Abstract
The androgen receptor (AR, NR3C4) is a nuclear receptor whose main function is acting as a transcription factor regulating gene expression for male sexual development and maintaining accessory sexual organ function. It is also a necessary component of female fertility by affecting the functionality of ovarian follicles and ovulation. Pathological processes involving AR include Kennedy’s disease and Klinefelter’s syndrome, as well as prostate, ovarian, and testicular cancer. Strict regulation of sex hormone signaling is required for normal reproductive organ development and function. Therefore, testing small molecules for their ability to modulate AR is a first step in identifying potential endocrine disruptors. We screened the Tox21 10K compound library in a quantitative high-throughput format to identify activators of AR using two reporter gene cell lines, AR β-lactamase (AR-bla) and AR-luciferase (AR-luc). Seventy-five compounds identified through the primary assay were characterized as potential agonists or inactives through confirmation screens and secondary assays. Biochemical binding and AR nuclear translocation assays were performed to confirm direct binding and activation of AR from these compounds. The top seventeen compounds identified were found to bind to AR, and sixteen of them translocated AR from the cytoplasm into the nucleus. Five potentially novel or not well-characterized AR agonists were discovered through primary and follow-up studies. We have identified multiple AR activators, including known AR agonists such as testosterone, as well as novel/not well-known compounds such as prulifloxacin. The information gained from the current study can be directly used to prioritize compounds for further in-depth toxicological evaluations, as well as their potential to disrupt the endocrine system via AR activation.
1. Introduction
The androgen receptor (AR, NR3C4) is a transcription factor which regulates male sexual development, while also maintaining accessory sexual organ function (Culig et al. 2002b. The structure of AR includes an N-terminal region which contains the activation function-1 (AF-1), a DNA-binding domain (DBD), a hinge region, and a ligand binding domain (LBD) which contains the ligand-regulated AF-2 (Culig et al. 2002b; Roy et al. 2001). AR is an evolutionarily conserved receptor and is closely related to the human glucocorticoid and progesterone receptors including even recognizing analogous DNA response elements. However, these receptors have a different hormone ligand specificity (Culig et al. 2002b). AR is the main transcription factor implicated in transmitting hormone signals inside the prostate gland (Culig et al. 2002a). As a key transcription factor regulating male sexual development (Pihlajamaa et al. 2015), altering regulation of this nuclear receptor causes abnormal development of the prostate (Wen et al. 2015).
AR activation can occur through direct or indirect pathways (Culig et al. 2002b; Roy et al. 2001). Direct AR activation occurs through a multi-step process. First, the unliganded receptor sequestered by heat shock proteins and immunophilins in the cytoplasm of the cell binds to a ligand through the LBD. This causes a conformational change allowing for the dissociation from the complex anchoring AR in the cytoplasm. Once free, AR homodimerizes and the nuclear localization signal amino acid sequence becomes exposed. The nuclear localization signal subsequently binds to importins, which then transport AR into the nucleus (DeFranco 1999; Roy et al. 2001). Once inside the nucleus, the ligand-receptor complex and its co-activators accumulate at sequence-specific nuclear foci (Roy et al. 2001; Tyagi et al. 2000). However, like its other nuclear receptor counterparts, AR can also be activated through multiple other pathways in an indirect manner (Davey and Grossmann 2016); direct ligand binding is not necessary.
Xenobiotic perturbation of AR has many possible adverse outcomes in humans. This includes multiple types of endocrine disruption such as changes in spermatogenesis and the synthesis of sex hormones (Cook et al. 1999). AR is a key driver of prostate cancer growth and AR expression and sensitivity have also been shown to increase in the androgen-responsive human prostatic carcinoma (LNCaP) cell line when grown in androgen-depleted medium (Culig et al. 2002a; Kokontis et al. 1994). Other studies have shown that AR also has an important role in the modulation of multiple additional cancer types including liver, kidney, and bladder, and is linked to hepatocellular hypertrophy (Chang et al. 2014; Fujimoto et al. 2012). Therefore, recognizing exogenous compounds and environmental chemicals which activate AR is critical in detecting endocrine disrupters and possible cancer modulators.
To quickly evaluate the effect of environmental chemicals on the endocrine system, quantitative high-throughput screening (qHTS) has become a useful way to detect modulation of relevant receptors (Hsu et al. 2014; Huang et al. 2014). As part of the Tox21 program, a federal collaboration among the National Institutes of Health, including the National Center for Advancing Translational Sciences and the National Toxicology Program at the National Institute of Environmental Health Sciences, the Environmental Protection Agency, and the Food and Drug Administration, we screened the Tox21 collection of ~10,000 environmental chemical and drug (Tox21 10K) samples (~8300 unique compounds) for their AR agonist potential. From our primary screening assay (Tox21 10K), we conducted follow-up studies including a confirmation screen, binding assay, and translocation assay to identify a number of compounds which activate the androgen receptor.
2. Materials and Methods
2.1. Compound Library
The Tox21 chemical library consists of approximately 10,000 (~8300 unique) small molecules (NCATS 2016; PubChem 2013), including pesticides, drugs, industrial chemicals, and food additives, commercially sourced by the NTP, NCATS, and EPA (Attene-Ramos et al. 2013). These compounds were selected based on multiple criteria, including environmental hazards or exposure concerns, compounds with properties conducive to HTS (molecular weight, volatility, solubility, logP), commercial availability, and cost. There were also 88 diverse compounds (Tox21–88) selected as internal controls and plated in duplicate on every library plate to assess reproducibility and determine positional plate effects (Huang et al. 2014). Currently available chemical quality control information is also available (NCATS 2016).
2.2. Cell Culture
GeneBLAzer® AR-UAS-bla GripTite™ (AR-bla) cells, obtained from Invitrogen (Carlsbad, CA), are derived from HEK293 cells and stably integrated a β-lactamase reporter gene under control of an upstream activator sequence (UAS). The UAS is regulated by a fusion protein consisting of the rat AR ligand-binding domain (LBD) fused to the DNA-binding domain of GAL4 which binds the UAS. The rat LBD is identical in sequence to the human LBD although there are amino acid differences in the included hinge region. Agonists of the AR result in binding of the fusion protein to the UAS leading to expression of beta-lactamase. AR-bla cells were cultured in DMEM high-glucose (Invitrogen) supplemented with 10% dialyzed FBS, 80 μg/mL zeocin, 80 μg/mL hygromycin and 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen). The cells were maintained at 37ºC under a humidified atmosphere and 5% CO2. The second cell line, AR MDA-kb2 luciferase-expressing (AR-luc), was obtained from ATCC (Manassas, VA). The AR-luc cell line (developed from the parental cell line MDA-MB-453 of human breast cancer cells (Wilson et al. 2002)) expresses firefly luciferase under control of the MMTV promoter that contains response elements for both glucocorticoid receptors (GR) and AR. AR-luc cells were cultured in L-15 Medium (ATCC) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). The cells were maintained at 37ºC under a humidified atmosphere and 0% CO2.
2.3. qHTS of AR β-lactamase reporter gene assay
AR-bla cells suspended in phenol red-free Opti-MEM medium containing 10% dialyzed FBS, were dispensed at 2,000 cells/6 μl/well in 1536-well black wall/clear bottom plates using a Thermo Scientific Multidrop Combi (Thermo Fisher Scientific Inc., Waltham, MA). After the assay plates were incubated at 37ºC/5% CO2 for 5 hours, 23 nL of the test compounds dissolved in DMSO, positive controls, or DMSO, were transferred to the assay plates using a Wako Pintool station (Wako Automation, San Diego, CA). The final compound concentration ranged from 0.8 nM to 76 μM over 15 concentrations. The plate format of the positive control (R1881) is as follows: Column 1: concentration-response titration of R1881 (Perkin Elmer, MA) from 0.2 pM to 3.83 μM; Column 2: 30 nM of R1881; Columns 3 & 4: top halves with 20 nM of R1881 and bottom halves with DMSO only. The plates were then incubated at 37ºC/5% CO2 for 16 hrs. One μL of LiveBLAzer™ B/G FRET substrate (Invitrogen, Carlsbad, CA) detection mix was added using a Flying Reagent Dispenser (FRD, Aurora Discovery, Carlsbad, CA). The assay plates were then incubated at room temperature (RT) for 2 hrs, after which fluorescence intensity was measured at 460 and 530 nm following excitation at 405 nm using an EnVision plate reader (Perkin Elmer, Shelton, CT). The assays were performed three times for each compound concentration and data was expressed as the ratio of 460 nm/530 nm emissions and analyzed as described later.
2.4. qHTS of AR-luciferase reporter gene assay
AR-luc cells suspended in L-15 medium containing 10% FBS, were dispensed at 3,000 cells/5 μL/well in 1536-well white tissue culture plates using the Multidrop Combi. After the assay plates were incubated at 37ºC/0% CO2 for 5 hrs, 23 nL of compounds dissolved in DMSO, positive controls, or DMSO were transferred to the assay plates by the Wako Pintool station. The final compound concentration ranged from 1 nM to 92 μM over 15 concentrations. The plate format for the positive control was as follows; Column 1: concentration-response titration of R1881 (Perkin Elmer, MA) from 0.3 pM to 4.6 μM; Column 2: R1881 in top half with 36 nM and bottom half with 10 nM; Columns 3 & 4: R1881 in top halves with 24 nM & 20 nM respectively; DMSO only in bottom halves. The assay plates were then incubated at 37ºC/0% CO2 for 16 hrs. After 5 μL of ONE-Glo™ Luciferase reagent (Promega, Madison, WI) was added using the FRD, the plates were incubated at RT for 30 min, after which luminescence intensity was measured by a ViewLux plate reader (Perkin Elmer). The assays were performed three times for each compound concentration and data were expressed as relative luminescence units and analyzed as described later.
2.5. qHTS Data Analysis
The qHTS data was analyzed according to the previous protocol (Huang et al. 2014). Briefly, raw plate reads for each titration point were first normalized relative to the positive control compound (agonist mode: R1881 = 100%) and DMSO-only wells (0%) as follows: % Activity=[(Vcompound-VDMSO)/(Vpos-VDMSO)] x 100, where Vcompound denotes the compound well values, Vpos denotes the median value of the positive control wells, and VDMSO denotes the median values of the DMSO-only wells. The data set was then corrected using the DMSO-only compound plates at the beginning and end of the compound plate stack by applying an in-house pattern correction algorithm (Wang and Huang 2016). The half maximum effective values (EC50) for each compound and maximum response (efficacy) values were obtained by fitting the concentration-response curves of each compound to a four-parameter Hill equation (Wang et al. 2010). Compounds designated as Class 1–4 according to the type of concentration–response curve observed (1.1, 1.2, 1.3, 1.4, 2.1, 2.2, 2.3, 2.4, and 3 for activators; 4 for inactive) (Huang et al. 2011; Inglese et al. 2006). Curve classes are heuristic measures of data confidence, classifying concentration–responses on the basis of efficacy, the number of data points observed above background activity, and the quality of fit. To facilitate analysis, each curve class was combined with an efficacy cutoff and converted to a numerical curve rank such that more potent and efficacious compounds with higher quality curves were assigned a higher rank (Huang et al. 2011). A detailed definition of curve rank is shown in Supplemental Table S2. For each reading, the activity outcome of a test compound was first categorized based on the average curve rank from the triplicate runs and the reproducibility calls. The final activity outcome of each compound was determined based on its multi-channel readout activity. For example, compounds were only considered active when they showed agonist activity in both the 460 nm channel and the FRET ratios. A compound was assigned as autofluorescent in the AR-bla assay and inconclusive when it was promiscuously active in the 460 nm readout of all the Tox21 bla assays or was active in the auto fluorescence counter screen at similar potencies as the ratiometric readout (i.e. AC50, auto fluor/AC50, ratio<3). Data reproducibility was categorized as active match, inactive match, inconclusive, and mismatch according to the previously described criterion (Huang et al. 2014). The 10K library was clustered based on structural similarity (Leadscope® fingerprints; Leadscope, Inc., Columbus, OH, USA) using the self-organizing map algorithm. Each cluster was evaluated for its enrichment of active agonists and significance of enrichment as determined by p-values from the Fisher’s exact test.
2.6. Fluorescence polarization AR competition binding assay
PolarScreen™ AR Competitor Assays, non-radioactive binding assays, were acquired from Life Technologies (Carlsbad, CA) and used to determine binding affinity of selected compounds to the AR-LBD. Fluormone AL Green, a fluorescent labeled AR ligand, was thawed on ice prior to the following experiments. The mixing of complete AR Green Screening buffer, containing 2 mM DTT, and the fluormone tracer was conducted to create a 4 nM tracer solution. Using a Mantis single-channel liquid dispenser (Formulatrix, Bedford, MA), we distributed 5 μL of the 4 nM tracer solution into each well of a 1536-well plate. Twenty-three nL of DMSO solution of each compound was added into the 1536-well plate using a Wako Pintool station, followed by addition of 5 μL of complete buffer to the first 16 wells of column one. In all other wells, 5 μL of 892 nM AR-LBD protein was added to create a final concentration of 446 nM of protein. After the assay plate was centrifuged (1,000 rpm for 1 minute) and incubated for 4 hours at RT, the fluorescence polarization signal (excitation at 480 nm and emission at 535 nm) was measured on the EnVision plate reader.
2.7. GFP-AR translocation assay and analysis
The 3108 cell line (a derivative of 3134 mouse mammary adenocarcinoma cell line) expresses green fluorescent protein (GFP)-tagged human full length AR (GFP-AR) from a chromosomal locus under control of the tetracycline-repressible promoter (Klokk et al. 2007; Stavreva et al. 2012). Experiments were performed in 96-well plates with a glass bottom. Cells were grown overnight in DMEM containing 10% charcoal stripped serum (Hyclone, Logan, UT) without tetracycline (allowing expression of the GFP-AR) at 10, 000 cells/well. Cells were treated with compounds at different concentrations, with the positive control (testosterone), or with vehicle control (DMSO) for 30 min at 37°C. Upon treatment, cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 15 min and washed three times with PBS. Cells were then stained with DRAQ5 (BioStatus Limited, Shepshed, GB) at a concentration of 1:5000 for 10 min and, after three washes with PBS, imaged either immediately on the Perkin Elmer Opera Image Screening System (Waltham, MA) or kept in PBS at 4°C for later imaging up to one month.
The Opera Screening platform was used for an automated confocal collection of images. A 40x water immersion objective lens, laser-illuminated Nipkow disk, and cooled CCD cameras were employed to digitally capture high-resolution confocal fluorescence micrographs (300 nm pixel size with 2×2 camera pixel binning). An image analysis pipeline was customized using the Columbus software (PerkinElmer) to automatically segment the nucleus using the DRAQ5 channel (excitation 640 nm, emission filter 690/70) and then construct a ring region (cytoplasm) around the nucleus for each cell in the digital micrographs. The pipeline measured the mean GFP-AR intensity in both compartments in the GFP channel (excitation 488 nm, emission filter 520/35). Translocation was calculated as a ratio of these intensities. Each value was further normalized to the value for the control (DMSO) sample on the same plate.
3. Results
3.1. qHTS performance and reproducibility
We conducted the primary assay by screening the Tox21 10K compound library in a qHTS platform, using AR-bla (LBD, partial receptor) and AR-luc (full receptor) cells, to identify environmental chemicals and drugs as possible AR agonists (see PubChem assay IDs 743036, 743053, 743040). R1881, the positive control for these assays, showed consistent activity throughout the screen with an EC50 of 1.06 ± 0.10 nM in the AR-bla assay and 0.10 ± 0.02 nM for the AR-luc assay. The assay performance statistics including signal to background (S/B) ratio, coefficient of variance (CV), and Z’ factor (Zhang et al. 1999) for the AR-bla assay were 1.85 ± 0.17, 4.82% ± 0.79, and 0.21 ± 0.11, respectively. The AR-luc assay had better performance statistics: 6.63 ± 0.55 for the S/B ratio, 8.59% ± 0.80 CV, and a Z’ factor of 0.68 ± 0.06.
To further assess the performance of the assay, we evaluated the reproducibility of the three copies of the 10K compound library tested independently, as well as the Tox21–88 compound array, which was plated in duplicate on every library plate. Following the primary 10K compound library screening, all compounds were categorized as follows: active, inactive, or inconclusive. The mismatches in activity were <0.5% in the 10K triplicate run and <1.5% in the Tox21–88 duplicates indicating robust assay performance (Table 1). In addition, the correlation of EC50 values (R2) between the duplicates that were active matches in the primary agonist screen for the AR-bla and AR-luc assays were 0.81 and 0.98, respectively (data not shown).
Table 1.
Assay performance using Tox21–88 duplicates and 10K triplicate runs.
| Assay Reproducibility | Active match (%) | Inactive match (%) | Inconclusive (%) | Mismatch (%) | EC50 fold change |
|---|---|---|---|---|---|
| 10 K triplicate run | |||||
| AR-bla | 5.36 | 86.88 | 7.44 | 0.30 | 1.77 |
| AR-luc | 4.14 | 93.65 | 2.20 | 0.00 | 1.36 |
| Tox21–88 | |||||
| AR-bla | 9.43 | 79.55 | 10.45 | 0.57 | 1.78 |
| AR-luc | 3.07 | 92.16 | 3.52 | 1.25 | 1.39 |
For each assay, the reproducibility was calculated for the Tox21–88 compounds (duplicates in each plate) and for the 10K library (three copies) with compounds plated in different well locations
3.2. Identification of potential AR agonists
The well-documented AR agonists, androstenedione, spironolactone, and testosterone, were identified in both AR-bla and AR-luc assays (Supplemental Figure 1). The activity of all active AR agonists, and some inactives from a list of AR reference compounds (Kleinstreuer 2016) further verified the biological relevance of our qHTS assays. The EC50 and efficacy of these reference compounds are listed in Table 2. All the compounds were identified correctly as a positive or negative hit in either the AR-bla or AR-luc assays.
Table 2.
Previously (Kleinstreuer 2016) identified AR reference compounds and their present activity.
| Chemical Name (CASRN) [Purity Rating] |
Structure | AR-bla Primary Assayc | AR-luc Primary Assayc | Known AR Activity |
|---|---|---|---|---|
| EC50 (μM) [Efficacy (%)a] |
EC50 (μM) [Efficacy (%)a] |
|||
| 4-Androstenedione (63–05-8) [A] |
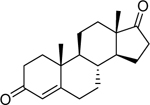 |
0.391 [190] |
0.0579 [113] |
Agonist |
| 5α-Dihydrotestosterone (521–18-6) [A] |
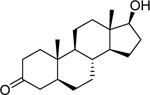 |
0.00628 [119] |
0.000926 [113] |
Agonist |
| Atrazine (1912–24-9) [A] |
 |
Inactiveb | Inactiveb | Inactive |
| Carbendazim (10605–21-7) [A] |
 |
Inactiveb | Inactiveb | Inactive |
| Cyproterone acetate (427–51-0) [A] |
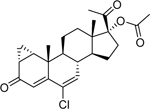 |
Inactiveb | 2.38 [107] |
Weak Agonist |
| Deltamethrin (52918–63-5) [A] |
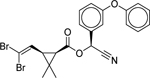 |
Inactiveb | Inactiveb | Inactive |
| Fenarimol (60168–88-9) [A] |
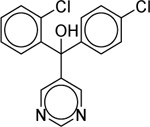 |
Inactiveb | Inactiveb | Inactive |
| Fluoxymestrone (76–43-7) [A] |
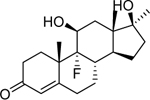 |
0.279 [163] |
0.0239 [162] |
Agonist |
| Flutamide (13311–84-7) [A] |
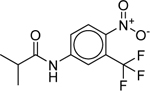 |
Inactiveb | Inactiveb | Inactive |
| Iprodione (36734–19-7) [A] |
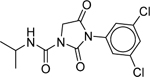 |
Inactiveb | Inactiveb | Inactive |
| Levonorgestrel (797–63-7) [A] |
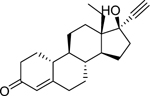 |
0.00152 [46.4] |
0.0119 [108] |
Agonist |
| Medroxyprogesterone acetate (71–58-9) [A] |
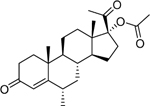 |
0.269 [84.4] |
0.0102 [274] |
Agonist |
| Methyltestosterone (58–18-4) [A] |
 |
0.00592 [156] |
0.00276 [88.1] |
Agonist |
| Methyltrienolone (R1881) (965–93-5) [A] |
0.00131 [131] |
0.000144 [110] |
Agonist | |
| Nandrolone (434–22-0) [A] |
 |
0.00265 [153] |
0.000934 [90.1] |
Agonist |
| Nilutamide (63612–50-0) [A] |
 |
Inactiveb | Inactiveb | Inactive |
| Norethindrone (68–22-4) [A] |
 |
0.00261 [60.6] |
0.0814 [106] |
Agonist |
| Norethindrone acetate (51–98-9) [A] |
 |
0.0264 [77.6] |
0.0191 [98.1] |
Agonist |
| Norethynodrel (68–23-5) [A] |
 |
0.00367 [71.5] |
0.0412 [94.2] |
Agonist |
| p,p’-DDT (50–29-3) [A] |
 |
Inactiveb | Inactiveb | Inactive |
| Prochloraz (67747–09-5) [A] |
 |
Inactiveb | Inactiveb | Inactive |
| Resveratrol (501–36-0) [A] |
 |
Inactiveb | Inactiveb | Inactive |
| Tamoxifen (10540–29-1) [A] |
 |
Inactiveb | Inactiveb | Inactive |
| Testosterone (58–22-0) [A] |
 |
0.00278 [179] |
0.000864 [105] |
Agonist |
| Trenbolone (10161–33-8) [A] |
 |
0.00297 [105] |
0.001 [122] |
Agonist |
% efficacy is based on the maximal efficacy produced by R1881 (positive control).
Compound was identified as inactive if the efficacy was below 30% of R1881’s activity.
Values are in mean ± SD format, where n=3.
Purity Rating: A = MW Confirmed, Purity > 90%.
Seventy-five compounds were selected from the primary screen based on potency (EC50 < 30 μM), efficacy (> 30% of R1881), purity (NCATS 2016), and structural class similarity to be re-examined using the same AR-bla and AR-luc assays (Supplementary Table 1). Out of the 75 compounds that were retested, 42 were confirmed as active in the AR-luc assay and 39 compounds were confirmed active in the AR-bla assay. Twenty-three of the 75 compounds were confirmed inactive in the AR-luc assay, while the AR-bla assay confirmed 16 compounds as inactive. Including confirmed actives and inactives, these values produced confirmation rates of 73% and 87% for the AR-bla and AR-luc assays, respectively. The most interesting 17 known and potential novel AR agonists are listed in Table 3. Five compounds were identified as potentially novel (or not well-known) AR activators: prulifloxacin, norethisterone enanthate, oxiglutathione, GSK232420A, and glycyrrhizin. All five showed an efficacy above 60% in at least one of the assay screens performed. Four of these potential novel compounds have an EC50 below 1 μM, and the other one has an EC50 below 10 μM in at least one of the assays. These 17 compounds were chosen as the best illustrations of AR agonists due to their potency, efficacy, curve quality, reproducibility, purity, and direct AR binding activity through the primary, confirmatory, and competitive binding assays (described below).
Table 3.
Potential novel and representative AR modulators identified through qHTS, clusters, binding, and translocation.
| Compound Name (CASRN) [Purity Rating] |
Chemical Structure | Cluster Number [assay] |
AR-blac,d EC50 (μM) [Efficacy (%)a] |
AR-lucc EC50 (μM) [Efficacy (%)a] |
FP Bindingc IC50 (μM) [Efficacy (%)a] |
AR Translocationc Fold Change [Concentration, nM] |
|---|---|---|---|---|---|---|
| 3,3’-Dichlorobenzidine dihydrochloride* (612–83-9) [A] |
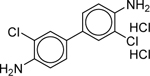 |
28.4 | Inactive | 27.49 ± 4.46 [109.29 ± 1.77] |
3.35 ± 0.63 [−64.20 ± 16.55] |
2.46 ± 0.115 [1000] |
| Drospirenone* (67392–87-4) [A] |
 |
36.26 [Both] |
0.018 ± 0.0015 [163.52 ± 31.25] |
4.39 ± 0.30 [86.22 ± 4.85] |
0.31 ± 0.056 [−68.07 ± 8.16] |
3.87 ± 0.463 [62.5] |
| Equilin* (474–86-2) [A] |
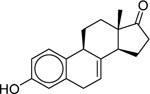 |
20.10 [luc] |
19.50 ± 0.00 [31.53 ± 3.88] |
6.31 ± 2.03 [89.27 ± 18.86] |
21.01 ± 3.41 [−89.77 ± 13.22] |
2.09 ± 0.0633 [62.5] |
| Ethynodiol diacetate* (297–76-7) [A] |
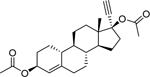 |
34.26 [Both] |
0.083 ± 0.045 [59.76 ± 6.90] |
0.23 ± 0.058 [51.25 ± 3.72] |
4.78 ± 0.86 [−53.19 ± 2.93] |
3.24 ± 0.202 [62.5] |
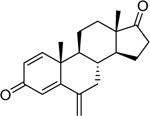
| Exemestane* (107868–30-4) [A] |
 |
37.26 [Both] |
0.61 ± 0.14 [215.30 ± 17.92] |
0.031 ± 0.0056 [108.39 ± 7.03] |
2.73 ± 1.59 [−110.10 ± 21.05] |
2.74 ± 0.134 [62.5] |
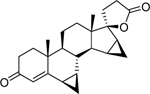
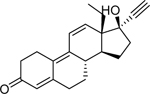
| Gestrinone* (16320–04-0) [A] |
 |
38.25 [Both] |
0.0034 ± 0.0029 [63.41 ± 14.86] |
0.0039 ± 0.0012 [125.33 ± 14.48] |
0.019 ± 0.0087 [−87.76 ± 4.33] |
9.34 ± 0.757 [62.5] |
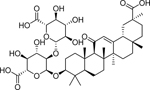
| Glycyrrhizin (1405–86-3) [A] |
 |
1.2 | 18.2 ± 12.2 [35.2 ± 4.13] |
9.48 ± 3.45 [88.5 ± 8.41] |
9.77 ± 2.36 [−50.4 ± 4.24] |
1.32 ± 0.0312 [1000] |
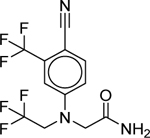
| GSK232420A (864283–48-7) [A] |
 |
41.11 | 5.87 ± 3.41 [38.03 ± 5.31] |
0.015 ± 0.0099 [96.57 ± 11.56] |
0.089 ± 0.045 [−71.27 ± 10.12] |
8.75 ± 0.765 [62.5] |
| Melengestrol acetate* (2919–66-6) [A] |
 |
38.26 [Both] |
Inactive° | 0.017 ± 0.012 [288.12 ± 44.83] |
1.17 ± 0.20 [−124.24 ± 3.58] |
2.57 ± 0.149 [62.5] |
| Methenolone enanthate* (303–42-4) [A] |
 |
36.26 [Both] |
0.036 ± 0.018 [72.47 ± 15.52] |
0.0031 ± 0.00021 [120.97 ± 12.18] |
1.20 ± 0.22 [−75.13 ± 17.71] |
6.41 ± 0.750 [62.5] |
| Methylcoumarin* (91–44-1) [A] |
 |
24.19 | 23.65 ± 1.54 [335.08 ± 9.51] |
27.43 ± 3.15 [88.69 ± 8.59] |
3.14 ± 2.97 [−60.70 ± 4.82] |
2.46 ± 0.0676 [1000] |
| Nandrolone phenpropionate* (62–90-8) [A] |
 |
36.26 [Both] |
0.010 ± 0.0018 [163.42 ± 1.14] |
0.0019 ± 0.00022 [133.58 ± 5.15] |
2.05 ± 0.33 [−67.56 ± 27.79] |
4.31 ± 0.449 [62.5] |
| Norgestimate* (35189–28-7) [A] |
 |
33.25 | 0.027 ± 0.0044 [70.90 ± 6.13] |
0.053 ± 0.00 [85.83 ± 4.25] |
2.72 ± 1.92 [−76.92 ± 32.06] |
6.29 ± 0.469 [62.5] |
| Norethisterone enanthate (3836–23-5) [A] |
 |
36.26 [Both] |
0.20 ± 0.013 [61.26 ± 3.79] |
0.23 ± 0.091 [61.59 ± 5.94] |
1.62 ± 0.19 [−83.23 ± 7.27] |
2.99 ± 0.295 [62.5] |
| Oxabolone cypionate* (1254–35-9) [Ac] |
 |
36.26 [Both] |
0.016 ± 0.0028 [137.70 ± 10.04] |
0.0048 ± 0.00055 [133.67 ± 9.16] |
2.49 ± 1.27 [−59.76 ± 14.45] |
6.20 ± 0.403 [62.5] |
| Oxiglutathione (27025–41-8) [A] |
 |
17.26 | 0.058 ± 0.0038 [99.95 ± 2.09] |
0.011 ± 0.0030 [105.83 ± 6.49] |
1.89 ± 0.24 [−95.59 ± 13.02] |
7.68 ± 0.685 [62.5] |
| Prulifloxacin (123447–62-1) [A] |
 |
37.1 [luc] |
1.26 ± 0.43 [134.90 ± 17.19] |
0.28 ± 0.019 [116.23 ± 8.01] |
5.23 ± 2.79 [−72.36 ± 7.67] |
3.66 ± 0.499 [62.5] |
% efficacy is based on the maximal efficacy produced by R1881.
These compounds have documented direct connections to AR as modulators.
Values are in mean ± SD format, where n=3.
Compound was identified as inactive if the efficacy was below 30% of R1881’s activity.
Purity Rating: A = MW Confirmed, Purity > 90%; Ac = Purity > 90%, Low concentration of sample.
3.3. Identification of structural clusters activating AR
From the primary screen, the entire Tox21 10K compound collection was grouped into 1,014 clusters based on structural similarity utilizing a self-organizing map algorithm (Kohonen 2006) such that each cluster contained chemicals belonging to the same structural class. As shown in Figure 1, each compound cluster was colored according to the significance of enrichment of AR activators in the cluster. Fourteen clusters (containing 409 compounds) were identified as significantly (p < 0.01) enriched with AR agonists in both AR-bla and AR-luc assays. There were 13 additional clusters (including 303 compounds) containing potential AR agonist scaffolds identified only in the AR-bla assay. There were another five clusters (containing 130 compounds) identified using the AR-luc assay, bringing the total amount to 32 clusters, representing a total of 842 compounds. All of the clusters enriched with AR agonists in both assays have a steroid like scaffold (k19.11, k20.11, k34.26, k36.26, k37.26, k38.25, k38.26, k39.25, k39.26, k40.25, k40.26, k41.26, k42.25, and k42.26), consistent with the well-documented modulation of AR through steroid hormones. Cluster k36.26 includes some known AR agonists on the reference list, such as testosterone and trenbolone (Table 2). This cluster also includes known AR modulators: nandrolone phenpropionate, drospirenone, methenolone enanthate, and oxabolone cypionate (Table 3).
Figure 1. Structural class heat map of AR agonist activity.
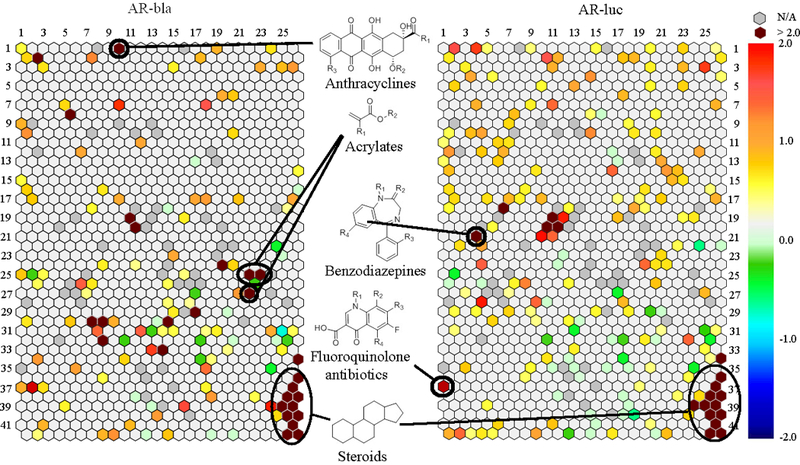
Every hexagon represents a class of structurally similar compounds. The color gradient is indicative of the enrichment of AR actives in that specific cluster [negative logarithmic scale of the p-value, -log(p-value)]. Each color represents a group of chemicals with similar scaffolds to activate AR in the AR-bla and AR-luc assays. Clusters with multiple actives in their class are closer to maroon, while clusters with no activity are a light grey color. Empty clusters with no available (N/A) compounds in them are darker grey in color.
The scaffolds of the five clusters that were active exclusively in the AR-luc assay contain anti-androgens (k18.6), steroids (k20.10 and k39.24), benzodiazepines (k21.4), and antibiotics (37.1). The anti-androgens and steroids include well-known modulators of AR such as androstenediol, dutasteride, and estrone. However, to our knowledge, the benzodiazepines oxazepam and nimetazepam have not been previously identified as AR agonists. In our study, oxazepam showed a moderate potency (EC50: 12.6 μM; efficacy: 58%). Nimetazepam had a much higher potency and efficacy of 4.4 μM and 470%, respectively, which was confirmed in the confirmation screen. A group of antibiotics, sparfloxacin (EC50: 20 μM; efficacy: 83%), orbifloxacin (EC50: 26 μM; efficacy: 59%), and prulifloxacin (EC50: 0.3 μM; efficacy: 116%), was also identified as potential novel AR agonists.
The active clusters identified in only the AR-bla assay include steroids (k2.2), anthracyclines (k1.10), acrylates (k25.22, k25.23, and k27.22), and flavins (k30.7) to name a few. Some of the anthracyclines, such as doxorubicin, were already known to have connections with AR (Ikeda et al. 2010). Conversely, carubicin has not been previously identified, to our knowledge; its EC50 and efficacy values were 2.1 μM and 197%, respectively.
3.4. Direct Binding of compounds to AR
To further determine the AR binding ability of these compounds, a fluorescent polarization binding assay was performed on a subset of the selected compounds. The top 17 compounds which had a binding efficacy greater than 20% and were not among the AR reference compounds are listed in Table 3 along with the primary screening and translocation data described later. The five potentially novel/not well-characterized AR modulators identified using the primary, confirmation, and binding assays are glycyrrhizin, GSK232420A, norethisterone enanthate, oxiglutathione, and prulifloxacin. Figure 2 illustrates three of these compounds and their AR modulating effects. GSK232420A, norethisterone enanthate, and prulifloxacin are able to bind to AR, with binding efficacies of 71%, 83%, and 72%, respectively; and potencies (IC50s) of 0.089 μM, 1.62 μM, and 5.23 μM, respectively.
Figure 2. Concentration-response curves alongside binding data of potentially novel AR agonists.
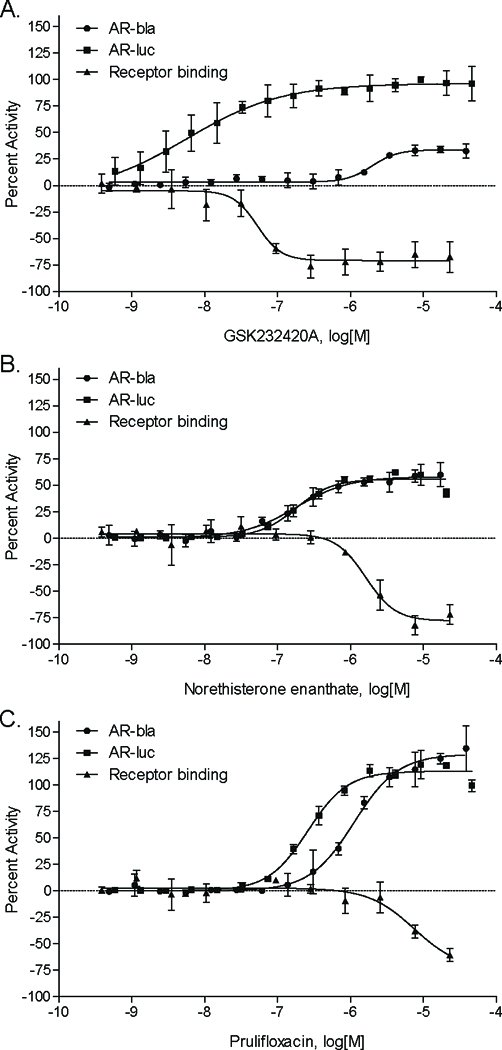
Fifteen point dilutions and receptor binding studies of, GSK232420A (A), Norethisterone enanthate (B), and prulifloxacin (C) were performed. Data were collected from follow-up studies, including the AR-bla confirmation assay, the AR-luc confirmation assay, and the receptor binding screen. They were expressed as mean ± SD from triplicate experiments for each assay.
The 75 compounds, chosen from the primary screen and tested in the confirmation screen were also compared between both reporter gene assays and the binding assay (Supplemental Table 1). Out of the 42 compounds confirmed as active AR agonists in the AR-luc assay, 23 had binding efficacies greater than 20%. When the 23 confirmed inactive compounds were tested, 2 were positive in the binding assay. This produced a 68% concordance rate between the AR-luc and binding assays. When comparing the AR-bla and binding assays, a 60% concordance rate was found. Twenty-one out of the 39 confirmed AR agonists bound to AR, while 12 out of the 16 confirmed inactive compounds did not bind. Out of the 27 agonists confirmed using both assays, 19 were identified as binding to AR. Two inactive compounds identified in both assays, did not bind. Therefore, when combining both the AR-luc and AR-bla assays, the overall concordance between the binding data and the reporter gene assay results was 72%.
3.5. GFP-AR translocation
In order to further investigate if these compounds are true AR agonists, a GFP-AR translocation assay was performed to examine this early step of AR activation. Results from the 3108 cell line, used to identify the presence of AR-translocators among the top 17 compounds, are shown in Table 3. Sixteen out of the 17 compounds significantly translocated AR into the nucleus (Figure 3A) in a concentration-dependent manner (Supplemental Figure 2). One exception was glycyrrhizin, which increased AR translocation by 1.32-fold, suggesting only slight activation. Pictures of the GFP-AR protein and nucleus staining, upon treatment of the cells with the three representative novel AR agonists identified previously (Figure 3B), clearly identify these three compounds as translocators of AR, confirming our binding and primary screening data.
Figure 3. Translocation of GFP-AR protein of top 17 compounds.
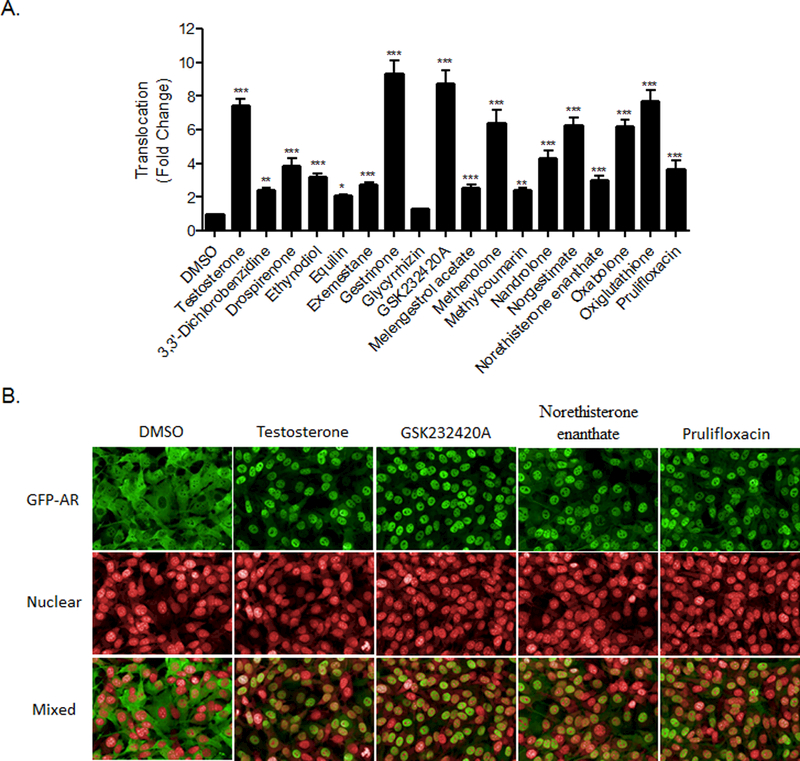
A mouse mammary adenocarcinoma cell line that expresses GFP-AR was treated with the vehicle control (DMSO), the positive control (Testosterone), or each of the top 17 potential and known agonists for 30 minutes, at concentrations indicated in Table 3. An automated image analysis of localization was performed and the mean GFP-AR intensity for both the nucleus and the cytoplasm was identified. The ratio of both intensities was calculated and the triplicate values were then normalized to the corresponding control (DMSO) values (A). Data represent the mean ± SD (n=3). **, P<0.01; ***, P<0.001. Representative micrographs of the GFP-AR, nucleus, and combined channels of the cells treated with 62.5 nM of the representative three potential novel compounds, as well as the positive and negative controls, are shown in B.
4. Discussion
The two qHTS assays, AR-bla and AR-luc, enabled us to identify novel and known AR agonist compounds. The AR is an important receptor in sexual development; dysregulation by endogenous or exogenous factors results in changes in the activity, amount, or polymorphisms of this receptor which may lead to adverse medical conditions including developmental diseases, such as Klinefelter syndrome or Kennedy’s disease, or cancer (Ran et al. 2015; Skakkebæk et al. 2014; Tanaka et al. 2012). Therefore, identifying environmental chemicals which might affect AR would be an interesting and necessary scientific discovery.
One structural cluster, k37.1, contained apparently novel AR agonists identified from the primary AR-luc qHTS assay. It consisted of fluoroquinolone antibiotics with two rings as their common scaffold (see Figure 1 for structure). Sparfloxacin, orbifloxacin, and prulifloxacin all displayed an efficacy of over 50% in the AR-luc assay. However, only prulifloxacin, a prodrug used mainly for urinary tract and community-acquired respiratory tract infections, was also identified as a strong binder to AR (Karageorgopoulos et al. 2013). It has very few reported side effects, and is therefore being assessed as an excellent choice of antibiotic (Prats et al. 2006). A rat fertility study found no reproductive or developmental effects with oral administration, and identified the No Observed Adverse Effect Level (NOAEL) to be 1000 mg/kg (Morinaga et al. 1996a). In a rabbit development study, a NOAEL of 30 mg/kg for pregnancy was found although the general toxicity NOAEL was 10 mg/kg (Morinaga et al. 1996b. Further studies may be needed to determine prulifloxacin’s in vivo modulation of the AR and whether it has adverse effects. It is also interesting to note that prulifloxacin was also active in the analogous estrogen receptor screens in our lab (data not shown).
The binding and translocation assays further characterized compounds as AR agonists or inactives identified from the primary and confirmation screens. Out of the 75 compounds tested, the binding assay had an overall 72% concordance rate with our primary screens. This 28% discrepancy is most likely due to the primary screens being cell-based, while the binding assay was biochemical. Previous reports have shown differences in results based on the type of assay performed (Shukla et al. 2011). AR can work through a few indirect pathways, including non-DNA binding-dependent or ligand independent activation (Davey and Grossmann 2016); this means the chemical does not need to bind directly to the receptor, nor does the AR complex need to directly bind to the DNA. Due to this indirect activation, a positive AR agonist identified in our primary and confirmation screen in the AR-luc assay could acquire negative results in the binding assay. Since AR-bla only uses a partial receptor, this indirect activation method would not apply. Any compound which has a discrepancy should be studied further to identify its true nature. The translocation assay helped to further examine potential AR agonists by identifying sixteen out of seventeen compounds to translocate AR from the cytoplasm into the nucleus, thus confirming this early step of AR activation. The combined use of all the assays performed here were able to identify a group of potentially novel AR agonists.
Along with prulifloxacin, there were four other compounds identified as potentially novel (or not well-known) AR agonists. The compound GSK232420A was used as a positive control androgen receptor agonist in a SARM drug development project (Handlon et al. 2016) (Supporting Information). However, there is little or no other information publicly available for this pharmaceutical compound.
Norethisterone enanthate is a progestin prodrug used in female contraception. (Adeyemi and Adekanle 2012). The parent drug released by esterase activity, norethisterone, is structurally highly related to the synthetic androgenic steroid nandrolone. Nandrolone undergoes rapid first-pass metabolism rendering it not orally bioavailable. Medical usage of nandrolone requires delivery as a prodrug through intramuscular injection (Wijnand et al. 1985). Norethisterone has previously been shown to have androgenic effects (Kuhl 2005). It has also demonstrated a decrease in circulating sex hormone-binding globulin (SHBG), consistent with androgenic effects (Kuhnz et al. 1997). High-dose side effects, such as hirsutism and voice changes, have also been seen, suggesting androgen activity (Jacobson 1962).
The natural product glycyrrhizin, the active component of licorice, was identified in this study as an activator of AR. This compound has multiple effects and is commonly used in traditional Asian medicine, such as an anti-inflammatory, antiviral, antitumor, immune system regulator, and can be used to treat certain symptoms of liver disease (Li et al. 2014). It has also been previously identified as a weak binder of AR (Tamaya et al. 1986). However, studies have been done which show licorice to modulate the metabolism and/or synthesis of androgen as well as imply glycyrrhizin inhibits the growth of androgen-dependent prostate cancer cells (Armanini et al. 2004; Hawthorne and Gallagher 2008). This conflicting data demonstrates the importance of further study of glycyrrhizin and how it affects the androgen pathway.
In conclusion, the two qHTS AR assays combined with confirmatory follow-up assays, including the binding assay and GFP-AR translocation study, enabled us to identify both known and potentially novel AR agonists. The AR-luc assay, which has the full length receptor, had a more robust assay performance, higher confirmation and reproducibility rates (87% confirmation and 0% mismatch rate) over the AR-bla assay (73% confirmation and 0.3% mismatch rate), better concordance with the binding assay, and higher accuracy in identifying known AR agonists. The AR-bla assay was also found to be less sensitive than the AR-luc assay. This may be due to additional coregulatory activity in the AR-luc full-length receptor that is lacking in the partial receptor (AR-bla). The AR cell line with the full-length receptor presents a more physiologically relevant condition compared to the AR cell line with only a partial receptor. Finally, as illustrated with several compounds characterized here that also had previously reported animal studies or human clinical data, adverse effects are a possibility but do not always follow. Further studies are needed on the novel compounds discovered to better understand their efficacy in vivo and potential for adverse effects.
Supplementary Material
Acknowledgements
This work was supported by the U.S. Environmental Protection Agency (Interagency Agreement #Y3-HG-7026–03) and the interagency agreement IAG #NTR 12003 from the National Institute of Environmental Health Sciences/Division of the National Toxicology Program to the National Center for Advancing Translational Sciences, National Institutes of Health. We also acknowledge the support of the NCI High Throughput Facility.
The views expressed in this paper are those of the authors and do not necessarily reflect the statements, opinions, views, conclusions, or policies of the U.S. EPA, the NIEHS, the National Center for Advancing Translational Sciences, the NIH, or the U.S. government. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
The authors declare they have no actual or potential competing financial interests.
References
- Adeyemi A and Adekanle D 2012. Progestogen-only injectable contraceptive: Experience of women in Osogbo, southwestern Nigeria. Annals of African Medicine 11, 27–31. [DOI] [PubMed] [Google Scholar]
- Armanini D, Mattarello MJ, Fiore C, Bonanni G, Scaroni C, Sartorato P and Palermo M 2004. Licorice reduces serum testosterone in healthy women. Steroids 69, 763–766. [DOI] [PubMed] [Google Scholar]
- Attene-Ramos MS, Miller N, Huang R, Michael S, Itkin M, Kavlock RJ, Austin CP, Shinn P, Simeonov A, Tice RR and Xia M 2013. The Tox21 robotic platform for the assessment of environmental chemicals – from vision to reality. Drug Discovery Today 18, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Lee SO, Yeh S and Chang TM 2014. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene 33, 3225–3234. [DOI] [PubMed] [Google Scholar]
- Cook JC, Klinefelter GR, Hardisty JF, Sharpe RM and Foster PMD 1999. Rodent Leydig Cell Tumorigenesis: A Review of the Physiology, Pathology, Mechanisms, and Relevance to Humans. Critical Reviews in Toxicology 29, 169–261. [DOI] [PubMed] [Google Scholar]
- Culig Z, Bartsch G and Hobisch A 2002. a. Interleukin-6 regulates androgen receptor activity and prostate cancer cell growth. Molecular and Cellular Endocrinology 197, 231–238. [DOI] [PubMed] [Google Scholar]
- Culig Z, Klocker H, Bartsch G and Hobisch A 2002b. Androgen receptors in prostate cancer. Endocrine-Related Cancer 9, 155–170. [DOI] [PubMed] [Google Scholar]
- Davey RA and Grossmann M 2016. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. The Clinical Biochemist Reviews 37, 3–15. [PMC free article] [PubMed] [Google Scholar]
- DeFranco D 1999. Regulation of steroid receptor subcellular trafficking. Cell Biochem Biophys 30, 1–24. [DOI] [PubMed] [Google Scholar]
- Fujimoto N, Inoue K, Yoshida M, Nishikawa A, Ozawa S, Gamou T, Nemoto K and Degawa M 2012. Estrogen and androgen receptor status in hepatocellular hypertrophy induced by phenobarbital, clofibrate, and piperonyl butoxide in F344 rats. The Journal of Toxicological Sciences 37, 281–286. [DOI] [PubMed] [Google Scholar]
- Handlon AL, Schaller LT, Leesnitzer LM, Merrihew RV, Poole C, Ulrich JC, Wilson JW, Cadilla R and Turnbull P 2016. Optimizing Ligand Efficiency of Selective Androgen Receptor Modulators (SARMs). ACS Medicinal Chemistry Letters 7, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne S and Gallagher S 2008. Effects of glycyrrhetinic acid and liquorice extract on cell proliferation and prostate-specific antigen secretion in LNCaP prostate cancer cells. Journal of Pharmacy and Pharmacology 60, 661–666. [DOI] [PubMed] [Google Scholar]
- Hsu C-W, Zhao J, Huang R, Hsieh J-H, Hamm J, Chang X, Houck K and Xia M 2014. Quantitative High-Throughput Profiling of Environmental Chemicals and Drugs that Modulate Farnesoid X Receptor. Scientific reports 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Sakamuru S, Martin MT, Reif DM, Judson RS, Houck KA, Casey W, Hsieh J-H, Shockley KR, Ceger P, Fostel J, Witt KL, Tong W, Rotroff DM, Zhao T, Shinn P, Simeonov A, Dix DJ, Austin CP, Kavlock RJ, Tice RR and Xia M 2014. Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Scientific reports 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Xia M, Cho M-H, Sakamuru S, Shinn P, Houck KA, Dix DJ, Judson RS, Witt KL, Kavlock RJ, Tice RR and Austin CP 2011. Chemical Genomics Profiling of Environmental Chemical Modulation of Human Nuclear Receptors. Environmental Health Perspectives 119, 1142–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Aihara K. i., Akaike M, Sato T, Ishikawa K, Ise T, Yagi S, Iwase T, Ueda Y, Yoshida S, Azuma H, Walsh K, Tamaki T, Kato S and Matsumoto T 2010. Androgen Receptor Counteracts Doxorubicin-Induced Cardiotoxicity in Male Mice. Molecular Endocrinology 24, 1338–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W and Austin CP 2006. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proceedings of the National Academy of Sciences of the United States of America 103, 11473–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson BD 1962. Hazards of norethindrone therapy during pregnancy. Am J Obstet Gynecol 84, 962–968. [PubMed] [Google Scholar]
- Karageorgopoulos DE, Maraki S, Vatopoulos AC, Samonis G, Schito GC and Falagas ME 2013. Antimicrobial activity of prulifloxacin in comparison with other fluoroquinolones against community-acquired urinary and respiratory pathogens isolated in Greece. European Journal of Clinical Microbiology & Infectious Diseases 32, 1417–1422. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer N, Ceger P, Browne P, Judson R, Allen D, Hamm J, and Casey W 2016. Development and Application of a Reference Database for Androgen Receptor Activity. 55th Annual Meeting of the Society of Toxicology, New Orleans, LA, USA. [Google Scholar]
- Klokk TI, Kurys P, Elbi C, Nagaich AK, Hendarwanto A, Slagsvold T, Chang C-Y, Hager GL and Saatcioglu F 2007. Ligand-Specific Dynamics of the Androgen Receptor at Its Response Element in Living Cells. Molecular and Cellular Biology 27, 1823–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen T 2006. Self-organizing neural projections. Neural Networks 19, 723–733. [DOI] [PubMed] [Google Scholar]
- Kokontis J, Takakura K, Hay N and Liao S 1994. Increased Androgen Receptor Activity and Altered c-myc Expression in Prostate Cancer Cells after Long-Term Androgen Deprivation. Cancer Research 54, 1566–1573. [PubMed] [Google Scholar]
- Kuhl H 2005. Pharmacology of estrogens and progestogens: influence of different routes of administration. Climacteric 1, 3–63. [DOI] [PubMed] [Google Scholar]
- Kuhnz W, Heuner A, Humpel M, Seifert W and Michaelis K 1997. In vivo conversion of norethisterone and norethisterone acetate to ethinyl etradiol in postmenopausal women. Contraception 56, 379–385. [DOI] [PubMed] [Google Scholar]
- Li J. y., Cao H. y., Liu P, Cheng G. h. and Sun M. y. 2014. Glycyrrhizic Acid in the Treatment of Liver Diseases: Literature Review. BioMed Research International 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga T, Fujii S, Furukawa S, Kikumori M, Yasuhira K, Shindo Y, Watanabe M and Sumi N 1996a. [Reproductive and developmental toxicity studies of prulifloxacin (NM441)(1)--A fertility study in rats by oral administration]. J Toxicol Sci 1, 171–185. [DOI] [PubMed] [Google Scholar]
- Morinaga T, Fujii S, Furukawa S, Kikumori M, Yasuhira K, Shindo Y, Watanabe M and Sumi N 1996b. [Reproductive and developmental toxicity studies of prulifloxacin (NM441)(3)--A teratogenicity study in rabbits by oral administration]. J Toxicol Sci 1, 207–217. [DOI] [PubMed] [Google Scholar]
- NCATS. (2016). “Tox21 Data Browser.” Retrieved November 17, 2016, from https://tripod.nih.gov/tox21.
- Pihlajamaa P, Sahu B and Jänne OA 2015. Determinants of Receptor- and Tissue-Specific Actions in Androgen Signaling. Endocrine Reviews 36, 357–384. [DOI] [PubMed] [Google Scholar]
- Prats G, Rossi V, Salvatori E and Mirelis B 2006. Prulifloxacin: a new antibacterial fluoroquinolone. Expert Review of Anti-infective Therapy 4, 27–41. [DOI] [PubMed] [Google Scholar]
- PubChem. (2013). “Tox21 Phase II compound collection.” Retrieved November 17, 2016, from https://www.ncbi.nlm.nih.gov/pcsubstance/?term=tox21.
- Ran F, Xing H, Liu Y, Zhang D, Li P and Zhao G 2015. Recent Developments in Androgen Receptor Antagonists. Archiv der Pharmazie 348, 757–775. [DOI] [PubMed] [Google Scholar]
- Roy AK, Tyagi RK, Song CS, Lavrovsky YAN, Ahn SC, Oh T-S and Chatterjee B 2001. Androgen Receptor: Structural Domains and Functional Dynamics after Ligand-Receptor Interaction. Annals of the New York Academy of Sciences 949, 44–57. [DOI] [PubMed] [Google Scholar]
- Shukla SJ, Sakamuru S, Huang R, Moeller TA, Shinn P, VanLeer D, Auld DS, Austin CP and Xia M 2011. Identification of Clinically Used Drugs That Activate Pregnane X Receptors. Drug Metabolism and Disposition 39, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skakkebæk A, Bojesen A, Kristensen MK, Cohen A, Hougaard DM, Hertz JM, Fedder J, Laurberg P, Wallentin M, Østergaard JR, Pedersen AD and Gravholt CH 2014. Neuropsychology and brain morphology in Klinefelter syndrome – the impact of genetics. Andrology 2, 632–640. [DOI] [PubMed] [Google Scholar]
- Stavreva DA, George AA, Klausmeyer P, Varticovski L, Sack D, Voss TC, Schiltz RL, Blazer VS, Iwanowicz LR and Hager GL 2012. Prevalent Glucocorticoid and Androgen Activity in US Water Sources. Scientific reports 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaya T, Sato S and Okada H 1986. Inhibition by plant herb extracts of steroid bindings in uterus, liver and serum of the rabbit. Acta Obstetricia et Gynecologica Scandinavica 65, 839–842. [DOI] [PubMed] [Google Scholar]
- Tanaka F, Katsuno M, Banno H, Suzuki K, Adachi H and Sobue G 2012. Current Status of Treatment of Spinal and Bulbar Muscular Atrophy. Neural Plasticity 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi RK, Lavrovsky Y, Ahn SC, Song CS, Chatterjee B and Roy AK 2000. Dynamics of Intracellular Movement and Nucleocytoplasmic Recycling of the Ligand-Activated Androgen Receptor in Living Cells. Molecular Endocrinology 14, 1162–1174. [DOI] [PubMed] [Google Scholar]
- Wang Y and Huang R 2016. Correction of Microplate Data from High-Throughput Screening In: Zhu H and Xia M (Eds), High-Throughput Screening Assays in Toxicology, Springer; New York, New York, NY, pp. 123–134. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jadhav A, Southal N, Huang R and Nguyen D-T 2010. A Grid Algorithm for High Throughput Fitting of Dose-Response Curve Data. Current Chemical Genomics 4, 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Chang H-C, Tian J, Shang Z, Niu Y and Chang C 2015. Stromal Androgen Receptor Roles in the Development of Normal Prostate, Benign Prostate Hyperplasia, and Prostate Cancer. The American Journal of Pathology 185, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnand HP, Bosch AM and Donker CW 1985. Pharmacokinetic parameters of nandrolone (19-nortestosterone) after intramuscular administration of nandrolone decanoate (Deca-Durabolin) to healthy volunteers. Acta Endocrinol Suppl 271, 19–30. [DOI] [PubMed] [Google Scholar]
- Wilson VS, Bobseine K, Lambright CR and Gray LE 2002. A Novel Cell Line, MDA-kb2, That Stably Expresses an Androgen- and Glucocorticoid-Responsive Reporter for the Detection of Hormone Receptor Agonists and Antagonists. Toxicological Sciences 66, 69–81. [DOI] [PubMed] [Google Scholar]
- Zhang J-H, Chung TDY and Oldenburg KR 1999. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening 4, 67–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


