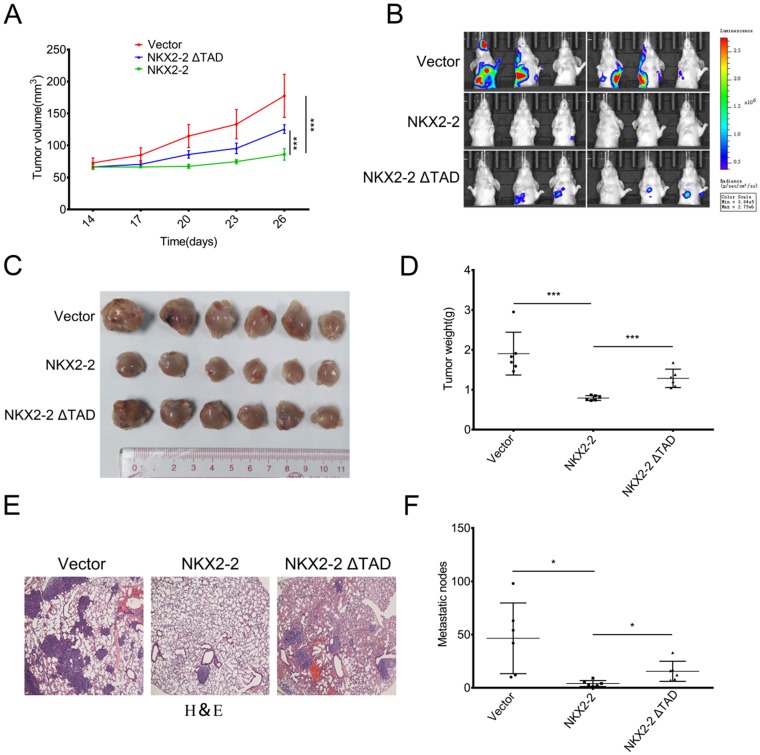
NKX2-2 overexpression inhibits the growth and metastasis of osteosarcoma in vivo
To study the effect of NKX2-2 on osteosarcoma development in vivo, osteosarcoma cells were xenografted into nude mice. Luciferase-expressing 143B (143B-luc) cells transduced with the vector control, NKX2-2 or NKX2-2 ΔTAD were orthotopically injected into right tibiae of the mice. Pulmonary metastasis was monitored longitudinally using bioluminescent imaging (BLI). The tumor volume was measured every three days beginning two weeks after injection. As shown in Fig. 4A, NKX2-2 overexpression in 143B-luc cells decreased the primary tumor growth compared with the vector control, while NKX2-2 ΔTAD lost its ability to inhibit tumor growth. Three weeks after the injection, lung metastasis was monitored weekly using bioluminescence imaging. Five weeks after the injection, the majority of the mice in the vector group developed lung metastases, whereas only one mouse in the NKX2-2-overexpressing group. The NKX2-2 ΔTAD group formed more lung metastasis than the NKX2-2 group, although still less than the vector group (Fig. 4B). Then we sacrificed all mice, all tumors were collected and weighed. Upon gross visualization of the primary 143B tumors, we observed a significant difference in tumor size between the vector, NKX2-2 and NKX2-2 ΔTAD groups (Fig. 4C and D). Lungs from all mice were simultaneously subjected to H&E staining. As shown in Fig. 4E and F, the NKX2-2 group exhibited much fewer metastatic nodules than the vector group, while NKX2-2 ΔTAD group showed a restoration of the nodule number.
Figure 4.
NKX2-2 negatively regulates the growth and metastasis of osteosarcoma in vivo. (A-F) Growth of primary tumors of 143B-luc cells transduced with the vector control, NKX2-2 or NKX2-2 ΔTAD that had been injected into the proximal tibia of nude mice (n = 6 animals/group). The mice were monitored for 35 days. The tumor volumes were monitored on day 14 and then every 3 days, as indicated. The data are presented as the mean tumor volumes ± SD from 6 mice (A). (B) The development of metastatic tumors in the lungs was monitored by measuring bioluminescence. Images of lung colonization in all mice 35 days after the injection are shown. (C and D) Xenografts were excised and weighed on day 35. Data represent as means ± SDs. ***P < 0.001 (E) Representative images of H&E staining of lungs from the indicated tumor-bearing nude mice are shown. (F) Results of the statistical analysis of the number of metastatic nodules in the lungs of nude mice are shown (error bars, SD; *P < 0.05).
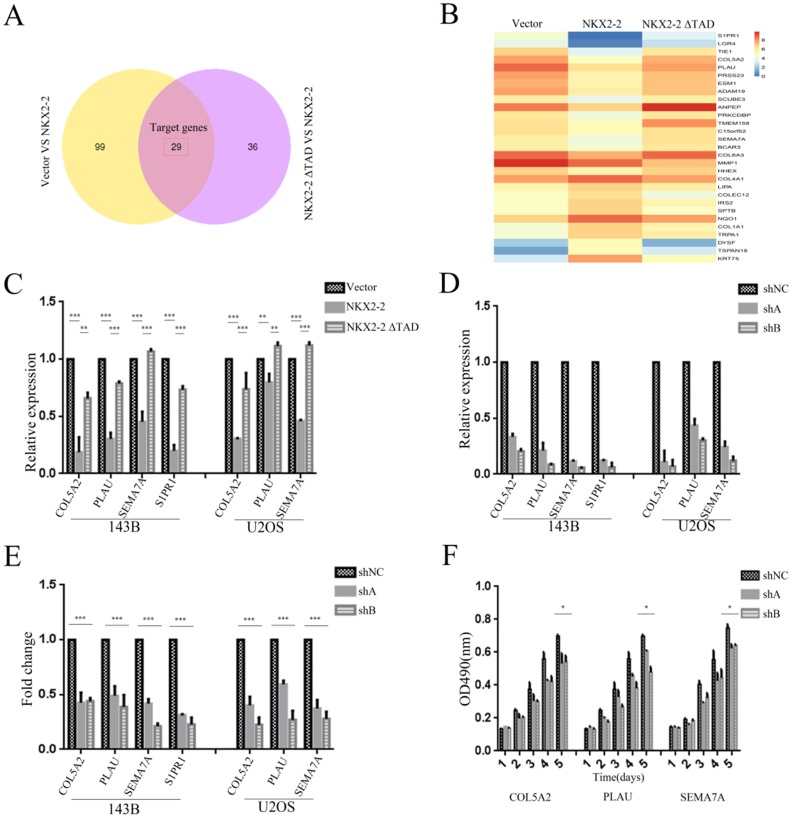
NKX2-2 exerts its tumor suppressor function partially through its downstream effectors COL5A2, PLAU, SEMA7A and S1PR1
We applied RNASeq to identify the potential functional downstream effectors of NKX2-2 and reveal the mechanism by which NKX2-2 regulates osteosarcoma proliferation and metastasis. The Venn diagram presented in Fig. 5A contains 29 genes shared by the vector versus NKX2-2 and NKX2-2 ΔTAD versus NKX2-2 comparisons. Gene expression profiles of the 29 candidate genes in the vector, NKX2-2 and NKX2-2 ΔTAD are displayed in Fig. 5B. Among these genes, several have been reported to be involved in tumor metastasis and proliferation. Collagen alpha-2 (V) chain (COL5A2), a key determinant in the assembly of tissue-specific matrices, has been reported to be associated with the malignancy status of colorectal cancer 27. Urokinase-type plasminogen activator (PLAU) converts plasminogen to plasmin and regulates cell adhesion and migration though cleavage of ECM and MMPs 28. Semaphorin-7A (SEMA7A) increases the migration of MC3T3 cells 29, and increased levels of SIPR1 in CD4 (+) T cells have been reported to promote STAT3 activation and JAK/STAT3-dependent Treg tumor migration 30. Therefore, we choose these four potential genes downstream of NKX2-2 to validate their functions in osteosarcoma metastasis and proliferation. First, we validated their expression levels in 143B and U2OS cells transduced with vector, NKX2-2 or NKX2-2 ΔTAD. Their relative expression levels were consistent with the RNASeq results, except that S1PR1 was not expressed in U2OS cells (Fig. 5C). Then we knocked down each of these genes with two separate shA and shB constructs and confirmed their knockdown efficiency by real-time PCR (Fig. 5D). Finally, the functional assay showed that knockdown of these genes dramatically inhibited the migration of both 143B and U2OS cells (Fig. 5E). A proliferation assay performed using the U2OS cell line showed a minor inhibitory effect of knockdown of these downstream genes (Fig. 5F). Based on these data, NKX2-2 may exert its tumor suppressor function by down-regulating the expression of its downstream effector genes COL5A2, PLAU, SEMA7A and S1PR1.
Figure 5.
Identification of the downstream effectors of NKX2-2 that mediates its tumor suppressive function. (A) The differentially expressed genes between the vector and NKX2-2 groups (left circle) and between the NKX2-2 ΔTAD and NKX2-2 groups (right circle) are displayed in a Venn diagram. The shared parts of two circles represented the target genes. (B) Gene expression profiles of target genes. Values are presented as log2 (fold changes). (C) Real-time qPCR analysis of COL5A2, PLAU, SEMA7A and S1PR1 expression in osteosarcoma cells transduced with the vector control, NKX2-2 or NKX2-2 ΔTAD. The graph of the qPCR data shows the average relative expression normalized to GAPDH from three replicates per condition. Data are representative of two independent experiments. (D) COL5A2, PLAU, SEMA7A and S1PR1 levels in osteosarcoma cells stably expressing shRNAs (shA and shB) were assessed by real-time qPCR as described in (C). (E) The migration of osteosarcoma cells transduced with the indicated shA, shB or shNC construct was determined as described in the “Methods”. n = 5 fields per replicate; 3 replicates per condition; data are representative of three independent experiments. (F) The proliferation of shA-, shB- or shNC-transduced U2OS was measured using MTT assays. The OD490 value was measured on the indicated day (error bars, SD; *P < 0.05; **P < 0.01; ***P < 0.001).
Discussion
In this study, a series of NKX genes suppressed the migration of osteosarcoma cells. Among these NKX genes, NKX2-2 was a bona fide suppressor of proliferation and metastasis both in vitro and in vivo. We also identified NKX2-2 downstream effectors that mediate its tumor suppressor function.
Aberrant expression of NKX genes may have an important role in tumorigenesis 12. However, few studies have examined the roles of NKX genes in osteosarcoma. NKX2-2 is required for cell fate decisions in the pancreatic islet and cell patterning in the ventral neural tube 31, 32. Subsequent studies have revealed its role in tumorigenesis. NKX2-2 is necessary for the oncogenic phenotype of Ewing's sarcoma, while its downregulation is correlated with increased tumor malignancy in glioblastoma 17. However, the role of NKX2-2 in osteosarcoma is largely unknown. Here, we showed for the first time that NKX2-2 functions as a tumor suppressor in osteosarcoma using both in vitro and in vivo experiments. In this study, although we focused on NKX2-2 and revealed its tumor suppressive function and mechanisms, other NKX genes including NKX2-1, 2-3, 2-5 and 6-2, which dramatically suppressed the migration of 143B cells, may also be involved in regulating osteosarcoma pathogenesis. Further studies examining the function and mechanism of other NKX genes in osteosarcoma are necessary and urgently needed.
NKX2-2 contains three highly conserved regions: the transcriptional activation domain (TAD), the transcriptional repression domain (TN) and the NK2-specific domain (SD) 26. Previous studies in the ventral neural tube and pancreas have shown that the TN domain of NKX2-2 is important for interaction with the transducin-like enhancer of split (Tle) proteins to regulate gene expression 33. The SD domain of NKX2-2 was reported to be important for enteroendocrine cell specification 34. However, little is known about the function of the TAD domain of NKX2-2. In our study, we analyzed three common functional domains of NKX2-2 and found that the TAD domain is of great importance for its tumor suppressive function. Also, we believed the existence of other unknown functional domains of NKX2-2 for its tumor suppressive function and this is worth further study.
The mechanisms regulating gene expression depend on the interplay between numerous transcription factors acting in concert to produce cell- and tissue-specific expression patterns 6. Much attention has been focused on transcription factors because of their crucial roles in tumorigenesis and metastasis. However, the potential use of transcription factors as direct therapeutic targets is a substantial challenge, because most of the transcription factors have been proven to be difficult to target directly 8. Although recent studies have revealed a number of potential avenues for targeting transcription factors, the classic and most frequently employed strategy is still to target the critical downstream effectors that mediate their functions. To identify the downstream effectors of NKX2-2 that mediate its tumor suppressor function, we identified a series of important effectors, including COL5A2, PLAU, SEMA7A and S1PR1 genes, using RNASeq and validated their functions. Knockdown of each of these four genes dramatically inhibited osteosarcoma metastasis and proliferation. Notably, the S1PR1 inhibitor ozanimod inhibited both migration and proliferation of osteosarcoma in vitro (data not shown); this work is still ongoing, but the results indicate that NKX2-2-S1PR1-ozanimod axis may be a promising therapeutic strategy for osteosarcoma.
In summary, our studies of NKX2-2 revealed new molecular mechanisms underlying osteosarcoma proliferation and metastasis and a series of downstream effectors were identified, which may represent new therapeutic strategies for treating osteosarcoma.
Acknowledgments
This work was supported by grants from Science and Technology Program of Guangzhou, China (Grant No.201508020102 to T. K.), the National Key Research and Development Program of China [2016YFC0904601 and 2016YFA0500304 to T. K.], the National Nature Science Foundation of China (NSFC) [81772922 to Y. W., 81502512 to D. L., 81530081 and 31571395 to T. K.], Guangdong Natural Science Foundation Team Project [2014A030312015 to T. K], the Special Key Project of Guangzhou Scientific Research [201607020038 to T. K], the Natural Science Foundation of Guangdong Province [2016A030310218 to W. Y.], and the Fundamental Research Funds for the Central Universities [16ykpy39 to W. Y.]
References
- 1.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13(8):480–91. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 2.Zambo I, Vesely K. [WHO classification of tumours of soft tissue and bone 2013: the main changes compared to the 3rd edition] Cesk Patol. 2014;50(2):64–70. [PubMed] [Google Scholar]
- 3.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Link MP, Goorin AM, Miser AW, Green AA, Pratt CB, Belasco JB. et al. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. N Engl J Med. 1986;314(25):1600–6. doi: 10.1056/NEJM198606193142502. [DOI] [PubMed] [Google Scholar]
- 5.Meyers PA, Healey JH, Chou AJ, Wexler LH, Merola PR, Morris CD. et al. Addition of pamidronate to chemotherapy for the treatment of osteosarcoma. Cancer. 2011;117(8):1736–44. doi: 10.1002/cncr.25744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ell B, Kang Y. Transcriptional control of cancer metastasis. Trends Cell Biol. 2013;23(12):603–11. doi: 10.1016/j.tcb.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418(6900):823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 8.Mees C, Nemunaitis J, Senzer N. Transcription factors: their potential as targets for an individualized therapeutic approach to cancer. Cancer Gene Ther. 2009;16(2):103–12. doi: 10.1038/cgt.2008.73. [DOI] [PubMed] [Google Scholar]
- 9.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14(11):722–35. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 11.Stanfel MN, Moses KA, Schwartz RJ, Zimmer WE. Regulation of organ development by the NKX-homeodomain factors: an NKX code. Cell Mol Biol (Noisy-le-grand) 2005;(Suppl 51):OL785–99. [PubMed] [Google Scholar]
- 12.Homminga I, Pieters R, Meijerink JP. NKL homeobox genes in leukemia. Leukemia. 2012;26(4):572–81. doi: 10.1038/leu.2011.330. [DOI] [PubMed] [Google Scholar]
- 13.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473(7345):101–4. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magee JA, Abdulkadir SA, Milbrandt J. Haploinsufficiency at the Nkx3.1 locus. A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell. 2003;3(3):273–83. doi: 10.1016/s1535-6108(03)00047-3. [DOI] [PubMed] [Google Scholar]
- 15.Kusy S, Gerby B, Goardon N, Gault N, Ferri F, Gerard D. et al. NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia. J Exp Med. 2010;207(10):2141–56. doi: 10.1084/jem.20100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR. et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9(5):405–16. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Muraguchi T, Tanaka S, Yamada D, Tamase A, Nakada M, Nakamura H. et al. NKX2.2 suppresses self-renewal of glioma-initiating cells. Cancer Res. 2011;71(3):1135–45. doi: 10.1158/0008-5472.CAN-10-2304. [DOI] [PubMed] [Google Scholar]
- 18.Zou CY, Wang J, Shen JN, Huang G, Jin S, Yin JQ. et al. Establishment and characteristics of two syngeneic human osteosarcoma cell lines from primary tumor and skip metastases. Acta Pharmacol Sin. 2008;29(3):325–32. doi: 10.1111/j.1745-7254.2008.00756.x. [DOI] [PubMed] [Google Scholar]
- 19.Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67(5):1979–87. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- 20.Liao D, Zhong L, Duan T, Zhang RH, Wang X, Wang G. et al. Aspirin Suppresses the Growth and Metastasis of Osteosarcoma through the NF-kappaB Pathway. Clin Cancer Res. 2015;21(23):5349–59. doi: 10.1158/1078-0432.CCR-15-0198. [DOI] [PubMed] [Google Scholar]
- 21.Su Y, Luo X, He BC, Wang Y, Chen L, Zuo GW. et al. Establishment and characterization of a new highly metastatic human osteosarcoma cell line. Clin Exp Metastasis. 2009;26(7):599–610. doi: 10.1007/s10585-009-9259-6. [DOI] [PubMed] [Google Scholar]
- 22.Kelly AD, Haibe-Kains B, Janeway KA, Hill KE, Howe E, Goldsmith J. et al. MicroRNA paraffin-based studies in osteosarcoma reveal reproducible independent prognostic profiles at 14q32. Genome Med. 2013;5(1):2. doi: 10.1186/gm406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diao C, Xi Y, Xiao T. Identification and analysis of key genes in osteosarcoma using bioinformatics. Oncol Lett. 2018;15(3):2789–94. doi: 10.3892/ol.2017.7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weekes D, Kashima TG, Zandueta C, Perurena N, Thomas DP, Sunters A. et al. Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene. 2016;35(22):2948. doi: 10.1038/onc.2015.420. [DOI] [PubMed] [Google Scholar]
- 25.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–51. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing's sarcoma. PLoS One. 2008;3(4):e1965. doi: 10.1371/journal.pone.0001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer H, Stenling R, Rubio C, Lindblom A. Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis. 2001;22(6):875–8. doi: 10.1093/carcin/22.6.875. [DOI] [PubMed] [Google Scholar]
- 28.Smith HW, Marshall CJ. Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol. 2010;11(1):23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 29.Delorme G, Saltel F, Bonnelye E, Jurdic P, Machuca-Gayet I. Expression and function of semaphorin 7A in bone cells. Biol Cell. 2005;97(7):589–97. doi: 10.1042/BC20040103. [DOI] [PubMed] [Google Scholar]
- 30.Priceman SJ, Shen S, Wang L, Deng J, Yue C, Kujawski M. et al. S1PR1 is crucial for accumulation of regulatory T cells in tumors via STAT3. Cell Rep. 2014;6(6):992–9. doi: 10.1016/j.celrep.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL. et al. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125(12):2213–21. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 32.Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL. et al. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398(6728):622–7. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- 33.Papizan JB, Singer RA, Tschen SI, Dhawan S, Friel JM, Hipkens SB. et al. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev. 2011;25(21):2291–305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross S, Garofalo DC, Balderes DA, Mastracci TL, Dias JM, Perlmann T. et al. The novel enterochromaffin marker Lmx1a regulates serotonin biosynthesis in enteroendocrine cell lineages downstream of Nkx2.2. Development. 2016;143(14):2616–28. doi: 10.1242/dev.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]