Abstract
Purpose:
To introduce a framework for rapid prototyping of MR pulse sequences.
Methods:
We propose a simple file format, called “TOPPE”, for specifying all details of an MR imaging experiment, such as gradient and radiofrequency (RF) waveforms and the complete scan loop. In addition, we provide a TOPPE file “interpreter” for GE scanners, which is a binary executable that loads TOPPE files and executes the sequence on the scanner. We also provide MATLAB scripts for reading and writing TOPPE files and previewing the sequence prior to hardware execution. With this setup, the task of the pulse sequence programmer is reduced to creating TOPPE files, eliminating the need for hardware-specific programming. No sequence-specific compilation is necessary; the interpreter only needs to be compiled once (for every scanner software upgrade). We demonstrate TOPPE in three different applications: k-space mapping, non-Cartesian PRESTO whole-brain dynamic imaging, and myelin mapping in the brain using inhomogeneous magnetization transfer.
Results:
We successfully implemented and executed the three example sequences. By simply changing the various TOPPE sequence files, a single binary executable (interpreter) was used to execute several different sequences.
Conclusion:
The TOPPE file format is a complete specification of an MR imaging experiment, based on arbitrary sequences of a (typically small) number of unique modules. Along with the GE interpreter, TOPPE comprises a modular and flexible platform for rapid prototyping of new pulse sequences.
INTRODUCTION
Implementing novel magnetic resonance (MR) pulse sequences on clinical scanners generally requires low-level, platform-dependent programming, which is technically challenging and time-consuming. The time from sequence design to hardware implementation can therefore be very long, and may be measured in weeks even with a skilled application scientist on staff. This presents a significant barrier to MR research, particularly for rapid prototyping of novel sequences.
To help overcome this barrier, several different programming environments have been proposed, involving various degrees of abstraction away from the physical hardware layer. KSFoundation (1) is a recently introduced application programming interface (API) for pulse programming on General Electric scanners (General Electric Healthcare, Waukesha, WI, USA), that “hides” many of the lower level programming tasks such as detailed timing specifications for individual sequence elements. Moving up one abstraction layer, SequenceTree (2) is a graphical programming interface for designing, visualizing, and simulating MR pulse sequences, that has been integrated with Siemens scanners (Siemens Medical Solutions, Erlangen, Germany) to allow direct execution on that vendor’s platform. Conceptually similar setups were introduced in (3) and (4), in which a sequence is designed within a graphical user interface (GUI) and exported to a GE or Siemens scanner for execution, respectively.
Although these programming environments can ease the burden of MR pulse programming, their use can nevertheless involve a significant learning curve. Here we introduce a further layer of abstraction, based on the idea of separating the sequence specification from both the scanner hardware and the software used to design the sequence. Specifically, we propose a simple sequence file format, which we call TOPPE (derived from “The End Of Pulse Programming”). In addition, we provide a TOPPE file “interpreter” for GE scanners, which is a binary executable that loads TOPPE files and executes the sequence on the scanner. With this setup, the task of the pulse sequence programmer is reduced to creating TOPPE files, eliminating the need for hardware-specific programming. No sequence-specific compilation is necessary; the interpreter only needs to be compiled once (for every scanner software upgrade).
We provide a MATLAB script for writing TOPPE files from a given set of arbitrary RF and gradient waveforms; the task of designing the waveforms in the first place is left to the programmer. In addition, we provide a library of complete TOPPE sequence examples that can be used as starting points for implementing new sequences.
TOPPE is similar to a recently introduced pulse sequence prototyping platform called Pulseq (5). Pulseq and TOPPE were created independently, but nevertheless share the same design philosophy: to clearly separate the sequence specification from scanner implementation. Like TOPPE, Pulseq merely specifies a file format for describing an MR imaging experiment, and does not restrict the software used to design the waveforms, or how their hardware interpreters are implemented. Indeed, our TOPPE GE interpreter was in fact the GE interpreter for Pulseq files used in (5) (on which JFN is a co-author), made possible by a simple file conversion from Pulseq to TOPPE. The Siemens and Bruker Pulseq interpreters demonstrated in (5) were developed independently by the Freiburg group. To our knowledge, TOPPE represents the only interpreter of Pulseq files for the GE platform. We provide additional discussion of TOPPE and Pulseq in Supporting Text S1.
In the following, we first describe the TOPPE file structure and sequence execution workflow. To demonstrate the broad applicability of TOPPE, we then present results for three different applications: (i) k-space trajectory and B0 (constant offset) phase measurements, (ii) T2*-weighted brain imaging using 3D stack-of-spirals echo-shifted MRI (PRESTO) (6, 7), and (iii) brain myelin imaging using inhomogeneous magnetization transfer (ihMT) imaging (8).
METHODS
TOPPE file structure and workflow
Figure 1 illustrates the three types of files required by TOPPE: (i) two or more ‘.mod’ files, (ii) one ‘modules.txt’ file, and (iii) one ‘scanloop.txt’ file. Throughout this manuscript, file names are typed in monospace font. Each .mod file specifies one unique pulse sequence segment, or module, contained in the planned MR experiment. In the example in Fig. 1, there are four unique modules: two different multi-dimensional RF excitations, one spiral-in readout with kz-encoding, and one gradient spoiler. The .mod files are the basic “building blocks” of the MR experiment, and can be shared (re-used) among different TOPPE sequences. Multiple RF and gradient waveform shapes (of equal length) can be contained in each .mod file; during sequence execution, the particular waveform to be played out is selected on-the-fly according to the corresponding entry in scanloop.txt (described below).
Figure 1:
Overview of the TOPPE file structure, example containing four unique modules: two 3D RF excitations, one spiral-in readout, and one gradient spoiler (a). Each module contains a set of arbitrary gradient waveforms and a complex RF waveform (if applicable), and is contained within a file with extension ‘.mod’. These modules are unique up to waveform scale factors, and to a rotation in the logical trans-verse (kxy) plane. The modules are listed in the file ‘modules.txt’ (b). (c) The file ‘scanloop.txt’ lists the order in which to execute the modules, and specifies all other dynamic sequence information. A few select columns are shown. Each row specifies the module number as listed in modules.txt, which of the waveforms in the .mod file to execute, RF and gradient waveform amplitudes, in-plane rotation angle (‘phi’), transmit and receive phase, a parameter ‘textra’ by which to extend module duration (for dynamic TR changes), and RF transmit frequency (for slice offsets).
The file modules.txt lists each .mod file contained in the MR experiment. In addition, modules.txt specifies module duration, and whether it is an RF excitation, data acquisition, or gradients-only module. In this way, RF (or “b1”) waveforms or data acquisition windows can be easily turned on or off, which is useful for, e.g., testing or gradient calibration purposes. If the module duration specified in modules.txt is less than the minimum achievable duration, the minimum duration is used.
Finally, scanloop.txt lists the order in which to execute the modules, and specifies waveform amplitudes and other dynamic information (see Fig. 1 caption for details). Waveform amplitudes are given in short integer (16-bit signed) format, such that the value 215 − 2 = 32766 corresponds to the full am plitude as specified in the corresponding .mod file. Unlike modules.txt, scanloop.txt can become very long for some acquisitions. For example, we use TOPPE for 3D fMRI with a scanloop.txt file containing 34,000 lines.
Once the TOPPE files have been created, they are loaded into a TOPPE binary executable (interpreter) that executes the sequence on the scanner (Fig. 2). The interpreter itself is “generic” in the sense that its source code is independent of the specifics of any one sequence; the interpreter merely loads the modules listed in modules.txt and executes them according to the instructions in scanloop.txt. Thus, the sequence specification (encapsulated in the TOPPE files) is strictly separated from sequence implementation (the interpreter).
Figure 2:
Overview of rapid prototyping of MR pulse sequences using TOPPE (continuing with example from Fig. 1). The elements within the dashed box show the novel contributions presented in this paper. After designing the RF and gradient waveforms (left), the pulse programmer uses the mat2mod.m script to write each sequence module to a file. The resulting .mod files are passed along with modules.txt and scanloop.txt, which must be created by the pulse programmer in any way he or she sees fit, to the TOPPE interpreter, a binary executable that executes the sequence on the (GE) scanner. With this setup, no sequence-specific source code compilation is required.
Creating .mod files
The task of designing the various RF and gradient waveforms that enter into each .mod file is left to the pulse programmer, who is free to choose any suitable method for this purpose. For example, in the examples presented below we designed slice-selective RF pulses using the Shinnar-Le Roux (SLR) algorithm (9), using John Pauly’s 0053LR MATLAB toolbox available at http://rsl.stanford.edu/research/software.html . We designed spiral readouts using Brian Hargreaves’ MATLAB code available at http://mrsrl.stanford.edu//~brian/vdspiral/.
Once the waveforms have been designed, the user simply converts each pulse (module) to a .mod file using the ‘mat2mod.m’ MATLAB script included in the TOPPE distribution (see Fig. 2). Also included are scripts for viewing .mod files, and for displaying the sequence in movie loop mode.
All TOPPE MATLAB cxode is made freely and publicly available under the GNU Library General Public License version 2.0, and can be downloaded via the TOPPE website, https://toppemri.github.io/ or from the TOPPE MATLAB repository, https://github.com/toppeMRI/matlab/.
This repository also contains complete sequence examples that can simply be downloaded and executed directly on the MR scanner. Also included are the sequence files for the examples presented here.
The TOPPE GE interpreter
We have developed a TOPPE file interpreter for GE scanners that is a binary executable compiled from a GE pulse sequence source file written in a C-like programming language called “EPIC”. To achieve a high degree of modularity and flexibility, the interpreter takes advantage of a convenient feature of EPIC, namely the ability to create separate modules by simply sectioning the code and loading all waveforms for a given module within a single section. For example, a section specifying a typical slice-selective RF excitation contains code for specifying three waveforms: RF magnitude (|b1|), RF phase (∠ b1), and z gradient waveforms. The TOPPE interpreter loops through the .mod files listed in modules.txt, and creates one section for each module. The modules are stored in an array whose length is dynamically allocated according to the total number of .mod files. We have determined empirically that at present, a maximum of 20 different .mod files can be loaded on our scanner. The ability to load up to 20 different modules, each capable of switching between multiple waveforms as described above, offers the sequence designer considerable flexibility.
Once the modules are loaded into memory, the scan is executed by simply stepping through the rows in scanloop.txt as described above. Hence, the notion of a “sequence repetition time”, or TR, does not strictly exist in TOPPE, except perhaps in the mind of the sequence designer. The modular design of the TOPPE interpreter makes the source code quite compact, but also introduces certain timing constraints. Specifically, the hardware imposes a small delay after the execution of a module, and in the case of data acquisition or RF excitation modules there is also a delay prior to module execution. As a result, modules are separated by ~.4–0.5 msec.
Source code for the TOPPE interpreter is posted on the GE MR online collaboration community, https://collaborate.mr.gehealthcare.com/docs/DOC-1247, and on Github, https://github.com/toppeMRI/psd/.
Imaging experiments
We performed imaging experiments on a GE Discovery MR750 3T scanner, using a 32-channel receive head array and body RF coil transmission. We reconstructed all images offline (in Matlab), since the TOPPE interpreter does not (at present) allow for online reconstruction at the MR console.
Measuring k-space and B0 errors
In non-Cartesian imaging, small deviations from the prescribed (nominal) gradients can have a large impact on image quality if not accounted for in reconstruction. We have implemented a “pencil-beam” measurement for measuring readout gradient (k-space) trajectories and B0 fluctuations for a 2D balanced spiral-in trajectory (Fig. 3(a)), based on the standard approach in (10). The advantage of pencil-beam measurements is that both gx and gy gradients can be played simultaneously, unlike “slice” measurements that make the assumption that independent measurements of k-space and B0 fluctuations along orthogonal directions add linearly. Acquisition parameters were: spiral-in readout of duration 11 msec; TR=1 sec; slice-selective SLR excitation and spin-echo pulses; pencil-beam width 1 mm. We excited a pencil-beam in each corner of a square of width 4 cm centered at iso-center. For each pencil-beam, we also acquired a reference acquisition with zero readout gradient amplitude, the phase of which was subtracted from the non-zero gradient measurement. By subtracting and adding the observed phase, measures of k-space and B0 phase offsets, respectively, are obtained. To achieve sufficient signal-to-noise ratio (SNR), we averaged the signals from twenty repetitions.
Figure 3:
k-space and B0 phase measurements for a balanced spiral-in readout, using pencil-beam excitations (the pulse sequence is shown in Supporting Fig. S1). (a) Measured (red) and prescribed (green) k-space trajectories are in excellent agreement. (b) Measured B0 (constant offset) phase.
Stack-of-spirals T2*-weighted imaging
Dynamic T2*-weighted non-Cartesian imaging is useful for, e.g., functional MRI, but requires custom pulse sequences. We implemented such a sequence using TOPPE, based on a 3D stack-of-spirals readout and echo-shifting (PRESTO) (Fig. 4(a)). Acquisition parameters were: spiral-in readout of duration 11 msec; flip angle 10o; 3.33 mm isotropic voxels; reconstructed image matrix size 72×72×54; acquisition bandwidth ±125 kHz (4μs sampling); 4 fully sampled reference frames followed by 432 six-fold undersampled frames; 3 spiral-in leafs per kz “platter”; TE/TR=32/18 msec; volume acquisition time2.9 msec.
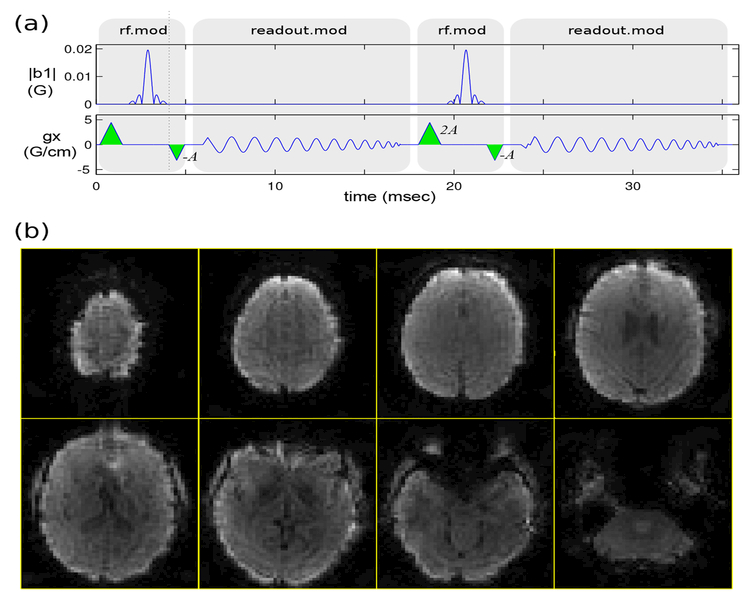
Figure 4:
Rapid whole-brain dynamic T2*-weighted imaging using 3D stack-of-spirals PRESTO. Each 72×72×54 image volume was acquired in 2.9 sec (fully sampled). (a) Pulse sequence diagram (gy, gz waveforms not shown). Echoes were shifted by one TR by placing trapezoidal gradients (green) with area −A and 2A just before and after the spiral-in readout, respectively, with the net gradient area A corresponding to two cycles of phase across the voxel along both x and y dimensions. (b) Reconstructed image volume for one time-frame (only eight of the 54 slices are shown), showing strong T2*-weighting as expected.
We demonstrate this acquisition by reconstructing a 3D image corresponding to one of the fully sampled reference frames using inverse FFT and nonuniform FFT (11) in the through- and in-plane dimensions, respectively, the latter implemented with the IRT MATLAB toolbox available at http://www.eecs.umich.edu/~fessler.
Myelin imaging using inhomogeneous magnetization transfer (ihMT)
Loss of myelin in the brain has long been implicated in Multiple Sclerosis and other disorders, however quantifying this loss on a per-voxel basis has remained a challenging task. Recently, a new method called “inhomogeneous magnetization transfer” (ihMT) was introduced (8), that is based on combining a series of MT-weighted images in a way that produces images that appear to only show myelin. We note, however, that there is currently no consensus regarding the best choice of MT preparation or data readout (e.g., fast spin-echo vs spoiled gradient-echo).
To demonstrate the use of TOPPE for this application, we implemented an ihMT spoiled gradient-echo (SPGR) steady-state sequence and imaged a healthy volunteer (Fig. 5(a)). Acquisition parameters were: 3D Cartesian (spin-warp) readout; flip angle 15o; voxel size 1×1×5 mm3; image matrix size 240×240×8; acquisition bandwidth ±31.25 kHz; TE/TR=3/40 msec. We acquired one image volume with no MT preparation, and three with off-resonance RF excitation at +10 kHz, −10 kHz, and at both positive and negative 10 kHz (cosine-modulated). The amplitude of the MT pulses were adjusted for equal RF power in all acquisitions. Inhomogeneous MT images were obtained as 100×(Im+10kHz+Im−10kHz−2×Im±10kHz)/(2×M0), where M0 is the image without MT preparation.
Figure 5:
Myelin imaging with inhomogeneous magnetization transfer (ihMT) in the brain of a healthy volunteer. (a) Pulse sequence diagram (gy, gz waveforms not shown). In each TR, an off-resonance MT pulse is followed by a conventional 3D spoiled gradient-echo (SPGR) readout. (b) Myelin maps in six adjacent slices. The skull and air space surrounding the head have been masked out.
RESULTS
Figures 3(a) and (b) show the observed k-space trajectory and B0 phase, respectively. The observed k-space is highly accurate apart from small anisotropic delays (not shown), however the B0 phase exhibits large swings with a periodicity matching the spiral trajectory (i.e., approximately 11 full cycles). In addition, we observe a slow, approximately linear, B0 drift. It is worth noting that in separate measurements using vertical slice excitations (not shown), we did not observe this drift.
Figure 4(b) shows a representative result from the stack-of-spirals PRESTO imaging experiment. Lower slices exhibit signal loss near the frontal sinus and ear canals which is evidence of strong T2*-weighting as desired.
Finally, Fig. 5(b) shows results from the ihMT experiment. These myelin maps are in qualitative agreement with ihMT maps in the literature (8), and reflect normal white matter anatomy as expected.
In addition to the examples presented here, we have used TOPPE in a variety of other projects, such as dual-echo steady-state (DESS) imaging (12) and suppression of ghost artifacts in dynamic RF-spoiled imaging (13). Supporting Figs. S2–S4 show other recent examples from our laboratory, and illustrate the flexibility of TOPPE for sequence execution.
DISCUSSION
We have found TOPPE to be a convenient framework for rapid implementation and testing of new pulse sequences in a research setting, particularly for non-oblique 3D imaging. For example, in TOPPE it is easy to loop through several image acquisitions with, e.g., different flip angles and/or TR, as is done in MR fingerprinting (14); modifying the scan loop is simply a matter of editing scanloop.txt. In addition, TOPPE can simplify the experimental workflow substantially by, e.g., combining several related scans into one long scan. Another benefit is that TOPPE makes it easy to ensure that an MR experiment is executed consistently from one scan session to the next, even if the scanner undergoes a software upgrade between sessions.
TOPPE is a framework for rapid sequence prototyping, and is not intended as a complete replacement for conventional pulse sequence programming. For example, the TOPPE GE interpreter does not allow sequence parameters such as in-plane shifts, slice thickness, or field-of-view to be specified programmatically based on scan prescription, which can be limiting outside of a research setting. In addition, adding vendor-supplied “canned” code for, e.g., navigator support, would require modifying the interpreter source code and recompiling, which partly defeats the purpose of coding in TOPPE in the first place. Also, as already mentioned, our TOPPE GE interpreter requires the module duration to be somewhat (~0.4–0.5 msec) longer than the waveform duration. TOPPE may therefore have limited applicability for sequences requiring very short TR such as balanced steady-state free precession (bSSFP), or overlapping waveforms across modules, e.g. for minimizing TE. Nevertheless, in these situations TOPPE may still be useful during the sequence design and validation stages, e.g., for exploring the feasibility of a novel sequence idea.
At present, the TOPPE GE interpreter does not contain built-in checks for gradient heating, peripheral nerve stimulation (PNS), or RF power deposition (SAR). Fortunately, our GE scanner monitors RF power deposition and will stop the scan if SAR limits are exceeded. It also monitors the temperature of the gradient bore. In addition, vendor (“stock”) sequences include gradient heating calculations as an additional check, whereas no such calculations are currently performed by our TOPPE GE interpreter. Real-time monitoring does not exist for PNS, and it is therefore the responsibility of the sequence designer to ensure that PNS limits are met.
Another limitation of TOPPE and related projects such as (5,15) is limited support for real-time decision making. For example, although a simple cardiac or respiratory trigger can be implemented, real-time decisions based on, e.g., heart rate or presence of arrhythmia, are not possible.
Although we have only implemented a TOPPE interpreter for GE scanners, we do not see this as a limitation since Pulseq and possibly related projects (e.g., (15)) promise to “bridge the gap” across vendor platforms. In particular, and as mentioned in Introduction, the TOPPE GE interpreter was used in (5) as the Pulseq interpreter for GE, after converting a Pulseq file to a corresponding set of TOPPE files (using the seq2ge.m script included in the TOPPE Matlab library). This made it possible to execute the same Pulseq scan definition on Siemens, Bruker, and GE scanners (5). We also provide a Matlab script (ge2seq.m) for converting TOPPE to Pulseq format, thus allowing sequence definitions created in TOPPE or Pulseq to be freely exchanged and executed across GE, Siemens, and Bruker scanners. We encourage other laboratories, or the vendors themselves, to develop Pulseq or TOPPE interpreters for other platforms.
CONCLUSION
We have proposed a file format for fully specifying an MR imaging experiment, along with a hardware interpreter for GE scanners. The resulting framework allows rapid prototyping of MR pulse sequences, without the need for sequence-specific source code compilation; indeed, we have demonstrated that completely different sequences can all be executed correctly, without even re-prescribing the TOPPE GE binary executable (driver).
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott Swanson, University of Michigan, for designing the ihMT experiment. We also thank Tianrui Lui, University of Michigan, for reconstructing the stack-of-spirals PRESTO data. Finally, we thank Michelle Karker, University of Michigan, for designing EPI waveforms with “random” phase-encoding.
REFERENCES
- 1.Skare S, Avventi E, Norbeck O, Ryden H. An abstraction layer for simpler EPIC pulse programming on GE MR systems in a clinical environment In Proc., ISMRM, 25th Annual Meeting. Honolulu, 2017; 3813. [Google Scholar]
- 2.Magland JF, Li C, Langham MC, Wehrli FW. Pulse sequence programming in a dynamic visual environment: SequenceTree. Magn Reson Med 2016;75:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debbins J, Gould K, Halleppanavar V, Polzin J, Radick M, Sat G, Thomas D, Trevino S, Haworth R. Novel software architecture for rapid development of magnetic resonance applications. Concepts Magn Reson 2002;15:216–237. [Google Scholar]
- 4.Honroth T, Cordes C, Archipovas S, von Samson-Himmelstjerna F, Günther M, Porter D. Advanced platform-independent MR prototyping: From EPI to Arterial Spin Labeling without code compilation In Proc., ISMRM, 25th Annual Meeting. Honolulu, 2017; 3824. [Google Scholar]
- 5.Layton KJ, Kroboth S, Jia F, Littin S, Yu H, Leupold J, Nielsen JF, Stöcker¨ T, Zaitsev M. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med 2017; 77:1544–1552. [DOI] [PubMed] [Google Scholar]
- 6.Moonen CT, Liu G, van Gelderen P, Sobering G. A fast gradient-recalled MRI technique with increased sensitivity to dynamic susceptibility effects. Magn Reson Med 1992;26:184–189. [DOI] [PubMed] [Google Scholar]
- 7.van Gelderen P, Duyn JH, Ramsey NF, Liu G, Moonen CT. The PRESTO technique for fMRI. Neuroimage 2012;62:676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varma G, Duhamel G, de Bazelaire C, Alsop DC. Magnetization transfer from inhomogeneously broadened lines: A potential marker for myelin. Magn Reson Med 2015;73:614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pauly J, Le Roux P, Nishimura D, Macovski A. Parameter relations for the Shinnar-Le Roux selective excitation pulse design algorithm. IEEE Trans Med Imag 1991;10:53–65. [DOI] [PubMed] [Google Scholar]
- 10.Duyn JH, Yang Y, Frank JA, van der Veen JW. Simple correction method for k-space trajectory deviations in MRI. J Mag Res 1998;132:150–3. [DOI] [PubMed] [Google Scholar]
- 11.Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Trans Sig Proc 2003;51:560–74. [Google Scholar]
- 12.Nataraj G, Nielsen JF, Fessler JA. Dictionary-free MRI parameter estimation via Kernel Ridge Regression In International Symposium on Biomedical Imaging (ISBI’17). Melbourne, 2017; 79. [Google Scholar]
- 13.Nielsen JF, Noll DC. Improved spoiling efficiency in dynamic RF-spoiled imaging by ghost phase modulation and temporal filtering. Magn Reson Med 2016;75:2388–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature 2013;495:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archipovas S, Honroth T, Cordes C, Günther M, Porter D. MRI Pulse Sequence Development Using Graphical User Interface Modules In Proc., ISMRM, 25th Annual Meeting. Honolulu, 2017; 1510. [Google Scholar]
- 16.Vannesjo SJ, Haeberlin M, Kasper L, Pavan M, Wilm BJ, Barmet C, Pruessmann KP. Gradient system characterization by impulse response measurements with a dynamic field camera. Magn Reson Med 2013;69:583–593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.