Abstract
Changes in gene regulation during differentiation are governed by networks of transcription factors. The Arabidopsis root endodermis is a tractable model to address how transcription factors contribute to differentiation. We used a bottom-up approach to understand the extent to which transcription factors required for endodermis differentiation can confer endodermis identity to a non-native cell-type. Our results show the transcription factors SHORTROOT and MYB36 alone have limited ability to induce ectopic endodermal features in the absence of additional cues. The stele-derived signaling peptide CIF2 stabilizes SHORTROOT-induced endodermis identity acquisition. The outcome is a partially impermeable barrier deposited in the sub-epidermal cell layer, which has a transcriptional signature similar to the endodermis. These results demonstrate other root cell-types can be forced to differentiate into endodermis and highlights a previously unappreciated role for receptor-kinase signaling in maintaining endodermis identity.
INTRODUCTION
Development of multi-cellular organisms requires tight spatiotemporal coordination and stabilization of cell differentiation. Transcription factors (TFs) are pivotal components of differentiation and can direct cell-identity acquisition in animals and plants 1–3. TF-regulated events that underlie differentiation are often simplified in the construction of gene regulatory networks (GRNs) 4. The Arabidopsis root endodermis has been a useful model for understanding differentiation and has a well-characterized GRN, as well as signature features of differentiation 5. Differentiated endodermis has lignified cell-wall impregnations that form the Casparian strip as well as developmentally delayed secondary cell-wall depositions of suberin lamellae 6,7. Both of these features contribute to the endodermis’ physiological role as a barrier to the external environment 6,7.
The key TFs of the endodermis GRN are SHORTROOT (SHR), SCARECROW (SCR) and MYB36. These TFs form a transcriptional cascade with SHR as the “master regulator” activating SCR and subsequently MYB36 5. SHR is expressed in the root vasculature (stele) and moves into adjacent cells where it initiates the asymmetric cell division that separates the endodermis from the cortex (collectively the ground tissue) 8. Both the scr and shr mutants contain only one layer of ground tissue, with shr being devoid of lignin deposition and scr having a sporadic lignification pattern 9,10. MYB36, which is downstream of SHR and SCR, regulates expression of the transmembrane CASPARIAN STRIP ASSOCIATED PROTEINS (CASPs) that mark the domain where the Casparian strip forms 11–13. Mutations in MYB36 result in delayed apoplastic barrier formation 11,12.
Proper endodermis barrier function also relies on signaling events downstream of the SHR cascade. The receptor-like kinase SCHENGEN3 (SGN3), which is regulated by SHR, seals the Casparian strip upon activation by the stele-derived ligand CASPARIAN STRIP INTEGRITY FACTOR 2 (CIF2), with some contribution from its homolog CIF1 14,15,16,17.
It remains unclear in what contexts the endodermal TFs SHR, SCR and MYB36 have the ability to confer endodermis cell identity. Ectopic expression of SHR or overexpression of MYB36 results in few cells with ectopic lignin deposition 8,12,18, however, these structures do not resemble a Casparian strip. Here, we show SHR in combination with CIF2 is sufficient for endodermal identity acquisition. We determine that the major transcriptional contributor to endodermis identity is SHR, define the developmental stages competent to respond to SHR and CIF2, and show endodermal identity acquisition is unstable in the absence of CIF2. We find that the CIF2 response does not depend on SCR and that the combination of SHR and CIF2 can induce identity acquisition in other root cell-types. Taken together, these results provide new insight into partially redundant pathways regulating the specification and stabilization of endodermis identity.
RESULTS
Endodermal TFs induce patchy ectopic Casparian strip-like structures:
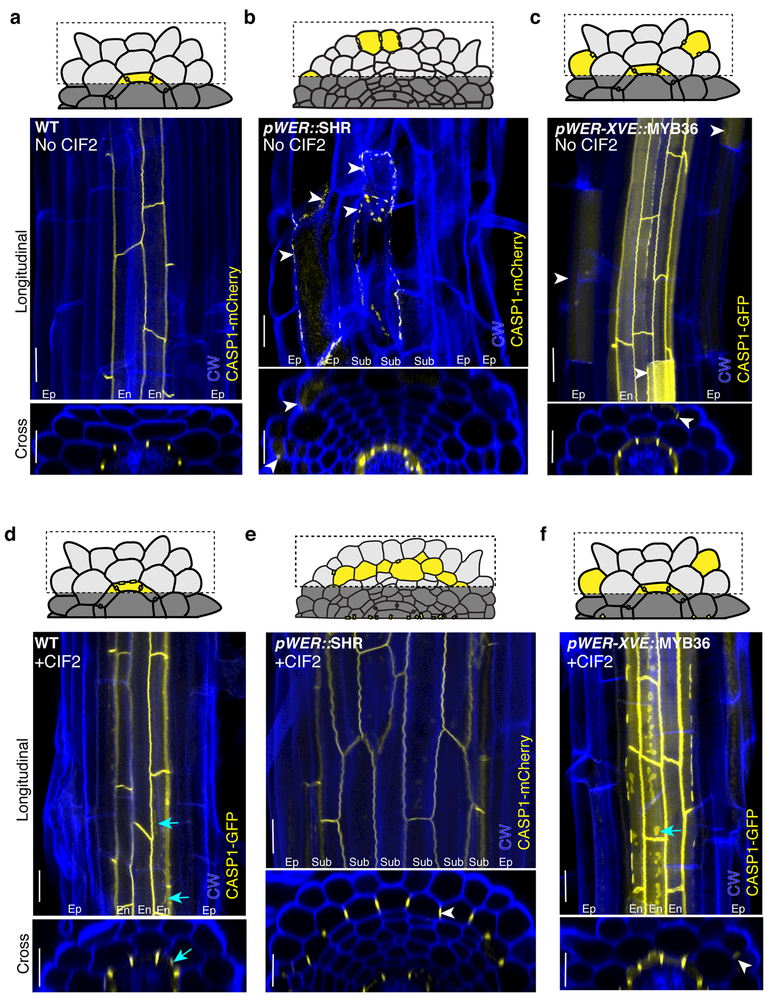
To determine if SHR, SCR and MYB36 are sufficient to form a Casparian strip, we individually expressed these TFs in the epidermis using the WEREWOLF (WER) promoter, known to confer expression primarily in the epidermis, in both constitutive and inducible forms 19,20. Expression of CASPs, and the deposition of lignin and suberin were used as markers for endodermal differentiation. We found that epidermis-expressed SHR induces stochastic expression of CASP1, lignin and suberin in a small subset of epidermal cells (Fig. 1A-B, FigS1A-D). Occasionally, CASP1 was detected in the supernumerary ground tissue layers that are present in these lines as a result of mis-expression of SHR 18 (the median percentage of sub-epidermal cells with ectopic CASP1 = 8.4%, interquartile range (IQR) = 0 – 10.8%; Fig. 1B). Epidermal expression of SCR was unable to induce CASP1, lignin or suberin in any cells (Fig. S1E-F). As was previously reported for lines driving MYB36 under a ubiquitous promoter 12, we found epidermis-expressed MYB36 also had limited ectopic CASP1 and lignin deposition in the epidermis, as well as suberin deposition (the median number of cells with ectopic CASP1 per epidermal view = 5, IQR = 3–8; Fig. 1C, Fig. S1G, Fig. S1H). We conclude that SHR and MYB36, but not SCR, are able to stochastically generate endodermal features in the epidermis.
Figure 1.
CIF2 stabilizes TF-induced endodermis identity acquisition. Maximum projections of confocal images of fixed and cleared roots stained with Calcofluor white (CW). Schematic of sub-epidermis projection views are represented above each image. Both longitudinal and cross section views are shown. a, CASP1 wild-type (Col-0, abbreviated as WT). b, CASP1 in pWER::SHR-GFP, median percentage of sub-epidermis with ectopic CASP1= 8.4%, IQR = 0 – 10.8%. c, pWER-XVE::MYB36-Venus plants, median number of epidermis cells with ectopic CASP1 in view = 5, IQR = 3 – 8. d, CASP1 in CIF2-treated wild-type (Col-0). e CASP1 in CIF2 treated pWER::SHR-GFP, median percentage of sub-epidermis with ectopic CASP1 = 90.4%, IQR = 87.2%−100%. f, CASP1 in CIF2 treated pWER-XVE::MYB36-Venus plants, median number of epidermis cells with ectopic CASP1 in view = 6, IQR = 5 – 8. White arrows indicate examples of ectopic CASP1. Cyan arrows in d and f indicate extra plaques of CASP1 in native endodermis as a result of CIF2 treatment. Scale bars = 20 μM. For a-b, d-e n=20 roots; for c, f n=17 roots. Mann Whitney test p-value (see methods) of b to e = <0.0001, and for c to f = 0.41 (not significant). Each experiment was repeated at least four times.
CIF2 and SHR induce Casparian strip-like structures:
The ectopic Casparian strip-like structures in the epidermis do not resemble the native Casparian strip. Instead, they have a “beads on a string” appearance similar to the Casparian strip found in the sgn3 receptor mutant (Fig. 1B,C) 14. It was previously reported that SGN3 expression is significantly down-regulated in shr but is relatively unaffected in scr or myb36 11,12,15. Recently, the peptide CIF2 was identified as a ligand for SGN3 and shown to play a key role in sealing the Casparian strip 16,17. CIF2 is produced in the stele and its expression is independent of SHR 15–17. We hypothesized that the “beads on a string” phenotype of the ectopic Casparian strip-like structures was a result of present but inactive SGN3, as CIF2 may not be able to reach the epidermal cells.
To test this hypothesis, we treated epidermis-expressed SHR seedlings with 100 nM CIF2. This treatment resolved the “beads on a string” CASP1 phenotype, and intriguingly, also induced expression of polarized CASP1 in most of the cells in the sub-epidermal layer (the median percentage of cells with ectopic CASP1 in the sub-epidermis = 90.4%, IQR = 87.2% - 100%; Fig. 1D-E). These cells also show deposition of lignin and suberin (Fig. S1I-L). In contrast, plants with epidermis-expressed SCR show no markers of endodermis differentiation even when treated with CIF2 (Fig. S1M-N). Ectopic Casparian strip-like structures in roots with epidermis-expressed MYB36 also have a “beads on a string” phenotype (Fig. 1C). Treatment with CIF2 did not seal ectopic Casparian strip-like structures, nor did it induce significantly more cells expressing CASP1 (the median number of cells with ectopic CASP1 per epidermal view = 6, IQR = 5–8; Fig S1). These results are consistent with previously published data showing MYB36 does not regulate SGN3 expression 11,12. We confirmed that the induction in epidermal-SHR plants is not due to SGN3 alone, since epidermis-expressed SGN3 in a sgn3–3 background was only capable of inducing rare patches of CASP1, even when treated with CIF2 (the median number of cells with ectopic CASP1 per epidermal view = 0, IQR = 0 – 0; without CIF2, median = 0, IQR = 0 – 1 with CIF2; Fig. S2).
The sub-epidermis has limited barrier function:
Epidermis-expressed SHR combined with CIF2 treatment results in an ectopic Casparian strip and suberin deposition suggesting that this structure might form an impermeable barrier. We tested the effectiveness of the sub-epidermis to act as a symplastic barrier by measuring fluorescein diacetate (FDA) penetration 21 and as an apoplastic barrier by propidium iodide (PI) penetration 22.The sub-epidermal layer in both pWER::SHR-GFP alone and pWER::SHR-GFP with CIF2 treatment had similar FDA penetration rates, suggesting that the suberin deposited in this layer has only a minor effect on symplastic flow (Video S1, S2). In contrast, PI penetration was partially inhibited in the sub-epidermal cell-layer (Fig S3), suggesting that the ectopic endodermal cell wall components form a limited barrier but that it is not as effective as the Casparian strip found in native endodermis.
The identity of the sub-epidermis before and after CIF2 treatment:
It has previously been proposed that the ectopic cell layers present in roots with mis-expression of SHR assume the identity of cortex 23. To clarify the identity of the supernumerary layers, we crossed seedlings expressing pWER::SHR-GFP to two cortex-associated fluorescent transcriptional markers, CO2 and 315.1 24. The mature cortex marker 315.1 is present before and after CIF2 treatment, although CIF2 treatment dampens its expression (the median percentage of 315-expressing cells in the sub-epidermis before CIF2 treatment = 82.1%, IQR = 73.9% - 87.9%; after CIF2 treatment = 36.4%, IQR = 26.5%−50.4%; Fig. 2A-C, Fig S4). In contrast, the meristematic cortex marker CO2 is usually absent from the sub-epidermis, even prior to CIF2 treatment (Fig. 2D-F). MYB36 and SGN3 are present in both epidermis and sub-epidermis of pWER::SHR-GFP roots, before and after CIF2 treatment (Fig. 2G-L). Combined, this suggests that in pWER::SHR-GFP plants, prior to CIF2 treatment, there are already transcriptional changes to the sub-epidermis that may prime it for endodermis identity acquisition.
Figure 2.

The identity of the sub-epidermis before and after CIF2 treatment. (A-L) Confocal images of fixed and stained roots. a, Cortex marker p315.1::erGFP in Col-0 (WT), b, pWER::SHR-GFP roots without CIF2 and c, with CIF2. Cortex marker pCO2::H2B-YFP in d Col-0, e pWER::SHR-GFP roots without CIF2 and f with CIF2. SGN3 in f Col-0, h pWER::SHR-GFP roots without CIF2 and i with CIF2. Maximum projections of MYB36 transcriptional reporter in j Col-0 (live sample), k pWER::SHR-GFP roots without CIF2 and l with CIF2. CW = Calcofluor white, PI = propidium iodide. White asterisks indicate epidermis cell files. Red asterisks indicate sub-epidermis cell file. Red arrowheads in g-l indicated ectopic expression. White arrowheads in g-l indicate endogenous expression. Scale bars = 20 μM. For a-l, n ≥ 10 roots. m, Schematic of the two genotypes and two treatments used for FACS. Cells with CASP1 (yellow) were enriched for by cell sorting. n, Heat map of the mean Index of cell identity value of three replicates from samples described in m compared to transcriptome from RootMap 3.0 (cell-types listed along bottom, see Table S2 for RootMap 3.0 libraries and values). Black asterisks indicate ICI p-value <0.05. Each experiment was repeated at least three times.
After CIF2 treatment, the sub-epidermis resembles an endodermis in terms of the presence of a Casparian strip and deposition of suberin. We asked if the identity of the sub-epidermal cell layer resembles an endodermis at the transcriptional level or simply produces a Casparian strip-like structure in a cortex-like cell. We performed RNA-seq on mCherry-positive cells isolated by FACS from wild-type plants (CASP1-mCherry) and from plants with epidermis-expressed SHR (pWER::SHR-GFP;CASP1-mCherry), grown in the presence or absence of CIF2 (Fig. 2M). Note that in all cases, cells from the endogenous endodermis contain CASP1 and are analyzed along with the ectopically expressing cells (Fig. 2M). Interestingly, presence of CIF2 alters relatively few genes compared to the untreated controls in both genotypes (Table S1). This suggests that CIF2 acts primarily in a post-transcriptional manner on cells already expressing CASP1.
To further explore the identity of the sub-epidermal cells, we utilized the Index of Cell Identity (ICI) tool to compare our expression profiles to those from high-resolution RNA-seq cell type-specific profiles (RootMap 3.0) 25–26. The ICI tool characterizes cell identity based on the weighted expression of marker genes defining different cell-types in the root 25 by calculating an “identity score” as opposed to an exclusive categorization into one cell type (e.g. there can be a significant score for both endodermis and cortex, Table S2). We found that the CASP1-expressing cells from all samples significantly associate with mature endodermis (E30) (Fig. 2N). In CASP1-expressing cells from epidermis-expressed SHR plants there is also a low, insignificant score for cortex identity (COR/315.1) (Fig. 2N). We also carried out a multi-dimensional scaling plot analysis comparing our expression profiles to those from RootMap 3.0 (Fig. S5) 26. The CASP1 cells from epidermis-expressed SHR with and without CIF2 treatment clustered closely with wild-type CASP1-positive cells and with the endodermis marker E30 (Figure S5).
While the FACs approach clarified the identity of the sub-epidermis, it did not capture changes in gene expression in the sub-epidermal cells that were not expressing CASP1 before CIF2 treatment. Based on the increase of CASP1, lignin and suberin in most of the sub-epidermal cells, we hypothesized that there are transcriptional changes in response to CIF2 that were not captured by FACS sorting for CASP1 expressing cells. To address the transcriptional effect of CIF2 on sub-epidermis cells, we carried out whole root RNA-seq analysis on pWER::SHR-GFP roots before and after CIF2 treatment and found that CIF2 treatment enriched for over 500 genes (Fig S6A-B, Table S3). These targets are expressed broadly across root cell types in RootMap 3.0 (Fig S6B). A subset of these are enriched in the endodermis, including several peroxidases, CASP-like transmembrane proteins and MYB15, which has a role in lignin deposition in defense 27(Table S3). There were also 35 genes depleted in response to CIF2, including a subset which are normally enriched in the cortex (Fig S6C). We then examined the combined effect of CIF2 and SHR in order to identify genes regulated in a non-additive manner (Table S3, Fig S6D, see Methods). This approach determined only 78 genes are regulated synergistically in epidermis-expressed SHR lines compared to wild-type, which suggests that pWER::SHR-GFP plants respond in a similar manner to CIF2 as wild-type plants (Fig S6D). Taken together, our results indicate that WER-driven expression of SHR generates a sub-epidermis that in the absence of CIF2 resembles the endodermis and is primed to respond to the addition of CIF2.
CIF2 maintains the sub-epidermal ectopic Casparian strip:
The sporadic nature of reprogrammed epidermal cells prior to CIF2 treatment suggested that expression of the downstream targets was unstable. To analyze the dynamics of downstream targets, we live-imaged CASP1-mCherry for 24 hours in plants with epidermis-expressed SHR. We found that the ectopic CASP1 intensity fluctuates over time (Video S3, Fig 3A, FigS7). Consistent with the stochastic expression of CASP1, SCR is also present in some but not all epidermal and sub-epidermal meristematic cells in epidermis-expressed SHR roots (the median percentage of cells with ectopic SCR in the epidermis and the sub-epidermis = 26.4%, IQR = 12.6% - 32.3%). This is in contrast to previous reports that SCR is only induced by epidermis-expressed SHR in the initial cells18 (Fig. S8). SCR expression is markedly different from MYB36 and SGN3 in epidermis-expressed SHR roots, which are present in most epidermal and sub-epidermal cells (Fig 2H-J).
Figure 3.
Dynamics of ectopic CASP1 expression in CIF2 treated and untreated samples. Frames from confocal live imaging over 24 hours of ectopic CASP1 (yellow channel) of a, pWER::SHR-GFP, no CIF2; b, pWER::SHR-GFP removed from CIF2 treatment; c, pWER::SHR-GFP with CIF2 added. Cyan arrows in a point to cells with fluctuating CASP1 intensity. Cyan arrows in b indicate cells losing CASP1 polarity. Cyan arrows in c indicate cells gaining CASP1 expression and polarity. Time elapsed is indicated in the bottom right. Scale bar = 25 μM, n ≥ 3 roots. Each experiment was repeated at least three times.
To determine if the SHR/CIF2 induction resulted in stabilized commitment to endodermal cell identity, we transferred five-day-old pWER::SHR-GFP seedlings germinated in the presence of CIF2 to plates without CIF2 for 24h. Under these conditions, the most mature (shootward) cells retained CASP1 expression (Fig. S9A). However, in a small region of the differentiated tissue closer to the meristem, CASP1 became diffuse and depolarized (Fig. S9B). Rootward of this region, which contains cells that were dividing prior to removal from CIF2, the root appears similar to the untreated control (Fig. S9C). To understand the dynamics of CASP1 in this unstable region, we live-imaged differentiated sub-epidermal cells of plants removed from CIF2 (Video S4, Fig 3B). A subset of cells loses previously established CASP1 in the sub-epidermis within this region (Video S4, Fig 3B). While CIF2 treatment on pWER::SHR-GFP plants substantially increased the number of cells expressing CASP1 in the sub-epidermis, it also reduced the number of cells expressing ectopic CASP1 in the epidermis (the median number of cells ectopic CASP1 in the epidermis per epidermal view = 3 cells, IQR = 2–6 with no CIF2 treatment; the median = 0, IQR = 0–2 with CIF2 treatment; Fig S10A). To determine the effect of CIF2 on the few cells expressing CASP1 in the epidermis of pWER::SHR-GFP plants, we imaged epidermal-SHR roots every three hours over a 24h treatment with CIF2. We found a median of 67.8% of cells retain CASP1 expression (Fig S10B), suggesting that CIF2 does not strongly affect established CASP1 in the epidermis.
CIF2 and SHR act at different developmental stages:
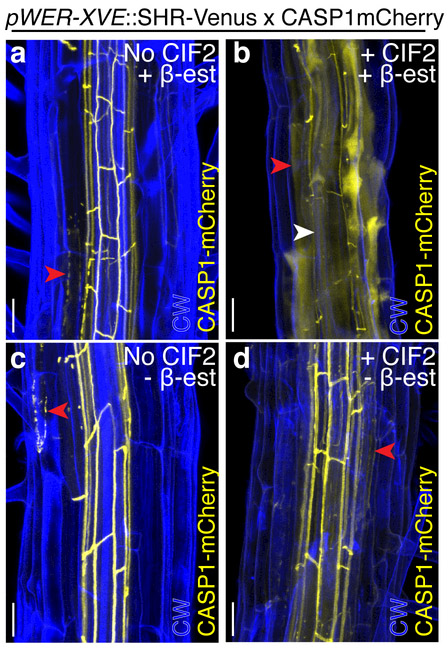
In order to investigate dynamics of the CIF2 response, we live-imaged epidermis-expressed SHR plants during CIF2 treatment. We found that cells already in the differentiation zone respond to CIF2 by expressing CASP1 in a polarized manner (Video S5, Fig 3C). Because the WER promoter confers expression primarily in early meristematic cells of the epidermis, we asked if SHR could alter the identity of cells when expressed later in the differentiation pathway. To this end, we examined plants expressing SHR under the GLABRA2 (GL2) and COBL9 promoters, which confer expression in non-hair cells of the transit-amplifying region of the meristem and in differentiated hair cells, respectively 28,29. It was previously reported that SHR driven by GL2 has some ectopic lignification in the epidermis18. Under our conditions, neither the COBL9 or GL2 driven SHR lines exhibited ectopic endodermal features, even with CIF2 treatment (Fig S11A-D). This suggests the window of competence for SHR to induce ectopic endodermal features in the epidermis is restricted to proximal meristem cells or stem cells. To further investigate the cell stage responsive to both CIF2 and SHR, we transiently induced SHR expression in the epidermis with an estradiol-inducible system (XVE) 20. After removing from β -estradiol for 24h, we treated plants with CIF2 for 24h. We found sub-epidermal cells in the mature regions of the plant that had been immature when treated with β-estradiol could produce ectopic Casparian strip-like structures after SHR induction and application of CIF2 (the median percentage of sub-epidermal cells = 83.3%, IQR = 80%−100%; for untreated, the median =14.3%, IQR = 6.6% - 27.8%, Fig 4A-B). These results support the conclusion that SHR must be present in the meristem or in stem cells to prime cells for endodermis identity acquisition, while CIF2 can act later in development. In the younger sections of the root not treated with β-estradiol, there were a few sporadic cells with ectopic Casparian strip-like structures (the median percentage of ectopic CASP1 in untreated = 6.2%, IQR = 0% - 11.1%; in CIF2 treated = 0%, IQR = 0% - 19.0%, Fig 4C-D). We reason that this is due to perdurance of β-estradiol after removing plants from the source rather than CIF2 since these structures are present in both CIF2-treated and untreated plants removed from β-estradiol (Fig 4C-D).
Figure 4.
SHR and CIF2 act at different stages of root development. Maximum projections of confocal images of stained and fixed portions of pWER-XVE::SHR-Venus x CASP1mCherry roots treated with B-estradiol a, without CIF2, median percentage of cells with ectopic CASP1 in sub-epidermis = 14.3%, IQR = 6.7% - 27.8%, and b, with CIF2, median percentage of cells with ectopic CASP1 in sub-epidermis = 83.3%, IQR = 80%−100%. c, Section of the same root in a not treated with β-estradiol, median percentage of cells with ectopic CASP1 in sub-epidermis = 6.3%, IQR = 0%−11.1%. d, Section of the same root in b that was not treated with β-estradiol, median percentage of cells with ectopic CASP1 in sub-epidermis = 0%, IQR = 0%−19.0%. Red arrowheads indicate examples of ectopic CASP1. White arrowheads in b indicate examples of endogenous CASP1. CW= Calcoflour white, scale bar = 20 μM. a, c n = 7 roots; b, d n = 11 roots. Mann Whitney test p-value of a to b is <0.0001, and c to d 0.55 (not significant). Experiment was repeated three times.
MYB36 and SGN3 but not SCR are required for induction of endodermal features:
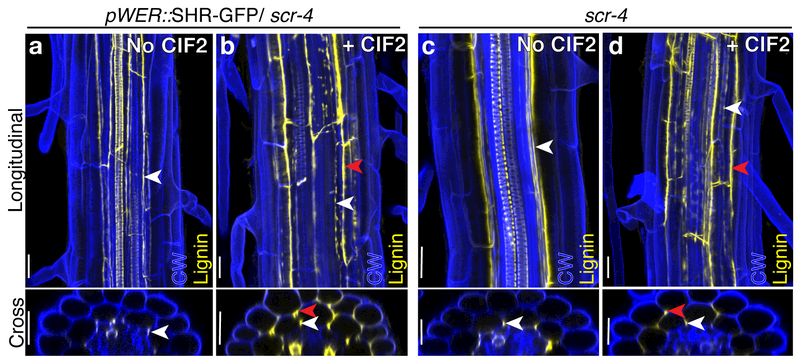
Neither epidermis-expressed MYB36 nor SGN3 is sufficient to induce a fully sealed ectopic Casparian strip in the presence of CIF2 (Fig 1C, F; Fig S2). By examining SHR/CIF2 induction in mutant backgrounds, we confirmed that both of these factors are required for ectopic Casparian strip induction (Fig S12). SCR was not sufficient to induce ectopic endodermal features (Fig S1E-F, M-N). We also tested if SCR is necessary for SHR-CIF2 Casparian strip induction. Plants with epidermis-expressed SHR in a scr-4 background lack the supernumerary cell divisions seen in a wild-type background and resemble scr-4 mutants containing a single mutant layer of ground tissue 18 (Fig. 5). In both scr-4 and pWER::SHR-GFP/scr-4 lines without CIF2, lignin is deposited on the inner edge of the mutant layer (Fig 5A, C). Upon CIF2 treatment, the mutant layer becomes excessively lignified, especially on the outer edges, in both pWER::SHR-GFP/scr-4 (a median percentage of 90.0%, IQR = 83.9%−100%) and scr-4 plants (a median percentage of 100%, IQR = 100%, Fig 5B,D). These results suggest SCR is dispensable for the response to CIF2 in formation of a Casparian strip.
Figure 5.
SCR is not required for a response to CIF2. Maximum projections of confocal images from fixed roots stained with Calcofluor white (CW) and Basic fuchsin for lignin (yellow channel), both longitudinal and cross section views. a, pWER::SHR-GFP/scr-4 plants without CIF2, median percentage of cells with lignin localization on the outer face of the mutant layer = 0%, IQR = 0% - 0%. b, pWER::SHR-GFP/scr-4 plants with CIF2, median percentage of cells with lignin localization on the outer face of the mutant layer = 90.0%, IQR = 83.9% - 100%. c, scr-4 plants without CIF2, no plants observed had cells lignified on the outer face of the mutant layer d, scr-4 plants with CIF2, all observed plants had all cells lignified on the outer face of the mutant layer. White arrowheads indicate lignin localized to the inner face of the mutant ground tissue layer. Red arrowheads in b and d indicate lignin localized to the outer face of the mutant ground tissue layer. Scale bar = 20 μM. a, b, n = 12 roots, c, d, n = 6 roots. Mann Whitney test p-value of a to b and c to d is <0.0001. Each experiment was repeated at least three times.
Movement of SHR from the epidermis does not explain endodermal identity acquisition:
The epidermal expression of SHR induces cortex-like supernumerary layers due to the movement of SHR within the epidermal stem cells 18. It was previously proposed that SHR cannot move from the epidermis in epidermis-expressed SHR lines based on GFP-localization18. However, we reasoned that a small amount of SHR, below the detection levels of the previous analysis, could move into the neighboring sub-epidermis and explain the formation of an ectopic Casparian strip in that cell layer. In order to measure SHR movement, we utilized a confocal scanning technique called paired correlation function (pCF)30. We were unable to detect movement of SHR from the epidermis to the sub-epidermis (comparable to non-moving control TMO5::3xGFP30, Fig S13A, see methods). Yet, interestingly, the pCF analysis revealed there are low levels of SHR present in the sub-epidermis, which were previously not visible (Fig. S13A, Table S4). We also generated a construct with a nuclear localization sequence (NLS-tag) to attempt to interfere with SHR movement. Similar to GFP-tagged SHR, there was no movement from epidermis to cortex in NLS-tagged lines (Fig S13A). The inducible NLS-Venus construct had low levels of SHR detectable in the sub-epidermis when treated for several days with β-estradiol (Fig S13B). Despite few supernumerary cell layers, these roots respond in a similar fashion to CIF2 treatment, making an ectopic Casparian strip in their sub-epidermal layer (the median percentage of cortex cells with ectopic CASP1 in CIF2 treated plants = 95.2%, IQR = 90.0%−100% compared to a median of 22.2%, IQR = 5.9%−33.3% in untreated; Fig S13C-D). The simplest explanation for these results is that SHR does not move from the epidermis but the WER promoter is active at very low levels in the cortex. Consistent with this, WER mRNA is present broadly in RootMap 3.026 (Fig S13E).
SHR and CIF2 can induce endodermis in ground tissue:
The lack of SHR movement from the epidermis suggested that the source of SHR for induction was from the sub-epidermis itself. To determine if SHR combined with CIF2 can change the identity of other root cell-types, we examined lines expressing SHR under the cortex promoter CO2 and endodermis promoter EN7. Both lines produce ectopic cell divisions, although not to the extent of the pWER::SHR-GFP lines (Fig 6). In pCO2::SHR-GFP lines, there are few cells with ectopic lignin (the median percentage of supernumerary cells with ectopic lignin in untreated = 14.3%, IQR = 12.5%−27.3%; Fig 6A). Upon CIF2 treatment, the number of cells with lignification increases substantially (a median percentage = 85.7%, IQR = 83.3% - 88.8%; Fig 6B). In pEN7::SHR-GFP lines, there is lignification of ectopic cell files, independent of CIF2 treatment (a median percentage of supernumerary cells with lignin = 77.5%, IQR = 66.7%−95% for untreated, a median of 100%, IQR = 100% for treated; Fig 6C-D). It is possible that the sealed, lignified domains in the absence of the CIF2 in these cell files are a result of the proximity of this layer to the endogenous source of CIF2. It could also be due to expression of SHR in the cortex-endodermal initial cell when driven by EN7, which is consistent with our results that SHR must be present in the meristem or stem cells to produce endodermal features ectopically. A non-mutually exclusive possibility is the presence of other factors in the endodermis allow SHR to more efficiently activate downstream targets. The latter has been previously proposed for lines expressing SHR in the endodermis8. In conclusion, the combination of cell-autonomously expressed SHR with CIF2 is able to alter the identity of cortex in its native context or in the context of ectopic ground-tissue layers.
Figure 6.
Ectopic SHR in the cortex and endodermis with and without CIF2 treatment. Maximum projections of confocal images from fixed roots stained with Calcofluor white (CW) and Basic fuchsin for lignin (yellow channel). a, pCO2::SHR-GFP without CIF2, median percentage of ectopic cells with lignin = 14.3%, IQR = 12.5% - 27.3%. b, pCO2::SHR-GFP with CIF2, median percentage of ectopic cells with lignin = 85.7%, IQR = 83.3% - 88.9%. c, pEN7::SHR-GFP without CIF2, median percentage of ectopic cells with lignin = 77.5%, IQR = 66.7% - 95.0%. Of cells with ectopic lignin, 100% are completely sealed. d, pEN7::SHR-GFP with CIF2, median percentage of ectopic cells with lignin = 100%, IQR = 100% - 100%. Of cells with ectopic lignin, all observed are completely sealed. Red arrowheads indicate examples of ectopic lignin. White arrowheads indicate examples of endogenous lignin. White asterisks indicate the native endodermis cell file. Red asterisks indicate ground tissue supernumerary cell files. Scale bar = 20 μM. a, b n=7 roots, c, d n=8 roots. Mann Whitney test p-value for a to b = 0.0012. For percentage of ectopic cells with lignin in c compared to d, Mann Whitney test p-value is 0.007 (significant); for median percentage of sealed ectopic lignin, Mann Whitney test p-value of c to d is >0.99 (not significant). Each experiment was repeated three times.
DISCUSSION
Our results establish a simple path to endodermis differentiation: the combination of the TF SHR and the peptide CIF2 is sufficient. By introducing components in a non-native context, we demonstrate that the SGN3/CIF2 pathway stabilizes endodermis differentiation, a process that was assumed to function far downstream from the “master regulator” SHR. The action of CIF2 and SHR support a model in which the primary differentiation cues for the endodermis emerge from the neighboring tissue, the stele. Additional support comes from our transcriptome data showing that SHR and CIF2 synergistically activate genes enriched in the stele (Fig S56D).
It is striking that acquisition of differentiated endodermis is so stable in the sub-epidermis and not in the epidermis. This could be because of a competing GRN present in the epidermis. Prior to CIF2 treatment, there is an obvious decrease in root hair density, which is relieved upon CIF2 treatment (Fig S14). Additionally, the formation of root hairs, a hallmark of differentiation in epidermal cells, was never observed in combination with ectopic CASP1.
Epidermis-expressed SHR can induce ectopic MYB36 and SGN3 in epidermal and sub-epidermal cells (Fig 2H-I, K-L). This is markedly different from ectopic SCR induced by pWER::SHR-GFP, which is present sporadically in the meristematic cells of the epidermis and sub-epidermis (Fig S8). The most parsimonious explanation for these results is that SHR regulates SCR, MYB36 and SGN3 using different mechanisms. These results were surprising since SCR was previously thought to be the link between SHR and MYB36 expression. However, here we show SCR is rarely present in the sub-epidermis and MYB36 is present in both the epidermis and sub-epidermis of pWER::SHR-GFP plants11(Fig S8). Our results suggest that SCR is not required for a general response to CIF2, which is consistent with published data showing SGN3 is expressed independently of SCR 15. We also show that neither SHR nor SGN3/CIF2 alone is sufficient to induce formation of an ectopic Casparian strip to the extent of the SHR/CIF2 combination. Consistent with these data, cif1/2, sgn3 mutants and myb36 mutants have some lignin assembly, while a myb36/sgn3 double mutant has no Casparian strip (Niko Geldner, personal communication). This suggests that the MYB36 and SGN3/CIF2 pathways have some functional redundancy in contributing to barrier formation and maintaining endodermis identity.
Our results show cells must express SHR in the meristem to be competent to respond to CIF2, and that they maintain that competence for at least several days. This further demonstrates the importance of both lineage and positional cues in plant cell differentiation 31. We also show cells no longer dividing or elongating have a narrow window in which they can reverse CASP1 localization. Since lignification occurs later in development, it could be that while the cells have established a CASP1 domain, they have not yet assembled lignin. To confirm that CIF2 doesn’t have a general role in polarizing CASP1, we treated roots over-expressing CASP1 with CIF2 and found no polarization outside the endodermis with or without CIF2 (Fig. S15). We propose that in the native endodermis, cell identity is initiated by SHR and stabilized by CIF2. The role of CIF2 in stabilization is only apparent with reduced levels of SHR as observed with ectopic expression of SHR.
A remaining question is, “Why is there only one layer rather than many layers of ectopic Casparian strip?” It is possible SHR must reach a critical threshold level to activate expression of SGN3 and MYB36, which occurs in the sub-epidermis but not in other supernumerary layers. It could also be that other factors involved in endodermis differentiation, such as the Indeterminate Domain/BIRD family, are expressed differently across the supernumerary ground tissue layers15,32. Another non-mutually exclusive possibility is that there is some paracrine signaling to inhibit neighboring cells from acquiring endodermis identity. This may also explain why we see few cells ectopically express CASP1 in the epidermis after CIF2 induction (Fig S10A). Finally, we note the similarity of the sub-epidermal layer with ectopic Casparian strip to the exodermis, a waterproof sub-epidermal layer present in most vascular plants, but absent in Arabidopsis 33. We speculate that homologs of these components may play a role in exodermis formation in other species.
MATERIALS AND METHODS:
Plant growth conditions:
Several plant growth conditions adjusting MS, sucrose, long day vs. 24h light, liquid and agar were tested and none alter the SHR-CIF2 induction phenotype. Seedlings were grown on ½ x MS, 1% sucrose plates with or without 100 nM CIF2 (Invitrogen), unless otherwise noted as a 24–72h treatment. For transgenic lines containing the XVE inducible promoter system 20 seedlings were germinated on MS 0.5% plates and transferred at day 2 plates containing 10µM β-estradiol. All plants were imaged at five days old.
Clearing, staining and imaging of fixed samples:
For examining CASP1 expression, five day-old roots were fixed in PFA and cleared in ClearSee as described in Kurihara et al. 201534 and cell walls were visualized with Calclofluor white as described in Ursache et al. 2018.35. For lines in a CASP1-GFP background, lignin localization was assessed by combining Basic Fuchsin and ClearSee and suberin localization was assessed by combining Nile red and ClearSee, both described in Ursache et al. 2018.35. Roots were imaged on a Zeiss 880 using a 40X or 20X objective. The following are the excitation and detection parameters: Calcofluor white ex 405 nm, em 425–475 nm; ex Basic Fuchsin/mCherry 561 nm, em 600–650 nm; ex Fluorol-Yellow/GFP 488nm, em 525–550 nm; ex Nile Red 561 nm, em 600–620 nm.
For examining lignin and suberin in lines in a CASP1-mCherry background, lignin was assessed by Basic Fuchsin staining as described in Malamy & Benfey 199736 and suberin was assessed by Fluorol Yellow staining as described in Barberon et al 201621. Basic Fuchsin-stained roots were imaged on a Zeiss 880 confocal using a plan apochromatic 40x objective and Fluorol-Yellow stained roots on a Leica DM5000 microscope using a 10X objective (excitation/emission parameters above). At least three biological replicates were acquired for each experiment.
All image analysis was carried out in ImageJ. For all image slices and projections, minimum signal for each channel was adjusted by measuring the intensity histogram of the background and removing the mean and two standard deviations from the signal. Brightness was adjusted for channels with dim signal to maximize range of display. Video S4 was drift corrected using Imaris software.
Live-Imaging of CASP1 dynamics without CIF2 and after CIF2 treatment:
At four days old, plants were removed from CIF2 or treated with CIF2 and imaged in a sterile chamber on a ½ x MS, 1% sucrose, 1% agar pad. Video S3 samples were carried out on a Zeiss 880 and Video S3 and 4 were on a Zeiss 510 (all with 40x objective). Excitation for CASP1mCherry was 561 nm (Zeiss 880) and 543 (Zeiss 510), emission was 600–650 nm. At least three biological replicates were acquired for each experiment.
Quantification of CASP1 intensity was carried out in ImageJ. Briefly, cells were outlined using the polygon tool and the raw integrated density was measured for each time point (see Table S5 for measurements). In a few rare cases, a time point was not recorded or laser power was diminished as determined by auto-fluorescence of root hairs. In these rare cases, the frame was omitted from analysis (see Table S8).
FDA and Barrier Assays:
FDA and barrier assays were carried out on five day-old plants as previously described in Barberon et al. 2016 21 and Alassimone et al. 2010 22.
FACS Sorting and RNA-sequencing:
Seeds were sterilized with 3% (vol/vol) sodium hypochlorite and 0.1% Tween for 5 min and rinsed five times in sterile water. Seeds were stratified for 48h and plated on sterile 1X MS, 1% sucrose plates with or without 100nM CIF2. Roots were harvested on day 5, digested and subjected to Fluorescence Activated Cell Sorting (FACS) as described previously37. Total RNA was isolated using RNAeasy Micro-kit (Qiagen). RNA integrity and quantity were assessed on Agilent Bioanalyzer system and QuBit, respectively (ThermoFisher Scientific). cDNA synthesis for library preparation was carried out using the SMART-Seq v4 Ultra Low Input RNA kit for Sequencing (Takara). The Low Input Library Prep Kit v2 (Takara) was used to prepare libraries for sequencing. Single-end reads were obtained using the Illumina HiSeq4000 platform at the Duke University Sequencing Core.
Whole Root RNA-sequencing:
Seeds were sterilized with 3% (vol/vol) sodium hypochlorite and 0.1% Tween for 5 min and rinsed five times in sterile water. Seeds were stratified for 48h and plated on sterile 1X MS, 1% sucrose plates with or without 100nM CIF2. Roots were harvested on day 5 and RNA was isolated using RNA-easy mini kit (Qiagen). RNA integrity and quantity were assessed on Agilent Bioanalyzer system and QuBit, respectively (ThermoFisher Scientific). mRNA libraries were prepared using KAPA stranded mRNA-seq kit (Roche). Single-end reads were obtained using the Illumina HiSeq4000 platform at the Duke University Sequencing Core. Three biological replicates were acquired for each experiment.
Sequencing Analysis:
All scripts for analysis can be found on github: https://github.com/cdrapek/Endodermis. Briefly, reads were aligned with TopHat (mapping results are available in Table S6). Counts were normalized using the Bioconductor EdgeR package 38. Differentially expressed gene lists (Table S1, S3) were identified using EdgeR GLM model 38. For FACS samples, results were filtered by logFC>0.3 and a P-value<0.01, with the exception of two comparisons which were also filtered by FDR<0.05 (pWER::SHR-GFP +CIF2 vs CASP1mCherry +CIF2; pWER::SHR-GFP vs CASP1-mCherry; Table S1). For whole root samples, results were filtered by logFC>0.3, FDR<0.05 (Table S3). For calculating combination effect of SHR and CIF2, we removed significant differences from CIF2 treatment in wild-type from CIF2 differences in epidermis-expressed SHR (contrasts on https://github.com/cdrapek/Endodermis). Multi-dimensional scaling analysis was carried out using the MDS-function in EdgeR to select samples pairwise from FACS libraries described in this manuscript and RootMap 3.0. For ICI analysis, marker Spec scores and ICI values were calculated as previously described 25. Briefly, Spec scores were calculated from RootMap FPKM RNA-seq data with the “getAllSpec” function and the default filters (medianfilter=0, cuts=FALSE, distshape=0). The index of cell identity (ICI) was calculated for each replicate using the “getIdentity” function. Values reported are for the normalized ICI score. Significance was determined from FDR corrected p-values (Table S2). Three biological replicates for each genotype and treatment were acquired.
Cell counting and statistical analysis:
A subset of randomly selected images from experiments were used for counting cells or calculating percentage of cells with ectopic features (all counts for all median and interquartile ranges are recorded in Table S7). Distribution of data was plotted using R. No experiment had mostly normal distribution, therefore, we used the Mann Whitney test to determine calculate p-value. Prism software was used for statistical analysis.
Paired Correlation Function:
Paired correlation function was carried out as previously described in Clark et al. 2016 30. Briefly, time series were carried out on a Zeiss 880 on PI-stained roots (to determine location of cell wall) with 40x objective using the following parameters for GFP and YFP: Images were analyzed using SimFCS software and statistical analysis was carried out as described in Clark et al. 2016 30. Arc of fluorescence over time across a cell wall was considered movement (movement index =1) and no crossing is considered no movement (movement index =0; see Table S4). Data from pSHR::SHR-GFP was previously generated as described in Clark et al. 201630.
Transgenic lines and cloning:
The following lines have been previously published: pWER::SHR-GFP, CASP1-mCherry, CASP1-GFP, SGN3-venus, pSCR::SCR-CFP, pCO2::SHR-GFP, pEN7::SHR-GFP, pGL2::SHR-GFP. For full list of primers used for cloning, see supplemental material Table S8. The 3Kb fragment upstream of the MYB36 start codon was fused to H2B and 3xYFP fragments using the multi-site gateway system. The inducible WER-XVE and COBL9-XVE promoter were previously generated as described in 20. The WER-XVE promoter was fused to MYB36, SCR, SGN3 or SHR cDNA entry vectors and fluorophore-terminator containing entry vectors using the multi-site gateway system into a Norflurazon-resistant destination vector. The SHR-mKate2 lines were generated by fusing SHR cDNA to mKate2 by In-Fusion technology (Takara) followed by insertion into pDONR221 vector by BP reaction. The WER-XVE promoter or COBL9-XVE promoter and SHR-mKate2 construct were assembled into the FastRed selection system destination vector using the Gateway Cloning system. For each transgenic line, three to twelve T1 lines containing a single-insertion were observed and a representative line was selected for follow up study.
Data availability:
All normalized CPM RNA-seq reads are recorded in Tables S1 and S3. All code used for analyzing mapped reads is available on GitHub (https://github.com/cdrapek/Endodermis)). Seed lines available upon request to corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS:
We thank the Duke University Genome Sequencing & Analysis Core for sequencing the RNA libraries. We thank Idan Efroni for assistance in the ICI pipeline and Guy Wachsmann for assistance in mapping RNA-seq reads. We are also grateful to Natalie Clark and Ross Sozzani for assistance with the pCF technique. We thank the Duke Light Microscopy Facility and Robertas Ursache for assistance in microscopy, members of the Benfey lab for comments on the manuscript, Veronica G. Doblas for assistance with CIF2 treatments, Heather Belcher and Jay Zhang for technical assistance, and Kim Gallagher for thoughtful discussion. This research was funded by grants from the NIH (R01-GM043778) and the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (GBMF3405) to PNB. N.G. was funded by an ERC Consolidator Grant (GA-N°: 616228 – ENDOFUN), an SNSF grant (31003A_156261). Additional support was provided to CD through an EMBO Short-term fellowship, to EES through startup funds from the University of Delaware, to PM by a FEBS Long-Term Fellowship, to TGA by an IEF Marie Curie fellowship
REFERENCES:
- 1.Takahashi K & Yamanaka S A decade of transcription factor-mediated reprogramming to pluripotency. Nature Publishing Group 1–11 (2016). doi: 10.1038/nrm.2016.8 [DOI] [PubMed] [Google Scholar]
- 2.Pillitteri LJ, Sloan DB, Bogenschutz NL & Torii KU Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Gallois J-L, Nora FR, Mizukami Y & Sablowski R WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes & Development 18, 375–380 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson EH, McClay DR & Hood L Regulatory gene networks and the properties of the developmental process. Proceedings of the National Academy of Sciences 100, 1475–1480 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drapek C, Sparks EE & Benfey PN Uncovering Gene Regulatory Networks Controlling Plant Cell Differentiation. Trends Genet. 33, 529–539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geldner N The endodermis. Annu. Rev. Plant Biol 64, 531–558 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Andersen TG, Barberon M & Geldner N Suberization-the second life of an endodermal cell. Current Opinion in Plant Biology 28, 9–15 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Nakajima K, Sena G, Nawy T & Benfey PN Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Helariutta Y et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Di Laurenzio L et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Liberman LM, Sparks EE, Moreno-Risueno MA, Petricka JJ & Benfey PN MYB36 regulates the transition from proliferation to differentiation in the Arabidopsisroot. Proceedings of the National Academy of Sciences 112, 12099–12104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamiya T et al. The MYB36 transcription factor orchestrates Casparian strip formation. Proceedings of the National Academy of Sciences 112, 10533–10538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roppolo D et al. A novel protein family mediates Casparian strip formation in the endodermis. Nature 473, 380–383 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Pfister A et al. A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. Elife 3, e03115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Risueno MA et al. Transcriptional control of tissue formation throughout root development. Science 350, 426–430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doblas VG et al. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355, 280–284 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Nakayama T et al. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355, 284–286 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Sena G, Jung JW & Benfey PN A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131, 2817–2826 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Lee MM & Schiefelbein J WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Siligato R et al. MultiSite Gateway-Compatible Cell Type-Specific Gene-Inducible System for Plants. PLANT PHYSIOLOGY 170, 627–641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barberon M et al. Adaptation of Root Function by Nutrient-Induced Plasticity of Endodermal Differentiation. Cell 164, 1–35 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Alassimone J, Naseer S & Geldner N A developmental framework for endodermal differentiation and polarity. Proceedings of the National Academy of Sciences 107, 5214–5219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S et al. A plausible mechanism, based upon SHORT-ROOT movement, for regulating the number of cortex cell layers in roots. Proceedings of the National Academy of Sciences 111, 16184–16189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady SM, Orlando DA, Lee JY, Wang JY & Koch J A high-resolution root spatiotemporal map reveals dominant expression patterns. Proceedings of the National Academy of Sciences (2007). doi: 10.1093/nar [DOI] [PubMed] [Google Scholar]
- 25.Efroni I, Ip P-L, Nawy T, Mello A & Birnbaum KD Quantification of cell identity from single-cell gene expression profiles. Genome Biol. 16, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Yamada M, Han X, Ohler U & Benfey PN High-Resolution Expression Map of the Arabidopsis Root Reveals Alternative Splicing and lincRNA Regulation. Developmental Cell 39, 508–522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chezem WR, Memon A, Li F-S, Weng J-K & Clay NK SG2-Type R2R3-MYB Transcription Factor MYB15 Controls Defense-Induced Lignification and Basal Immunity in Arabidopsis. The Plant Cell 29, 1907–1926 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masucci JD et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Brady SM, Song S, Dhugga KS, Rafalski JA & Benfey PN Combining expression and comparative evolutionary analysis. The COBRA gene family. PLANT PHYSIOLOGY 143, 172–187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark NM, Hinde E, Winter CM, Fisher AP & Crosti G Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. Elife (2016). doi: 10.7554/eLife.14770.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheres B Plant cell identity. The role of position and lineage. PLANT PHYSIOLOGY 125, 112–114 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long Y et al. Arabidopsis BIRD Zinc Finger Proteins Jointly Stabilize Tissue Boundaries by Confining the Cell Fate Regulator SHORT-ROOT and Contributing to Fate Specification. The Plant Cell 27, 1185–1199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enstone DE, Peterson CA & Ma F Root endodermis and exodermis: structure, function, and responses to the environment. J Plant Growth Regul (2002). doi: 10.1007/s00344-003-0002-2 [DOI] [Google Scholar]
- 34.Kurihara D, Mizuta Y, Sato Y & Higashiyama T ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ursache R & Geldner N A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. The Plant Journal [DOI] [PubMed] [Google Scholar]
- 36.Malamy JE & Benfey PN Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Birnbaum K et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Meth 2, 615–619 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All normalized CPM RNA-seq reads are recorded in Tables S1 and S3. All code used for analyzing mapped reads is available on GitHub (https://github.com/cdrapek/Endodermis)). Seed lines available upon request to corresponding author.