Abstract
A quantitative adverse outcome pathway (qAOP) consists of one or more biologically based, computational models describing key event relationships linking a molecular initiating event (MIE) to an adverse outcome. A qAOP provides quantitative, dose-response and time-course predictions that can support regulatory decision-making. Herein we describe several facets of qAOPs, including (a) motivation for development, (b) technical considerations, (c) evaluation of confidence, and (d) potential applications. The qAOP used as an illustrative example for these points describes the linkage between inhibition of cytochrome P450 19A aromatase (the MIE) and population-level decreases in the fathead minnow (FHM; Pimephales promelas). The qAOP consists of three linked computational models for: (a) the hypothalamicpitutitary-gonadal axis in female FHMs, where aromatase inhibition decreases the conversion of testosterone to 17β-estradiol (E2), thereby reducing E2-dependent vitellogenin (VTG; egg yolk protein precursor) synthesis, (b) VTG-dependent egg development and spawning (fecundity), and (c) fecundity-dependent population trajectory. While development of the example qAOP was based on experiments with FHMs exposed to the aromatase inhibitor fadrozole, we also show how a toxic equivalence (TEQ) calculation allows use of the qAOP to predict effects of another, untested aromatase inhibitor, iprodione. While qAOP development can be resource-intensive, the quantitative predictions obtained, and TEQ-based application to multiple chemicals, may be sufficient to justify the cost for some applications in regulatory decision-making.
Introduction
A vision for toxicity testing in the 21st century that was laid out by the US National Research Council (1) focuses on the assessment of critical mechanistic endpoints involved in the induction or early progression toward overt toxicity, rather than direct observation of toxic effects themselves (2). The adverse outcome pathway (AOP) framework has developed as a systematic approach for describing the scientifically credible basis for linking a toxicant-induced molecular initiating event (MIE: interaction of a chemical with a biomolecule in the body of an organism that causes a perturbation in its biology) to an adverse outcome (AO) considered relevant to risk assessment (generally defined at the individual or population level) (3). Efforts are underway to populate an AOP knowledgebase (AOP-KB; aopkb.org) with AOPs structured and described according to a key set of principles and internationally-harmonized guidance (4, 5). This includes defining a set of measurable biological changes, termed key events (KEs), that reflect progression from the initial biological perturbation (MIE) to a specific AO and describing both the biological plausibility and empirical evidence that supports a causal relationship between the KEs (4). Thus, AOPs represent a critical bridge to link pathway-specific bioactivities (i.e., mechanistic endpoints) to the apical hazards that are generally considered relevant to regulatory decision-making.
For the purposes of describing an AOP, an underlying assumption is that the severity of perturbation at the MIE, in terms of dose and duration, is sufficient to drive the pathway to its final AO. Adaptive, compensatory, and repair mechanisms are assumed to be overwhelmed. Thus, qualitative AOPs provide a fundamentally hazard-based assessment framework. They provide an indication of what hazards can plausibly, and based on available evidence, be connected with a particular perturbation of normal biology. However, they do not necessarily define the probability or severity of the AO that can be expected under a specified exposure scenario. Consequently, while many AOPs may have immediate utility as tools for hazard identification, hypothesis-driven testing, and prioritization, most are not appropriate for quantitative risk assessment.
The term quantitative AOP (qAOP) refers to a loosely defined, but relatively advanced stage in the progression of AOP development and description. At this stage, quantitative understanding of the relationships underlying transition from one KE to the next, as well as critical factors that can modulate those relationships, are sufficiently well defined to allow quantitative prediction of the probability or severity of the AO occurring for a given activation of the MIE (4, 6). Information concerning the quantitative understanding of what defines the transition from one KE in an AOP to the next is thus captured and included (where possible) in the KE relationship descriptions (7). That quantitative understanding may take many forms, depending on the extent of the available, relevant data. In the case of a relatively limited dataset containing little or no or dose-response and time-course information, the relationship between adjacent KEs may be as simple as a linear regression equation linking an upstream with an immediately downstream KE. With richer datasets, reflecting fuller dose-response and time-course designs, the quantitative understanding may be encoded into sophisticated biologically based models that simulate complex, non-linear, dynamics that can result from feedback loops, adaptive and compensatory responses, stochastic influences, interactions with other pathways, and/or influences of external or internal modulating factors. Whatever form they take, quantitative understanding of the KE relationships encompassed in an AOP description can facilitate a broader spectrum of applications (6). Consequently, there is interest in developing the quantitative understanding and description of AOPs to the extent that regulatory needs warrant and resources allow.
The goal of this paper is to provide an introduction to the concept of qAOPs. We seek to define the key attributes of a qAOP. The development and application of a qAOP are illustrated for an example AOP linking aromatase inhibition to reproductive dysfunction in fish and potentially other oviparous vertebrates (8). The importance of defining the relative confidence in the qAOP for support of decisions in different regulatory contexts is also described. While by no means comprehensive or illustrative of the many forms a qAOP may take, the fundamental concepts introduced here are intended to help inform the practice of qAOP development as an important component in the transition to the predictive toxicology paradigm envisioned by the NRC (1).
qAOP – an example
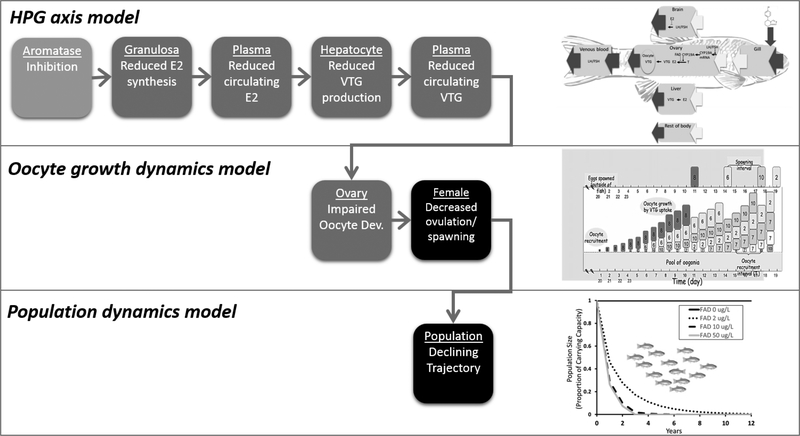
An AOP linking the MIE of aromatase inhibition to the AO of reproductive impairment (Fig. 1; 8) was one of the first described in accordance with OECD guidance and entered into the AOP-Wiki (https://aopwiki.org/aops/25). The AOP consists of eight key events reflecting the synthesis and circulation of 17β-estradiol (E2; Fig. 1, KEs 2, 3), synthesis and circulation of the egg yolk precursor vitellogenin (VTG; Fig. 1, KEs 4, 5), and uptake of VTG into developing oocytes (KE6) to support ovulation and spawning (KE7) as is required for a stable or increasing population trajectory (KE8). While the description found the AOP-Wiki lays out compelling scientific evidence linking inhibition of aromatase enzyme activity to reproductive impacts considered relevant for risk assessment and management, the qualitative description does not address the question of whether exposure to concentration “X” of aromatase inhibitor “Y” for duration “Z” should be expected to cause a significant reduction in cumulative fecundity or population trajectory. However, in the case of this AOP, a series of computational models that capture critical elements of the biological systems dynamics that underlie transition from one KE in the pathway to the next were developed independently and then coupled together as a qAOP.
Figure 1.
The aromatase inhibition qAOP. This qAOP consists of 3 linked models: fathead minnow HPG axis (9), fathead minnow oocyte growth dynamics (12), and fathead minnow population dynamics (14). The HPG axis model predicts plasma VTG concentration as a function of inhibition of aromatase. The oocyte model takes plasma VTG as its input and predicts fecundity (egg production). The population dynamics model takes fecundity as its input and predicts population dynamics.
qAOP first component - HPG axis model:
In the case of the aromatase inhibition AOP, feedback responses along the hypothalamic-pituitary-gonadal (HPG) endocrine axis and associated compensatory responses strongly influence the dose-response, time-course behaviors underlying KEs 1–5 (Fig. 1). Cheng et al. (9) described a computational model of aromatase catalyzed conversion of testosterone (T) to E2 and subsequent stimulation of VTG production and its dynamic regulation via endocrine feedback and VTG transport mechanisms. The model was calibrated using data from laboratory experiments where fathead minnows (Pimephales promelas) were exposed to the aromatase inhibitor fadrozole (fadrozole hydrochloride, CAS # 102676–31-3). A regulatory circuit coded in the model senses decreased levels of E2 associated with aromatase inhibition and in response upregulates ovarian aromatase activity. This regulatory circuit allows partial or even full recovery of plasma E2 during exposure to the aromatase inhibitor, depending on the degree of ongoing aromatase inhibition in a manner consistent with experimental observations (10). Also, when exposure to the inhibitor ends, the induced aromatase activity transiently generates a level of E2, greater than that in controls, that eventually returns to normal. A second regulatory circuit controls the rate of VTG uptake into the ovary to maintain ovarian VTG levels when the blood level of VTG varies. This circuit describes negative regulation of a VTG transporter, with the transporter being upregulated when ovarian VTG levels fall (9).
qAOP second component - Oocyte growth dynamics model:
A second independent model that captures the relationship between circulating plasma VTG and oocyte development and spawning (Fig. 1; KEs 5–7) is termed the oocyte growth dynamics model (OGDM) (11, 12). Measured or model-predicted plasma VTG concentrations are used as inputs to the OGDM. Oocyte growth is driven primarily by the absorption of VTG into the oocyte following first-order kinetics while accounting for water and other molecules that are also absorbed and contribute to its growth. When oocytes reach a critical volume (0.52 μL based on Leino et al. (13) for mature fathead minnow oocytes) spawning occurs (for more details see Li et al. [11]). The OGDM predictions include daily spawning that can be used to calculate average fecundity, the number of eggs per spawn and the number of spawns per female (i.e., Fig. 1 KE 7) for comparison with experimental results.
qAOP third component - Population model:
The third model incorporated into the qAOP is a density dependent logistic matrix model developed by Miller and Ankley (14), and previously applied in population modeling studies across multiple scenarios, including species (15, 16, 17, 18). The model uses inputs that include estimates of effects on vital rates (for example, alterations occurring to age-specific fecundity), a life table for the organism of interest, which is used to construct the Leslie projection matrix, and an estimate of carrying capacity. Output from the model is in the form of density dependent population trajectories. Further, model output can be expressed invariant of carrying capacity by plotting population size proportional to carrying capacity at each time step, as opposed to evaluation based on absolute numbers. In demonstrating model output in this manner, a value of 1.0 represents a population at carrying capacity and values between 0 and 1.0 represent a population below the carrying capacity threshold.
By coupling these three models together it becomes feasible to take a measure of a chemicals’ potency as an aromatase inhibitor, relative to fadrozole, and simulate dose-response and time-course behaviors for a given exposure scenario. The models can provide read-outs of the expected dose-response/time-course profile of most KEs along the pathway. This allows for in silico predictions of the probability or severity of a defined AO for a wide range of exposure scenarios and chemicals. While this is just one example of the many forms a qAOP may take, we use the example to illustrate attributes and considerations that generalize to the development and application of qAOPs more broadly.
Attributes of a qAOP
Quantitative AOPs share attributes of other types of biologically based models including biologically based dose-response models, toxicodynamic models, systems biology models, and population models. These kinds of models focus on biological determinants of response, motivated by the realization that the more accurately this biology is described, the more accurate the predictions provided by the models will be. However, one attribute of qAOPs that differentiates them from other types of biologically based models is that they align, specifically, with KEs defined in an AOP. That is to say, the model (or assembly of models) that makes up a qAOP is able to provide a read-out of condition or state at each KE in the pathway (e.g., Fig. 1). Likewise, ideally, a qAOP construct should be designed to accept measured or predicted values associated with any KE in the pathway and provide a simulated output for all downstream KEs (including the AO) in that pathway. In this way, a qAOP serves as a computational tool for translating or extrapolating from mechanistic measurements of an upstream KE to a predicted probability or severity of an AO. In addition, as with other biologically based models, a qAOP serves as a tool for hypothesis generation and verification in which key model outputs align with measurable biological parameters.
A second, important attribute of a qAOP construct is that it is not chemical or stressor-specific. AOPs are intended to describe a sequence of events, connecting the MIE with the AO, that will occur regardless of the specific identity of the chemical that perturbs a MIE or an intermediate KE (4). Thus, even if the qAOP is parameterized and/or validated based on data for a specific chemical or stressor, it should be generalizable to a broad range of stressors capable of eliciting perturbations captured in the AOP. In the case of the aromatase inhibition qAOP described above, the models were parameterized based on experiments with fadrozole. However, the assumption is that the qAOP can be applied to other chemicals by adjusting for their relative potency at the MIE and the absorption, distribution, metabolism, and elimination (ADME) properties that dictate how much of the chemical can reach the target. Thus, while chemical specific potency and ADME considerations need to be factored in for accurate prediction, the qAOP construct itself should be developed in a way that facilitates broad application across chemicals acting on a common biological target.
Like AOPs, qAOPs are pragmatic simplifications of biological complexity. A tremendous amount of known biology is essentially embedded in the KE relationships that link one measurable “check point” in the pathway to the next. This could include, for example, numerous steps in a signal transduction cascade linking activation of a G protein-coupled receptor to its effects on gene transcription. Feedback loops, compensation and repair processes etc. can also lie in the biology connecting one measurable KE to the next. While the qualitative AOP description is written in a manner that assumes such compensatory processes have been overwhelmed, a qAOP may need to represent specific aspects of this underlying biology in order to make reasonably accurate predictions of dose-response and time-course behaviors. Consequently, a qAOP model will often incorporate more biology than the AOP it aligns with, as reflected in the HPG axis model described above that incorporates endocrine feedback and VTG transport processes that are not represented as KEs in the AOP (Fig. 1). Nonetheless, as with an AOP, a qAOP need not provide a detailed representation of every aspect of the biology in question but, rather, should only be as complex as needed to provide useful predictions. For example, the HPG axis model can simulate the feedback responses that elicit empirically observed compensatory behaviors without incorporating the specific—and elaborate—biology associated with sensing of E2 concentrations in the hypothalamus, control of gonadotropin release from the pituitary, binding of gonadotropins to membrane bound G-protein coupled receptors on the surface of specific cell types within the ovary, resulting signal transduction and activation of transcription factors, etc. It uses a simplified formulation in which E2 directly influences gonadotropins which in turn directly influence aromatase expression. Such simplification is particularly useful when collecting the data needed to parameterize a more biologically detailed model is impractical. Thus, in the same sense that an AOP is a simplification of complex biology, a qAOP should be a parsimonious simulation of that biology that accounts for the data with a fairly straightforward (and in many cases knowingly simplified) explanation.
Finally, where feasible, qAOPs should employ the modular construction utilized for AOPs to help facilitate the eventual construction of qAOP networks. For example, Ankley et al. (3) presented a network of three AOPs that converge on a common KE of reducing hepatic VTG production in fish. For the three AOPs forming the network, the KEs upstream of reduced hepatic VTG differ. In one case, the MIE involves antagonism of estrogen receptor (ER) signaling in the liver (https://aopwiki.org/aops/30). A second case involves aromatase inhibition in the ovary and reductions in circulating estrogen concentrations (the AOP depicted in Fig. 1). The third case involves negative feedback presumably originating in the brain leading to decreased gonadal steroid production and subsequent reductions in circulating estradiol concentrations (https://aopwiki.org/aops/23). Rather than build an independent qAOP model for the entire sequence of KEs in each AOP, it would be useful to build components that can be coupled together as needed. In the present case, the HPG axis model applies specifically to the aromatase inhibition AOP. However, the OGDM and Population models would apply to all three AOPs.
Practical Considerations for qAOP Construction
As noted previously, a qAOP at its simplest could consist of a regression equation based on limited MIE, KE, and AO data. However, in moving from the qualitative characteristics of an AOP intended for hazard identification to qAOPs intended to predict quantitative dynamics of dose-response and time-course, capturing both the adaptive capabilities of the biological system of interest and the effects of important modulating factors become important. For example, adaptation can alter, in a time-dependent manner, the quantitative characteristics of the KE relationships, and thus the overall quantitative MIE-AO relationship. Regulatory motifs such as feedback and feedforward loops can create complex, time-dependent, dynamics as a function of the magnitude and duration of the input stimuli (19). These dynamics cannot be adequately simulated based on the two-dimensional data, but rather require characterization of more complex dose-time response surfaces, which allow for the shape of the dose-response curve to change with time. Thus, characterization of KE relationships that support identification of adaptive responses should involve experimental designs incorporating multiple time-points and doses (e.g., 9, 20, 21). Development of the HPG axis model included in the aromatase inhibition qAOP was based on exactly those kinds of studies (10, 22). Likewise, modulating factors may alter the shape of those surfaces dramatically. Thus, experimentation to define how key modulating variables alter those surfaces may also be important.
While the modeling itself becomes more complex as more biological detail, variables, and additional data are included, perhaps the more significant consideration is the kind of experimental designs required to obtain data that support the modeling. While experimental characterization of dose-time-response surfaces is informative, it is time-consuming and expensive and so it is important to consider “value added” in terms of the investment. There will generally be a correlation between the amount and types of relevant data collected and the level of confidence in the AOP and qAOP. Model predictions change as KE relationships are calibrated against progressively more complete datasets (reflecting experimental designs that increasingly capture dose-response and time-course behaviors). As datasets become more comprehensive, the resultant changes in model predictions should become smaller, suggesting greater confidence in the predictions and, therefore, reduced uncertainty. In that respect, an iterative process in which models are developed based on available data, predictions from those models are tested, and then the models refined based on the results and any model short-comings is often a pragmatic way to guide the investment of experimental resources to allocate to qAOP development.
Much of the “art” of qAOP development lies in deriving models of sufficient detail to provide reasonably accurate predictions across a wide range of scenarios while at the same time incorporating sufficient abstraction or simplification to make the model development and verification tractable from a resource investment perspective. There are no “hard” rules that define the appropriate level of abstraction or detail to include in a qAOP. Indeed, the answer to that question will often depend on the intended application and the associated degree of certainty that application requires. Nonetheless, there are a few guiding principles to keep in mind.
First, AOPs and qAOPs are intended to be chemical agnostic. While it is recognized that chemical-specific differences in potency and ADME properties are important determinants that should be factored into qAOP-based predictions, they should be handled external to the qAOP construct itself. Relative potencies and key chemical-specific determinants of internal dose at the MIE should be entered as input parameters to the qAOP model(s), not embedded into the models themselves. This allows the reproducible biological responses to perturbation of the MIE to be modeled in a generalizable manner that can flexibly tailored on a case by case for chemical-specific predictions.
Second, qAOPs need to be practical. Both AOPs and qAOPs are intended to facilitate predictive toxicology. They arise from the need to more effectively utilize our understanding of biology to predict effects that we cannot afford to measure, either due to the costs and time involved, or ethical and practical considerations (e.g., the inability to conduct in vivo toxicity studies in humans, endangered species, organisms that cannot be held/reared in captivity, etc.). While significant investment may be needed to develop and test a qAOP construct, the goal is to produce models with broad practical application. Just as a qAOP model tailored to a single chemical is not the intent in developing a qAOP, likewise, qAOPs tailored to just a small subset of an AOP’s applicability domain is similarly undesirable. In this respect, when developing the qAOP, it is desirable to do so with an aim toward encompassing or generalizing to as much of the applicability domain of the associated AOP as possible. With regard to the aromatase inhibition qAOP example, this means that although the model was initially built on data for a single fish species (FHM) exposed to one model aromatase inhibitor (fadrozole), additional efforts are focused on identifying the key species-specific and chemical-specific parameters to modify in the model(s) to allow the construct to be more broadly generalized. This has already been demonstrated for the population model component of the qAOP, where life-table parameters for different species can easily be inserted into the model structure to facilitate its use for other species (17). In order to make this practical, the parameter changes needed to tailor the model for different species, sexes, life-stages, etc. that fall within the applicability domain of the AOP should be as few in number as possible, ideally be parameters that are easily measured or estimated using cost effective methods, and should be easy to change within the model structure, without need for specialized expertise in coding, etc. (e.g., through development of a graphical user interface where key model parameters could be input by a user).
As described by Ankley et al. (3), qAOPs require data across the full range of biological organization from molecular up through individuals and, for ecological effects, populations. Parameter values can often be set to measured values from existing literature or, when necessary, can be obtained through targeted experimental work. Nonetheless, there are usually some parameters whose values must be obtained indirectly by optimizing the fit of model predictions to data. Informal optimization, where parameter values are adjusted manually until a visually good fit to data is obtained, is valuable for learning about model behavior. Formal optimization (e.g., 23), used to minimize a least squares or some other form of cost function, has the advantage of being reproducible, which is not necessarily true of informal approaches. In practice, the parameter values appearing in a “final” version of a qAOP will include both those measured in the laboratory and obtained through formal optimization. Optimization procedures along with sensitivity analyses may also be useful for identifying critical parameters that can be altered to tailor the model for different species or scenarios to broaden its applicability domain.
Evaluation of Confidence in qAOP Predictions
qAOPs are members of a larger class of computational, biologically-motivated models developed with the goal of improving quantitative understanding of the relationship between toxicant exposure and health-related outcomes. Other members of the class include PBPK models and biologically based dose-response (BBDR) models (24, 25). Lau et al. (24) state that “Biologically based dose–response (BBDR) modeling represents a novel approach for quantitative assessment of health risk by incorporating pharmacokinetic and pharmacodynamic characteristics of a chemical and by relating the immediate cellular responses to a cascade of aberrant biological actions that leads to detectable adverse outcomes.” The US EPA Guidelines for Carcinogen Risk Assessment (26) state that “toxicodynamic models” of a chemical agent’s mode of action are the preferred approach for analysis of cancer risk. Thus, although the AOP concept was only recently formally described (3), the essential operative features of AOPs and qAOPs and their potential for support of regulatory decision-making have been recognized for some time.
As computational models, qAOPs tend to be structurally complex and parameter-rich. Crump et al. (27) expressed concern that development of BBDR (and by extension, qAOP) models inevitably introduces uncertainties such that any use of the model to predict low dose risk is “…unlikely to be fruitful in reducing uncertainty in quantitative estimates of human risk from low-level exposures…” The theoretical justification for qAOP development is simply that (a) the quantitative relationship between the MIE and the AO is determined by the relevant biology and (b) the more accurate the description of this biology in the qAOP, the more accurate its predictions.
The concern stated by Crump et al. (27) is therefore only valid to the degree that there are problems with either the technical aspects of model development or with assumptions about model structure and parameterization. Quality control for technical aspects of model development is important and has been thoroughly addressed in numerous publications (e.g., 28, 29). A biologically based model structure that is complex relative to more empirical or statistical models can, at first glance, appear to be introducing complexity and hence uncertainty. However, it is important to recognize that the biologically based model is explicitly representing biology that is implicit in the simpler models. For example, a benchmark dose (BMD) model describes dose-response data empirically, without specific reference to any of the underlying biology that determines the shape of the dose-response curve. A biologically based model would explicitly describe at least some of the mechanistic basis determining the dose-response behavior, and so makes explicit the mechanism that is implicit in the BMD model. Thus, there is no introduction of uncertainty per se, only a transition from an implicit to an explicit representation of the actual complexity of the system.
The validity of assumptions regarding model structure and parameterization does require evaluation by experts, since incorrect specification of the biology is of course a source of error. If experts cannot agree on a preferred specification, then the more conservative specification among multiple alternatives, predicting more risk, can be chosen. For example, Conolly et al. (25) used this conservative option approach to address specific uncertainties about biological structure by building risk conservatism into the overall dose-response behavior of a BBDR model for formaldehyde carcinogenicity.
In addition to the selection of risk-conservative options, uncertainties identified during model development and evaluation can be addressed through targeted experimentation. The toolbox available for studying biological systems has evolved dramatically in recent years. Examination of toxicological mechanisms (and the structures of AOPs) are today constrained not so much by limitations in laboratory technologies, as by funding and resource constraints and by our ability to synthesize and understand the oncoming flood of data. Thus, uncertainties in toxicological mechanisms, and in the computational models that represent these mechanisms, are addressable to the extent that resources and motivation allow. In addition, statistical analyses can be used to evaluate alternative model formulations and simplifications through, e.g., model reduction and identification of correlated variables (30, 31) or parameter sensitivity analyses (32). Difficulties in communicating the complexity of models can be addressed by using standardized methods of model description and illustration such as the AOP wiki (aopwiki.org) and the OECD Effectopedia project (www.effectopedia.org) where graphical depictions of pathways are linked to model components.
Application Examples
Application Example 1: Comparing qAOP Simulation Results to Empirical Data for a 21 Day Continuous Exposure to Fadrozole
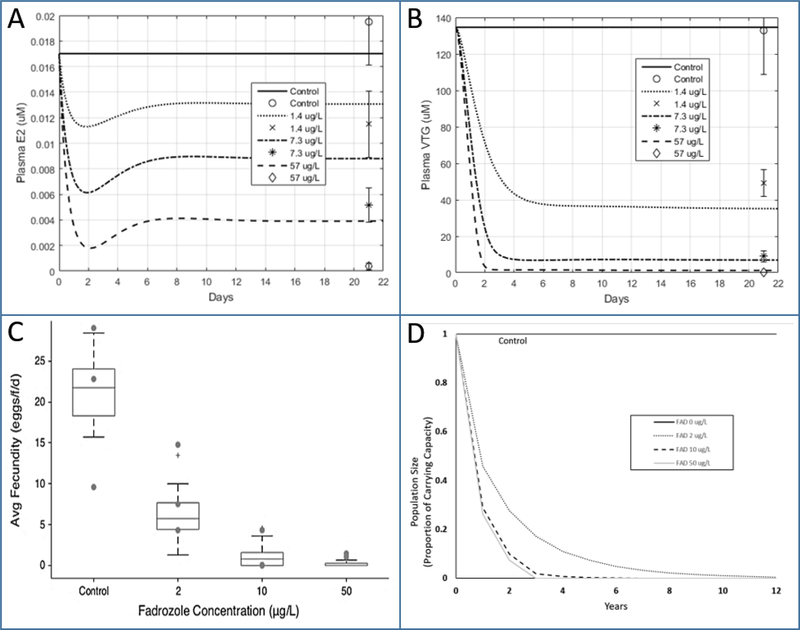
In this example, the qAOP (Fig. 1) was applied to predict the dose-response, time-course behaviors for four KE along the AOP for an exposure scenario modeled after the experimental design of Ankley et al. (33). In that experiment, FHM were exposed in a group spawning design to fadrozole continuously for 21 days at nominal concentrations of 0, 2, 10, and 50 μg/L (actual reported concentrations were 0, 1.4, 7.3 and 57 μg/L). Plasma E2 and VTG concentrations were measured at 21 days, while cumulative fecundity was reported before and during fadrozole exposure. While the qAOP was developed based on data from dose-response, time-course studies in fathead minnows exposed to fadrozole (10, 22), the data from the Ankley et al. study (33) were not used for model development. Thus, this case study provided opportunity to test the qAOP predictions against an empirical data set that falls firmly within the narrowest applicability domain of the qAOP (same species and chemical used in model development).
Ankley et al. (33) reported their data as mean ± SEM. HPG axis model simulations of the plasma E2 data fell within these bounds for control and 1.4 μg/L but over-predicted the data at 7.3 and 57 μg/L (Fig. 2A). These simulations of the 21-day E2 time-course also show the adaptive changes associated with upregulation of aromatase that occur during the first few days of continuous exposure. The basis for the adaptive behavior is fully described in Cheng et al. (9). Simulations of the plasma VTG data fell with the error bounds at all doses except for 1.4 μg/L, where the data were slightly under-predicted (Fig. 2B). These are reasonably good results, given that the E2 and VTG data were not used for development of the HPG axis model.
Figure 2.
Fadrozole 21 day, continuous exposure study. HPG axis model (9) predictions after 21 days of continuous exposure to fadrozole at concentrations of 0, 1.4, 7.3, and 57 μg/L (41). The time sequence data are plotted for plasma E2 (A) and plasma VTG (B). Boxplots of averaged fecundity (12) are plotted against fadrozole concentration (C). Fathead minnow population size (14) resulting from exposure to fadrozole in comparison to control (D). Data in Figs. 2A and2B are shown as mean ± SEM.
The OGDM was used to predict effects on fecundity using plasma VTG inputs provided by the HPG axis model. We followed the methods used by Watanabe et al. (21) for simulating group spawning design studies and performed a total of 600 simulations for each treatment (150 groups of four female FHM). Average fecundity, eggs per spawn and spawns per female were calculated (Fig. 2C). Watanabe et al. (21) showed that, using measured plasma VTG concentrations, the OGDM simulated reproduction metrics very well (i.e., average fecundity [eggs/female/day], eggs per spawning per female, and cumulative fecundity). Given this predictive accuracy, in this case study the accuracy of the OGDM predictions will track with the good accuracy of the plasma VTG predictions provided by the HPG axis model (Fig. 2B).
Predictions provided by the OGDM of daily spawning for each simulated fish over a 42-day period were provided as inputs to the population dynamics model. For each exposure concentration, the population model was executed over a 10-year simulation period and results were recorded using an annual time step (Fig. 2D). Population-level outcomes were not empirically tested (33). Thus, the accuracy of the predicted population impacts cannot be directly evaluated. They do, however, provide some insight into potential population significance of the reductions in cumulative fecundity observed, based on relevant life-table parameters for the species. The results suggest that, at even the lowest concentration tested, 1.4 μg/L, continuous exposure would exceed that adaptive capability of the fathead minnow and result in a dramatic population decline, absent other off-setting demographic parameters (e.g., decreased mortality rates resulting from less competition for habitat and food resources)..
Application Example 2: Derivation of Response-Response Relationships
At present, the aromatase inhibition qAOP relies on three independent models that span the KEs in the AOP. While the models are described and cited as part of the KE relationship descriptions in the AOP-Wiki (https://aopwiki.org/aops/25), they are not necessarily readily accessible or simple to run for a naïve user, nor do they align one-to-one with the KERs in a modular fashion. One way to enhance the accessibility and use of the quantitative understanding afforded by the models is to generate response-response functions for each KE relationship along the AOP. These could be entered into the AOP descriptions in the form of parameters for a regression equation describing key response-response relationships. Thus, the second application case study illustrates how the aromatase inhibition qAOP models were used to develop quantitative response-response relationship information that could be directly input to the AOP-Wiki and easily be used by consumers of that information.
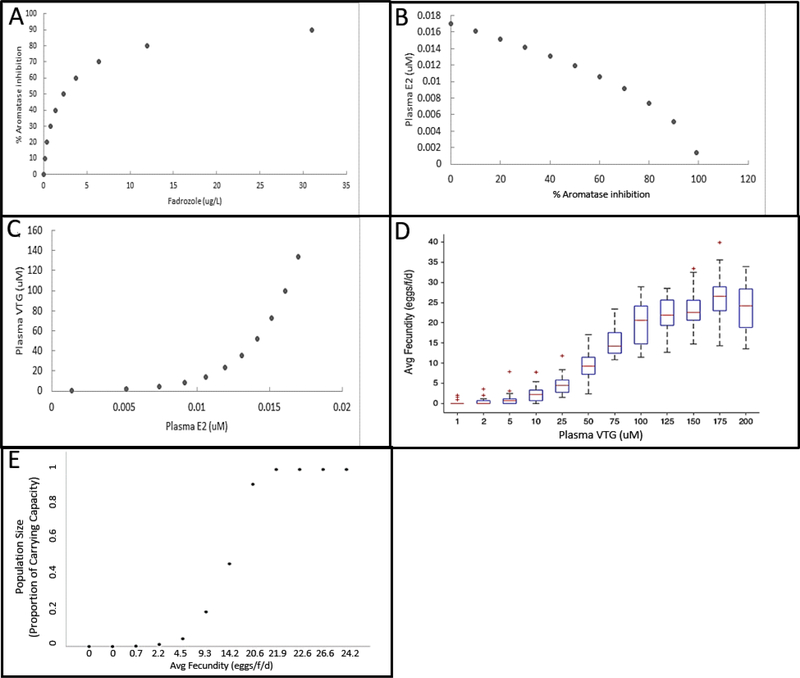
As noted in Application Example 1, during the first few days of continuous exposure to an aromatase inhibitor, the regulatory circuits encoded in the HPG axis model attempt to compensate for the effects of aromatase inhibition and can achieve varying degrees of recovery of plasma E2 and VTG, depending on the extent of the aromatase inhibition (Figs. 2A, 2B). As accommodation completes, the model reaches a new, albeit stressed, steady state. Under this condition, we generated response-response plots for aromatase inhibition as a function of fadrozole concentration (Fig. 3A), plasma E2 as a function of aromatase inhibition (Fig. 3B), plasma VTG as a function of plasma E2 (Fig. 3C), fecundity as a function of plasma VTG (Fig.3D) and FHM population size as a function of fecundity (Fig. 3E).
Figure 3.
Response-response predictions. With the qAOP (9, 12, 14) at steady state, response-response predictions are plotted. The MIE of aromatase inhibition is plotted against fadrozole concentration (A). The KE of plasma E2 concentration is plotted against percent aromatase inhibition (B). The KE of plasma VTG is plotted against plasma E2 level (C). The KE of average fecundity is plotted against plasma VTG concentration (D). The adverse outcome of fathead minnow population size is plotted as a function of fecundity (E).
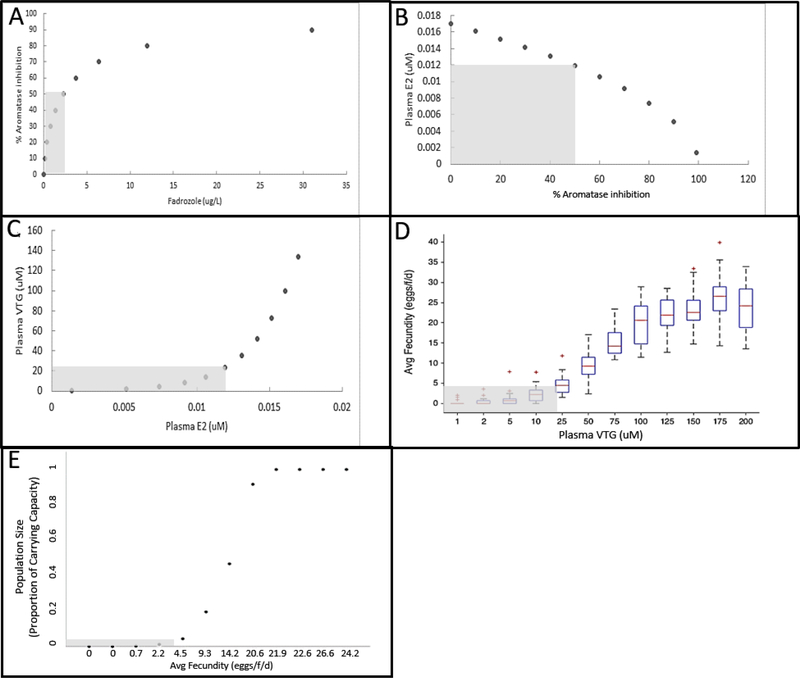
These response-response plots allow visual, semi quantitative estimation of changes in KEs and the AO without having to actually run the computer models comprising the qAOP (Fig. 4). For example, read-across of the response-response plots shows that female FHM exposure to 2.3 μg/L fadrozole was predicted to inhibit aromatase activity by 50% (Fig. 4A), reduce plasma E2 by 30% (Fig. 4B), plasma VTG by about 80% (Fig. 4C), average fecundity by about 80% (Fig. 4D), and a FHM population size by 90% (Fig. 4E). For a more quantitative approach, regression parameters associated with a non-linear fit to each response-response curve could be used to precisely estimate the value for any given point along each curve. Thus, the response-response functions generated represent an easily transferrable means to characterize the quantitative relationships among the KEs represented in the AOP and allow for simple, steady state predictions of outcome. While this approach is more limited than the predictions that can be achieved using the full models, it still has considerable utility for first tier applications where a steady state prediction is sufficient to support a decision. Similarly, the approach could be very useful in settings where limited KE data are available (e.g., plasma steroid or VTG concentrations from field monitoring studies), and there is a desire to rapidly translate these data into potential risks at the individual or population levels.
Figure 4.
Read-across of response-response plots to illustrate rapid evaluation of the qAOP-predicted effects of aromatase inhibition on key events and adverse outcome (9, 12, 14). In this example, exposure to 2.3 μM fadrozole causes 50% inhibition of aromatase (A), which in turn results in a decrease of plasma E2 to 0.012 μM (B), which in turn results in a decrease of plasma VTG to about 20 μM (C), which in turn results in a decrease of average fecundity to about 4 eggs/day (D), which in turn results in a decrease of fathead minnow population to about 5% of carrying capacity (E).
Application Example 3: Estimating a Benchmark Dose for an Untested Chemical
As noted previously, qAOP models are intended to be chemical agonistic and, ideally, generalizable to a broad range of chemicals that can activate the MIE. As a final application example, we show how the qAOP developed based on data for fadrozole can be applied to predict an estimated BMD for an untested chemical. Specifically, a toxic equivalence (TEQ) calculation (34) was used with the HPG axis model to predict the effect of exposure to the aromatase inhibitor iprodione on plasma E2.
Iprodione, a dicarboximide fungicide, is widely used in landscape maintenance and on ornamental and agricultural plants (35, 36). Although most monitoring studies have not detected iprodione in the environment, one study at golf courses in Japan found a surface water concentration of 1 μg/L (37). ToxCast data (38) were used to calculate a fadrozole toxicity equivalent factor (TEF) for iprodione. The 50% maximum effect concentrations (EC50) of iprodione for aromatase inhibition was measured in the cell-free NVS ADME hCYP19A1 assay and this EC50 was in turn used to calculate its fadrozole TEF (0.03). This TEF was then used to derive the TEQ concentration of fadrozole equivalent to 1 μg/L iprodione (0.03 μg/L). Read across of the response-response analysis, as described above, predicts that continuous exposure to 1 μg/L iprodione (as 0.03 μg/L fadrozole) would result in negligible inhibition of aromatase (< 0.001% as calculated by the HPG axis model) and, subsequently, negligible effects on plasma E2, plasma VTG, average fecundity or FHM population status (Fig. 3).
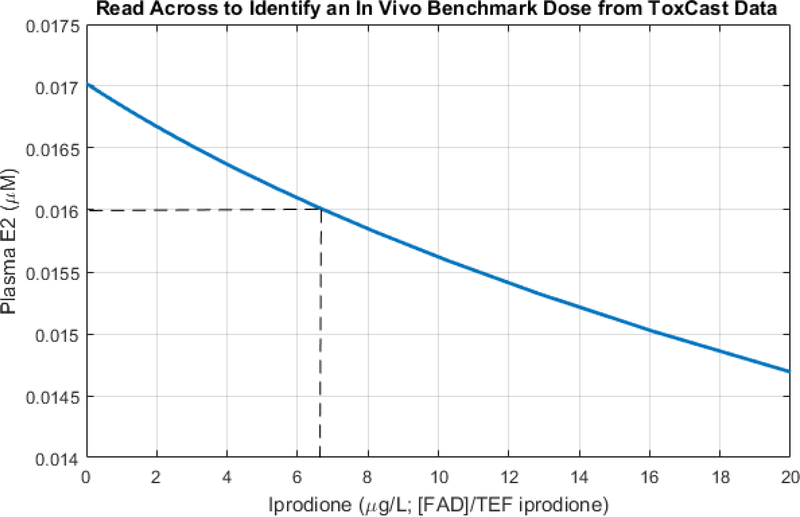
If we assume that a 20% reduction in the fathead minnow population upon continuous exposure to iprodione would be an endpoint of regulatory interest, we can use read across of the response-response plots to semi-quantitatively estimate the associated BMD exposure to iprodione. Using Fig. 3, read-across associates the 20% reduction in fathead minnow population with a plasma E2 level of about 0.016 μM and a fadrozole exposure below 1 μg/L, which is equivalent to an iprodione exposure of below 1 μg/L fadrozole / TEFiprodione, or less than 33 μg iprodione/L. The HPG axis model was then used to more precisely identify the BMD exposure to iprodione as between 6 and 7 μg/L (Fig. 5).
Figure 5.
Use of the qAOP (9, 12, 14) to semi-quantitatively predict a BMD for iprodione. A 20% decrease in fathead minnow population was considered to be a possible endpoint of regulatory interest. Read across (Fig. 4) of the qAOP was then used to identify a predicted plasma E2, continuous exposure to iprodione, as fadrozole equivalents, to identify a BMD assoicated with the 20% population decline (0.016 μM). The current plot, obtained with the HPG axis model, indicates that this level of plasma E2 is associcated with a continuous exposure to iprodione of between 6 and 7 μg/L.
It should be noted that, in addition to aromatase inhibition, iprodione appears to also inhibit androgen synthesis, possibly through inhibition of steroidogenic CYPs upstream of aromatase. Specifically, iprodione reduces serum testosterone levels and ex vivo testicular testosterone production, and inhibits male pubertal development in rats (35, 36, 39). Confidence in the predicted effects of iprodione using the aromatase inhibition qAOP described above should be evaluated in this context. The ToxCast program has evaluated iprodione in 41 assays involving various CYPs. Of these 41 assays, 26 were reported to have nonzero EC50s, and of these 26, the lowest reported EC50 was for the NVS ADME CYP19A1 aromatase inhibition assay (0.413 μM). The EC50 values for the other CYPs ranged from 1.84 to 21.3 μM. While this is not an exhaustive evaluation of possible interactions of iprodione with components of the HPG axis, these data do suggest that, while iprodione at higher concentrations is likely to interact with multiple CYPs, it is somewhat selective for aromatase at lower, arguably more environmentally-relevant concentrations.
Likewise, it should also be noted that in this simple example, we have only adjusted for iprodione’s relative potency compared to fadrozole. For a more sophisticated analysis, one would also want to take into consideration uncertainties associated with the ToxCast EC50s and potential differences in ADME between the two compounds. ToxCast EC50s are calculated based on the applied (unmeasured) concentration in the in vitro assays. Potential loss of chemical to, for example, the wall of the culture dish, is a source of uncertainty if not evaluated when EC50 values are used in computational models of in vivo biology, as with our qAOP. Armitage et al. (40) developed a computational model that illustrates how loss to the walls of the culture dish and other such aspects of in vitro experiments can have quantitatively significant effects on experimental results. ADME differences between chemicals are also a potential source of uncertainty. While data are available showing that water and fathead minnow plasma concentrations of fadrozole are similar (41), this equivalence may not hold for other compounds, such as iprodione, that inhibit aromatase. Lack of data addressing ADME is thus also a source of uncertainty when a TEF calculation is used to derive an equivalent concentration of fadrozole for input to the HPG axis model. However, while keeping these concerns in mind, the iprodione example illustrates conceptually how the chemical agonistic qAOP can be applied to predict a toxicological outcome or effect concentration for a chemical which has not been characterized in vivo. It is also worth reiterating that the biologically based modeling approach used for qAOP development leads to explicit identification of potential sources of uncertainty in qAOP predictions, thereby supporting targeted experimentation to address the uncertainties.
qAOPs and 21st Century Toxicology
The goal of “21st century” regulatory toxicology is to achieve a greater coverage of the chemical universe while utilizing fewer resources (time, money, animals). Critical to this is the ability to make reliable predictions of potential adverse effects of untested chemicals on individuals (or populations) based on rapid, inexpensive assessments of their ability to interact with biological systems. This type of information may be derived from computational models, in vitro assays, or short-term in vivo tests with pathway-specific endpoints. The critical role of the AOP framework in this paradigm is translation, in a causal manner, of these resource-efficient estimates of chemical activity into responses relevant to risk assessment. The qAOP is a natural evolution of this process in the context of quantitative predictions of AOs such that, theoretically, estimates of risk can be generated without any long-term animal testing. In this paper, we provide the example of a qAOP that can utilize easily-collected measures of chemical inhibition of a key, rate-limiting steroidogenic enzyme (aromatase) to predict reductions in egg production (fecundity) and, subsequently, population size of fish. This was achieved through linking three discreet models describing different components of the AOP, from the MIE (aromatase inhibition) through five intermediate KEs, to impacts of regulatory interest (fecundity, population size).
There are several potential applications of qAOPs, including some directly relevant to existing regulatory activities. For example, the qAOP described herein would be of immediate utility to the US EPA endocrine disruptor screening program (EDSP), a legislatively-mandated effort to identify and assess potential human health and ecological risks of chemicals that interact with specific endocrine systems (http://www.epa.gov/endo/). Chemicals of concern include those that perturb the HPG axis through direct activation or antagonism of estrogen or androgen receptors, or through indirect effects - usually inhibition - of enzymes involved in sex steroid synthesis, including aromatase (http://www.epa.gov/endo/). The EDSP is charged with evaluating around 10,000 chemicals for potential endocrine toxicity; it is anticipated that a critical step in the evaluation process will be the use of high throughput in vitro data from the US EPA ToxCast program to identify chemicals with the potential to interact with components of the HPG axis (42). To date, roughly 2,000 chemicals have been assessed for inhibition of aromatase activity in one of the core ToxCast assays (NVS ADME hCYP19A1). A total of 168 chemicals have been identified as exhibiting some degree of aromatase inhibition in this assay, with EC50 values ranging from 0.3 nM to 16.3 μM. As illustrated above for iprodione, potency data for the various aromatase inhibitors can be converted into TEQ values (relative to fadrozole) and compared to predicted (or, when available, measured) concentrations of the chemicals in relevant aquatic environments to determine potential aromatase inhibition. This resultant estimate can be directly incorporated into the computational models comprising the qAOP to produce quantitative predictions of possible effects on reproduction in individuals, and fish population size in the field.
In addition to this type of prospective application of qAOPs, there are opportunities for use in assessments of existing impacts of environmental contaminants. An example of this recently was provided by Miller et al. (28), who sought to use reproductive endocrine data from field-collected fish (white sucker, a large cyprinid species indigenous to the Great Lakes) to predict population status of the species at a site impacted by a pulp and paper mill plant. Miller et al. (28) employed a portion of the basic AOP construct used in this paper to link empirical steroid and fecundity information collected over the course of several years at the pulp mill and reference sites in northern Lake Superior, and used the resultant regression relationships as input to a species-specific population model. From this, they could use steroid data to make predictions of population trends of white sucker at the impacted site under different possible mitigation scenarios, such as decreased effluent discharge or enhanced treatment. The ability to quantitatively forecast the effects of remedial activities on extant populations using easily-collected data (steroid production in this instance) would be of great utility to efforts, such as the Great Lakes Restoration Initiative, focused on recovery of contaminated sites (43).
Use of the response-response approach described above avoids the need to run the computational models, instead allowing rapid, semi quantitative visual screening for effects at the individual and population levels. Further, this approach is applicable to the predictive assessment of multiple aromatase inhibitors that might occur as a mixture, through an estimate of “total” aromatase inhibition based on summation of the derived TEF values.
Developing qAOPs from a MIE, through multiple intermediate KEs, to AOs at the individual and/or population levels can require a significant resource investment. For example, initial toxicological work underlying the aromatase inhibition qAOP described herein was published 15 years ago (33); in the intervening years, the core team involved in the work has conducted a large amount of additional research critical to the final qAOP (9, 10, 12, 22, 25, 29, 41, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53). While efficiency certainly will increase as experience grows, it is not a reasonable expectation that there will be many qAOPs in the near term. Rather, qAOP development efforts would be best focused on a few high-visibility/impact MIEs or AOs of concern from a regulatory perspective. A good example of this are endocrine pathways of interest to the EDSP in the US and other international regulatory organizations (54). In fact, three of four qAOPs explored as short case examples in a recent workshop on the topic were focused on endocrine pathways associated with steroid synthesis (the current analysis), estrogen receptor activation, and thyroid signaling (6). The fourth example qAOP from that workshop analysis also addressed a high-visibility issue relative to a legislated need to reduce animal use in terms of safety testing for chemicals potentially causing skin sensitization (6). Other logical targets in terms of qAOP development in the near term include relatively complete qualitative AOPs archived, for example, in the AOP wiki (aopwiki.org), that have undergone a weight-of-evidence evaluation using approaches as described by Becker et al. (55), and rated as “strong” in the context of their intended use. These types of AOPs reflect relatively rich biological and toxicological knowledge bases likely amenable to modeling without the need for extensive collection of new data. For example, AOPs focused on adverse effects associated with Ah receptor activation may be logical candidates for qAOP development in the near future (aopwiki.org; 55).
It is worth noting that, at any point in time, a qAOP reflects the current understanding of KEs and KE relationships. As knowledge relevant to the qAOP increases, either through research specifically targeted to address data gaps or model uncertainties, or through activities in the broader scientific community, the qAOP can be correspondingly updated. Thus, qAOPs can be thought of as tools that ride the moving crest of scientific understanding, organize the relevant information, and identify the potential health risk implications of toxicant exposures.
Acknowledgements
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army or the U.S. EPA.
References
- 1.National Research Council. 2007. Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: The National Academies Press; DOI 10.17226/11970. [DOI] [Google Scholar]
- 2.Krewski D; Acosta D Jr.; Andersen M; Anderson H; Bailar JC 3rd; Boekelheide K; Brent R; Charnley G; Cheung VG; Green S Jr.; Kelsey KT; Kerkvliet NI; Li AA; McCray L; Meyer O; Patterson RD; Pennie W; Scala RA; Solomon GM; Stephens M; Yager J; Zeise L Toxicity testing in the 21st century: A vision and a strategy. J. Toxicol. Environ. Health B Crit. Rev 2010, 13, 51138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ankley GT; Bennett RS; Erickson RJ; Hoff DJ; Hornung MW; Johnson RD; Mount DR; Nichoils JW; Russom CL; Schmieder PK; Serrano JA; Tietge JE; Villeneuve DL 2010. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem 2010, 29, 730–741. [DOI] [PubMed] [Google Scholar]
- 4.Villeneuve DL; Crump D; Garcia-Reyero N; Hecker M; Hutchinson TH; LaLone CA; Landesmann B; Lettieri T; Munn S; Nepelska M; Ottinger MA; Vergauwen L; Whelan M Adverse outcome pathway (AOP) development I: Strategies and principles. Toxicol. Sci 2014a, 142, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Organization for Economic Co-operation and Development. 2016. Users’ Handbook supplement to the Guidance Document for developing and assessing Adverse Outcome Pathways, OECD Series on Adverse Outcome Pathways, No. 1, OECD Publishing, Paris: DOI 10.1787/5jlv1m9d1g32-en. [DOI] [Google Scholar]
- 6.Wittwehr C; Aladjov H; Ankley G; Bryne H; de Knecht J; Heinzle E; Klambauer G; Landesmann B; Luijten M; MacKay C; Maxwell G; Meek B; Paini A; Perkins E; Sobanski T; Villeneuve D; Waters K; Whelan M How adverse outcome pathways can aid the development of computational prediction models for regulatory toxicology. Toxicol. Sci 2017, 155, 326–336, doi: 10.1093/toxsci/kfw207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villeneuve DL; Crump D; Garcia-Reyero N; Hecker M; Hutchinson TH; LaLone CA; Landesmann B; Lettieri T; Munn S; Nepelska M; Ottinger MA; Vergauwen L; Whelan M Adverse outcome pathway development II: Best practices. Toxicol. Sci 2014b, 142, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villeneuve D Adverse outcome pathway on aromatase inhibition leading to reproductive dysfunction (in fish) In OECD Series on Adverse Outcome Pathways, No. 4, OECD Publishing: Paris: 2016. DOI 10.1787/5jlsv05mx433-en. [DOI] [Google Scholar]
- 9.Cheng WY; Zhang Q; Schroeder A; Villeneuve D; Ankley GT; Conolly R Computational modeling of plasma vitellogenin alterations in response to aromatase inhibition in fathead minnows. Toxicol. Sci 2016, 154, 78–89. [DOI] [PubMed] [Google Scholar]
- 10.Villeneuve DL; Mueller ND; Martinovic D; Makynen EA; Kahl MD; Jensen KM; Durhan EJ; Cavallin JE; Bencic D; Ankley GT Direct effects, compensation, and recovery in female fathead minnows exposed to a model aromatase inhibitor. Environ. Health Perspect 2009, 117, 624–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li ZH; Villeneuve DL; Jensen KM; Ankley GT; Watanabe KH A computational model for asynchronous oocyte growth dynamics in a batch-spawning fish. Can. J. Fish Aquat. Sci 2011b, 68, 1528–1538. [Google Scholar]
- 12.Watanabe KH; Mayo M; Jensen KM; Villeneuve DL; Ankley GT; Perkins EJ Predicting fecundity of fathead minnows (Pimephales promelas) exposed to endocrine disrupting chemicals using a MATLAB®-based model of oocyte growth dynamics. PLoS One, 2016, 11, e0146594 DOI 10.1371/journal.pone.0146594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leino RL; Jensen KM; Ankley GT Gonadal histology and characteristic histopathology associated with endocrine disruption in the adult fathead minnow (Pimephales promelas). Environ. Toxicol. Pharmacol 2005, 19, 85–98. [DOI] [PubMed] [Google Scholar]
- 14.Miller DH; Ankley GT Modeling impacts on populations: Fathead minnow (Pimephales promelas) exposure to the endocrine disruptor 17beta-trenbolone as a case study. Ecotoxicol. Environ. Saf 2004, 59, 1–9. [DOI] [PubMed] [Google Scholar]
- 15.Miller DH; Jensen KM; Villeneuve DE; Kahl MD; Makynen EA; Durhan EJ; Ankley GT Linkage of biochemical responses to population-level effects: A case study with vitellogenin in the fathead minnow (Pimephales promelas). Environ. Toxicol. Chem 2007, 26, 521–527. [DOI] [PubMed] [Google Scholar]
- 16.Ankley GT; Miller DH; Jensen KM; Villeneuve DL; Martinovic D Relationship of plasma sex steroid concentrations in female fathead minnows to reproductive success and population status. Aquat. Toxicol 2008, 88, 69–74. [DOI] [PubMed] [Google Scholar]
- 17.Miller DH; Tietge JE; McMaster ME; Munkittrick KR; Xia X; Ankley GT Assessment of status of white sucker (Catostomus commersoni) populations exposed to bleached kraft pulp mill effluent. Environ. Toxicol. Chem 2013, 32, 1592–1603. [DOI] [PubMed] [Google Scholar]
- 18.Miller DH; Tietge JE; McMaster ME; Munkittrick KR; Xia X; Griesmer DA; Ankley GT Linking mechanistic toxicology to population models in forecasting recovery from chemical stress: A case study from Jackfish Bay, Ontario, Canada. Environ. Toxicol. Chem 2015, 34, 1623–1633. [DOI] [PubMed] [Google Scholar]
- 19.Eungdamrong NJ; Iyengar R Modeling cell signaling networks. Biol. Cell 2004, 96, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ankley GT; Villeneuve DL Temporal changes in biological responses and uncertainty in assessing risks of endocrine-disrupting chemicals: insights from intensive time-course studies with fish. Toxicol. Sci 2015, 144, 259–275. [DOI] [PubMed] [Google Scholar]
- 21.Gillies K; Krone SM; Naler JJ; Schultz IR A computational model of the rainbow trout hypthalamous-pituitary-ovary-liver axis. PLoS Comput. Biol 2016, DOI: 10.1371/journal.pcbi.1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villeneuve DL; Breen M; Bencic DC; Cavallin JE; Jensen KM; Makynen EA; Thomas LM; Wehmas LC; Conolly RB; Ankley GT Developing predictive approaches to characterize adaptive responses of the reproductive endocrine axis to aromatase inhibition: I. Data generation in a small fish model. Toxicol. Sci 2013, 133, 225–233. [DOI] [PubMed] [Google Scholar]
- 23.Nelder JA; Mead R 1965. A simplex method for function minimization. Computer J. 1965, 7, 308–313. [Google Scholar]
- 24.Lau C; Andersen ME; Crawford-Brown DJ; Kavlock RJ; Kimmel CA; Knudsen TB; Muneoka K; Rogers JM; Setzer RW; Smith G; Tyl R Evaluation of biologically based dose-response modeling for developmental toxicity: a workshop report. Regul. Toxicol. Pharmacol 2000, 31, 190–199. [DOI] [PubMed] [Google Scholar]
- 25.Conolly RB; Kimbell JS; Janszen D; Schlosser PM; Kalisak D; Preston J; Miller FJ 2004. Human respiratory tract cancer risk of inhaled formaldehyde: dose-response preictions derived from biologically-motivated modeling of a combined rodent and human dataset. Toxicol. Sci 2004, 82, 279–296. [DOI] [PubMed] [Google Scholar]
- 26.Guidelines for Carcinogen Risk Assessment; U.S. Environmental Protection Agency: Washington, DC, 2005; EPA/630/P–03/001F. [Google Scholar]
- 27.Crump KS; Chen C; Chiu WA; Louis TA; Portier CJ; Subramaniam RP; White PD 2010. What role for biologically based dose-response models in estimating low-dose risk? Environ. Health Perspect. 2010, 118, 585–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volker G; Augusiak J; Focks A; Frank BM; Gabsi F; Johnston ASA; Liu C; Martin BT, Meli M; Radchuk V; Thorbek P; Railsback SF; Towards better modelling and decision support: Documenting model development, testing, and analysis using TRACE. Ecological Modeling 2014, 280, 129–139. [Google Scholar]
- 29.McLanahan ED; El-Masri HA; Sweeny LM; Kopylev LY; Clewell HJ; Wambaugh JF; Schlosser PM Physiologically based pharmacokinetic model use in risk assessment – Why being published is not enough. Toxicol. Sci 2012, 126, 5–15. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Fernandez M, Rehberg M, Kremling A, Banga JR. 2013. Simultaneous model discrimination and parameter estimation in dynamic models of cellular systems. BMC Syst. Biol 2013, 7, 76 DOI 10.1186/1752-0509-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P; Vu QD Identification of parameter correlations for parameter estimation in dynamic biological models. BMC Syst. Biol 2013, 7, 91 DOI 10.1186/1752-0509-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saltelli A, Chan K, Scott EM, Eds. Sensitivity Analysis: Gauging the Worth of Scientific Models; Wiley: New York, 2000. ISBN: 978–0-471–99892-1. [Google Scholar]
- 33.Ankley GT; Kahl MD; Jensen KM; Hornung MW; Korte JJ; Makynen EA; Leino RL Evaluation of the aromatase inhibitor fadrozole in a short-term reproduction assay with the fathead minnow (Pimephales promelas). Toxicol. Sci 2002, 67, 121–130. [DOI] [PubMed] [Google Scholar]
- 34.Safe SH Hazard and risk assessment of chemical mixtures using the toxic equivalency factor approach. Environ. Health Perspect 1998, 106, Suppl. 4, 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning T Endocrine disrupting chemicals: A review of the state of the science. Australasian J. Ecotoxicol 2005, 11, 1–52. [Google Scholar]
- 36.Mnif W; Hassine AI; Bouaziz A; Bartegi A; Thomas O; Roig B Effect of endocrine disruptor pesticides: A review. Int. J. Environ. Res. Public Health 2011, 8, 2265–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T; Kondo H; Yaguchi K; Maki T; Suga T Estimation of leachability and persistence of pesticides at golf courses from point-source monitoring and model to predict pesticide leaching to groundwater. Environ. Sci. Technol 1998, 32, 920–929. [Google Scholar]
- 38.Richard AM; Judson RS; Houck KA; Grulke CM; Volarath P; Thillainadarajah I; Yang C; Rathman J; Martin MT; Wambaugh JF; Knudsen TB; Kancherla J; Mansouri K; Patlewicz G; Williams AJ; Little SB; Crofton KM; Thomas RS Toxcast chemical landscape: Paving the road for 21st century toxicology. Chem. Res. Toxicol 2016, 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- 39.Reregistration Eligibility Decision (RED) IPRODIONE; U.S. Environmental Protection Agency: Washington, DC, 1998; EPA738-R-98–019. [Google Scholar]
- 40.Armitage JM; Wania F; Arnot JA; Application of mass balance models and the chemical activity concept to facilitate the use of in vitro toxicity data for risk assessment. Environmental Science Technology 2014, 48, 9770–9779. [DOI] [PubMed] [Google Scholar]
- 41.Villeneuve DL; Garcia-Reyero N; Martinovic-Weigelt D; Li Z; Watanabe KH; Orlando EF; Lalone CA; Edwards SW; Burgoon LD; Denslow ND; Perkins EJ; Ankley GT A graphical systems model and tissue-specific functional gene sets to aid transcriptomic analysis of chemical impacts on the female teleost reproductive axis. Mutat. Res 2012, 746, 151–162. [DOI] [PubMed] [Google Scholar]
- 42.Environmental US Protection Agency Endocrine Disruptor Screening Program (EDSP); https://www.epa.gov/endocrine-disruption/endocrine-disruptor-screening-program-edspoverview.
- 43.Ekman DR; Ankley GT; Blazer VS; Collette TW; Garcia-Reyero N; Iwanowicz LR; Jorgensen ZG; Lee KE; Mazik PM; Miller DH; Perkins EJ; Smith ET; Tietge JE; Villeneuve DL Biological effects-based tools for monitoring impacted surface waters in the Great lakes: A multi-agency program in support of the Great Lakes Restoration Initiative. Environ. Practice 2013, 15, 409–426.; DOI: 10.1017/S1466046613000458. [DOI] [Google Scholar]
- 44.Ankley GT; Bencic DC; Cavallin JE; Jensen KM; Kahl MD; Makynen EA; Martinovic D; Mueller NO; Wehmas LC; Villeneuve DL Dynamic nature of alterations in the endocrine system of fathead minnows exposed to the fungicide prochloraz. Toxicol. Sci 2009, 112, 344–353. [DOI] [PubMed] [Google Scholar]
- 45.Ankley GT; Cavallin JE; Durhan EJ; Jensen KM; Kahl MD; Makynen EA; Thomas LM; Wehmas LC; Villeneuve DL A time-course analysis of effects of the steroidogenesis inhibitor ketoconazole on components of the hypothalamic-pituitarygonadal axis of fathead minnows. Aquat. Toxicol 2012, 114–115, 88–95. [DOI] [PubMed] [Google Scholar]
- 46.Ankley GT; Jensen KM; Durhan EJ; Makynen EA; Butterworth BC; Kahl MD; Villeneuve DL; Linnun A; Gray LE; Cardon M; Wison VS Effects of two fungicides with multiple modes of action on reproductive endocrine function in the fathead minnow (Pimephales promelas). Toxicol. Sci 2005, 86, 300–308. [DOI] [PubMed] [Google Scholar]
- 47.Ankley GT; Jensen KM; Kahl MD; Korte JJ; Makynen EA Description and evaluation of a short-term reproduction test with the fathead minnow (Pimephales promelas). Environ. Toxicol. Chem 2001, 20, 1276–1290. [PubMed] [Google Scholar]
- 48.Ankley GT; Jensen KM; Kahl MD; Makynen EA; Blake LS; Greene KJ; Johnson RD; Villeneuve DL Ketoconazole in the fathead minnow (Pimephales promelas): reproductive toxicity and biological compensation. Environ. Toxicol. Chem 2007, 26, 1214–1223. [DOI] [PubMed] [Google Scholar]
- 49.Breen M; Villeneuve DL; Ankley GT; Bencic DC; Breen MS; Watanabe KH; Lloyd AL; Conolly RB 2013. Developing predictive approaches to characterize adaptive responses of the reproductive endocrine axis to aromatase inhibition: II. Computational modeling. Toxicol. Sci 2013, 133, 234–247. [DOI] [PubMed] [Google Scholar]
- 50.Breen M; Villeneuve DL; Ankley GT; Bencic D; Breen MS; Watanabe KH; Lloyd AL; Conolly RB Computational model of the fathead minnow hypothalamicpituitary-gonadal Axis: Incorporating protein synthesis in improving predictability of responses to endocrine active chemicals. Comp. Biochem. Physiol. C Toxicol. Pharmacol 2016, 183–184, 36–45. [DOI] [PubMed] [Google Scholar]
- 51.Li Z; Kroll KJ; Jensen KM; Villeneuve DL; Ankley GT; Brian JV; Sepúlveda MS; Orlando EF; Lazorchak JM; Kostich M; Armstrong B; Denslow ND; Watanabe KH A computational model of the hypothalamic: pituitary: gonadal axis in female fathead minnows (Pimephales promelas) exposed to 17α-ethynylestradiol and 17β-trenbolone. BMC Syst. Biol 2011a, 5, 63; DOI 10.1186/1752-0509-5-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shoemaker JE; Gayen K; Garcia-Reyero N; Perkins EJ; Villeneuve DL; Liu L; Doyle FJ 3rd. Fathead minnow steroidogenesis: in silico analyses reveals tradeoffs between nominal target efficacy and robustness to cross-talk. BMC Syst. Biol 2010, 4, 89 DOI 10.1186/1752-0509-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe KH; Li Z; Kroll KJ; Villeneuve DL; Garcia-Reyero N; Orlando EF; Sepulveda MS; Collette TW; Ekman DR; Ankley GT; Denslow ND A computational model of the hypothalamic-pituitary-gonadal axis in male fathead minnows exposed to 17 alpha-ethinylestradiol and 17 beta-estradiol. Toxicol. Sci. 2009, 109, 180–192. [DOI] [PubMed] [Google Scholar]
- 54.Organization for Economic Co-operation and Development. 2010. Workshop report on OECD countries activities regarding testing, assessment and management of endocrine disrupters. OECD Environment, Health and Safety Publications, Series on Testing and Assessment, Number 118 Paris, France. [Google Scholar]
- 55.Becker RA; Ankley GT; Edwards SW; Kennedy SW; Linkov I; Meek B; Sachana M; Segner H; Van Der Burg B; Villeneuve DL; Watanabe KH; BartonMacLaren TS Increasing scientific confidence in adverse outcome pathways: Application of tailored Bradford-Hill considerations for evaluating weight-of-evidence. Regul. Toxicol. Pharmacol 2015, 72, 514–537. [DOI] [PubMed] [Google Scholar]