Abstract
The ALK gene fusion has been identified as a new driver gene in non–small cell lung cancer (NSCLC). It includes the EML4-ALK rearrangement as a recurring event that renders the tumor sensitive to ALK tyrosine kinase inhibitor crizotinib. In addition, several other fusion partners to ALK kinase domain (eg, TFG, KLC1, and KIF5B) have been identified in NSCLC. However, clinical data relevant to response in lung cancer harboring these rare ALK translocations are not fully available. A nonsmoking Chinese male originally diagnosed with “stage Ib lung adenocarcinoma” showed metastases in regional lymph nodes, pleura, and bone 1 year after surgery. The patient refused invasive tissue biopsy, and chemotherapy was administrated, which failed as a first- and second-line treatment. We then identified a rare fusion gene of ALK and Striatin (STRN) using next-generation sequencing (NGS)–based circulating tumor DNA (ctDNA) analysis. The NGS of the patient’s originally paraffin-embedded surgical tumor samples also indicated the fusion. Reverse transcription–polymerase chain reaction and Sanger sequencing further confirmed the results. The STRN-ALK involves the fusion of exon 3 of STRN retaining a coiled-coil domain to exon 20 of ALK containing a kinase domain. The patient was treated with crizotinib and showed excellent clinical, radiographic, and molecular response. Repetitive dynamic ctDNA analysis revealed that the fraction of molecular alterations in plasma was closely associated with response to crizotinib treatment. This is the first clinical evidence involving advanced NSCLC due to a rare STRN-ALK fusion and has been effectively treated with crizotinib.
Abbreviations and Acronyms: ctDNA, circulating tumor DNA; MRI, magnetic resonance imaging; NGS, next-generation sequencing; NSCLC, non–small cell lung cancer; STRN, striatin; TKI, tyrosine kinase inhibitor
Crizotinib, a multitargeted ALK/ROS1/MET tyrosine kinase inhibitor (TKI), represents the standard treatment for patients with locally advanced or metastatic ALK-rearranged non–small cell lung cancer (NSCLC).1 EML4-ALK is the most common ALK fusion gene in NSCLC and encodes a cytoplasmic chimeric protein with constitutive kinase activity.2 In addition, several other fusion partners to ALK kinase domain (eg, TFG, KLC1, and KIF5B) have been identified in NSCLC,3 indicating that ALK-rearranged NSCLC is a molecularly heterogeneous subset of lung cancer. Because of the paucity of cases reported, the treatment of patients with NSCLC harboring these rare ALK translocations including the clinical behavior, histopathologic characteristics, and ALK TKI response is not fully understood. Here, we describe the discovery of a rare striatin (STRN)-ALK fusion in a Chinese male patient diagnosed with stage IV lung adenocarcinoma using next-generation sequencing (NGS)–based circulating tumor DNA (ctDNA) profiling, and also the first clinical evidence suggesting an excellent response to crizotinib via STRN-ALK translocation in NSCLC.
Case
The patient is a 59-year-old Chinese male never-smoker who was diagnosed with adenocarcinoma of the right lung at the local hospital and received operative treatment in March 2013. The postoperative pathological stage was pT2aN0M0 Ib.4 In August 2015, he was diagnosed with metastatic disease of regional lymph nodes by thoracic scan and treated with 2 cycles of chemotherapy including platinum combined with taxanes at the local hospital. In addition to symptomatic progression, the tumor developed both mediastinal and pleural lesions (Figure 1A). He was referred to our institution for further medical help. The serum carcinoembryonic antigen level was 77.28 ng/mL (normal, <5 ng/mL). Emission computed tomographic bone scanning showed multisite abnormal radioactivity. Lumbar magnetic resonance imaging (MRI) confirmed bone metastases at the L5 vertebral body. Additional imaging examination in other regions showed no tumor burden. Because the patient refused invasive procedures, real-time pathological evaluation and molecular phenotyping were not obtained. Thus, right lung adenocarcinoma of postoperative metastases (including regional lymph nodes, right pleura, and bone) was clinically diagnosed. Pemetrexed-based chemotherapy was started as per our protocol. Bisphosphonate was synchronously used to inhibit bone metastases. The cough improved, and a computed tomography scan after 3 cycles of chemotherapy showed stable disease. The sixth cycle of chemotherapy was administered in February 2016, at which time our patient’s cough aggravated and tumor progressed as evidenced by imaging examination (Figure 1B).
Figure 1.
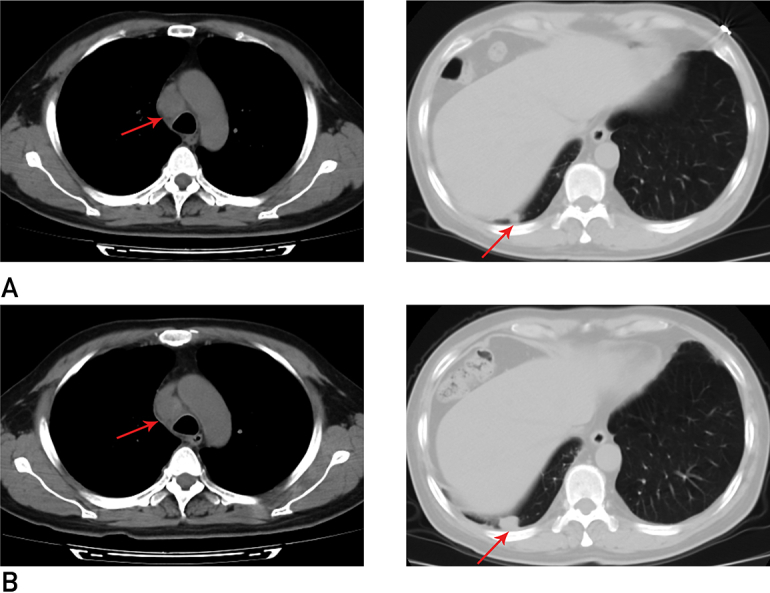
Imaging characteristics of the patient during chemotherapy. A, Chest CT scan after first-line chemotherapy showed enlarged paratracheal lymph nodes in the mediastinum and a nodule on the right pleura. B, Chest CT scan after 6 cycles of second-line chemotherapy showed tumor progression. CT = computed tomography.
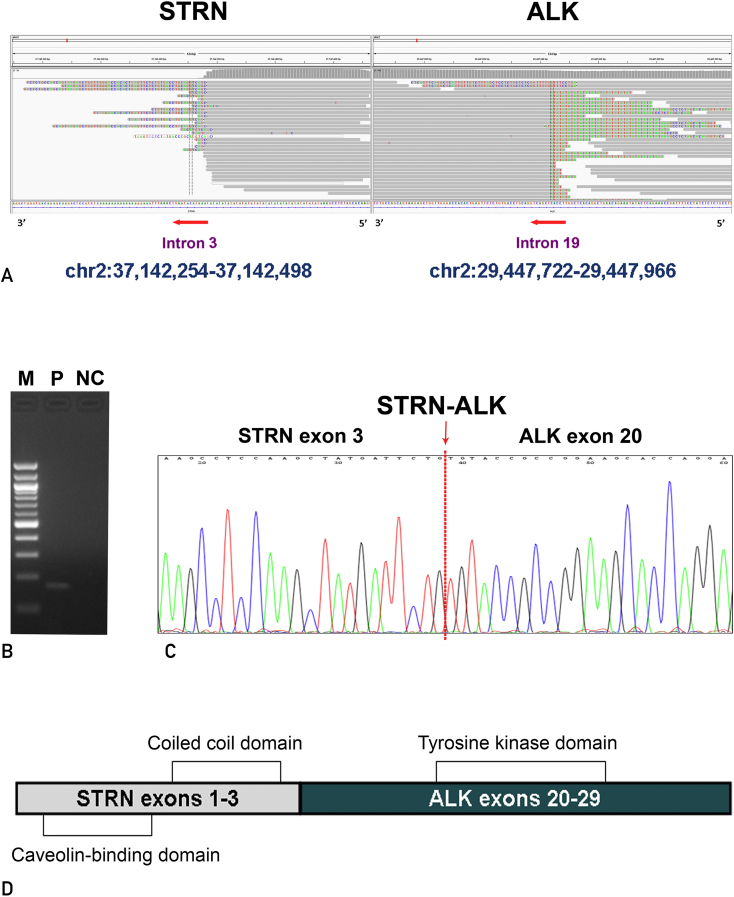
In the absence of standard treatment in this setting, we submitted the patient’s plasma for commercially available cell-free ctDNA analysis in consultation with him to uncover therapeutic targets. Targeted NGS of ctDNA (Geneseeq Biotechnology Inc) identified no aberrations of the EGFR, ROS-1, MET, HER-2, RET, RAS, or BRAF gene. However, we found STRN-ALK fusion at a 1% mutant allele frequency, MYC gene amplification, and R181C mutation of TP53 gene. The results of ctDNA revealed an atypical fusion. Therefore, we further screened the patient’s previous paraffin-embedded surgical samples and confirmed the results by NGS of the tumor tissues (Figure 2A). Reverse transcription–polymerase chain reaction successfully amplified cDNA products including a fusion point from tumor samples (Figure 2B). Sanger sequencing revealed the fusion of exon 3 of STRN retaining a coiled-coil domain to exon 20 of ALK containing a kinase domain (Figure 2C and D).
Figure 2.
Identification of STRN-ALK fusion in the patient’s paraffin-embedded surgical samples. A, Paired-end sequencing data from tumor tissue samples indicated somatic intrachromosomal STRN-ALK fusion as demonstrated by Integrative Genomics Viewer program. B, Confirmation of the STRN-ALK fusion by polymerase chain reaction. The expected product size is 166 bp. C, DNA sequence chromatograms show the conjoined regions at the DNA sequence level of the STRN-ALK fusion gene. D, Schematic diagram of the predicted domains of the STRN-ALK fusion protein. M = marker (100-bp ladder); NC = negative control; P = paraffin-embedded tissue sample.
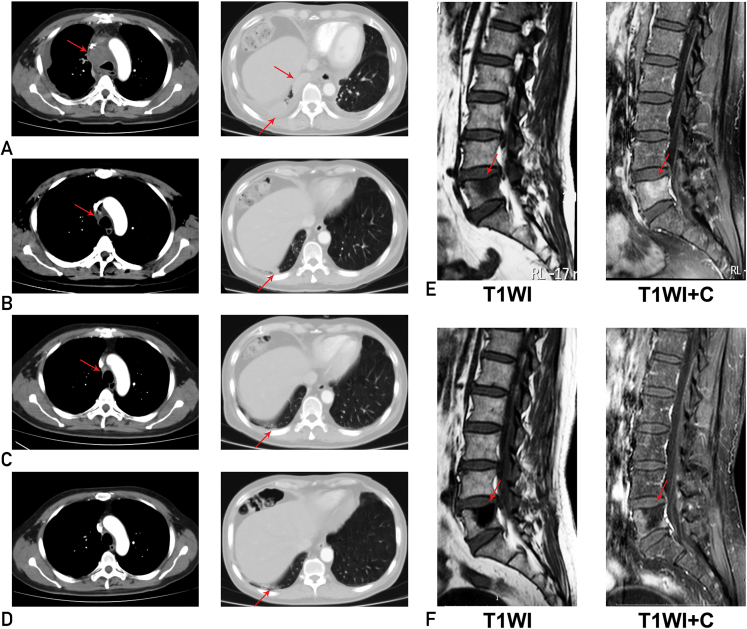
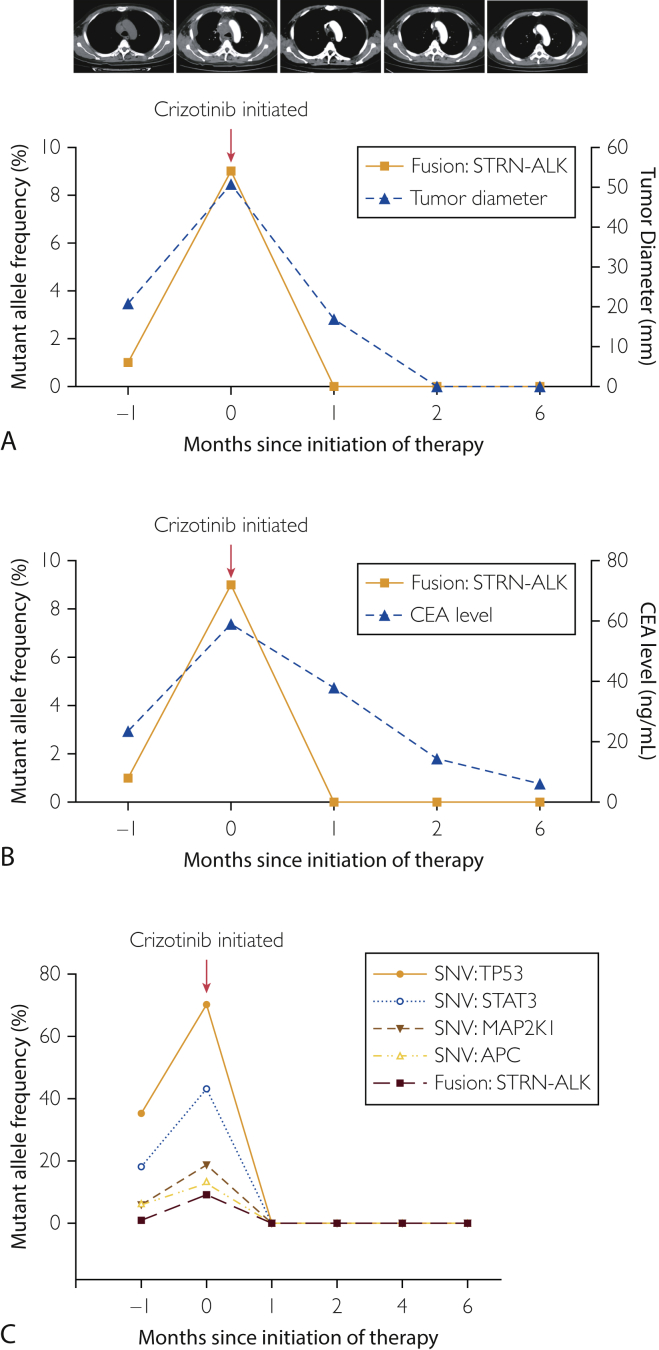
On the basis of molecular findings, we recommended crizotinib therapy, which was not accepted promptly. Shortly thereafter, the patient developed worsening cough, chest distress, and dyspnea. Scans demonstrated significant disease progression involving a 54 × 51 mm mass in the mediastinum, 2 nodules on the right pleura (28 × 19 mm and 43 × 13 mm, respectively), and bilateral pleural effusion and inflammation, especially on the right side (Figure 3A). The ctDNA analysis revealed an abundance of STRN-ALK fusion increasing to 9% mutant allele frequency (Figure 4). He began oral intake of crizotinib (250 mg twice daily) and this treatment ameliorated his clinical symptoms significantly and promptly. Scans demonstrated significant tumor shrinkage of the target lesions after 1 month (Figure 3B), complete response after 2 months (Figure 3C), and sustained response after 6 months (Figure 3D). Compared with the baseline lumbar MRI (Figure 3E), MRI at 2 months after therapy showed a restricted clear edge of metastatic bone lesions (Figure 3F). Repeated ctDNA analysis showed undetectable STRN-ALK after 1 month of crizotinib therapy and complete remission maintenance after 6 months (Figure 4). Dynamic ctDNA analyses revealed that the fraction of tumor-related molecular alterations in the plasma was closely associated with tumor burden (Figure 4A) and carcinoembryonic antigen level (Figure 4B), accurately reflecting the real-time tumor response to crizotinib therapy. Currently, the patient is still receiving crizotinib treatment with good tolerance.
Figure 3.
Imaging characteristics of the patient during crizotinib treatment. Chest CT-enhanced scans were shown before (A) and 1 month (B), 2 months (C), and 6 months (D) after initiation of crizotinib therapy, demonstrating dramatic shrinkage of tumor lesions. E, Lumbar MRI after first-line therapy revealed an apparent enhancement of lesion at the L5 vertebral body after intensification, indicating the existence of bone metastases. F, Lumbar MRI after treatment with crizotinib for 2 months showed limited scope for reinforcement. MRI = magnetic resonance imaging.
Figure 4.
Noninvasive detection and monitoring of ctDNA using targeted NGS. Dynamic changes in tumor burden (A) and carcinoembryonic antigen (B) in response to crizotinib treatment were closely correlated with alterations in fractional abundance of STRN-ALK in plasma. C, Concordance between different reporters (SNVs and fusion) in response to crizotinib. APC = adenomatous polyposis coli; CEA = carcinoembryonic antigen; ctDNA = circulating tumor DNA; MAF = mutant allele frequency; MAP2K1 = mitogen-activated protein kinase kinase 1; NGS = next-generation sequencing; SNV = single nucleotide variant; STAT3 = signal transducer and activator of transcription 3; TP53 = tumor protein 53.
Discussion
Postprogression tumor biopsies remain essential to both clinical care and research efforts. However, a single biopsy may not fully reflect the whole tumor biology. Under certain clinical conditions, tumor samples are not available (eg, tumor location, patient refusal for invasive procedures as in the present case). Plasma genotyping of ctDNA has the potential to facilitate rapid noninvasive diagnosis while avoiding the inherent shortcomings of tissue genotyping and repeat biopsies. Newman et al5 have shown that the levels of ctDNA are significantly correlated with tumor volume and provide earlier response assessment than radiographic approaches. Furthermore, the diagnostic ability and specificity of plasma droplet digital polymerase chain reaction to identify specific populations with distinct targets has recently been confirmed by investigators from the Harvard Medical School.6 These studies provide clinical support for ctDNA genetic profiling as a valuable tool in the care of cancer patients, where treatment is driven by genetic status.
ALK gene fusion is a unique molecular subtype occurring in 3% to 7% of all the patients with NSCLC,7, 8 including recurring EML4-ALK rearrangement more prevalent in young, male, and nonsmoking patients diagnosed with adenocarcinoma.9, 10 In the patients with adenocarcinoma carrying wild-type EGFR and KRAS in Taiwan and mainland China, the incidence of such fusion is as high as 34%.11 The patient with adenocarcinoma in the present case is a middle-aged Asian male without smoking history. He refused invasive procedures throughout the disease course; thus, the real-time pathological evaluation and molecular phenotyping could not be performed. Moreover, when further screening the paraffin slices of the resected lung cancer tissues of the patient archived at the local hospital at the first diagnosis, we found that the quality of the samples was not suitable for fluorescence in situ hybridization analysis according to pathologists’ assessment. In the absence of a standard recommendation for noninvasive treatment, we recommended targeted NGS-based ctDNA profiling upon progression of second-line chemotherapy. The molecular genotyping of this patient was negative for EGFR or KRAS mutations, ROS1 rearrangements, or MET amplifications and positive for STRN-ALK fusion and TP53 mutations, which was clearly consistent with the features of ALK-positive NSCLC. This rare fusion was further confirmed by NGS of paraffin-embedded surgical tumor samples, indicating that the ALK gene fusion was an early event.
STRN-ALK fusion in NSCLC was first described in 2013 by Majewski et al12 using kinome-centred RNA sequencing. Subsequently, this fusion type was also discovered in cancers of thyroid,13, 14 kidney,15 and colon.16 The STRN-ALK fusion was reported to involve intrachromosomal translocation of exons 1 to 3 of STRN to exons 20 to 29 of ALK within the short arm of chromosome 2 (2p22.2 and 2p23, separated by ∼7.7 Mb).12, 13, 14, 15, 16 The fusion site of this patient was consistent with previous studies (Figure 2). To date, several other rare fusion partners of ALK (eg, TFG, KLC1, and KIF5B) have also been identified in NSCLC.3 ALK fusions involving different partners or even different fusion points with the same partner demonstrate differential sensitivity to the structurally different ALK inhibitors.17, 18 Both in vivo and in vitro studies indicate that STRN-ALK fusion in thyroid carcinoma responded to ALK TKI crizotinib.13, 19 However, clinical data associated with response in lung cancer are lacking. Crizotinib treatment for NSCLC represents third-line therapy in the present case. However, the patient showed excellent clinical, radiographic, and molecular response, indicating that the tumor was driven by the STRN-ALK gene. Thus, we provide proof of principle that crizotinib should be considered in advanced NSCLC harboring STRN-ALK rare fusion, as it has been shown for EML4-ALK.
Conclusion
By using powerful NGS-based ctDNA profiling in a clinical setting, we were able to detect a rare STRN-ALK rearrangement in a patient with advanced NSCLC. To our knowledge, this is the first case to be described in the literature of STRN-ALK-positive NSCLC treated effectively with crizotinib. However, whether this finding represents an inherent property of this fusion protein or unique clinicopathologic characteristics in patients carrying this fusion remains to be investigated. Moreover, the patient’s durable response to crizotinib and even future resistance mechanisms need further follow-up. To clearly understand the potential of crizotinib in tumors bearing an aberration of the STRN-ALK, the prevalence of this rearrangement needs to be assessed in a larger group.
Acknowledgments
We thank Ye-Wei Xia, MD, and Xian Zhang, MD, PhD, of Geneseeq Technology Inc for technical assistance.
Footnotes
Grant Support: The work was supported by grants 81402514 and 81572458 from the National Natural Science Foundation of China.
Potential Competing Interests: The authors report no competing interests.
References
- 1.Kazandjian D., Blumenthal G.M., Chen H.Y., et al. FDA approval summary: crizotinib for the treatment of metastatic non-small cell lung cancer with anaplastic lymphoma kinase rearrangements. Oncologist. 2014;19(10):e5–e11. doi: 10.1634/theoncologist.2014-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soda M., Choi Y.L., Enomoto M., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 3.Katayama R., Lovly C.M., Shaw A.T. Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: a paradigm for precision cancer medicine. Clin Cancer Res. 2015;21(10):2227–2235. doi: 10.1158/1078-0432.CCR-14-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edge S., Byrd D.R., Compton C.C., eds, et al. Springer; New York: 2011. AJCC Cancer Staging Manual. [Google Scholar]
- 5.Newman A.M., Bratman S.V., To J., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacher A.G., Paweletz C., Dahlberg S.E., et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2(8):1014–1022. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koivunen J.P., Mermel C., Zejnullahu K., et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14(13):4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y., Zhang Y., Li Y., et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer. 2014;84(2):121–126. doi: 10.1016/j.lungcan.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Shaw A.T., Yeap B.Y., Mino-Kenudson M., et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27(26):4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gainor J.F., Varghese A.M., Ou S.H., et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19(15):4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S.G., Kuo Y.W., Chang Y.L., et al. EML4-ALK translocation predicts better outcome in lung adenocarcinoma patients with wild-type EGFR. J Thorac Oncol. 2012;7(1):98–104. doi: 10.1097/JTO.0b013e3182370e30. [DOI] [PubMed] [Google Scholar]
- 12.Majewski I.J., Mittempergher L., Davidson N.M., et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230(3):270–276. doi: 10.1002/path.4209. [DOI] [PubMed] [Google Scholar]
- 13.Kelly L.M., Barila G., Liu P., et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci U S A. 2014;111(11):4233–4238. doi: 10.1073/pnas.1321937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perot G., Soubeyran I., Ribeiro A., et al. Identification of a recurrent STRN/ALK fusion in thyroid carcinomas. PLoS One. 2014;9(1):e87170. doi: 10.1371/journal.pone.0087170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusano H., Togashi Y., Akiba J., et al. Two cases of renal cell carcinoma harboring a novel STRN-ALK fusion gene. Am J Surg Pathol. 2016;40(6):761–769. doi: 10.1097/PAS.0000000000000610. [DOI] [PubMed] [Google Scholar]
- 16.Yakirevich E., Resnick M.B., Mangray S., et al. Oncogenic ALK fusion in rare and aggressive subtype of colorectal adenocarcinoma as a potential therapeutic target. Clin Cancer Res. 2016;22(15):3831–3840. doi: 10.1158/1078-0432.CCR-15-3000. [DOI] [PubMed] [Google Scholar]
- 17.Heuckmann J.M., Hölzel M., Sos M.L., et al. ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res. 2011;17(23):7394–7401. doi: 10.1158/1078-0432.CCR-11-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heuckmann J.M., Balke-Want H., Malchers F., et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res. 2012;18(17):4682–4690. doi: 10.1158/1078-0432.CCR-11-3260. [DOI] [PubMed] [Google Scholar]
- 19.Godbert Y., Henriques de Figueiredo B., Bonichon F., et al. Remarkable response to crizotinib in woman with anaplastic lymphoma kinase-rearranged anaplastic thyroid carcinoma. J Clin Oncol. 2015;33(20):e84–e87. doi: 10.1200/JCO.2013.49.6596. [DOI] [PubMed] [Google Scholar]