Abstract
Human pluripotent stem cells are considered to be ideal cell sources for regenerative medicine, but their clinical and industrial application is hindered by their tumorigenic potential. Previously we have identified a pluripotent stem cell-specific lectin rBC2LCN recognizing podocalyxin as a cell surface ligand. More recently, podocalyxin was found to be a soluble ligand of rBC2LCN that is secreted specifically from human pluripotent stem cells into cell culture media. Taking advantage of this phenomenon, we have previously developed a sandwich assay targeting the soluble podocalyxin using rBC2LCN as a capturing probe and another lectin rABA as an overlay probe to detect human pluripotent stem cells residing in cell therapy products derived from human pluripotent stem cells. A drawback to this, however, was that cell culture media containing fetal bovine serum was found to cause a substantial background signal to the sandwich assay. To reduce the background and increase the sensitivity, we screened different overlay probes to detect the soluble podocalyxin. Among them, an anti-keratan sulfate monoclonal antibody called R-10G showed the highest sensitivity and provided a low background signal to fetal bovine serum. The established sandwich assay using rBC2LCN and R-10G was proved to be powerful, which allowed the high-sensitive detection of human induced pluripotent stem cells residing among clinical-grade cardiomyocytes and neural stem cells, both derived from human induced pluripotent stem cells. The developed method has a possibility to be a standard technology to detect human induced pluripotent stem cells resided in various types of cell therapy products.
Keywords: Pluripotent stem cells, Tumorigenicity, Regenerative medicine, Glycan, Lectin
Highlights
-
•
A nondestructive method was developed to detect undifferentiated cells.
-
•
The developed method is applicable to hiPSC-derived cardiomyocytes.
-
•
The developed method is applicable to hiPSC-derived neural stem cells.
1. Introduction
Human pluripotent stem cells (hPSCs), such as embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), hold enormous potential as cell sources for cell therapy products (CTPs) due to their ability for infinite self-renewal and differentiation into any cell type [1], [2]. Several clinical trials using hPSC-derived cell products such as hESC-derived oligodendrocyte progenitor cells [3], hESC-derived retinal pigment epithelium (RPE) [4], and hiPSC-derived RPE [5] have been conducted. Clinical trials using hiPSC-derived cardiomyocytes and neural stem cells (hNSCs) are also planned to treat severe heart failure [6] and spinal cord injury [7], respectively. While clinical applications of hPSC-based CTPs are moving forward, there is a major safety concern that residual undifferentiated cells in hPSC-derived CTPs could form teratoma in recipients. Therefore, establishment of a method to detect such cells is keenly sought for the safety assessment of hPSC-derived CTPs. However, there has been no internationally recognized guideline for the testing of tumorigenicity in hPSC-derived CTPs [8], [9], [10].
An in vitro teratoma formation assay is the sole method used to directly assess the tumorigenic potential of undifferentiated cells. However, this assay is laborious and time-consuming, resulting in more practical in vitro assays such as flow cytometry and quantitative real-time PCR being proposed [11]. Recently, Tano et al. reported a novel approach for the direct and simple detection of trace numbers of hPSCs using an efficient hPSC culture method [12].
Previously, we performed comprehensive glycome analyses of a large number of hPSCs using high-density lectin microarray and found an hPSC-specific lectin designated rBC2LCN (recombinant N-terminal domain of BC2L-C lectin derived from Burkholderia cenocepacia) [13]. rBC2LCN recognizes a mucin-type O-glycan, H type3 structure (Fucα1-2Galβ1-3GalNAc) that is heavily displayed on podocalyxin, a sialomucin containing a mucin domain with >100 putative O-glycosylation sites [14], [15]. Recently, rBC2LCN-positive podocalyxin was found to be secreted into cell culture media by undifferentiated hPSCs [16]. Taking advantage of this phenomenon, a nondestructive and quantitative sandwich assay (termed GlycoStem) targeting the soluble rBC2LCN-positive podocalyxin was developed to detect hPSCs residing in CTPs using cell culture supernatants. In this method, hPSC cell culture media was reacted with rBC2LCN immobilized on a microtiter plate and the captured rBC2LCN-positive podocalyxin was detected with another lectin, rABA. The rBC2LCN-rABA sandwich assay allowed the detection of undifferentiated cells resided in transplanting cells. However, the assay appeared to show background to fetal bovine serum (FBS) included in cell culture media. This drawback would limit the applicability of the assay to regenerative medicine.
Here, we screened alternative overlay probes and found that an anti-keratan sulfate monoclonal antibody, called R-10G, provides a low background signal to FBS-containing media. Using the established rBC2LCN-R-10G sandwich assay, termed GlycoStem-HP, we analyzed the number of undifferentiated hPSCs residing among in hPSC-derived CTPs, such as hiPSC-derived neural stem cells (hNSCs) and hiPSC-derived cardiomyocytes. The developed method is highly practical and should contribute to the safety of stem cell-based cell therapy.
2. Methods
2.1. Ff-I01 hiPSCs, fetal hNSCs, and hiPSC-derived hNSCs
This study was conducted in accordance with the principles of the Helsinki Declaration, and approval to use human neural stem/progenitor cells derived from neural tissues (fetal hNSCs) and hiPSCs generated from human primary somatic cells was obtained from the Ethics committee of Osaka National Hospital (No. 110, No. 146 and No. 150). Ff-I01 hiPSCs generated from human adult peripheral blood mononuclear cells by plasmid vectors [17] were obtained from the Center for iPS Cell Research and Application (CiRA), Kyoto University and maintained on cell culture plates coated with iMatrix-511 (Nippi) in StemFit®AK03 media (Ajinomoto) [18]. hiPSC-derived hNSCs were induced using SFEBq [19] and propagated by the neurosphere culture technique [20]. Fetal hNSCs (oh-NSC-7-fb) were also expanded using the same method [20]. hNSCs were cultured in DMEM/F-12 (1:1; Sigma–Aldrich) with EGF (20 ng/ml; PeproTech Inc.), FGF2 (20 ng/ml; PeproTech), leukemia inhibitory factor (10 ng/ml; Millipore), B27 supplement (final 2%; Life Technologies), heparin (5 μg/ml; Sigma–Aldrich), and HEPES (final 15 mM, Nacalai Tesque). To generate cell culture supernatants, semi-confluent hiPSCs (five days after last passage), fetal-derived hNSCs (eleven days after last passage), or hiPSC-derived hNSCs (eleven days after last passage) were transferred to new culture medium. After 24 h incubation, the medium was collected and centrifuged for 5 min at 1400 ×g to remove cellular debris. Aliquots of the conditioned medium were stored at −80 °C until analyzed.
2.2. 201B7 hiPSCs
201B7 hiPSCs were cultured in 2.5 mL of mTeSR1 (STEMCELL Technologies), TeSR-E8 (STEMCELL Technologies), StemSure hPSC (Wako), and MEF-CM on 6 cm dishes coated with Matrigel (BD Biosciences) [1], [21]. 253G4 hiPSCs were cultured in 2.5 mL of mTeSR1 (STEMCELL Technologies) on 6 cm dishes coated with Matrigel (BD Biosciences) [1], [21]. After 24 h culture, the medium was collected and centrifuged at 1400 ×g for 10 min to remove cell debris. Aliquots of the conditioned medium were stored at −80 °C until analyzed. MEF-CM is the cell culture supernatant of Mitomycin C-treated mouse embryonic fibroblasts (MEF) cultured overnight in DMEM Ham's F12 HEPES+ (ThermoFisher SCIENTIFIC), 20% KSR (ThermoFisher SCIENTIFIC), 1× MEM NEAA (ThermoFisher SCIENTIFIC), 100 μM 2-mercaptoethanol (Wako), Penicillin–Streptomycin (Wako), and 5 ng/mL bFGF (Wako).
2.3. 253G1 cells and cardiomyocyte differentiation
253G1 hiPSCs were differentiated into cardiomyocytes according to a previously described protocol with minor modifications [22]. Cell culture supernatants were centrifuged at 190 ×g for 1 min and stored at −80 °C until analyzed. Just before the analysis, cell culture supernatants were centrifuged again at 1400 ×g for 10 min to completely remove cell debris.
2.4. Lectin and antibody
The N-terminal domain (1–156 aa) of BC2L-C identified from B. cenocepacia and the full-length (1–143 aa.) of Agaricus bisporus agglutinin were inserted into the pET27b bacterial vector between the NdeI and XhoI restriction sites, generating rBC2LCN-pET27b and rABA-pET27b. The plasmid was transformed into Escherichia coli BL21 CodonPlus (DE3)-RIL competent cells for expression. The transformed E. coli was cultured in LB medium containing 10 μg/mL of kanamycin at 37 °C until the OD600 reached 0.4. Expression of rBC2LCN and rABA was induced by the addition of 1 mM IPTG at 20 °C for 24 h. The E. coli cells were harvested by centrifugation at 4450 ×g for 30 min and lyzed by sonication in PBSET (6 mM Na2HPO4·12H2O, 1.4 mM KH2PO4, 140 mM NaCl pH 7.0, 1 mM EDTA, 0.1% Triton X-100) containing a protease inhibitor cocktail (Nacalai tesque). After centrifugation at 24,910 ×g for 30 min, supernatants were applied onto l-fucose-Sepharose (for rBC2LCN) or GlcNAc-Sepharose (for rABA) and the bound recombinant lectins were eluted with 0.2 M l-fucose (for rBC2LCN) or 0.2 M GlcNAc (for rABA) in PBSE (6 mM Na2HPO4·12H2O, 1.4 mM KH2PO4, 140 mM NaCl pH 7.0, 1 mM EDTA). The purified lectins were finally dialyzed against PBS. The protein concentration was measured by BCA protein assay (Thermo Scientific) and the purity was analyzed by electrophoresis using 17% XV pantera MP Gel (DRC). rBC2LCN was labeled with biotin using Biotin Labeling Kit – NH2 (Dojindo, Cat#: LK03). rABA, R-10G (Wako, Cat#: 011-25811), SSEA3 (Millipore, Cat#: MAB4303), SSEA4 (Millipore, Cat#: MAB4304), Tra-1-60 (Millipore, Cat#: MAB4360), Tra-1-81 (Millipore, Cat#: MAB4381), and anti-podocalyxin pAb (R&D, Cat#: AF1658) were labeled with horseradish peroxidase using Peroxidase Labeling Kit-NH2 (Dojindo). Tra-1-60 (BD, Cat#: 560380) and cTNT (Thermo SCIENTIFIC, Cat#: MS-295-P) were used for flow cytometry analysis.
2.5. GlycoStem-HP
Biotin-labeled rBC2LCN (15 ng) diluted in PBS (Takara) was immobilized on streptavidin-coated plates (SUMITOMO BAKELITE, BS-X7603) at room temperature for 1 h. After washing 5 times with 200 μL of wash buffer (PBS containing 0.1% Triton X-100), 50 μL of cell culture media were allowed to react at room temperature for 1 h. After washing, 50 μL of HRP-labeled R-10G was overlaid for 1 h at room temperature. After washing 6 times, 100 μL of TMB solution (Wako, Cat#: 208-17371) were then added and developed for 30 min at room temperature. The reaction was stopped by 100 μL of 1 N HCL and detected at OD450, OD450-650, or OD450-620.
3. Results
3.1. GlycoStem shows background to FBS-containing media
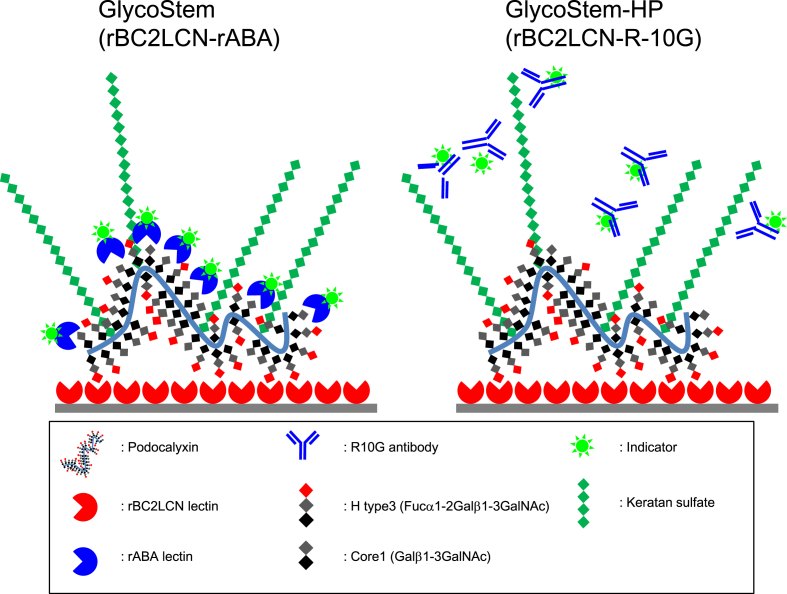
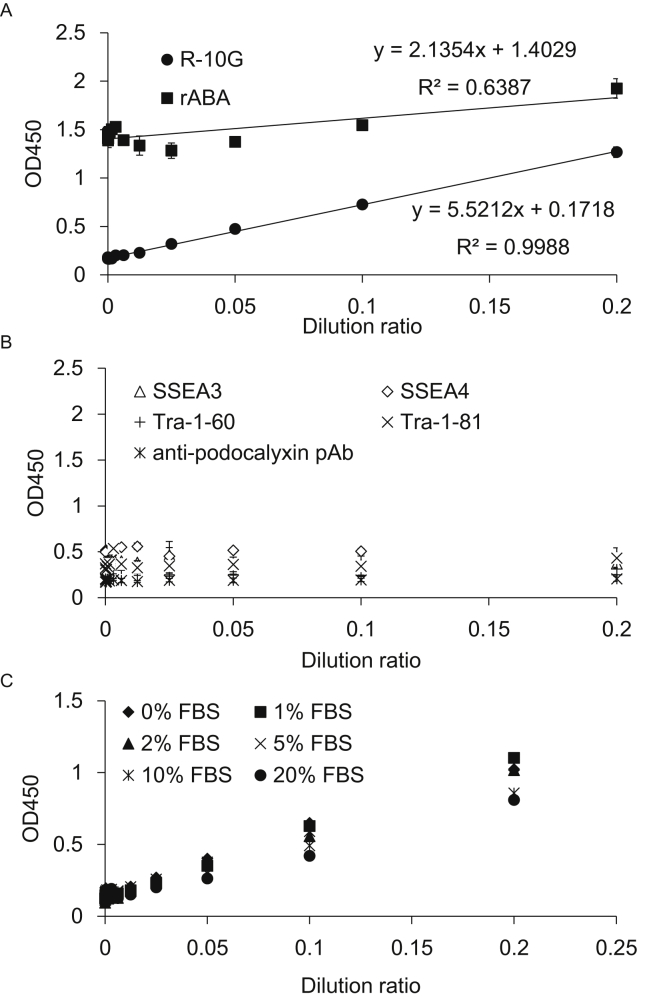
Previously, we developed a sandwich assay using rBC2LCN and rABA lectins to detect of hiPSCs using cell culture supernatants as illustrated in Fig. 1 left [16]. Supernatants of hiPSC cultures were incubated with rBC2LCN immobilized on a microtiter plate. rBC2LCN recognizes an H type3 epitope (Fucα1-2Galβ1-3GalNAc) heavily displayed on podocalyxin secreted from hPSCs. The captured rBC2LCN-positive podocalyxin was then detected with rABA, which recognizes core1 (Galβ1-3GalNAc), a precursor of H type3. The rBC2LCN-rABA sandwich assay (termed GlycoStem) allowed quantitative detection of hiPSCs (201B7 and 253G4) and hESCs (H1) cultured in different types of cell culture medium, including Nutristem, ReproFF, MEF-CM, mTeSR1, and StemSure hPSC [16]. The lower limit of detection (LLOD) varied depending on the type of cell culture medium, ranging from 478 to 4753 cells/mL. However, it appeared to show a heightened background signal to FBS-containing media. To confirm this, supernatants from 201B7 hiPSC cultures were serially diluted with Dulbecco's Modified Eagle Medium (DMEM) containing 2% FBS, incubated with rBC2LCN immobilized on a microtiter plate, and overlaid with horseradish peroxidase (HRP)-labeled rABA (Fig. 1 left). As shown in Fig. 2A (filled squares), the rBC2LCN-rABA sandwich assay was >1 at OD450 for DMEM containing 2% FBS and had a low correlation coefficient (R2 = 0.6387), indicating that the assay is not applicable to FBS-containing media.
Fig. 1.
Schematic representation of the principle of GlycoStem and GlycoStem-HP. Podocalyxin carrying H type3 specifically secreted from hPSCs is captured by hPSC-specific lectin rBC2LCN immobilized on a microplate plate. The rBC2LCN-positive podocalyxin is detected with either HRP-labeled rABA recognizing core1 (GlycoStem, left) or HRP-labeled R-10G recognizing keratan sulfate displayed on podocalyxin (GlycoStem-HP, right).
Fig. 2.
Development of GlycoStem-HP. (A) Cell culture supernatants of 201B7 hiPSCs were serially diluted with DMEM containing 2% FBS and reacted with rBC2LCN immobilized on a microtiter plate. The captured rBC2LCN-positive podocalyxin was detected with HRP-labeled R-10G or rABA. (B) The captured rBC2LCN-positive podocalyxin was detected with HRP-labeled SSEA3, SSEA4, Tra-1-60, Tra-1-81, or anti-podocalyxin pAb. Absorbance at OD450 was measured. Data shown are the mean ± SD of triplicate samples. (C) Effect of FBS on GlycoStem-HP. Cell culture supernatants of 201B7 hiPSCs were serially diluted with DMEM containing different percentages (0–20%) of FBS and reacted with rBC2LCN immobilized on a microtiter plate. The captured rBC2LCN-positive podocalyxin was overlaid with HRP-labeled R-10G. Absorbance at OD450 was measured. Data shown are means of triplicate samples.
3.2. Development of GlycoStem-HP
To reduce the background signal seen with FBS-containing media, we searched for alternative overlay probes to detect the captured rBC2LCN-positive podocalyxin, which shows no or low background to FBS-containing media. For this purpose, antibodies specific to hPSCs such as SSEA3, SSEA4, Tra-1-60, Tra-1-81, anti-podocalyxin polyclonal antibody (pAb), and R-10G monoclonal antibody (mAb) were screened as overlay probe candidates [23], [24], [25]. Among them, R-10G mAb was found to exhibit low background and provide a high correlation coefficient (R2 = 0.9988) [25] (Fig. 2A, filled circles), whereas other antibodies provided only low signals to the cell culture supernatants of 201B7 hiPSCs (Fig. 2B). R-10G has been reported to recognize a type of keratan sulfate that lacks oversulfated structures displayed on podocalyxin expressed in hPSCs [25] (Fig. 1 right). Supernatants from 201B7 hiPSC cultures were then serially diluted with DMEM containing varying percentage of FBS (0–20%) and analyzed by the rBC2LCN-R-10G sandwich assay. As shown in Fig. 2C, the sandwich assay had a low background signal to DMEM containing as much as 20% FBS. A linear regression curve could be obtained (Fig. 2C, filled circles), although the signals were slightly decreased compared to DMEM without FBS (Fig. 2C, filled diamonds). These results demonstrate that the rBC2LCN-R-10G sandwich assay, termed GlycoStem-high performance (GlycoStem-HP), is capable of detecting undifferentiated hPSCs even in the presence of FBS (Fig. 1 right).
3.3. Lower limit of detection of GlyoStem-HP
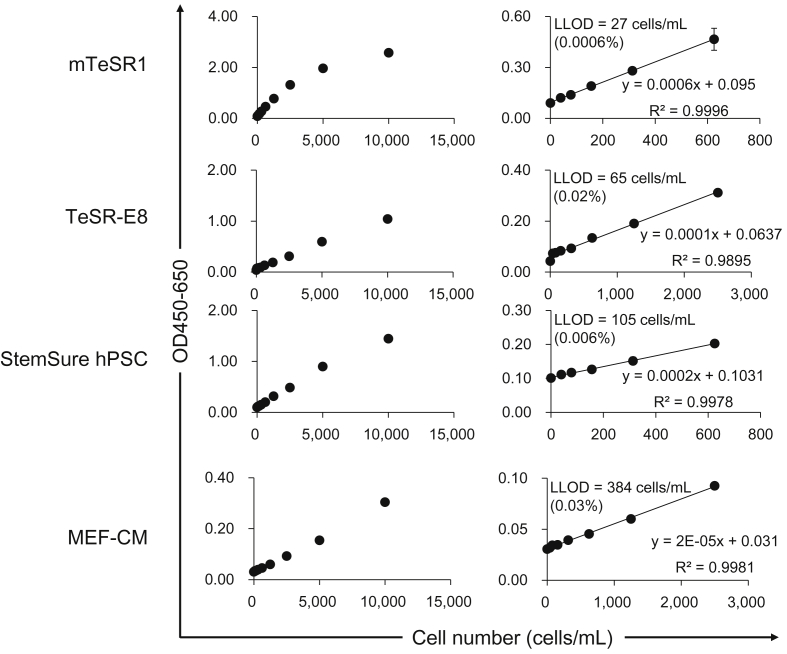
We next determined the lower limit of detection (LLOD) of the developed system using cell culture supernatants from 201B7 hiPSCs cultured in different types of cell culture medium. The value of LLOD was calculated for each medium as the mean plus 3.3-fold the standard deviation of the measurement of the negative control medium (Fig. 3) [11]. The LLOD values for mTeSR1, TeSR-E8, StemSure hPSC media, and mouse embryonic fibroblast-conditioned media (MEF-CM) were determined to be 27, 65, 105, and 384 cells/mL, respectively, indicating that the method could detect 0.0006, 0.02, 0.006, and 0.03% of 201B7 hiPSCs, respectively. The previous method (GlycoStem) gave 3792, 623, and 3775 cells/mL as the LLOD for mTeSR1, StemSure hPSC, and MEF-CM, respectively [16], indicating that the novel method (GlycoStem-HP) is 5.9 ˜ 140-fold more sensitive than the previous method.
Fig. 3.
Sensitivity of GlyoStem-HP. Cell culture supernatants of 201B7 hiPSCs cultured in mTeSR1, TeSR-E8, StemSure hPSC, MEF-CM media were serially diluted using the corresponding cell culture media and analyzed by GlycoStem-HP. Absorbance at OD450-OD650 was measured. Data are shown as the mean ± SD of triplicate samples.
3.4. Monitoring the number of hiPSCs during cardiomyocyte differentiation
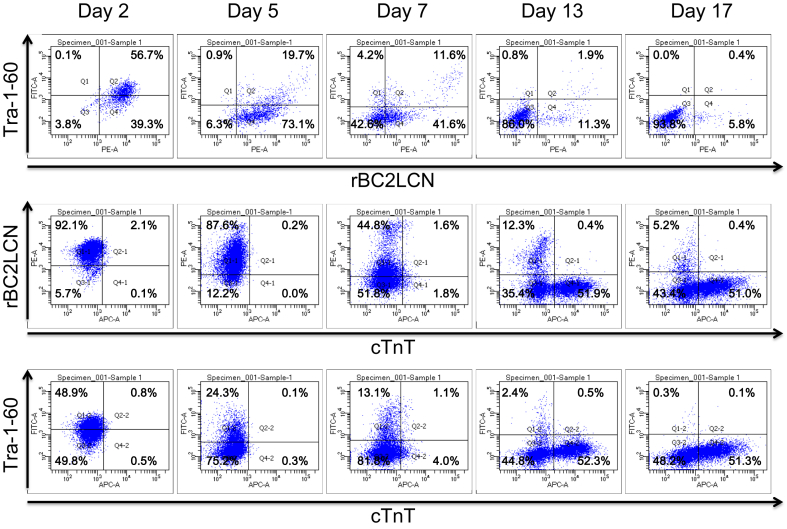
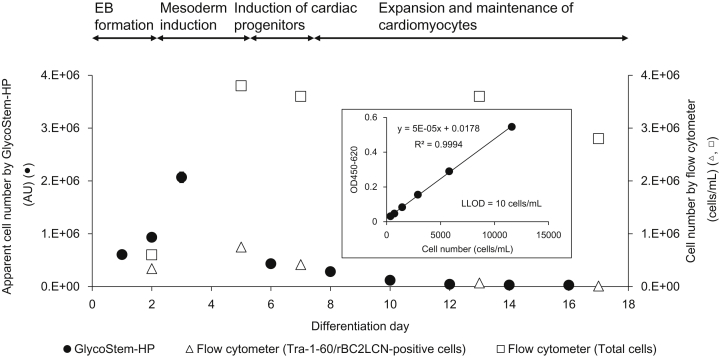
Using the developed system, we aimed to monitor the number of undifferentiated hiPSCs during the differentiation into cardiomyocytes, a procedure that is planned for clinical trials at Osaka University to treat severe heart failure [6]. 253G1 hiPSCs were differentiated into cardiomyocytes according to a previous report with minor modification [22] and the states of differentiation were analyzed by flow cytometry. As shown in Fig. 4, cardiac troponin T (cTnT)-positive and Tra-1-60/rBC2LCN-negative cells gradually increased in number. In contrast, the number of Tra-1-60/rBC2LCN-positive hiPSCs gradually decreased. After 17 days of differentiation, 0.4% of Tra-1-60/rBC2LCN-positive hiPSCs was detected. The apparent number of undifferentiated hiPSCs during the differentiation process was then monitored by GlycoStem-HP (Fig. 5). A standard curve was generated using the cell culture supernatants of 253G4 hiPSCs cultured for 24 h in mTeSR1, and the apparent cell number was calculated by the following formula: apparent cell number = ((OD450-620) − 0.0178)/5 × 10−5. The cell number divided by the volume of culture medium (mL) estimated by GlycoStem-HP was expressed as an “arbitrary unit (AU)”. As shown in Fig. 5, the apparent cell number of hiPSCs obtained by GlycoStem-HP increased from day 1 to day 3 (Fig. 5, filled circles) due to the increase of the total cell number (Fig. 5, open squares). After day 3, the apparent cell number of hiPSCs gradually decreased, which agrees well with the actual cell number of Tra-1-60/rBC2LCN-positive hiPSCs obtained by flow cytometry (Fig. 5, open triangles). After 16 days of differentiation, the apparent number of hiPSCs was calculated to be 1.78 × 104 AU (Fig. 5, filled circles), which is consistent with the number of Tra-1-60/rBC2LCN-positive hiPSCs at 17 days of differentiation obtained by flow cytometry (1.12 × 104 cells/mL, Fig. 5, open triangles). These results demonstrate that the number of hiPSCs estimated by GlycoStem-HP reflects the actual number of hiPSCs determined by flow cytometry. Therefore, GlycoStem-HP is applicable to the quantitative monitoring of undifferentiated cells during differentiation.
Fig. 4.
Differentiation of hiPSCs into cardiomyocytes. 253G1 hiPSCs were differentiated into cardiomyocytes for 17 days. After 2, 5, 7, 13, 17 days, cells were recovered, stained with anti-Tra-1-60, anti-cTNT, and BC2LCN, and analyzed by flow cytometry.
Fig. 5.
Monitoring of the number of hiPSCs during cardiomyocyte differentiation using GlycoStem-HP. Supernatants from 253G1 hiPSC cultures during cardiomyocyte differentiation were analyzed by GlycoStem-HP. The apparent cell number was calculated from the linear equation obtained from the standard curve generated using cell culture supernatants of 253G4 hiPSCs and expressed as AU (closed circles). The number of total cells (open squares) and Tra-1-60/rBC2LCN-positive hiPSCs (open triangles) determined by flow cytometry are also shown.
3.5. Detection of hiPSCs residing among hiPSC-derived hNSCs using GlycoStem-HP
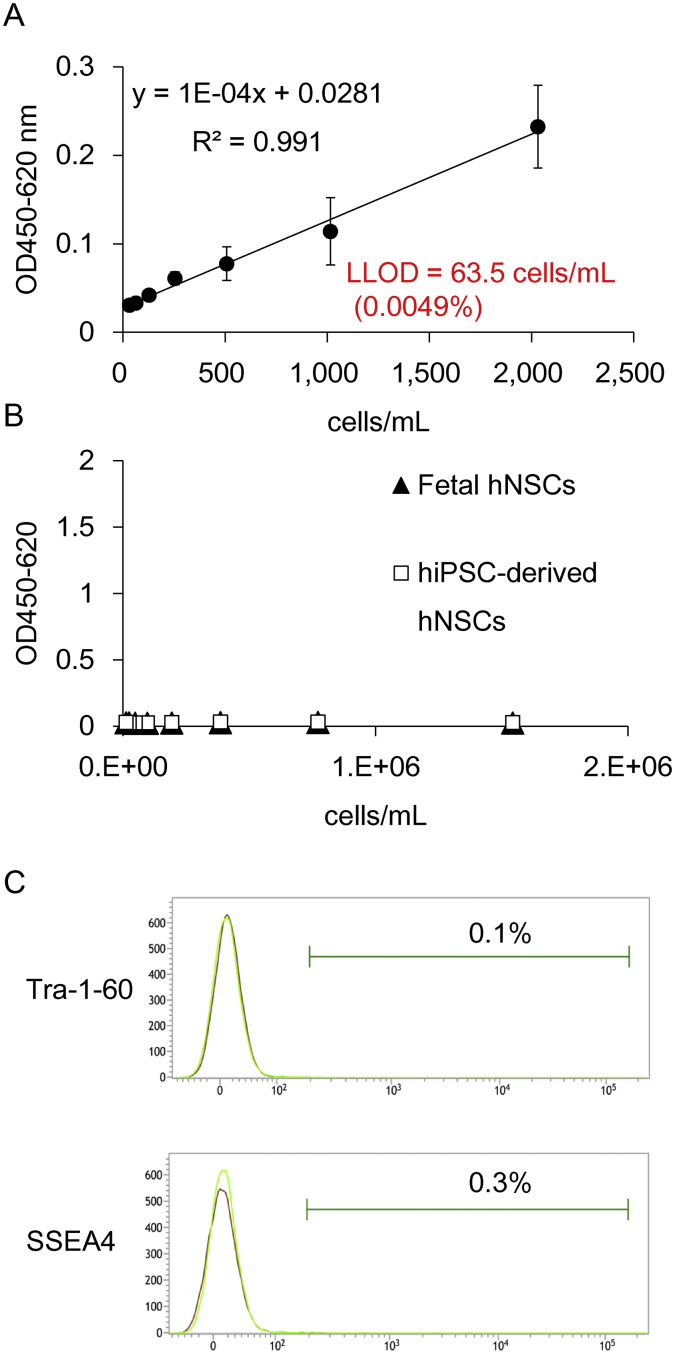
We next applied GlycoStem-HP to detect residual undifferentiated hiPSCs in hiPSC-derived hNSCs, which are planned to be used to treat spinal cord injury in patients at Keio University [7]. For this purpose, we first analyzed the cell culture supernatants of clinical grade of hiPSCs, Ff-I01 hiPSCs (2.6 × 106 cells), cultured in 2 mL of StemFit®AK03 medium to generate a standard curve. As shown in Fig. 6A, a linear regression curve could be generated with a high correlation coefficient (R2 = 0.991). The LLOD value obtained was 63.5 cells/mL, which corresponds to 0.0049% of hiPSCs. We then used GlycoStem-HP to analyze supernatants obtained from cultured fetal hNSCs. As shown in Fig. 6B, GlycoStem-HP gave no signal to fetal hNSCs even at 1.5 × 106 cells/mL (closed triangle), indicating that GlycoStem-HP is highly specific to hiPSCs, and not to other cell types such as hNSCs. No GlycoStem-HP signal was observed for hiPSC-derived hNSCs (open squares) indicating that less than the LLOD value (0.0049%) of hiPSCs is contaminated in hiPSC-derived hNSCs. Consistently, Tra-1-60/SSEA4-positive hiPSCs were not detected by flow cytometry in hiPSC-derived hNSCs (Fig. 6C).
Fig. 6.
Detection of fetal hNSCs and hiPSC-derived hNSCs. (A) Supernatants from Ff-I01 hiPSC cultures were serially diluted with PBS and analyzed by GlycoStem-HP. (B) Cell culture supernatants of fetal hNSCs and hiPSC-derived hNSCs were serially diluted with PBS and analyzed by GlycoStem-HP. Absorbance at OD450-620 was measured. Data are shown as the mean ± SD of triplicate samples. (C) hiPSC-derived hNSCs were stained with Tra-1-60 and SSEA4 and analyzed by flow cytometry.
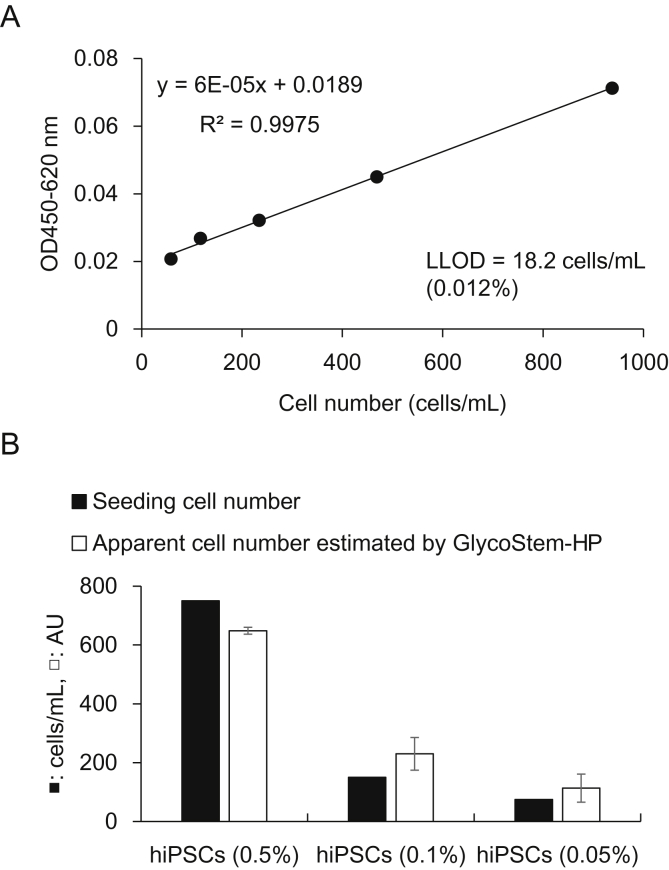
We then assessed whether the system can be used to detect hiPSC in a mixed cell culture. The conditioned medium obtained from 24 h culture of Ff-I01 hiPSCs (1.5 × 105 cells) was serially diluted with StemFit®AK03 medium in order to generate a standard curve. As shown in Fig. 7A, a linear regression curve was obtained with a high correlation coefficient (R2 = 0.9975). The LLOD value was 18.2 cells/mL, which corresponds to 0.012% of hiPSCs. hiPSC-derived hNSCs (1.5 × 105 cells) were then cultured in the presence of 750 (corresponding to 0.5% hiPSCs), 150 cells (0.1% hiPSCs), or 75 cells (0.05% hiPSCs) of Ff-I01 hiPSCs in 1 mL of cell culture media for 24 h. The cell culture supernatants were recovered and then analyzed by GlycoStem-HP. As shown in Fig. 7B, the estimated cell numbers obtained by GlycoStem-HP were 648, 230, and 113 cells/mL, respectively, which agreed well with the number of seeding hiPSCs (750, 150, 75 cells/mL). Altogether, these results demonstrate that GlycoStem-HP is applicable for the detection of undifferentiated cells residing among hiPSC-derived hNSCs.
Fig. 7.
Detection of undifferentiated hiPSCs residing among hiPSC-derived hNSCs. (A) A standard curve was generated using the conditioned medium obtained from 24 h culture of Ff-I01 hiPSCs (1.5 × 105) serially diluted with StemFit®AK03 medium. (B) hiPSC-derived hNSCs (1.5 × 105 cells) were then cultured in the presence of 750 (corresponding to 0.5% hiPSCs), 150 cells (0.1% hiPSCs), or 75 cells (0.05% hiPSCs) of Ff-I01 hiPSCs in 1 mL of cell culture media for 24 h. The cell culture supernatants were recovered and then analyzed by GlycoStem-HP. The seeding cell number (cells/mL) and the apparent cell number (AU) of hiPSCs estimated by GlycoStem-HP were shown in filled and open bar graphs, respectively. Data of GlycoStem-HP (open bar graph) are shown as the mean ± SD of triplicate samples.
4. Discussion
Quantitative and sensitive methods to detect residual undifferentiated hPSCs are required to evaluate the tumorigenicity of CTPs during cell manufacturing processes. In this aspect, Kuroda et al. evaluated three conventional methods to detect residual undifferentiated hiPSCs; soft agar colony formation, flow cytometry, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) [11]. Among the three methods, they concluded that qRT-PCR targeting the LIN28 gene was the most sensitive assay, which could detect 0.002% of residual undifferentiated hiPSCs in RPE cells induced from hiPSCs, while the LLOD values determined for soft agar colony formation and flow cytometry were estimated to be 1% (500 hiPSCs in 5 × 104 RPE) and 0.1% (50 hiPSCs in 5 × 104 RPE cells), respectively. However, the LIN28 expression level did not decrease during the differentiation of hiPSCs into human mesenchymal stem cells (hMSCs) [12]. The expression of LIN28 was suggested to be derived from partially differentiated cells without the ability of tumor formation, but not from intact hiPSCs. Other hPSC markers such as NANOG and OCT3/4 were also detected even in partially differentiated cells. Thus, it is not straightforward to determine the presence of residual hPSCs with the ability of tumor formation simply by qRT-PCR. In this sense, Tano et al. recently reported a direct method to detect residual hPSCs using a highly efficient cell culture system [12]. The method allowed detection of even 0.01–0.001% of hPSCs spiked into hMSCs and human neurons, although it takes approximately one week for results to be obtained by this system. Furthermore, all of the methods require a significant number of invaluable transplant cells for the analysis. This makes it difficult to undertake continuous monitoring of the differentiation state of cells.
In this aspect, we previously developed a nondestructive and quantitative method, termed GlycoStem, to detect hPSCs using cell culture supernatants [16]. One of the biggest advantages of this assay is that it requires cell culture supernatants, and not cells that would otherwise be used for transplant. However, the previous GlycoStem was found to show a background signal to FBS-containing media, which would limit the applicability of the system. In this report, we describe improvements in the method that have been achieved by changing the overlay probe from rABA lectin to R-10G mAb to detect rBC2LCN-positive podocalyxin secreted from hPSCs. This newly developed sandwich assay, GlycoStem-HP, allowed the detection of hiPSCs even in the presence of a high concentration of FBS (˜20%). The obtained LLOD values ranged from 27 to 384 cells/mL. This indicates that the developed assay permits the detection of 0.0006–0.03% of residual hiPSCs. The different LLOD values are due to the variable amounts of rBC2LCN-positive podocalyxin secreted from hiPSCs, which may also change depending on the type of cell culture medium used.
As shown in Fig. 3, Fig. 5, the absorbance is highly correlated with the cell number of undifferentiated cells with R2 > 0.98. However, the values obtained by the test could vary depending on the degree of undifferentiation/differentiation of human pluripotent stem cells [16]. For example, fully undifferentiated cells and partially differentiated cells might secrete different amounts of podocalyxin. In this case, the values could represent “the apparent cell number” of undifferentiated cells obtained by the test. Therefore, the cell number estimated by the test was expressed as an “arbitrary unit (AU)”, but not “cells/mL” in Fig. 5.
The high background signal to FBS seen with previous sandwich assay, which employed rBC2LCN and rABA (GlycoStem), may be due to the presence of glycoproteins in FBS, which are also detected by the rBC2LCN and rABA sandwich assay. GlycoStem-HP gave little or no background signal, even in the presence of high percentage levels of FBS, indicating that the assay is more specific to rBC2LCN-positive podocalyxin secreted from hPSCs.
Importantly, GlycoStem-HP allowed for the number of residual undifferentiated hiPSCs to be monitored during cardiomyocyte differentiation. Furthermore, it gave no signals to fetal hNSCs, although these cells were reported to express some hPSC marker genes [26]. This indicates that the assay is applicable to detecting undifferentiated hiPSCs in hiPSC-derived hNSCs. Indeed, 0.05% of hiPSCs spiked in hiPSC-derived hNSCs could be successfully detected. The practicality of GlycoStem-HP makes it highly applicable to monitoring the number of undifferentiated cells included in hNSC and cardiomyocyte populations.
Tumorigenicity is one of the biggest concerns for hPSC-derived CTPs that are transplanted into patients. However, as a tumorigenicity test has not been established [8], a combination of methods such as flow cytometry, qRT-PCR, and a cell culture system such as GlycoStem-HP should contribute to the testing of strategies to combat tumorigenicity in hPSC-derived hCTPs. Translation of this technology to a clinical setting will help make regenerative medicine a safe option for the treatment of a range of diseases.
Conflict of interest
The authors declare no competing financial interests.
Acknowledgements
Human iPS cell lines 201B7 (HPS0063) and 253G1 (HPS0002) were obtained from the RIKEN Bioresource Center. Ff-I01 hiPSCs was obtained from Center for iPS Cell Research and Application (CiRA), Kyoto University. This work was supported in part by JSPS KAKENHI Grant Number 25712039.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
References
- 1.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Strauss S. Geron trial resumes, but standards for stem cell trials remain elusive. Nat Biotechnol. 2010;28:989–990. doi: 10.1038/nbt1010-989. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz S.D., Hubschman J.P., Heilwell G., Franco-Cardenas V., Pan C.K., Ostrick R.M. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 5.Kamao H., Mandai M., Okamoto S., Sakai N., Suga A., Sugita S. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawamura M., Miyagawa S., Fukushima S., Saito A., Miki K., Ito E. Enhanced survival of transplanted human induced pluripotent stem cell-derived cardiomyocytes by the combination of cell sheets with the pedicled omental flap technique in a porcine heart. Circulation. 2013;128:S87–S94. doi: 10.1161/CIRCULATIONAHA.112.000366. [DOI] [PubMed] [Google Scholar]
- 7.Okano H., Nakamura M., Yoshida K., Okada Y., Tsuji O., Nori S. Steps toward safe cell therapy using induced pluripotent stem cells. Circ Res. 2013;112:523–533. doi: 10.1161/CIRCRESAHA.111.256149. [DOI] [PubMed] [Google Scholar]
- 8.Kawamata S., Kanemura H., Sakai N., Takahashi M., Go M.J. Design of a tumorigenicity test for induced pluripotent stem cell (iPSC)-derived cell products. J Clin Med. 2015;4:159–171. doi: 10.3390/jcm4010159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanemura H., Go M.J., Shikamura M., Nishishita N., Sakai N., Kamao H. Tumorigenicity studies of induced pluripotent stem cell (iPSC)-derived retinal pigment epithelium (RPE) for the treatment of age-related macular degeneration. PLoS One. 2014;9:e85336. doi: 10.1371/journal.pone.0085336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey A.M. Balancing tissue and tumor formation in regenerative medicine. Sci Transl Med. 2012;4:147fs28. doi: 10.1126/scitranslmed.3003685. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda T., Yasuda S., Kusakawa S., Hirata N., Kanda Y., Suzuki K. Highly sensitive in vitro methods for detection of residual undifferentiated cells in retinal pigment epithelial cells derived from human iPS cells. PLoS One. 2012;7:e37342. doi: 10.1371/journal.pone.0037342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tano K., Yasuda S., Kuroda T., Saito H., Umezawa A., Sato Y. A novel in vitro method for detecting undifferentiated human pluripotent stem cells as impurities in cell therapy products using a highly efficient culture system. PLoS One. 2014;9:e110496. doi: 10.1371/journal.pone.0110496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tateno H., Toyota M., Saito S., Onuma Y., Ito Y., Hiemori K. Glycome diagnosis of human induced pluripotent stem cells using lectin microarray. J Biol Chem. 2011;286:20345–20353. doi: 10.1074/jbc.M111.231274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tateno H., Matsushima A., Hiemori K., Onuma Y., Ito Y., Hasehira K. Podocalyxin is a glycoprotein ligand of the human pluripotent stem cell-specific probe rBC2LCN. Stem Cells Transl Med. 2013;2:265–273. doi: 10.5966/sctm.2012-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerjaschki D., Sharkey D.J., Farquhar M.G. Identification and characterization of podocalyxin–the major sialoprotein of the renal glomerular epithelial cell. J Cell Biol. 1984;98:1591–1596. doi: 10.1083/jcb.98.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tateno H., Onuma Y., Ito Y., Hiemori K., Aiki Y., Shimizu M. A medium hyperglycosylated podocalyxin enables noninvasive and quantitative detection of tumorigenic human pluripotent stem cells. Sci Rep. 2014;4:4069. doi: 10.1038/srep04069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okita K., Hong H., Takahashi K., Yamanaka S. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat Protoc. 2010;5:418–428. doi: 10.1038/nprot.2009.231. [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa M., Taniguchi Y., Senda S., Takizawa N., Ichisaka T., Asano K. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep. 2014;4:3594. doi: 10.1038/srep03594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Kanemura Y., Mori H., Kobayashi S., Islam O., Kodama E., Yamamoto A. Evaluation of in vitro proliferative activity of human fetal neural stem/progenitor cells using indirect measurements of viable cells based on cellular metabolic activity. J Neurosci Res. 2002;69:869–879. doi: 10.1002/jnr.10377. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 22.Dubois N.C., Craft A.M., Sharma P., Elliott D.A., Stanley E.G., Elefanty A.G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kannagi R., Cochran N.A., Ishigami F., Hakomori S., Andrews P.W., Knowles B.B. Stage-specific embryonic antigens (SSEA-3 and -4) are epitopes of a unique globo-series ganglioside isolated from human teratocarcinoma cells. EMBO J. 1983;2:2355–2361. doi: 10.1002/j.1460-2075.1983.tb01746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schopperle W.M., DeWolf W.C. The TRA-1-60 and TRA-1-81 human pluripotent stem cell markers are expressed on podocalyxin in embryonal carcinoma. Stem Cells. 2007;25:723–730. doi: 10.1634/stemcells.2005-0597. [DOI] [PubMed] [Google Scholar]
- 25.Kawabe K., Tateyama D., Toyoda H., Kawasaki N., Hashii N., Nakao H. A novel antibody for human induced pluripotent stem cells and embryonic stem cells recognizes a type of keratan sulfate lacking oversulfated structures. Glycobiology. 2013;23:322–336. doi: 10.1093/glycob/cws159. [DOI] [PubMed] [Google Scholar]
- 26.Kim J.B., Greber B., Arauzo-Bravo M.J., Meyer J., Park K.I., Zaehres H. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–653. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]