Summary
To facilitate understanding of human cardiomyocyte (CM) subtype specification, and the study of ventricular CM biology in particular, we developed a broadly applicable strategy for enrichment of ventricular cardiomyocytes (VCMs) derived from human embryonic stem cells (hESCs). A bacterial artificial chromosome transgenic H9 hESC line in which GFP expression was driven by the human ventricular-specific myosin light chain 2 (MYL2) promoter was generated, and screened to identify cell-surface markers specific for MYL2-GFP-expressing VCMs. A CD77+/CD200− cell-surface signature facilitated isolation of >97% cardiac troponin I-positive cells from H9 hESC differentiation cultures, with 65% expressing MYL2-GFP. This study provides a tool for VCM enrichment when using some, but not all, human pluripotent stem cell lines. Tools generated in this study can be utilized toward understanding CM subtype specification, and enriching for VCMs for therapeutic applications.
Keywords: cardiac differentiation, human embryonic stem cells, ventricular cardiomyocytes, cell-surface marker signature
Highlights
-
•
Generation of an H9 MYL2-GFP-expressing reporter cell line for ventricular cardiomyocytes
-
•
Identified a cell-surface signature, CD77+/CD200− that enriches for hESC-derived VCMs
-
•
CD77+/CD200− enriches for VCMs for some, but not all, human stem cell lines
-
•
Tools generated in this study will advance understanding of human VCM biology
In this article, Evans and colleagues generated an H9 BAC transgenic reporter cell line and performed a flow cytometry screen to identify a cell-surface signature specific for MYL2-GFP-expressing VCMs. The cell-surface signature, CD77+/CD200−, facilitated isolation of a nearly pure hESC-derived CM population, enriched for VCMs. VCM enrichment was achieved when using some, but not all, human pluripotent stem cell lines. Tools generated in this study serve to advance our understanding of CM subtype specification, commitment, and maturation.
Introduction
Human embryonic stem cell (hESC)-derived cardiomyocytes (CMs) serve as an in vitro system to understand human CM lineage development, for cardiac disease modeling, drug discovery, toxicity, and regenerative medicine (Habib et al., 2008, Braam et al., 2009, Braam et al., 2010, Moretti et al., 2013). Existing differentiation protocols generate mixed cardiovascular (CM, smooth muscle cell, fibroblast, and endothelial cell) and CM (atrial, ventricular, and nodal) populations of varying yields (He et al., 2003, Yang et al., 2008, Kattman et al., 2011, Burridge et al., 2014), and potentially contain contaminating and undesired cell types that could markedly affect basic and clinical applications of hESC-derived CMs (Habib et al., 2008, Braam et al., 2009).
Methodologies have been developed that enrich for CMs or different CM subtypes (Mummery et al., 2012, Talkhabi et al., 2016). Previous studies have engineered hESC lines to express fluorescent reporters or antibiotic resistance elements driven by cardiac- or atrial- or ventricular-specific promoters to enrich for cardiac progenitors or CMs, or CM subtypes by fluorescence-activated cell sorting (FACS) or drug selection (Bernstein and Hyun, 2012, Den Hartogh and Passier, 2016). However, a major drawback of this approach is that genetic manipulation of hESCs precludes use of derivatives in downstream clinical applications. To overcome this, some cell-surface markers for human CMs have been identified, including SIRPA (signal-regulatory protein-α/CD172a) (Dubois et al., 2011, Elliott et al., 2011) and VCAM1 (vascular cell adhesion molecule 1/CD106) (Elliott et al., 2011, Uosaki et al., 2011), which distinguish stem cell-derived CMs from non-CMs using flow cytometry. These proteins, however, are not exclusively expressed by CMs, and are only useful for identifying CMs at certain stages of differentiation. Although progress has been made in directing CMs toward a specific phenotype (Zhang et al., 2011, Karakikes et al., 2014), cell-surface markers suitable for sorting subpopulations of CMs have not yet been established.
Here, we identified a CD77+/CD200− cell-surface signature that can be utilized to enrich for hESC-derived ventricular cardiomyocytes (VCMs). We generated a transgenic H9 hESC reporter line in which GFP expression was driven by ventricular-specific myosin light chain 2 (MYL2) (Chuva de Sousa Lopes et al., 2006) regulatory sequences (promoter/enhancers) derived from a MYL2 bacterial artificial chromosome (BAC), and performed a flow cytometry screen. MYL2-GFP-expressing cells (and CD77+/CD200−-sorted populations) displayed structural, molecular, and functional properties of VCMs.
Results
Generation of an H9 MYL2-GFP BAC Transgenic Reporter Cell Line
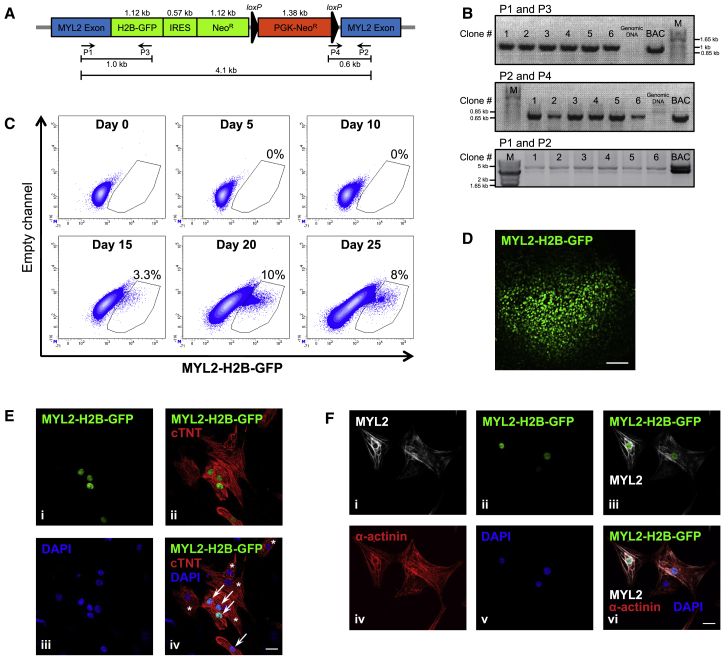
An H9 hESC BAC transgenic reporter cell line was generated by introducing a targeting construct containing a histone2B-GFP-IRES-neomycin resistance gene cassette (H2B-GFP-IRES-NeoR) integrated in-frame to the ATG start site of the cardiac ventricle-specific human MYL2 gene, encoding ventricular MYL2 (Figure 1A). An additional PGK-neomycin resistance (PGK-NeoR) gene cassette enabled selection of positive clones by G418 antibiotic treatment following electroporation of the BAC targeting vector into wild-type H9 hESCs. Based on the limited activity of a short MYL2 promoter (Huber et al., 2007, Bizy et al., 2013), a BAC was used so that GFP expression might more closely mimic that of endogenous MYL2. Genomic integration of the BAC construct in G418-resistent clones was verified by PCR (Figure 1B). Pluripotency of each transgenic clone was confirmed by immunofluorescence and flow cytometric analysis of intracellular and cell-surface stem cell markers, respectively (Figures S1A and S1B). Karyotype analyses indicated normal diploid chromosomes (Figure S1C).
Figure 1.
Generation of an H9 MYL2-GFP BAC Transgenic Reporter Cell Line
(A) A schematic representation of the BAC targeting vector containing: a histone2B-GFP-IRES-neomycin resistance gene cassette (H2B-GFP-IRES-NeoR) integrated in-frame to the ATG start site of the cardiac ventricle-specific human MYL2 gene, and a PGK-neomycin resistance (PGK-NeoR) gene cassette encoding G418 resistance flanked by loxP sites (black triangles). The predicted sizes of the PCR products using primers (P) 1–4 to detect genomic integration are indicated.
(B) RT-PCR amplification of a 1-kb band (P1 and P3) and a 0.6-kb band (P2 and P4), and long-range PCR amplification of a specific 4.1-kb band (P1 and P2) confirmed BAC construct integration in G418-resistent clones nos 1–6. BAC DNA was used as a positive control. Genomic DNA from non-transfected wild-type H9 hESCs was used as a negative control. M, marker. See Figure S1 for verification of H9 MYL2-GFP BAC reporter line pluripotency and cardiac differentiation capacity.
(C) Flow cytometric quantification of MYL2-H2B-GFP expression over the time course of embryoid body cardiac differentiation (day 0–25). Numbers indicate the percentage of GFP+ cells (see Figure S2A for cell gating strategy). Data are representative of a minimum of three independent clones.
(D) Immunofluorescence analysis (MYL2-H2B-GFP) of a day 25 embryoid body derived from H9 MYL2-GFP hESCs. Scale bar, 100 μm. The image is representative of a minimum of three independent clones.
(E) Immunofluorescence analysis of hESC-derived cardiomyocytes after 25 days of cardiac differentiation. MYL2-H2B-GFP-positive(+) cells (i) were positive for cardiac troponin T (cTNT) (ii). (iii) DNA stained with DAPI. (iv) Merged image. Arrows point to MYL2-H2B-GFP+/cTNT+ cardiomyocytes. Asterisks indicate cTNT+ cardiomyocytes not expressing MYL2-H2B-GFP. Scale bar, 20 μm. Images are representative of a minimum of three independent clones.
(F) Immunostaining for MYL2 (i) shows exclusive expression in MYL2-H2B-GFP+ hESC-derived cardiomyocytes after 25 days of cardiac differentiation (ii and iii). MYL2-H2B-GFP+/MYL2+ cardiomyocytes were positive for α-actinin (vi), but not all α-actinin+ cells (iv) were MYL2+ cardiomyoctes. (v) DNA stained with DAPI. Scale bar, 20 μm. Images are representative of a minimum of three independent clones.
To monitor ventricular specification, H9 MYL2-GFP hESCs were differentiated into CMs as embryoid bodies (EBs) (Noseda et al., 2011) (Figure S1D). Beating EBs appeared between days 7 and 10 of cardiac differentiation, coincident with the upregulation of the cardiac-specific genes: cardiac troponin I (TNNI3), cardiac troponin T (TNNT2), and the MYL2 atrial isoform (MYL7) (Figure S1E). Expression of endogenous MYL2 appeared around day 15 of differentiation (Figure S1E), which closely paralleled MYL2-H2B-GFP-positive (MYL2(-H2B)-GFP+) detection by flow cytometry (Figure 1C), and nuclear MYL2-H2B-GFP epifluorescence (Figures 1D–1F; Video S1). The proportion of the MYL2-GFP+ population increased over time, reaching a variable maximum between days 20 and 25 (Figures 1C and S2A). Regardless of cardiac differentiation efficiency, the proportion of CMs expressing MYL2-GFP was similar (Figure S1F). Immunofluorescence analysis of cell populations derived from dissociated EBs after 25 days of cardiac differentiation demonstrated the presence of both cardiac troponin T-positive (cTNT+)/MYL2-H2B-GFP+ and cTNT+/MYL2-H2B-GFP-negative (MYL2-GFP−) populations (Figure 1E). Immunostaining for MYL2 revealed that expression was exclusive to MYL2-H2B-GFP+/sarcomeric α-actinin-positive CMs (Figure 1F).
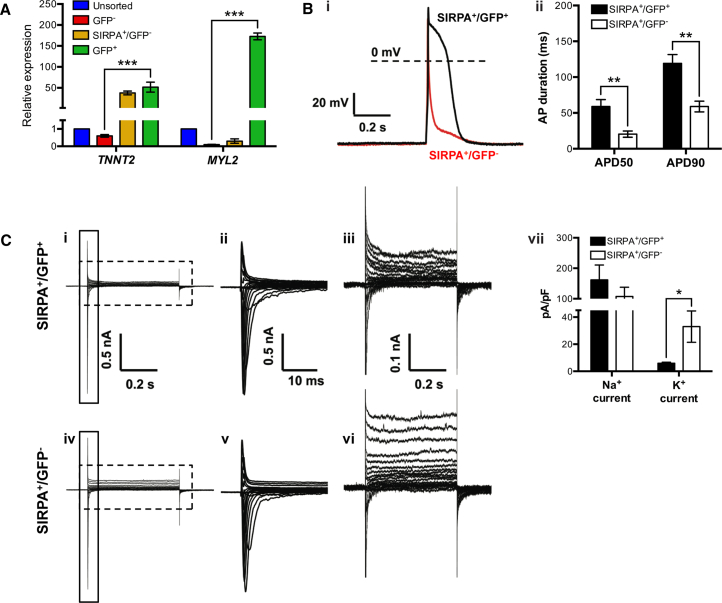
To further characterize the H9 MYL2-GFP reporter cell line, we purified MYL2-GFP+ cells by FACS to assess ventricular phenotype (Figure 2). As cardiac differentiation efficiency varied between experiments, we enriched for CMs by SIRPA-bright (SIRPA+) selection (Figure S2B) (Dubois et al., 2011). Cell-sorting gates were conservatively chosen based on corresponding isotype control and/or H9 parental cells at the same stage of cardiac differentiation (Figure S2). qRT-PCR analysis showed that day 25 MYL2-GFP+-sorted cells were significantly enriched for both TNNT2 (cTNT) and MYL2 compared with MYL2-GFP− cells (Figure 2A). Electrophysiological assessment revealed action potentials from SIRPA+/MYL2-GFP+ and SIRPA+/MYL2-GFP− cells to be characteristic of ventricular and atrial CMs, respectively (Figure 2B). Furthermore, differences in inward sodium (Na+) and outward potassium (K+) currents in SIRPA+/MYL2-GFP+ cells compared with SIRPA+/MYL2-GFP− cells, as measured by whole-cell voltage clamp (Figure 2C), were consistent with previous observations in ventricular and atrial CMs isolated from human myocardium (Varro et al., 1993).
Figure 2.
Characterization of H9 MYL2-GFP hESC-Derived Ventricular Cardiomyocytes
(A) qRT-PCR analysis of TNNT2 and MYL2 gene expression in unsorted, MYL2-GFP-negative (GFP−)-, SIRPA-bright/MYL2-GFP-negative (SIRPA+/GFP−)-, and MYL2-GFP-positive (GFP+)-sorted fractions of dissociated hESC-derived embryoid bodies at day 25 of cardiac differentiation (see Figure S2 for cell gating strategy). Data were normalized to corresponding ACTB expression, and are relative to the unsorted hESC-derived cell population. Error bars, mean ± SD. ∗∗∗p ≤ 0.001 by Student's t test (n = 3 technical replicates). Data are representative of a minimum of three independent biological replicates.
(B) Electrophysiological characterization of SIRPA+/GFP+- and SIRPA+/GFP−-sorted fractions of dissociated hESC-derived embryoid bodies at days 25–40 of cardiac differentiation (see Figure S2B for cell gating strategy). (i) Representative triggered action potential (AP) traces recorded from SIRPA+/GFP+ and SIRPA+/GFP− cardiomyocytes using whole-cell current-clamp technique. (ii) Quantification of AP duration (APD) at different times of repolarization (50% and 90%) for SIRPA+/GFP+ (n = 21) and SIRPA+/GFP− (n = 18) cardiomyocytes. Error bars, mean ± SEM. ∗∗p ≤ 0.01 by Student's t test. mV, millivolts; s, seconds. Data are representative of a minimum of three independent biological replicates.
(C) Representative whole-cell currents recorded from (i) SIRPA+/GFP+ (upper panel) and (iv) SIRPA+/GFP− (lower panel) cells, with an enlarged view of the solid boxed area (ii and v), and the dashed boxed area (iii and vi) to the right of the corresponding trace. (vii) Quantification of maximum inward Na+ currents, and maximum sustained K+ currents of SIRPA+/GFP+ (n = 21) and SIRPA+/GFP− (n = 18) cardiomyocytes. Error bars, mean ± SEM. ∗p ≤ 0.05 by Student's t test. Data are representative of a minimum of three independent biological replicates. nA, nanoampere; s, seconds; ms, milliseconds; pA/pF, picoampere per picofarad.
Taken together, these data document generation of an H9 MYL2-GFP BAC transgenic reporter hESC line that allowed for identification and purification of hESC-derived ventricular-like CMs based on MYL2 expression.
Human ESC-Derived VCMs Express a Specific Cell-Surface Marker Signature
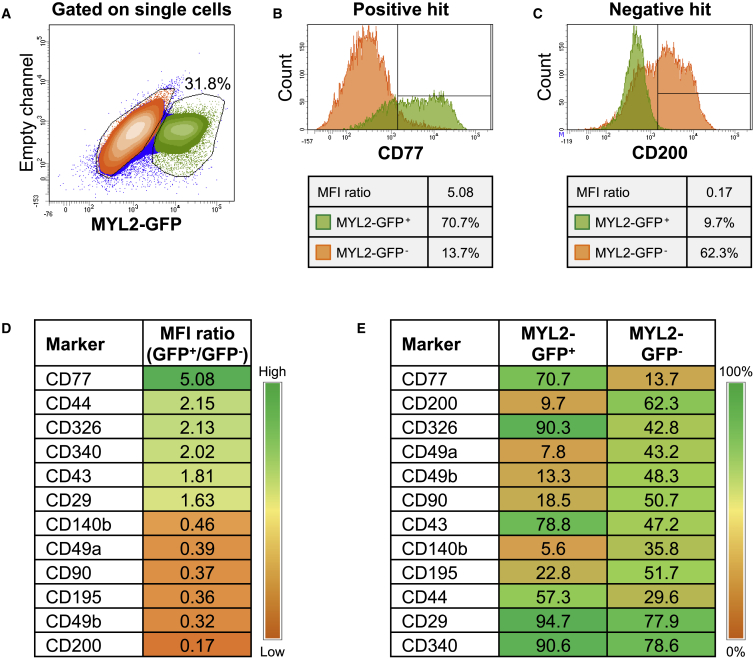
To identify a cell-surface marker signature that could specifically identify hESC-derived VCMs, we performed an unbiased flow cytometry-based screen using a panel of 242 known anti-human monoclonal antibodies on cell populations generated from the H9 MYL2-GFP reporter cell line after 25 days of cardiac differentiation in an EB culture system. Screening allowed for simultaneous interrogation of both MYL2-GFP+ and MYL2-GFP− populations for expression of 242 surface makers (Figure 3A).
Figure 3.
Identification of Positive and Negative Cell-Surface Markers Specific to hESC-Derived Ventricular Cardiomyocytes
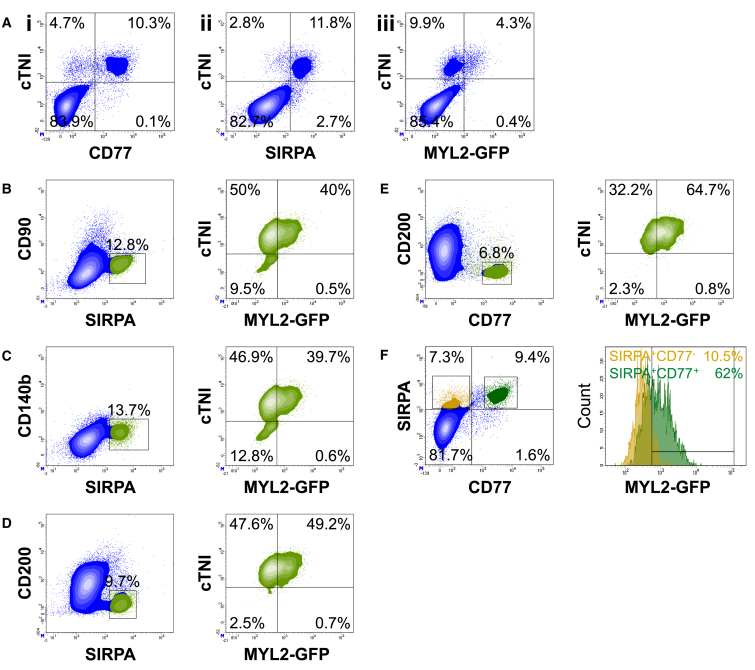
(A) After exclusion of debris and doublets based on light scatter properties, single cells were analyzed for the expression of MYL2-GFP. The gating strategy for the detection of MYL2-GFP+ cells was defined using parental H9 cells as a control (see Figure S2A). GFP+ and GFP− populations, represented in green and orange, respectively, were interrogated for the expression of each one of the 242 surface markers included in the BD Lyoplate screening panel.
(B and C) Representative example of positive and negative hits, (B) CD77 and (C) CD200, identified based on median fluorescence intensity (MFI) ratio and differential frequency (%) within GFP+ and GFP− subpopulations. See Figure S3 for other potential marker candidates.
(D and E) Representative heatmaps depicting the highest-ranked positive or negative hits based on (D) MFI ratio or (E) differential frequency (%) within GFP+ and GFP− populations (see Table S1 for heatmap of complete list of 242 markers).
To define positive and negative hits, markers were ranked based on expression intensity and differential frequency within MYL2-GFP+ and MYL2-GFP− populations (Figures 3B–3E). To identify markers enriching for MYL2-GFP+ cells (positive hit) or MYL2-GFP− cells (negative hit), data were expressed as a ratio of median fluorescence intensity (MFI), whereby the MFI value of MYL2-GFP+ cells stained with an individual antibody, and detected with an Alexa Fluor 647 (AF647)-conjugated second-step antibody, was quantified in relation to the MFI value of MYL2-GFP− cells in each individual antibody well (Figure 3D). A high MFI ratio indicated a positive hit, whereas a low ratio suggested a negative hit. The full heatmap with the MFI ratio for each antibody tested is reported in Table S1. We furthermore considered the differential expression of cell-surface markers in terms of frequency within the MYL2-GFP+ and MYL2-GFP− populations (Figure 3E) to narrow down the list of potential hits. For example, CD340 had a relatively high MFI ratio of 2.02; however, the differential expression was poor (Figures 3D, 3E, and S3). Although 90.6% of all MYL2-GFP+ cells were found to express CD340, it was also detected in 78.6% of MYL2-GFP− cells.
Taking both intensity and differential expression into consideration, we identified CD77 (also known as globotriaosylceramide; Gb3), as a potential VCM-specific marker (Figure 3B). With an MFI ratio of 5.08, CD77 also had a high differential expression pattern, with 70.7% of the MYL2-GFP+, and only 13.7% of the MYL2-GFP− population expressing the cell-surface marker. From the screen, we also identified CD200 (also known as OX-2 membrane glycoprotein) as a negative marker of hESC-derived VCMs (Figure 3C). Less than 10% of MYL2-GFP+ cells were shown to express CD200, whereas over 60% of MYL2-GFP− cells stained positively for this marker, which also had the lowest MFI ratio of 0.17 of all candidate cell-surface marker hits. As an internal control to demonstrate the accuracy of the screen in defining a cell-surface signature, antibodies including anti-CD90 (THY1 [Kisselbach et al., 2009]), and anti-CD140b (PDGFR-β [Lindahl et al., 1997]), did not mark MYL2-GFP+ cells (Figure S3). Thus, hESC-derived VCMs could be identified based on the positive and negative expression of select cell-surface markers.
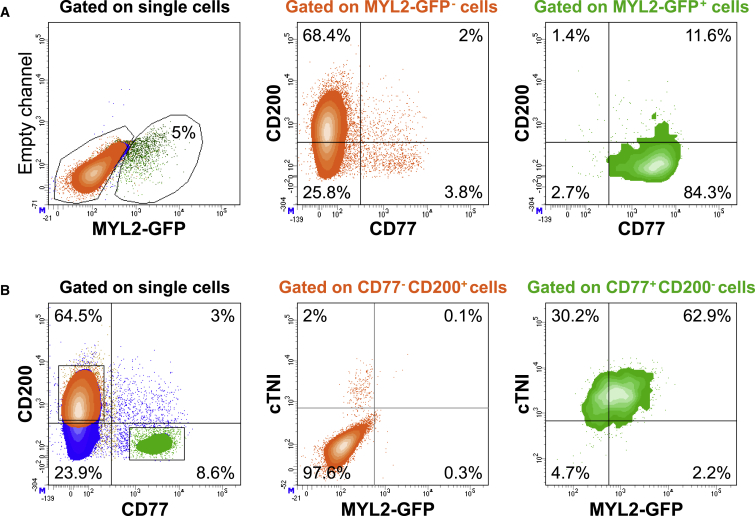
Validation of hESC-Derived VCM Cell-Surface Marker Signature
We postulated that the combination of CD77-positive (CD77+) and CD200-negative (CD200−) selection could be used as a cell-surface signature for enriching a population of hESC-derived VCMs from a heterogeneous cell population. Multi-color antibody panels were designed and validated to optimize signal resolution (data not shown). When gating on the MYL2-GFP− EB-derived population at day 25 of differentiation, the majority of cells resided within the CD77−/CD200+ subpopulation (Figure 4A). It is unclear whether the very low numbers of CD77+ cells present in the MYL2-GFP− fraction represent true CD77+ non-VCMs, or VCMs yet to express MYL2. The majority of the MYL2-GFP+ gated population, however, were CD77+/CD200−, with >95% of MYL2-GFP+ VCMs expressing CD77 (Figure 4A), thus validating the hits identified from the screen.
Figure 4.
Validation of hESC-Derived Ventricular Cardiomyocyte Cell-Surface Marker Signature
Multi-color flow cytometry was used to simultaneously analyze the expression of surface markers CD77 and CD200, the intracellular cardiomyocyte marker, cardiac troponin I (cTNI), and GFP at day 25 of cardiac differentiation. Data are representative of a minimum of three independent biological replicates.
(A) MYL2-GFP-positive (green) and -negative (orange) populations were defined within single cells, after exclusion of doublets and debris (left panel). The simultaneous expression of CD200 and CD77 was analyzed within the GFP− population (middle panel, orange) and the GFP+ population (right panel, green). See Figure S4 for co-expression of CD77 or CD200 with other marker candidates.
(B) CD77 and CD200 expression was first analyzed within the single-cell population (left panel). The expression of cTNI and MYL2-GFP was then analyzed within the CD77−/CD200+ population (middle panel, orange) and CD77+/CD200− population (right panel, green).
When using the reverse strategy, gating on the CD77−/CD200+ population, >97% of cells were negative for cardiac troponin I (cTNI), and virtually no CD77−/CD200+ cells were MYL2-GFP+ (Figure 4B). Conversely, when gating for CD77+/CD200−, 95% ± 2.7% of cells were cTNI+, and 63.8% ± 1.3% were cTNI+/MYL2-GFP+ VCMs (Figures 4B and 5E), providing further validation for CD77+/CD200− as a cell-surface marker signature of hESC-derived CMs. The minor proportion of CD77+/CD200− cells that were not MYL2-GFP+, might be more immature VCMs not yet expressing MYL2, or represent other CM lineages. Co-expression of other positive hits from the cell-surface marker screen was also evaluated (Figure S4A); however, using additional positive markers for the cell-surface signature did not improve enrichment of the MYL2-GFP+ VCM population (data not shown).
Figure 5.
CD77 Expression Highly Correlates with hESC-Derived Ventricular Cardiomyocyte Phenotype
All analysis is of the same population. Data are representative of a minimum of three independent biological replicates.
(A) Correlation of cTNI and (i) CD77, (ii) SIRPA, or (iii) MYL2-GFP in hESC-derived populations at day 25 of cardiac differentiation by flow cytometry.
(B–E) Representative plots depicting cTNI/MYL2-GFP cell populations (right panel, green plots) at day 25 of cardiac differentiation, after gating on different cell populations defined by distinct surface marker signatures: (B) SIRPA+/CD90−; (C) SIRPA+/CD140b−; (D) SIRPA+/CD200−; and (E) CD77+/CD200− (left panel, boxed area).
(F) Left panel: SIRPA+/CD77− (yellow) and SIRPA+/CD77+ (green) cell populations were defined within single cells. Right panel: histograms depicting MYL2-GFP expression within SIRPA+/CD77− and SIRPA+/CD77+ cell populations.
Expression of CD77 was mutually exclusive of cell-surface markers negatively expressed by MYL2-GFP+ cells (Figure S4B). These markers, which include CD31, CD49a, CD49b, CD90, CD140b, CD141, and epidermal growth factor receptor, are expressed by endothelial cells, fibroblast cells, smooth muscle cells, and other non-CMs that correlated with CD200 expression (Figure S4B). These data demonstrated that CD77+/CD200− could be used to isolate a nearly pure CM population enriched for VCMs.
Enrichment of VCMs from hESC-Derived Populations with a Cell-Surface Signature
Co-staining of CD77 and cTNI by flow cytometry established a strong correlation of the two markers (Figure 5Ai), confirming that CD77 was specifically expressed within the CM lineage in differentiated populations generated from hESCs. SIRPA also displayed clear co-expression with cTNI in our cultures at day 25 of cardiac differentiation (Figure 5Aii). To determine whether the expression of CD77 on the cell surface of hESC-derived CMs can be used to enrich for VCMs, combinations of known CM marker signatures were compared together with cell-surface markers identified in this study (Figures 5B–5F). Use of the H9 MYL2-GFP reporter cell line allowed for the interrogation of VCM enrichment in populations gated blindly based upon the expression of positive and negative cell-surface markers. Importantly, virtually all MYL2-GFP+ cells (4.3% of the total starting cell population) co-expressed cTNI, and made up ∼30% of the cTNI+-CM population in the differentiation culture (Figure 5Aiii). Cells expressing the previously identified stem cell-derived CM marker, SIRPA (Dubois et al., 2011), not included in the Lyoplate Human Cell Surface Marker Screening Panel, were positively selected for while excluding cell types expressing CD90 (Figure 5B) or CD140b (Figure 5C), surface markers specific for non-CMs (Lindahl et al., 1997, Kisselbach et al., 2009, Dubois et al., 2011). Gating for SIRPA+/CD90− and SIRPA+/CD140b− subpopulations resulted in 90% and 86.6% of cells staining positive for cTNI by intracellular flow cytometry, respectively, with up to 40% of cells expressing MYL2-GFP (Figures 5B and 5C). Enrichment of MYL2-GFP+ cells was improved using CD200 as a negative selection marker, with ∼50% of the 96.8% cTNI+-CM-enriched SIRPA+/CD200− population expressing MYL2-GFP (Figure 5D).
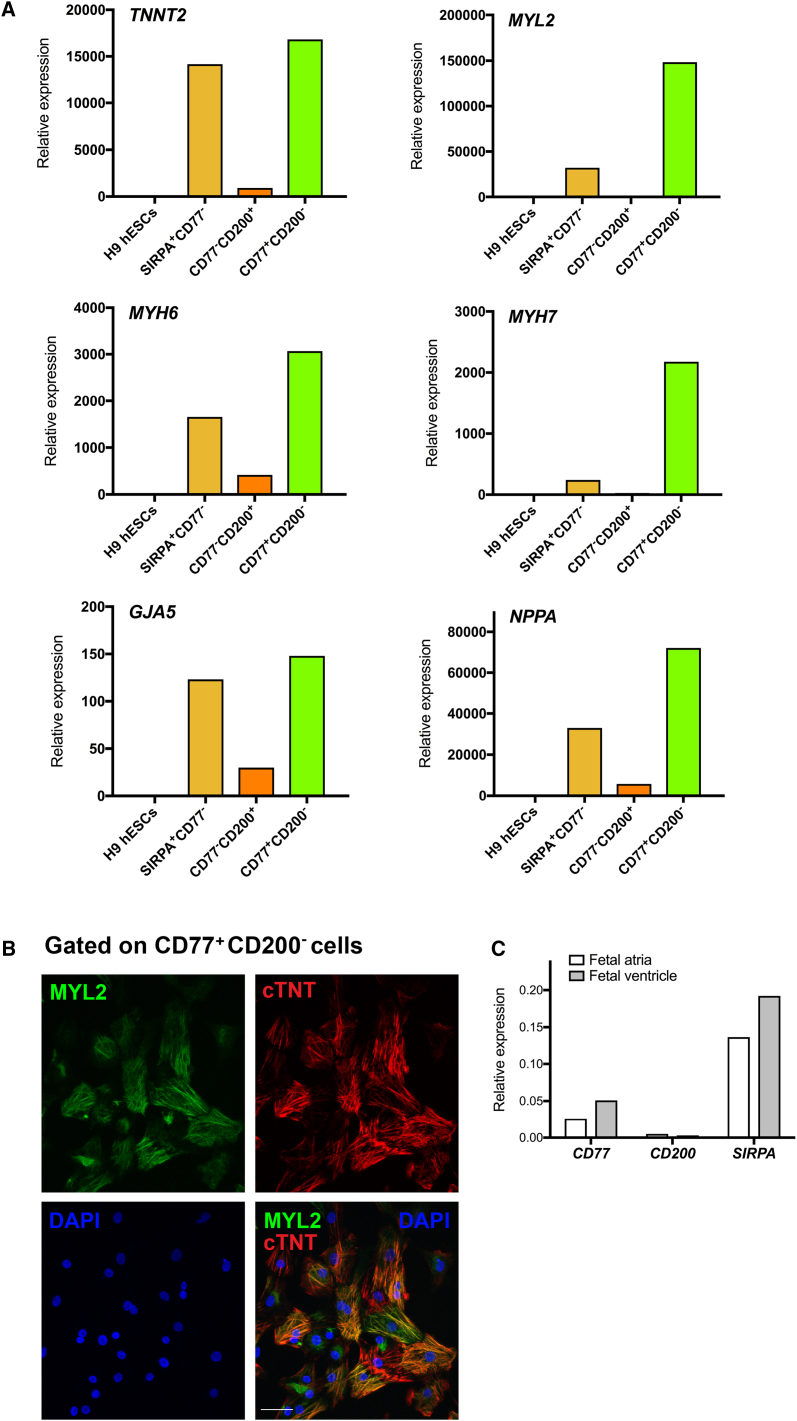
However, when selecting for CD77+ cells together with CD200− cells, MYL2-GFP+ VCMs were enriched from 4.3% in the starting total cell population to 64.7% in the CD77+/CD200− gated population (Figures 5Aiii and 5E). The CD77+/CD200− cell-surface marker signature enriched for a nearly pure cTNI+ CM population (Figure 5E). Furthermore, the CD77+ population was a subset of SIRPA+ cells selectively enriched for MYL2-GFP+ VCMs (Figure 5F). These data demonstrated that, although SIRPA allows for the isolation of populations highly enriched for CMs, CD77 further enriched for VCMs. For confirmation, molecular analyses revealed that CD77+/CD200− cells sorted following 25 days of cardiac differentiation expressed an extensive panel of cardiac markers, including TNNT2, MYL2, MYH6, MYH7, GJA5, and NPPA (Figure 6A). To ensure that the cell-surface signature was effective not only in our MYL2-H2B-GFP transgenic H9 cell line, but also for parental H9 cells, we performed immunocytochemical analysis of parental H9 hESC-derived CD77+/CD200−-sorted cells with an anti-MYL2 antibody after 25 days of EB cardiac differentiation, and further verified the high proportion of VCMs in these cultures (Figure 6B). CD77 also discriminated between atrial and ventricular cells in human hearts, as CD77 expression was significantly higher in human fetal ventricular tissue than atrial (Figure 6C).
Figure 6.
Validation of Ventricular Cardiomyocyte Enrichment from hESC-Derived Populations with a Cell-Surface Signature
(A) qRT-PCR analysis of TNNT2, MYL2, MYH6, MYH7, GJA5, and NPPA gene expression in SIRPA+/CD77−-, CD77−/CD200+-, and CD77+/CD200−-sorted fractions of dissociated hESC-derived embryoid bodies at day 25 of cardiac differentiation. Data were normalized to corresponding ACTB expression, and are relative to H9 hESC-derived embryoid bodies at day 0. Data are representative of a minimum of three independent biological replicates.
(B) Immunofluorescence analysis of H9 hESC-derived CD77+/CD200−-sorted cells after 25 days of cardiac differentiation. Cardiomyocytes were stained with cTNT. DNA was stained with DAPI. Scale bar, 50 μm. Images are representative of a minimum of three independent biological replicates.
(C) qRT-PCR analysis of CD77, CD200, and SIRPA gene expression in human 15-week fetal atrial and ventricular tissue. Data were normalized to corresponding GAPDH expression.
To determine whether the cell-surface marker signature identified in this study could be accurately used to define a stage of cardiac differentiation, we monitored expression of CD77, CD200, SIRPA, and CD90 over the time course of differentiation (Figure S5). The expression pattern of the cell-surface signature (CD77+/CD200−/SIRPA+/CD90−) showed a strong correlation with the emergence of MYL2-GFP+ VCMs (Figure S5A). We also compared the use of the cell-surface protein VCAM1 with our CD77+/CD200− cell-surface signature in the identification of CMs. Antibodies to VCAM1 are effective at identifying CMs from cardiac differentiation cultures from days 9 to 14 (Elliott et al., 2011, Uosaki et al., 2011). We performed EB cardiac differentiation of the parental H9 cell line and harvested cells for FACS at day 12 of differentiation. Subsequent immunocytochemical analysis of VCAM1-sorted cells demonstrated that antibodies to VCAM1 were able to yield populations of 92.4% ± 11.9% (n = 5 fields of view/n = 4 biological replicates) cTNT+ cells (Figure S5E). At day 12 of differentiation, MYL2+ cells were not observed, consistent with above studies showing that MYL2 gene and protein expression was first evident around day 15 of differentiation (Figures S1 and 1C). These results were in contrast to day 25 of differentiation, when no VCAM1 expression was detectable on cell populations generated from the H9 MYL2-GFP reporter cell line (Table S1). These results confirmed previous findings that VCAM1 is effective for identifying CMs at relatively early stages of cardiac differentiation (Elliott et al., 2011, Uosaki et al., 2011), emphasizing the use of the CD77+/CD200− cell-surface signature for enrichment of differentiated VCMs.
To evaluate the robustness of CD77 as a cell-surface marker to enrich for VCMs in other hESC lines, we analyzed both RUES2- and H1 ESC-derived CMs for MYL2 expression, both before and after sorting for CD77+/CD200−. Following 25 days of EB cardiac differentiation, selecting for CD77+/CD200− cells resulted in a significant fold enrichment of ventricular (MYL2-expressing) CMs in both RUES2 and H1 hESC lines (Figure S6A). We further analyzed expression of CD77 in CMs derived from two independent human wild-type induced pluripotent stem cell (iPSC) lines by flow cytometric analysis (Figure S6B). However, putative CM populations in both iPSC lines exhibited little to no CD77 expression, for reasons that are not clear, but may reflect negligible presence of VCMs in the tested cultures.
We also investigated whether the CD77+/CD200− cell-surface signature would be effective for enrichment of VCMs derived from monolayer cultures differentiated according to a two-dimensional monolayer protocol (Burridge et al., 2014). Using this protocol, expression of the MYL2-H2B-GFP transgene occurred at approximately day 40 of differentiation (data not shown), much later than observed with the EB protocol (Figure 1C). Accordingly, cells were cultured until day 60 (when MYL2-H2B-GFP+ cells were widely evident). Sorting for CD77+/CD200− resulted in a 2- to 5-fold enrichment of VCMs from monolayer cultures (Figure S6C), somewhat less than that obtained from EB cultures.
Together, the findings from these studies demonstrated the use of cell sorting with the cell-surface marker signature CD77+/CD200− for enrichment of VCMs when using some, but not all, PSC lines.
Discussion
Despite continuous improvements in efficiency and reproducibility, current differentiation methodologies generate heterogeneous populations of cardiovascular cells and/or CMs with varying degrees of maturity, necessitating further refinements to maximize use of hESC-derived CMs in certain basic and clinical applications (Habib et al., 2008, Braam et al., 2009).
FACS is the method of choice when very high purity of a desired cell population is required, when the target cell population expresses a very low level of the identifying surface marker, or when cell populations require separation based on differential marker frequency (Basu et al., 2010). Using an H9 MYL2-GFP BAC transgenic reporter cell line, we screened a panel of >240 known commercially available antibodies and identified CD77 as a surface protein expressed on hESC-derived VCMs. CD77, also known as Gb3, is a glycolipid characteristically expressed on Burkitt lymphoma cells and a subset of B lymphocytes (Nudelman et al., 1983, Mangeney et al., 1991). Interestingly, the gene encoding the enzyme responsible for the synthesis of CD77/Gb3, CD77/Gb3 synthase (Gb3S or α1,4-galactosyltransferase), is strongly expressed in normal human heart tissue (Kojima et al., 2000).
Using the cell-surface marker signature, CD77+/CD200−, we isolated populations consisting of >97% CMs from differentiating hESCs, with 65% being VCMs. CM identity was verified by flow cytometry, immunocytochemistry, and qRT-PCR. Furthermore, CD77+/CD200− selection enabled CM enrichment of >97% (65% MYL2-GFP+) from a cell source comprising only ∼15% cTNI+ CMs (∼4% MYL2-GFP+). Regardless of differentiation efficiency, the CD77+/CD200− population always strongly correlated with the MYL2-GFP+ population, highlighting the use of CD77+/CD200− selection in enriching for VCMs, even in the setting of low differentiation efficiencies. Enrichment was effective using both EB and monolayer cardiac differentiation protocols.
Another advantage to using the cell-surface marker signature CD77+/CD200− to isolate a nearly pure CM population enriched for ventricular cells is the ease of gating the population for cell-sorting applications. We found SIRPA expression to be a continuum, which made accurate gating of a true CM population a challenge without the use of additional negative markers or by fixing and performing intracellular cTNI staining. Even so, gating for SIRPA+/CD90− or SIRPA+/CD140b− populations led to the collection of some (10%–13%) non-CM contaminants, and the overall population of cTNI+ cells contained more MYL2-GFP− than MYL2-GFP+ cells. Using CD200 as a negative selection marker together with SIRPA allowed for easy gating of a more highly enriched pan-CM population. However, staining for CD77 and CD200 expression allowed for robust gating of a clearly separated CD77+/CD200− population rich in CMs of ventricular phenotype, when compared with other known cell-surface marker combinations. Enrichment for VCMs using CD77+/CD200− selection was effective not only in our MYL2-H2B-GFP transgenic H9 cell line, but also for parental H9 cells, and RUES2 and H1 hESC lines. One important caveat is that in contrast to our results using the hESC lines (RUES2, H1, and H9), flow cytometric analysis of CD77 expression in putative CM (SIRPA+/CD200−) populations derived from two human wild-type iPSC lines exhibited little to no CD77 expression, although CD77 was expressed in human fetal ventricular tissue. Future analysis of other hESC and iPSC lines and/or use of RNA probes to test CD77 expression in these lines will further address the use of CD77 as a marker.
In summary, identification of CD77 as a CM marker will facilitate access to highly enriched populations of VCMs derived from hESCs to enable development of high-throughput pharmacological assays, and generation of defined populations for regenerative therapies. Moreover, tools generated in this study could serve to improve and standardize cardiac differentiation methodologies that specifically enrich for VCMs, and advance our understanding of CM subtype specification, commitment, and maturation.
Experimental Procedures
Generation of the H9 MYL2-GFP BAC Transgenic Reporter Cell Line
The reporter construct, encoding two transcriptional units, a 1.12-kb GFP cDNA, 570-bp IRES promoter, and a 1.12-kb NeoR gene cassette, and a 1.38-kb PGK promoter and a NeoR gene cassette, were generated using standard cloning techniques, and was recombineered into a human BAC vector. The GFP cDNA was integrated in-frame to the ATG state site of the endogenous MYL2 locus. BAC DNA was purified using a NucleoBond Xtra Maxi endotoxin-free kit (Macherey-Nagel) according to the manufacturer's instructions. Electroporation was performed as described previously (Costa et al., 2007), with modifications as detailed. In brief, H9 (WA09) hESCs (WiCell Research Institute) were cultured on BD Matrigel (BD Biosciences) matrix-coated tissue culture plates to 70% confluence. Cells were harvested using 0.05% trypsin-EDTA (Thermo Fisher Scientific) and resuspended in hESC maintenance medium supplemented with 10 mM Y-27632 (Sigma-Aldrich). Purified BAC DNA (50 μg) was added to 1 × 107 hESCs in a 4-mm cuvette, cells were electroporated using the parameters 320 V and 200 μF (Gene Pulser Xcell, Bio-Rad), and immediately transferred to Matrigel-coated plates in hESC maintenance medium with 10 mM Y-27632. Medium was replaced daily and G418 (Corning) selection was applied 3 days after electroporation (2 days at 50 μg/mL, 2 days at 100 μg/mL, and 150 μg/mL thereafter). Surviving clones were picked, expanded, and verified by RT-PCR (Figure 1B). Of the six clones shown, nos 1–3 were used predominantly throughout the study. The data presented throughout the manuscript is representative of a minimum of three independent H9 MYL2 clones. Further, upon excision of the PGK-NeoR gene cassette, no difference in MYL2-GFP expression was observed (data not shown).
Maintenance and Cardiac Differentiation of hESCs
All procedures and use of human-derived stem cell lines were approved by the Institutional Review Board at the University of California, San Diego.
H9 MYL2-GFP hESCs and H9 control hESCs were maintained on Matrigel-coated plates in irradiated mouse embryonic fibroblast (MEF) (GlobalStem)-conditioned hESC medium consisting of DMEM/F-12 supplemented with 20% (v/v) KnockOut Serum Replacement, 100 μM non-essential amino acids, 2 mM L-glutamine, 55 μM β-mercaptoethanol (all from Thermo Fisher Scientific), and 20 ng/mL human basic fibroblast growth factor (PeproTech). Cells were passaged as small colonies, following dissociation with 1 mg/mL dispase (Thermo Fisher Scientific).
Prior to differentiation, hESCs were cultured on MEFs in hESC medium for 2–3 days to 90% confluence. EBs were differentiated to the cardiovascular lineage as described previously (Yang et al., 2008, Dubois et al., 2011, Kattman et al., 2011), with modifications as detailed and as shown in Figure S1D. At the indicated time points, EBs were harvested and dissociated to single cells using previously described methods (Dubois et al., 2011). In brief, EBs were washed and incubated in dissociation solution consisting of 1 mg/mL collagenase type II (Worthington) in Hank's balanced salt solution (Corning) with 5 mM HEPES (Corning), overnight at room temperature (RT) with gentle shaking. On the following day, an equal amount of dissociation solution supplemented with 1 mg/mL BSA (Thermo Fisher Scientific), 0.1 mM EGTA (Sigma-Aldrich), and 10 mM taurine (Sigma-Aldrich) was added to the cell suspension, and EBs were pipetted gently to dissociate the cells. After dissociation, cells were centrifuged (200 × g for 5 min), filtered (100 μm cell strainer), and used for analysis.
RT-PCR, Long-Range PCR, and Real-Time qPCR
Total RNA was isolated using an RNeasy Micro or Mini Kit (QIAGEN) according to the manufacturer's instructions. RNA (100 ng to 1 μg) was reverse transcribed into cDNA using random hexamers or oligo(dT) with Superscript III First-Strand Synthesis System (Thermo Fisher Scientific). RT-PCR, and long-range PCR, to confirm BAC DNA integration (Figure 1B), were performed using MyTaq Red Mix (Bioline) and a MasterAmp Extra-Long PCR Kit (Epicentre) used as described previously (Frazer et al., 2007), respectively. Bands were analyzed by agarose gel electrophoresis. qRT-PCR was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad) using IQ SYBR green supermix (Bio-Rad). All qRT-PCR reactions were performed in triplicate. Expression levels were normalized to β-actin (ACTB) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and assessed using the comparative change in threshold cycle (Ct) method. After normalization, samples were plotted relative to hESC-derived EBs at day 0 or unsorted hESC-derived cell populations at day 25 of cardiac differentiation. Primers were designed using the NCBI Primer-BLAST software tool and are listed in Table S2. Human 15-week fetal atrial and ventricular tissue was purchased from Novogenix Laboratories, and human atrial and ventricular tissue was acquired from the University of California San Diego. RNA from human tissue was isolated using an RNeasy Fibrous Tissue Mini Kit (QIAGEN).
Immunocytochemistry
H9 MYL2-GFP BAC transgenic clones or single hESC-derived CMs after 25 days of cardiac differentiation were plated on 35-mm Matrigel-coated plates in MEF-conditioned hESC or differentiation media, respectively, and cultured for 3 days. Cells were washed with PBS (Corning), fixed with 4% paraformaldehyde (Electron Microscopy Services) for 15 min at RT, and permeabilized with 0.2% Triton X (Sigma-Aldrich) for 10 min at RT. After three washes with PBS, cells were blocked in 2% fish skin gelatin (Sigma-Aldrich) for 1 hr at RT, then incubated with 1:100 mouse anti-cTNT (Abcam, ab8295), 1:200 rabbit anti-MYL2 (Proteintech, 10906-1-AP), and/or 1:100 mouse anti-sarcomeric α-actinin (Sigma-Aldrich, A7811), in blocking solution overnight at 4°C. After staining, cells were washed three times with PBS, and, when necessary, were incubated with fluorescently conjugated secondary antibodies: 1:400 donkey anti-mouse Alexa Fluor 555, 1:400 donkey anti-rabbit Alexa Fluor 488, or 1:400 donkey anti-rabbit Alexa Fluor 647 (all Thermo Fisher Scientific) in blocking solution for 1–2 hr at RT in the dark. Cells were washed again as above, and nuclei were stained with DAPI (Sigma-Aldrich) diluted in PBS for 5 min at RT. Coverslips were attached using mounting medium (Dako), and the standing edge of each plate was removed using a scalpel that had been passed through a flame. The plate base was then glued to a glass slide, and imaged with an Olympus FluoView FV1000 Confocal Microscope.
Cell-Surface Antibody Screening
After 25 days of differentiation, a single-cell suspension of H9 MYL2-GFP hESC-derived cells was obtained by overnight collagenase II digestion, as described, and resuspended in BD FACS Pre-Sort Buffer (BD Biosciences) with the addition of 10 kU/mL DNase II, penicillin, and streptomycin. Cells were then stained using the BD Biosciences BD Lyoplate Human Cell Surface Marker Screening Panel following the included product protocol, and as described previously (Yuan et al., 2011). In brief, cells were dispensed into 96-well round-bottom plates (BD Biosciences) at 1 × 106 cells per well. Antibodies in the BD Lyoplate Screening Panel Plates were reconstituted with 1× PBS and added to the corresponding wells of the plates containing the single-cell suspension. Cells were stained live on ice for 20 min, washed with Pre-Sort Buffer containing DNase II, penicillin, and streptomycin, and stained with the appropriate AlexaFluor 647-conjugated IgG secondary antibody for 20 min in the dark. Cells were washed again and resuspended in Pre-Sort Buffer containing DNase II, penicillin, and streptomycin. Cells were then analyzed on a BD FACSCanto II system with a High Throughput Sampler reader (BD Biosciences). Data were analyzed with BD FACSDiva software (BD Biosciences) to determine the expression of each of the antibodies screened within the MYL2-GFP+ and MYL2-GFP− subpopulations. The median values of the antigen screened were first normalized to their appropriate isotype control. A ratio was then generated between MYL2-GFP+ and MYL2-GFP− cells using the normalized median values. Any antigen that showed a ratio of greater than 1.5, or less than 0.5, was then further analyzed. Microsoft Excel 2007 was used for the generation of heatmaps. The full heatmap with the MFI ratio for each antibody tested is reported in Table S1.
Multi-Color Flow Cytometry
Single-cell suspensions obtained at the indicated times of cardiac differentiation were resuspended in BD Stain Buffer (BD Biosciences) and stained with different combinations of antibodies against cell-surface markers (antibodies are detailed in Table S3). Cells were stained for 20 min at RT in the dark. For the detection of intracellular cTNI, cells were fixed and permeabilized after the surface marker stain using a BD Cytofix/Cytoperm Solution Kit (BD Biosciences). In brief, cells were incubated with fixation/permeabilization solution for 20 min at RT, then washed twice with BD Perm/Wash Buffer and incubated with AlexaFluor anti-cTNI antibody for 20 min at RT in the dark. Following antibody staining, cells were washed with, and resuspended in, BD Stain Buffer and analyzed on a BD LSRFortessa (BD Biosciences). Parental H9 cells and isotype controls were used to define the cytometric gates.
Cell Sorting
A single-cell suspension was obtained by overnight collagenase II digestion, as described, washed once in PBS, and then resuspended in Pre-Sort Buffer containing DNase II, penicillin, and streptomycin. Cells were stained on ice with fluorochrome-conjugated antibodies against CD77, CD200, and/or CD172a (SIRPA) (antibodies are detailed in Table S3) in the necessary combinations for sorting the various immunophenotypes as detailed. Cells were washed again and resuspended in Pre-Sort Buffer containing DNase II, penicillin, and streptomycin. Cells were then sorted using a BD FACSAria III Cell Sorter (BD Biosciences) using a 100-μm nozzle at an approximate pressure of 20 psi. Cells were collected in differentiation medium before preparation for qRT-PCR, immunocytochemistry, and electrophysiology analysis, as described.
Electrophysiology
Sorted SIRPA+/GFP+ and SIRPA+/GFP− hESC-derived CM populations (Figure S2B) were plated on 35-mm Matrigel-coated plates in differentiation media and cultured for 3 days. Only spontaneously beating cells were used for data collection. The presence/absence of GFP was confirmed by microscopic observation. Single-cell patch-clamp recordings were performed using an Axopatch 200B Amplifier (Axon Instruments) and digitized using a Digidata 1440A Digitizer (Axon Instruments), as described previously (Ouyang et al., 2005). Data acquisition and analysis were performed using Clampex 10.2 and Clampfit 10.2 software (Axon Instruments), respectively. Action potentials were recorded under whole-cell current-clamp conditions, and whole-cell Na+ and K+ currents were recorded under voltage-clamp conditions. The resistance of the patch electrodes ranged from 4 to 6 MΩ. The standard external solution contained: 150 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 10 mM glucose (pH 7.4 adjusted with NaOH). The intracellular pipette solution contained: 5 mM NaCl, 150 mM KCl, 1 mM MgCl2, 2 mM EGTA, 1 mM MgATP, and 10 mM HEPES (pH 7.2 adjusted with KOH). All experiments were performed at RT (20°C–22°C).
Statistical Analysis
In all quantitation experiments, results are expressed as mean ± SD or SEM. Statistical differences between two or more sets of data were determined using an unpaired Student's t test or one-way ANOVA, respectively. A value of p ≤ 0.05 was considered statistically significant. All statistical calculations were performed using GraphPad Prism 6 software.
Author Contributions
J.V., M.C., J.G.V., N.E., C.T.C., R.J.D., N.C.C., J.C.M., J.C., and S.M.E. were responsible for the experimental design. J.V., E.N.F., M.C., A.D.W., K.P., J.G.V., K.O., C.L., P.v.V., M.Z., J.M.H., D.C.D., J.D.G., and J.C.M. performed the experimental work. All authors assisted in the data analysis and interpretation and provided experimental advice. J.V., M.C., N.C.C., J.C.M., J.C., and S.M.E. wrote the paper.
Acknowledgments
We thank Andrea Nguyen and Dennis Sasaki (BD Biosciences) and the University of California San Diego (UCSD) Human Embryonic Stem Cell Core Facility for their assistance with cell sorting, and the UCSD School of Medicine Microscopy Shared Facility (support grant NS047101). Karyotype analysis was performed by Molecular Diagnostic Services (San Diego, California). This work was supported by funding from the NIH to K.A.F. and E.D.A. (HL107442) to N.C.C. and S.M.E. (HL128773) and to J.C. (HL123626, HL130295, HL137957), and from the California Institute for Regenerative Medicine (CIRM) to E.D.A. (TR305687) to J.C., and S.M.E. J.C. is an American Heart Association Endowed Chair. J.V., P.v.V., and D.C.D. were supported by CIRM fellowships (TG2-01154). K.O. was supported by the National Key Basic Research Program of China (2013CB531200), the National Science Foundation of China (31370823, 91439130), and the Shenzhen Peacock Project (KQJSCX20170330155020267). D.C.D. and J.D.G. were supported by an NIH pre-doctoral training grant (T32 GM008666).
Published: August 16, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, three tables, and one video and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.07.007.
Contributor Information
Ju Chen, Email: juchen@ucsd.edu.
Sylvia M. Evans, Email: syevans@ucsd.edu.
Supplemental Information
References
- Basu S., Campbell H.M., Dittel B.N., Ray A. Purification of specific cell population by fluorescence activated cell sorting (FACS) J. Vis. Exp. 2010 doi: 10.3791/1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H.S., Hyun W.C. Strategies for enrichment and selection of stem cell-derived tissue precursors. Stem Cell Res. Ther. 2012;3:17. doi: 10.1186/scrt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizy A., Guerrero-Serna G., Hu B., Ponce-Balbuena D., Willis B.C., Zarzoso M., Ramirez R.J., Sener M.F., Mundada L.V., Klos M. Myosin light chain 2-based selection of human iPSC-derived early ventricular cardiac myocytes. Stem Cell Res. 2013;11:1335–1347. doi: 10.1016/j.scr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam S.R., Passier R., Mummery C.L. Cardiomyocytes from human pluripotent stem cells in regenerative medicine and drug discovery. Trends Pharmacol. Sci. 2009;30:536–545. doi: 10.1016/j.tips.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Braam S.R., Tertoolen L., van de Stolpe A., Meyer T., Passier R., Mummery C.L. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4:107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes S.M., Hassink R.J., Feijen A., van Rooijen M.A., Doevendans P.A., Tertoolen L., Brutel de la Riviere A., Mummery C.L. Patterning the heart, a template for human cardiomyocyte development. Dev. Dyn. 2006;235:1994–2002. doi: 10.1002/dvdy.20830. [DOI] [PubMed] [Google Scholar]
- Costa M., Dottori M., Sourris K., Jamshidi P., Hatzistavrou T., Davis R., Azzola L., Jackson S., Lim S.M., Pera M. A method for genetic modification of human embryonic stem cells using electroporation. Nat. Protoc. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- Den Hartogh S.C., Passier R. Concise review: fluorescent reporters in human pluripotent stem cells: contributions to cardiac differentiation and their applications in cardiac disease and toxicity. Stem Cells. 2016;34:13–26. doi: 10.1002/stem.2196. [DOI] [PubMed] [Google Scholar]
- Dubois N.C., Craft A.M., Sharma P., Elliott D.A., Stanley E.G., Elefanty A.G., Gramolini A., Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.A., Braam S.R., Koutsis K., Ng E.S., Jenny R., Lagerqvist E.L., Biben C., Hatzistavrou T., Hirst C.E., Yu Q.C. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Eskin E., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Habib M., Caspi O., Gepstein L. Human embryonic stem cells for cardiomyogenesis. J. Mol. Cell Cardiol. 2008;45:462–474. doi: 10.1016/j.yjmcc.2008.08.008. [DOI] [PubMed] [Google Scholar]
- He J.Q., Ma Y., Lee Y., Thomson J.A., Kamp T.J. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ. Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- Huber I., Itzhaki I., Caspi O., Arbel G., Tzukerman M., Gepstein A., Habib M., Yankelson L., Kehat I., Gepstein L. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J. 2007;21:2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- Karakikes I., Senyei G.D., Hansen J., Kong C.W., Azeloglu E.U., Stillitano F., Lieu D.K., Wang J., Ren L., Hulot J.S. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl. Med. 2014;3:18–31. doi: 10.5966/sctm.2013-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kisselbach L., Merges M., Bossie A., Boyd A. CD90 expression on human primary cells and elimination of contaminating fibroblasts from cell cultures. Cytotechnology. 2009;59:31–44. doi: 10.1007/s10616-009-9190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y., Fukumoto S., Furukawa K., Okajima T., Wiels J., Yokoyama K., Suzuki Y., Urano T., Ohta M., Furukawa K. Molecular cloning of globotriaosylceramide/CD77 synthase, a glycosyltransferase that initiates the synthesis of globo series glycosphingolipids. J. Biol. Chem. 2000;275:15152–15156. doi: 10.1074/jbc.M909620199. [DOI] [PubMed] [Google Scholar]
- Lindahl P., Johansson B.R., Leveen P., Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277:242–245. doi: 10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- Mangeney M., Richard Y., Coulaud D., Tursz T., Wiels J. CD77: an antigen of germinal center B cells entering apoptosis. Eur. J. Immunol. 1991;21:1131–1140. doi: 10.1002/eji.1830210507. [DOI] [PubMed] [Google Scholar]
- Moretti A., Laugwitz K.L., Dorn T., Sinnecker D., Mummery C. Pluripotent stem cell models of human heart disease. Cold Spring Harb. Perspect. Med. 2013;3 doi: 10.1101/cshperspect.a014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery C.L., Zhang J., Ng E.S., Elliott D.A., Elefanty A.G., Kamp T.J. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ. Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noseda M., Peterkin T., Simoes F.C., Patient R., Schneider M.D. Cardiopoietic factors: extracellular signals for cardiac lineage commitment. Circ. Res. 2011;108:129–152. doi: 10.1161/CIRCRESAHA.110.223792. [DOI] [PubMed] [Google Scholar]
- Nudelman E., Kannagi R., Hakomori S., Parsons M., Lipinski M., Wiels J., Fellous M., Tursz T. A glycolipid antigen associated with Burkitt lymphoma defined by a monoclonal antibody. Science. 1983;220:509–511. doi: 10.1126/science.6836295. [DOI] [PubMed] [Google Scholar]
- Ouyang K., Wu C., Cheng H. Ca(2+)-induced Ca(2+) release in sensory neurons: low gain amplification confers intrinsic stability. J. Biol. Chem. 2005;280:15898–15902. doi: 10.1074/jbc.C500026200. [DOI] [PubMed] [Google Scholar]
- Talkhabi M., Aghdami N., Baharvand H. Human cardiomyocyte generation from pluripotent stem cells: a state-of-art. Life Sci. 2016;145:98–113. doi: 10.1016/j.lfs.2015.12.023. [DOI] [PubMed] [Google Scholar]
- Uosaki H., Fukushima H., Takeuchi A., Matsuoka S., Nakatsuji N., Yamanaka S., Yamashita J.K. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS One. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varro A., Nanasi P.P., Lathrop D.A. Potassium currents in isolated human atrial and ventricular cardiocytes. Acta Physiol. Scand. 1993;149:133–142. doi: 10.1111/j.1748-1716.1993.tb09605.x. [DOI] [PubMed] [Google Scholar]
- Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yuan S.H., Martin J., Elia J., Flippin J., Paramban R.I., Hefferan M.P., Vidal J.G., Mu Y., Killian R.L., Israel M.A. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Jiang J., Han P., Yuan Q., Zhang J., Zhang X., Xu Y., Cao H., Meng Q., Chen L. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.