Abstract
Background
The aim of this study was to observe the expression of microRNA-222 (miR-222) and matrix metalloproteinase inhibitor 3 (TIMP3) in non-small cell lung cancer (NSCLC) and discuss their significance.
Methods
A total of 230 patients with NSCLC were enrolled in the observation group during the operation. Ninety-eight normal adjacent tissues were used as the control group. Two groups of miR-222 and TIMP3 were detected by in situ hybridization and immunohistochemistry. The distribution of miR-222 and TIMP3 in A549/H358/PC9 cells was observed by immunofluorescence. Chi-squared and Spearman correlation tests were used to analyze the relationship among miR-222, TIMP3 expression, and clinicopathological parameters of NSCLC. Kaplan–Meier and Cox proportional hazards regression were used to analyze the prognostic impact of miR-222 and TIMP3.
Results
Immunohistochemistry showed that the expression of miR-222 in lung cancer tissue was significantly higher, but TIMP3 was lower than that in normal lung tissue (P = 0.0001 for the former and P = 0.0002 for the latter). Meanwhile, miR-222 and TIMP3 were mainly distributed in the cytoplasm. Among them, cTIMP3 accounted for 70.29% (72/101), cmiR-222 for 59.35% (92/155), 14.85% for nTIMP3 (15/101), and 18.06% for nmiR-222 (28/155). There was a significant difference in distribution (both P < 0.0001). The expression of miR-222 and TIMP3 were negatively correlated in lung cancer tissues (r = −0.43, P = 0.0219). With the progression of clinical stage, the positive intensity of cTIMP3 showed a decreasing trend, while the cmiR-222 showed a reverse trend (the former P = 0.0024 and the latter P < 0.0001). In the Kaplan–Meier prognostic analysis, we found that the high expression of cTIMP3 could predict a better prognosis (P = 0.0040), whereas cmiR-222 was the opposite (P = 0.0016). Multivariate analysis shows that both can be used as independent factors.
Conclusion
TIMP3 expression in lung cancer is relatively low and has a negative correlation with lung cancer staging and prognosis, suggesting that it may play a defensive function in the development of lung cancer, while miR-222 has the opposite effect, and the expression of both proteins is negatively correlated, suggesting that in lung cancer progresses, both proteins may play some role together.
Keywords: lung cancer, tumor markers, A549/h358/pc9, biological mechanism, immunofluorescence
Introduction
TIMP31,2 is a protein with a molecular weight of 24 kD. It exists in a wide range of tissue cytoplasm and extracellular matrix and participates in many biological reactions. Studies3,4 have shown that TIMP3 has a specific inhibitory effect on matrix metalloproteinases (MMPs) and binds to MMPs through a 1:1 non-covalent binding pattern, which inactivates the latter, thereby protecting the extracellular matrix from MMPs degradation and remodeling, therefore suggesting that TIMP3 has the effect of inhibiting tumor growth and invasion. Studies have shown that low expression of TIMP3 is observed in both hepatocarcinoma (HCC) and HCC cell lines, suggesting that the latter is closely related to HCC.
miRNAs are a class of non-coding,5–7 single-stranded, small RNA molecules that inhibit the translation or degradation of target mRNA by completely or incompletely binding to the 3′ non-coding region of the target gene, thereby regulating gene expression at the post-transcriptional level. Data6,7 have shown that up to 60% of human total genes are regulated by microRNA post-transcriptional regulation, and numerous studies7 have confirmed that microRNAs play an important role in regulating cell proliferation, differentiation, cell cycle, and apoptosis and then affect the development of tumors. Studies3,8 showed that microRNA-222 (miR-222) and miR-221 are highly homologous, and their genes are located in the P11.3 region of the X chromosome, in the region of about 1 kb, and the two clusters are distributed. A number of studies8–14 have confirmed that miR-222 expression levels are significantly higher in various tumor tissues than in normal orthotopic tissues. This phenomenon is found in breast cancer, bladder cancer, colon cancer, glioblastoma, pancreatic cancer, etc. In addition, upregulation of miR-222 expression is also positively correlated with increased tumor grade, poor prognosis, and low expression in tumors with good differentiation, low grading, and good prognosis. In our previous study, we found that miR-222 and TIMP3 are negatively correlated in lung cancer expression, which may mean that both may have common pathways in the development of lung cancer. This study analyzed a large number of lung cancer tissue samples and followed up a large number of data to explore the relationship between the two and lung cancer characteristics and prognosis, to lay the foundation for further exploration of the relationship between the two.
Methods
From June 2010 to May 2014, histology samples of 230 patients aged 34–76 years were collected in the First Affiliated Hospital of Sun Yat-Sen University and were pathologically diagnosed with non-small cell lung cancer (NSCLC). All patients did not undergo chemoradiation before surgery. During the operation, tumor tissue was taken from the observation group, and 98 normal tissues with a distance of >5 cm from the tumor tissue were taken from the control group. Follow-up data included tumor staging (according to the TNM staging guidelines of the eighth edition), gender, smoking index, and overall survival prognosis.
Ethical approval and written informed consent
All procedures performed in studies involving human participants were approved by the ethics committee of the First Affiliated Hospital, Sun-Yat Sen University. Written informed consent was obtained from all participants included in the study.
Detection of miR-222 and TIMP3 using in situ hybridization and immunohistochemistry
In situ hybridization
Conventional section dewaxing, gradient ethanol rehydration. The slices were washed with 3% pepsin at room temperature for 15–20 minutes and again washed with 4% formaldehyde for 10 minutes. The pre-hybridization was performed at 55°C for 2 hours, and then the slices were hybridized with digoxigenin-labeled miR-222 probe overnight at 55°C. The next day, the slices were washed thoroughly with different concentrations of saline sodium citrate buffer. The blocking solution was closed for 30 minutes. The slices were incubated with biotinylated mouse anti-digoxin at room temperature for 2 hours. The slices were incubated with the strept avidin-biotin complex solution at room temperature for 30 minutes. 3,3-N-Diaminobenzidine coloration, hematoxylin re-stained nuclei, fully washed, neutral gel seal preservation.
Immunohistochemistry
TIMP3 was detected using the EliVision™ (Fuzhou Maxin Biological Technology Development Co., Ltd, Fuzhou, People’s Republic of China) Plus two-step method. Conventional section dewaxing, gradient ethanol rehydration. The slices were autoclaved on demand, and after cooling, they were washed with PBS. Primary antibody (50 µL) was added to the slices and incubated overnight at 4°C. They were washed with PBS. Biotin-labeled secondary antibody (50 µL) was added to the slices, incubated for 10 minutes at room temperature, and washed with PBS. Streptavidin-peroxidase solution (50 µL) was added dropwise, incubated at room temperature for 10 minutes, and washed with PBS. DAB coloration, hematoxylin stained nuclei, fully washed, neutral gel seal preservation. The brownish yellow particles indicate positive result.
Reading assessment
The three pathologists performed the readings respectively. Under a 400-fold microscope, 10 slides were randomly selected, and 100 cancer cells were selected for each visual field. A semi-quantitative method was used to calculate the proportion of positive cells by multiplying the fraction of staining intensity by the percentage of positive cells to obtain the final score (Immunohistochemistry [IHC] score: 1–16), where -, 1–4; +, 5–8; ++, 9–12; and +++, 13–16. The proportion of positive cells was calculated according to the following method: ≤20%, 1; 20%–50%, 2; 50%–70%, 3; and >70%, 4. The staining intensity was as follows: negative, 1; weak, 2; media, 3; and strong, 4.
Immunofluorescence assay
Culture medium H358, A549, and PC9 cells (purchased from Shanghai Tongpai Biological Technology Co., Ltd., Shanghai, People’s Republic of China) were cultured in a routine culture medium to make cell slides and perform fluorescence immunoassay experiments. In the culture plate, slides of cells that had been climbed were washed three times with PBS for 3 minutes each time; the slides were fixed with 4% paraformaldehyde for 15 minutes; slides were washed with PBS for three times, each time for 3 minutes; 0.5% Tri-ton X-100 (prepared in PBS) is permeable for 20 minutes at room temperature. The slides were immersed in PBS for three times, 3 minutes each time. The blotting paper was blotted to dry PBS. Normal goat serum was added dropwise onto the slides and blocked at room temperature for 30 minutes. The blotting paper was blotted off the blocking solution and was not washed. A sufficient amount of the blocking solution was added to each slide. The primary antibody (miR-222 was added with digoxigenin-labeled miR-222 probe, hybridized overnight at 55°C) and anti-TIMP3 (1:200) were placed in a wet box and incubated overnight at 4°C, and PBS was used instead of primary antibody in negative control group; fluorescent secondary antibody was added: phosphate buffered saline with Tween-20 (PBST) was diluted three times, 3 minutes each time. Absorb the excess liquid on the blotting paper and add the diluted secondary antibody (CY3 mouse anti-digoxin, CY3 mouse anti-IgG, 1:100, Abcam, Cambridge, UK), in a wet box, incubate at 20°C–37°C for 1 hour, PBST dip 3 times for 3 minutes each time, re-stain nucleus: Incubate DAPI in dark for 5 minutes, stain the specimen, and wash 4 times with PBST for 5 minutes. The liquid on the slide was blotted dry with absorbent paper, and the slide was sealed with a sealing liquid containing anti-fluorescence quencher. The images were then observed under a fluorescence microscope.
Statistical methods
Data were analyzed using GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Count data were expressed as rates, and chi-squared tests were used for comparison between groups. Spearman test was used for correlation analysis, and Kaplan–Meier and Cox proportional hazards regression were used for univariate and multivariate analysis. P < 0.05 was considered statistically significant. “*” indicates “P < 0.01”, “**” indicates “P < 0.001”, and “***” indicates “P < 0.0001”.
Results
Expression and relationship of miR-222 and TIMP3 in lung cancer
IHC results showed that the expression of miR-222 in lung cancer tissue was significantly higher, but TIMP3 was lower than normal lung tissue (P = 0.0001 for the former and P = 0.0002 for the latter), and miR-222 and TIMP3 were mainly distributed in the cytoplasm. Cytoplasm TIMP3 accounted for 70.29% (72/101) of TIMP3 positive, cmiR-222 for 59.35% (92/155), nTIMP3 for 14.85% (15/101), and nmiR-222 for 18.06% (28/155), and there are significant differences (both P < 0.0001, cytoplasm vs nucleus). In the immunofluorescence (IF) experiment, this result was further confirmed by observing the distribution of miR-222 and TIMP3 in A549/H358/PC9 cells. By Spearman correlation analysis, there was a close negative correlation between the two protein expressions (Figures 1 and 2 and Table 1).
Figure 1.
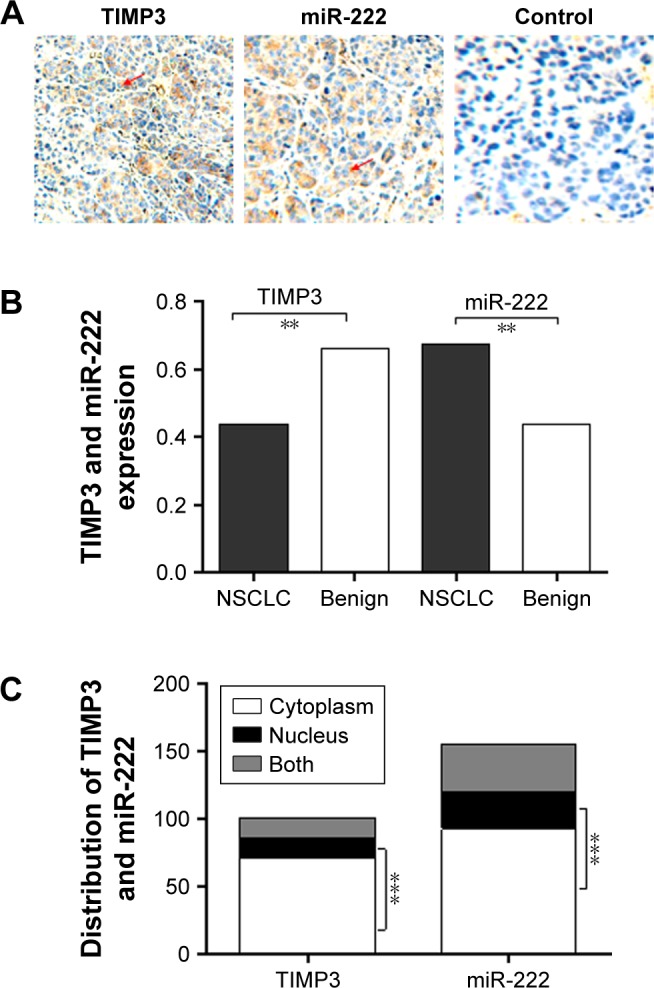
Expression and distribution of TIMP3 and miR-222 in lung cancer.
Notes: The results of the IHC test were as follows: (A) PBS was used as a negative control, and the microscope was observed ×200 times. The protein expression score was evaluated according to the method. (B) The expression of miR-222 in lung cancer tissues was significantly higher, while that of TIMP3 was lower than that in normal lung tissues. (C) TIMP3 and miR-222 were mainly distributed in the cytoplasm (arrow). Scale bar = 50 µm; magnification ×20. **P<0.001; ***P<0.0001.
Abbreviations: IHC, Immunohistochemistry; miR-222, microRNA-222; NSCLC, non-small cell lung cancer; TIMP3, matrix metalloproteinase inhibitor 3.
Figure 2.
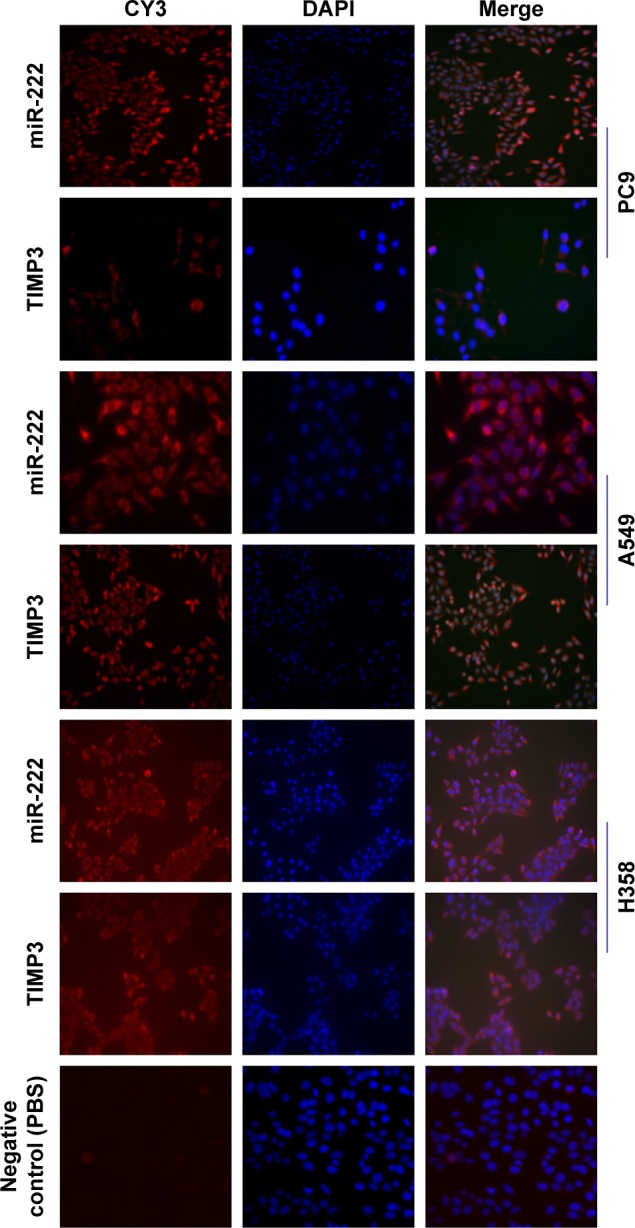
The distribution of TIMP3 and miR-222 in H358, A549, and PC9 cells was observed by IF.
Notes: A549, PC9, and H358 cells results are shown; PBS was used as primary antibody for negative control. Through IF experiments, it was observed that TIMP3 and miR-222 are mainly distributed in the cytoplasm. Scale bar = 20 µm; magnification ×40.
Abbreviations: IF, immunofluorescence; miR-222, microRNA-222; TIMP3, matrix metalloproteinase inhibitor 3.
Table 1.
Relationship between both proteins expression and TNM stage
| TIMP3 | P-value | miR-222 | P-value | |||||
| − | + | − | + | |||||
| NSCLC | 129 | 101 | 0.0002 | 75 | 155 | 0.0001 | ||
| Benign | 33 | 65 | 55 | 43 | ||||
| Spearman correlation S(r) between expression of TIMP3 and miR-222 | ||||||||
| Patients (n) | r | P-value | ||||||
| TIMP3 vs miR-222 | 230 | −0.430 | 0.0219 | |||||
| cTIMP3 | cmiR-222 | |||||||
| IHC score | I | II | III | IV | I | II | III | IV |
| − | 25 | 38 | 35 | 46 | 41 | 44 | 9 | 9 |
| + | 15 | 18 | 6 | 4 | 5 | 3 | 8 | 10 |
| ++ | 10 | 12 | 4 | 3 | 8 | 10 | 13 | 12 |
| +++ | 5 | 6 | 2 | 1 | 9 | 9 | 17 | 23 |
| Positive (%) | 54.54 | 48.64 | 25.53 | 14.81 | 34.92 | 33.33 | 80.85 | 83.33 |
| χ2 | 25.55 | 55.66 | ||||||
| P-value | 0.0024 | <0.0001 | ||||||
Abbreviations: IHC, Immunohistochemistry; miR-222, microRNA-222; TIMP3, matrix metalloproteinase inhibitor 3.
Relationship among cytoplasmic miR-222, TIMP3, and clinical features of lung cancer
The IHC results were scored and analyzed based on the assessment results. There was no significant relationship between the positive expression of cmiR-222 and cTIMP3 and sex, cancer type, smoking index, and age. It was only related to the pTNM stage (IA–IIB vs IIIA–IV, both P < 0.0001). Meanwhile, it was found that with pTNM stage progressively, the intensity of TIMP3 expression was significantly decreased, and the number of patients with positive expression decreased, while miR-222 showed the opposite change (Table 2).
Table 2.
Correlation of cytoplasmic TIMP3 and miR-222 with clinicopathologic parameters
| Groups | cTIMP3
|
P-value | cmiR-222
|
P-value | ||
|---|---|---|---|---|---|---|
| − | + | − | + | |||
| Gender | 1.0000 | 1.0000 | ||||
| Male | 71 | 42 | 51 | 62 | ||
| vFemale | 73 | 44 | 52 | 65 | ||
| Age (years) | 0.4139 | 0.5074 | ||||
| <55 | 68 | 46 | 54 | 60 | ||
| ≥55 | 76 | 40 | 49 | 67 | ||
| Smoking index | 0.5866 | 0.5960 | ||||
| <400 | 71 | 46 | 50 | 67 | ||
| ≥400 | 73 | 40 | 53 | 60 | ||
| Histological type | 0.8914 | 0.2316 | ||||
| SCC | 78 | 45 | 60 | 63 | ||
| AD | 66 | 41 | 43 | 64 | ||
| TNM staging | <0.0001 | <0.0001 | ||||
| IA–IIB | 63 | 66 | 85 | 44 | ||
| IIIA–IV | 81 | 20 | 18 | 83 | ||
Abbreviations: AD, adenocarcinoma; miR-222, microRNA-222; SCC, squamous cell carcinoma; TIMP3, matrix metalloproteinase inhibitor 3.
Analysis relation of cytoplasmic miR-222, TIMP3, and prognosis
The univariate and multivariate analyses were performed using the positive and negative cytoplasmic miR-222 and TIMP3 as indicators. It was found that the positive expression of cytoplasmic miR-222 suggested a poor prognosis (P = 0.0016, R = 2.230), whereas the positive expression of cytoplasmic TIMP3 showed better results (P = 0.0040, R = 2.092), and Cox proportional hazards regression analysis showed that both proteins can be used as independent prognostic factors (Figure 3 and Table 3).
Figure 3.
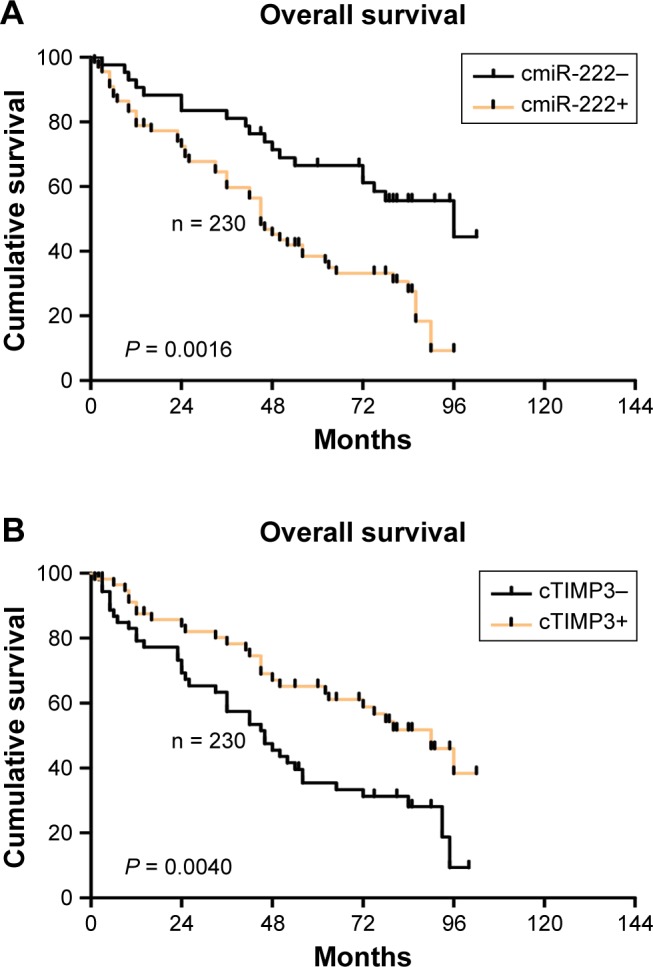
Analysis of cytoplasmic TIMP3 and miR-222 prognostic functions.
Notes: The univariate and multivariate analyses were performed using the positive and negative cytoplasmic miR-222 and TIMP3 as indicators. It was found that the positive expression of cytoplasmic miR-222 suggested a poor prognosis (P = 0.0016, R = 2.230) (A), whereas the positive expression of cytoplasmic TIMP3 showed better results (P = 0.0040, R = 2.092) (B).
Abbreviations: miR-222, microRNA-222; TIMP3, matrix metalloproteinase inhibitor 3.
Table 3.
Kaplan–Meier and Cox multivariate proportional hazard analysis
| Factors | Univarivate analysis
|
Multivariate analysis
|
||
|---|---|---|---|---|
| Log-rank | P-value | Hazard ratio (95%) | P-value | |
| Age (years) | 2.014 | 0.1302 | ||
| <55 | ||||
| ≥55 | ||||
| cTIMP3 | 8.301 | 0.0040 | 2.092 (1.266–3.456) | 0.0263 |
| Negative | ||||
| Positive | ||||
| cmiR-222 | 9.953 | 0.0016 | 2.230 (1.355–3.671) | 0.0112 |
| Negative | ||||
| Positive | ||||
| P-stage | 6.425 | 0.0321 | 2.715 (1.702–7.358) | 0.0323 |
Abbreviations: miR-222, microRNA-222; TIMP3, matrix metalloproteinase inhibitor 3.
Discussion
NSCLC is the most common histological type of lung cancer,15,16 and its 5-year survival rate is low.11 NSCLC includes both adenocarcinoma and squamous cell carcinoma. Up to 75% of lung cancer patients have had local and/or distant metastases at the time of diagnosis. Therefore, the search for effective, non-invasive biomarkers for prognosis is of great significance for the treatment of NSCLC.
Studies have shown that miRNAs are dysregulated in various tumor tissues and exert oncogene-like or tumor suppressor-like effects by regulating the expression of oncogenes or tumor suppressor genes.17–19 Upregulation of miR-222 expression is associated with poor prognosis of pancreatic cancer; miR-222 is highly expressed in bladder cancer and is associated with clinical stage, tumor diameter, and prognosis of bladder cancer, as well as in gastric cancer-related studies, similar results was obtained.20–22 Multiple miRNAs are closely related to the development of NSCLC and help to assess tumor progression. TIMP33,4,23 is a pro-apoptotic protein involved in the proliferation and invasion of various tumor cells. TIMP3 is downregulated in endometrial cancer tissues and participates in the migration and invasion of endometrial cancer cells. It is downregulated in gastric cancer tissues and is associated with clinical stage and lymph node metastasis.
The expression and distribution of miR-222 and TIMP3 in NSCLC tissues were observed by IHC. The positive expression rate of miR-222 in 230 cases of NSCLC was 67.39%, which was significantly lower than that in benign lung tissues; of which the simple cytoplasmic expression positive rate was 59.35% (92/155), while the positive expression rate in the cell nucleus alone was 18.06% (28/155); the former was significantly higher than the latter, and the positive rate of both cytoplasm and nucleus was 22.58% (35/155). While the expression of TIMP3 in lung cancer tissue is contrary to that of miR-222, the positive rate of lung cancer tissue is lower than that of normal lung tissue, and it is also mainly distributed in the cytoplasm, which is significantly higher than that in the nucleus. This result is consistent with our IF results and shows that in three types of cells (A549/H358/PC9), miR-222 and TIMP3 are mainly distributed in the cytoplasm. Further analysis showed that as the grade of tumor pTNM increased, the expression level of TIMP3 showed a significant downward trend, while that of miR-222 was the opposite. These results suggest that the functional characteristics of TIMP3 in lung cancer may be defensive and that miR-222 has cancer-promoting function. The conclusions of our abovementioned literature are consistent,24–27 and its underlying mechanism may be consistent with the above, and it exerts its effects in the cytoplasm. However, there is an interesting phenomenon in this study. The expression of TIMP3 is negatively correlated with the expression of miR-222. The two expressions may have a tendency to control each other in the opposite direction. This phenomenon has also been observed in our prognostic analysis. The cytoplasmic TIMP3 anti-cancer- and miR-222 cancer-promoting functions were further discovered. We found that the cytoplasmic TIMP3 positive has a good prognosis, with the median survival time being significantly longer than that of the negative ones, whereas the cytoplasmic miR-222 is the opposite. This means that TIMP3 may play a role in the defense mechanism and miR-222 play a related mechanism. At present, this research has not been confirmed by other studies; only we have observed, and further experiments in cells and animals were needed.
Conclusion
This study investigated the distribution of TIMP3 and miR-222 in the cytoplasm by analyzing a large number of clinical specimens and repeated experiments with three cells (A549, H358, and PC9). Through the analysis, we found that the former may have a good prognosis, and there is a negative correlation with the latter, and its effect mechanism may be related to the latter. These require further studies.
Acknowledgments
This study was supported by National Nature Science Foundation of China (NSFC; grant number: 81501964).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15(12):1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Descot A, Oskarsson T. The molecular composition of the metastatic niche. Exp Cell Res. 2013;319(11):1679–1686. doi: 10.1016/j.yexcr.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 3.Hadler-Olsen E, Winberg JO, Uhlin-Hansen L. Matrix metalloproteinases in cancer: their value as diagnostic and prognostic markers and therapeutic targets. Tumour Biol. 2013;34(4):2041–2051. doi: 10.1007/s13277-013-0842-8. [DOI] [PubMed] [Google Scholar]
- 4.Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27(31):5287–5297. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 8.Hui AB, Shi W, Boutros PC, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89(5):597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 9.Zheng C, Yinghao S, Li J. MiR-221 expression affects invasion potential of human prostate carcinoma cell lines by targeting DVL2. Med Oncol. 2012;29(2):815–822. doi: 10.1007/s12032-011-9934-8. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Zhang Y, Zhang H, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 11.Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25(5):387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Chou CK, Chen RF, Chou FF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20(5):489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 13.Sun K, Wang W, Zeng JJ, et al. MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacol Sin. 2011;32(3):375–384. doi: 10.1038/aps.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felicetti F, Errico MC, Bottero L, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68(8):2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 15.Pietras RJ, Márquez DC, Chen HW, et al. Estrogen and growth factor receptor interactions in human breast and non-small cell lung cancer cells. Steroids. 2005;70(5–7):372–381. doi: 10.1016/j.steroids.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H, Altieri DC, Lu CD, Cd L, et al. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58(22):5071–5074. [PubMed] [Google Scholar]
- 17.Kim MK, Jung SB, Kim JS, et al. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Arch. 2014;465(4):463–471. doi: 10.1007/s00428-014-1640-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Xu L, Yang Z, et al. MicroRNA-10b indicates a poor prognosis of non-small cell lung cancer and targets E-cadherin. Clin Transl Oncol. 2015;17(3):209–214. doi: 10.1007/s12094-014-1213-7. [DOI] [PubMed] [Google Scholar]
- 19.Xu T, Liu X, Han L, et al. Up-regulation of miR-9 expression as a poor prognostic biomarker in patients with non-small cell lung cancer. Clin Transl Oncol. 2014;16(5):469–475. doi: 10.1007/s12094-013-1106-1. [DOI] [PubMed] [Google Scholar]
- 20.Quintavalle C, Garofalo M, Zanca C, et al. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPµ. Oncogene. 2012;31(7):858–868. doi: 10.1038/onc.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong M, Yang P, Hua F. MiR-191 modulates malignant transformation of endometriosis through regulating TIMP3. Med Sci Monit. 2015;21:915–920. doi: 10.12659/MSM.893872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel R, Naishadham D, Jemal A, Statistics C. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 23.Cruz-Munoz W, Khokha R. The role of tissue inhibitors of metalloproteinases in tumorigenesis and metastasis. Crit Rev Clin Lab Sci. 2008;45(3):291–338. doi: 10.1080/10408360801973244. [DOI] [PubMed] [Google Scholar]
- 24.Garofalo M, di Leva G, Romano G, Michela G, Gianpiero DL, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Hen Y, Zaman MS, Deng G, et al. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostrate cancer. Cancer Prev Res. 2011;4:76–86. doi: 10.1158/1940-6207.CAPR-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin YJ, Kim JH. The role of EZH2 in the regulation of the activity of matrix metalloproteinases in prostate cancer cells. PLoS One. 2012;7(1):e30393. doi: 10.1371/journal.pone.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu CH, Peng KL, Kang ML, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2(3):568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]


