Abstract
Schizophrenia is a severe psychiatric disorder of complex etiology. Immune processes have long been proposed to contribute to the development of schizophrenia, and accumulating evidence supports immune involvement in at least a subset of cases. In recent years, large-scale genetic studies have provided new insights into the role of the immune system in this disease. Here, we provide an overview of the immunogenetic architecture of schizophrenia based on findings from genome-wide association studies (GWAS). First, we review individual immune loci identified in secondary analyses of GWAS, which implicate over 30 genes expressed in both immune and brain cells. The function of the proteins encoded by these immune candidates highlight the role of the complement system, along with regulation of apoptosis in both immune and neuronal cells. Next, we review hypothesis-free pathway analyses which have so far been inconclusive with respect to identifying immune pathways involved in schizophrenia. Finally, we explore the genetic overlap between schizophrenia and immune-mediated diseases. Although there have been some inconsistencies across studies, genome-wide pleiotropy has been reported between schizophrenia and Crohn’s disease, multiple sclerosis, rheumatoid arthritis, systemic lupus erythematosus, type 1 diabetes, and ulcerative colitis. Overall, there are multiple lines of evidence supporting the role of immune genes in schizophrenia. Current evidence suggests that specific immune pathways are involved—likely those with dual functions in the central nervous system. Future studies focused on further elucidating the relevant pathways hold the potential to identify novel biomarkers and therapeutic targets for schizophrenia.
Keywords: schizophrenia, genetic, genome-wide association study, immune, inflammation, autoimmune, pleiotropy, pathway analysis
Immune Hypothesis of Schizophrenia
Immune processes have long been considered a potential initiating insult in schizophrenia.1 Accumulating evidence from epidemiological2 and clinical3–6 studies suggests that immune disturbances are present in at least a subset of patients with schizophrenia, garnering further support for this immune hypothesis.7 Advances in neuroimmunology have provided new insights into the mechanisms by which the immune system influences brain development and function,8–11 generating diverse hypotheses about the potential role of the immune system in schizophrenia. Neuronal autoantibodies,12 priming of microglia,13 and altered T-cell distribution14 have all been proposed as immune mechanisms predisposing to the disease.
One approach to refine these hypotheses is via human genetic association studies; such studies identify genetic variants associated with schizophrenia case status, thereby providing a window into the biological processes that cause the disease. Here, we review the immunogenetic architecture of schizophrenia. The focus is on evidence from genome-wide association studies (GWAS), given that GWAS (1) are currently the largest studies available in schizophrenia and therefore have the greatest statistical power, and (2) allow direct comparison to other diseases with available GWAS data. Importantly, GWAS provide an incomplete picture of immunogenetic variation—common single nucleotide polymorphisms (SNPs) are well captured, but rare and complex structural variants are not. When GWAS findings are driven by rare or complex structural variants, sequencing studies can help identify the causal variant(s) and shed new light on disease biology.15 The utility of sequencing studies is particularly relevant for immune genes, which are enriched for complex genetic variation due to strong pathogen-driven selective pressure.16 For instance, targeted sequencing in a region of association identified by schizophrenia GWAS identified complex structural variation at complement component 4 (C4) as a risk factor for the disease.17 Subsequent functional studies revealed increased C4 expression was associated with schizophrenia, and implicated synaptic pruning as a potential biological mechanism by which C4 variants contribute to the disease.17 Thus, sequencing studies hold the potential to move beyond regions of association identified in schizophrenia GWAS toward the identification of causal immune variants and their functional roles. Moving forward, the integration of sequencing data will be an important step to further understand the immunogenetic architecture of schizophrenia and other human diseases.
Association of Immune Genes in Schizophrenia
Major Histocompatibility Complex Genes
The extended major histocompatibility complex (xMHC) region is a 7.7 Mb stretch of DNA on the short arm of chromosome 6 (25.7–33.4 Mb) containing over 400 genes.18 There is extensive linkage disequilibrium (LD) across the xMHC, posing a challenge for studies of genetic variants in this region.19 Important immune genes in the xMHC region include the highly polymorphic human leukocyte antigen (HLA) genes, as well as genes encoding tumor necrosis factor family (TNF, LTA, LTB) and complement components (BF, C2, C4A, C4B). There are also many nonimmune genes in the xMHC region, including candidates for central nervous system disorders such SYNGAP1, MOG, NOTCH4, histones, and tRNAs.
In light of a suspected immune-related etiology, more than 60 hypothesis-driven candidate gene studies of functional HLA variants have been conducted over the years.20 However, the sample sizes of these candidate gene studies were modest (less than 300 cases), and the implicated HLA variants were inconsistent. Overall, there was some evidence that DQB1*0602 and DRB1*04 alleles may protect against schizophrenia in a systematic analysis by Wright et al.20 In European populations, in more recent hypothesis-free GWAS of schizophrenia, SNP alleles in the xMHC region have consistently shown the strongest association (reviewed by Kodavali et al21).
Fine-mapping of the xMHC revealed 3 independent associations across the region17,22 in a large Psychiatric Genomics Consortium (PGC) schizophrenia GWAS28 of over 35000 cases and 46000 controls (referred to here as the ‘PGC2’ study).17,22 Interestingly, HLA variants do not appear to drive the xMHC28 region association in schizophrenia (in contrast to many immune-mediated diseases).17,22 The strongest association with schizophrenia corresponds to an extended class I region association spanning ~2 Mb which remains to be further localized.17,22 The second region of association in the xMHC corresponds to C4, and has been well elucidated in genetic and functional work.17 Genetic variation at the C4 locus containing C4A and C4B is complex, with both structural and copy number variation influencing C4 expression. Genetic variants conferring increased C4 expression were associated with schizophrenia in the PGC GWAS.17 Furthermore, C4 knockout mice demonstrated significantly reduced synaptic pruning,17 suggesting that increased expression of C4 in schizophrenia may contribute to disease via excessive synaptic pruning. The third region of association in the xMHC is just downstream of the extended class II region.17,22 The strongest associated SNP in this region (rs210133) was identified near BAK1 by Sekar et al17 when accounting for the extended class I region association and genotype-predicted C4 expression. In conditional analysis using GWAS data alone without accounting for C4 expression, the strongest associated SNP (rs9461856) was approximately 140 kb upstream within SYNGAP1.22 While these SNPs are not in significant LD (r2 = .15 based on 1000 Genomes Phase 3 CEU European reference panel),23 it is notable that conditional analyses using 2 different approaches provided converging evidence for an independent third region of association in the extended class II region. Interestingly BAK1 encodes a pro-apoptotic mitochondrial protein required to prevent autoimmunity,24 while SYNGAP1 encodes a postsynaptic density protein previously implicated in schizophrenia25 and other neurodevelopmental disorders.26,27
Overall, robust evidence has emerged for the immune gene C4 as a susceptibility gene in schizophrenia.17 Follow-up studies combining genetic and functional approaches, similar to those undertaken by Sekar et al,17 are needed to delineate the underlying causal variants for the extended class I and extended class II major histocompatibility complex (MHC) associations in schizophrenia.
Non-MHC genes
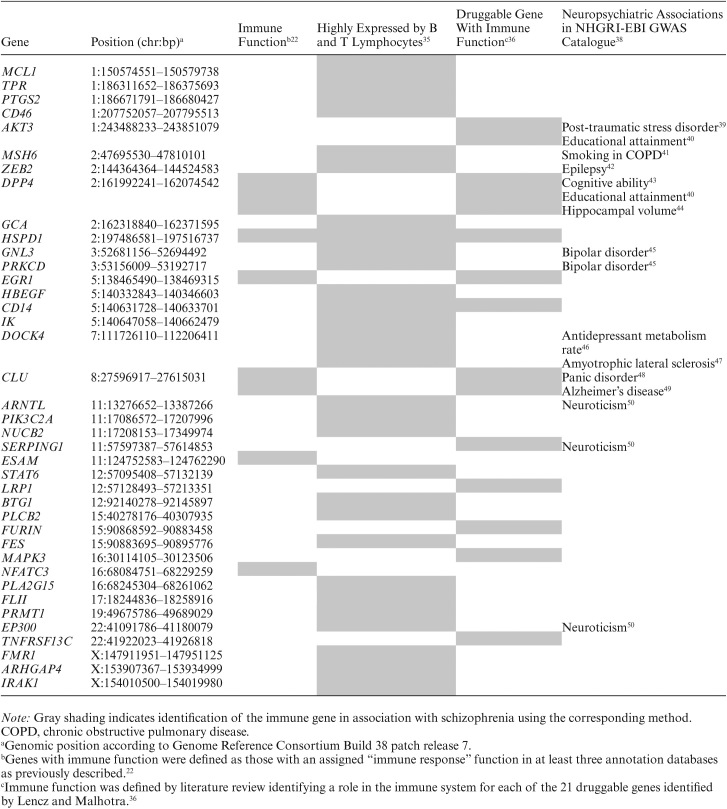
Outside of the xMHC region, over 100 loci reached genome-wide significance (P < 5 × 10−8) in the PGC2 GWAS.28 Follow-up investigations to identify the causal variants and/or genes in these regions are currently underway. Among the genome-wide significant variants in the PGC2 GWAS, our group highlighted 6 SNPs near immune genes CLU, DPP4, EGR1, ESAM, HSPD1, and NFATC3.22 All 6 of these immune candidates are expressed in human brain tissue,29 and have established roles in the brain30–34 in addition to their roles in immunity. An integrative analysis of the PGC and other schizophrenia GWAS incorporating eQTL and pathway analysis identified variants annotated to 28 non-MHC genes highly expressed in B lymphocytes and T lymphocytes (Box 1).35 Another study identified 18 genes located in genome-wide significant regions of the SCZ-52 GWAS which encoded proteins targeted by approved drugs, and an additional 21 genes whose products were targeted by drugs currently in clinical trials.36 Among these genes, 11 have known immune functions and are expressed by immune cells in addition to brain: AKT3, CD14, CLU, DPP4, EGR1, FURIN, HSPD1, LRP1, MAPK3, SERPING1, and TNFRSF13C (Human Protein Atlas available from www.proteinatlas.org, accessed March 16, 2018).37 Overall, these 3 studies22,35,36 identified 39 candidate immune genes for schizophrenia outside of the MHC region (table 1). Notably, HSPD1 was identified as a top schizophrenia gene in all 3 independent secondary analyses of schizophrenia GWAS data22,35,36 while CD14,35,36DPP4,22,36CLU,22,36 and EGR122,36 were identified in 2 independent secondary analyses (table 1).
Table 1.
Schizophrenia Immune Gene Candidates
Box 1. Schizophrenia susceptibility genes expressed in B and T lymphocytes
ARHGAP4, ARNTL, BTG1, CD14, CD46, DOCK4, EP300, FES, FLII, FMR1, GCA, GNL3, HBEGF, HSPD1, IK, IRAK1, MCL1, MSH6, NUCB2, PIK3C2A, PLA2G15, PLCB2, PRKCD, PRMT1, PTGS2, STAT6, TPR, and ZEB2.
There is accumulating evidence of shared genetic risk across schizophrenia and other neuropsychiatric phenotypes.51–55 Given this genetic sharing, it is notable that 12 of the 39 non-MHC immune candidate gene regions identified in previous studies22,35,36 have been associated with other neuropsychiatric traits in the NHGRI-EBI GWAS Catalogue38 including bipolar disorder, anxiety, neuroticism, cognitive abilities, and amyotrophic lateral sclerosis (table 1). In addition to these brain-related phenotypes, schizophrenia immune candidate gene regions have been implicated in a number of immune and inflammatory phenotypes (supplementary table 1). While these findings raise the possibility that certain immune genes implicated in schizophrenia also contribute to other brain- and immune-related phenotypes, there are some important caveats. Firstly, not all of the immune candidate genes reviewed here achieved genome-wide significance in the PGC2 GWAS (supplementary table 1). Thus, there may be some false positive findings, and further validation of these regions of association in larger samples is needed. Secondly, the immune genes reviewed here are located in broad regions of association containing additional genes (supplementary table 1). Until fine-mapping and functional investigations are completed to identify the causal variant(s) in each region, it remains unclear whether the association signals are driven by variants in the candidate immune genes or other variants in the region. Finally, although the same genomic region containing an immune gene of interest is associated with schizophrenia and other traits (table 1), the underlying causal genes or polymorphisms may be different (supplementary table 1). For instance, a susceptibility locus on chromosome 11 containing SERPING1—an inhibitor of the complement system—has been implicated in both neuroticism (rs10896636; Pdiscovery = 4.52 × 10−10, Preplication = .02)50 and schizophrenia (rs9420; P = 2.24 × 10−9).28 Although these SNPs are in high LD (r2 = .90 based on 1000 Genomes Phase 3 CEU European reference panel)23 the nearest gene to rs10896646 is not SERPING1, but instead ZDHHC5—a gene that has been shown to regulate hippocampal-dependent learning in ZDHHC5-deficient mice.56 Thus, before any conclusions about shared immune pathways across neuropsychiatric phenotypes can be made, future investigations must confirm a causal role for immune candidate genes in each phenotype of interest. Nevertheless, available data suggest that some immune genes may play a role in predisposing to schizophrenia specifically (eg, HSPD1, NFATC3, TNFRSF13C), while others may contribute to a number of different neuropsychiatric and immune-related phenotypes (eg, AKT3, DPP4, CLU).
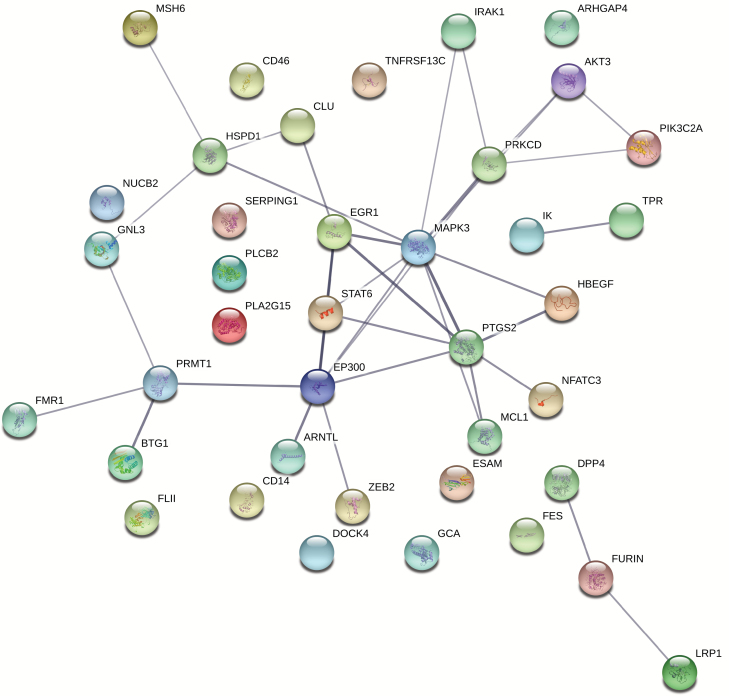
Keeping in mind the important limitations described above, we performed exploratory protein–protein interaction (PPI) analysis of previously identified schizophrenia candidate immune genes (table 1) using STRING, an online tool providing functional association of proteins based on evidence from a variety of data sources.57 We included functional interactions with total scores above 0.400 (medium confidence) based on evidence from 3 different scores: textmining, experiments, and co-expression. Our PPI analysis highlighted the roles of both the adaptive and innate immune response (see figure 1 for details of the PPI analysis, and supplementary table 2 for complete list of enriched biological pathways and gene members). Interestingly, regulation of cell death—including neuronal death—was a recurring biological process in the PPI network.
Fig. 1.
Functional protein association network for immune candidates implicated in schizophrenia. The 39 immune gene candidates outside of the xMHC region identified in this review were included in a PPI analysis using STRING.57 We included functional interactions with total scores above 0.400 (medium confidence) based on evidence from 3 different scores: textmining, experiments, and co-expression. Line thickness connecting the nodes indicates the strength of data support for a functional interaction between those proteins. Large nodes have known 3D structure, small nodes do not.
Current findings suggest that immune genes implicated in schizophrenia may be involved in both brain- and immune-related phenotypes, and may have dual roles in immunity and central nervous system (CNS) function (eg, regulation of cell death). The co-expression of these candidate immune genes in CNS is perhaps unsurprising given the vast transcriptome of immune cell types; an estimated 76% of all human proteins are expressed by at least one type of immune tissue (Human Protein Atlas available from www.proteinatlas.org, accessed March 16, 2018).37 Thus, further studies are needed to clarify whether specific immune pathways or types of immune cells may play a role in schizophrenia. For example, in autism large-scale RNA sequencing identified a gene co-expression network implicating dysregulated microglial responses in the disease.58 Given that our understanding of the genetic architecture of schizophrenia remains incomplete, it will be important to revisit these analyses in the future as new discoveries are made.
Immune Pathways in Schizophrenia
Schizophrenia is a polygenic disorder involving thousands of genetic risk variants.59 To identify the biological processes implicated by these risk variants, genetic pathway analyses evaluate whether groups of functionally related genes (eg, calcium channels) are jointly associated with the disorder. Several pathway analysis approaches have been developed and applied to schizophrenia, each with their own set of advantages and limitations.19 One major disadvantage of these methods is that they rely on existing research to assign biological functions to a gene set or variant set to define a pathway, typically based on known gene functions. To overcome this barrier, the recently developed Data-driven Expression-Prioritized Integration for Complex Traits (DEPICT) method integrates gene co-expression data with existing knowledge of the biological functions of genes.60 Unlike traditional pathway analyses where understudied genes with unknown functions are left out of the study, these genes are assigned a function in DEPICT based on similar expression to other genes with known function.60
Analyses of early schizophrenia GWAS suggested a role for immune pathways. For instance, in gene-set enrichment analysis of the Genetic Association Information Network (GAIN) schizophrenia GWAS (N = 1158 cases and 1378 controls), 3 of the 7 overrepresented pathways were related to the immune system (TGF-β, TNFR1, and TOB1 pathways).61 A meta-analysis of 18 schizophrenia GWAS including the ISC, SGENE, MGS, CATIE, and Swedish samples (N = 11185 cases and 10768 controls) also reported enrichment of immune pathways (antigen processing, T-cell adhesion molecules, and translocation to immunological synapse pathways).62 However, both of these pathway analyses were undertaken without special consideration of the xMHC region.61,62 Their findings must therefore be interpreted in light of the fact that extensive LD in the xMHC region can bias standard enrichment approaches and lead to false-positive results,19 particularly given the strong association of SNPs in this region with schizophrenia.
The Network and Pathway Analysis Subgroup of the PGC performed a rigorous analysis of 19752 pathways obtained from 6 sources (KEGG, GO, PANTHER, TARGETSCAN, Reactome, OMIM), testing for enrichment of these pathways in 5 psychiatric disorders—including schizophrenia, major depressive disorder (MDD), bipolar disorder (BP), attention deficit hyperactivity disorder (ADHD), and autism spectrum disorder (ASD)—by combining the results of 5 different pathway analysis methods (FORGE, MAGENTA, ALIGATOR, INRICH, and set screen testing). No pathway enrichment was detected in the schizophrenia sample (N = 9379 cases and 7736 controls), whether including or excluding the xMHC region.63 After pooling data across adult psychiatric disorders (schizophrenia, MDD, BP), TGF-β signaling and B-cell activation pathways were significantly enriched.63
In the primary analysis of the SCZ-52 GWAS by the PGC (N = 35476 cases and 46839 controls), all pathway analyses excluded the xMHC region and no immune pathway enrichment was observed.28 These analyses were not as comprehensive as that of the Network and Pathway Analysis Subgroup; a total of 9016 pathways obtained from 7 sources (KEGG, GO, PANTHER, Reactome, BioCarta, MGI, NCI) were tested for enrichment using 2 different methods (ALIGATOR and INRICH).28 Genome-wide significant schizophrenia associations were enriched at enhancers (defined by histone H3K27ac signal) active in CNS and, interestingly, B-lymphocytes.28 However, follow-up analyses by Finucane et al64 using Stratified LD Score Regression (sLDSC) to analyze all SNPs in the SCZ-52 GWAS, rather than restricting to the set of genome-wide significant SNPs, only found enrichment of CNS enhancers (defined by H3K27ac, H3K4me1, H3K4me3, or H3K9ac signal) with no enrichment of immune cell enhancers. In contrast, autoimmune diseases such as Crohn’s disease and rheumatoid arthritis showed enrichment of immune cell enhancers.64 Using sLDSC, our group tested for enrichment of SNPs capturing variation in genes with known immune and CNS function; we observed significant enrichment of the CNS genes in schizophrenia, but no significant enrichment of immune genes.22 In contrast, immune genes were enriched among 5 autoimmune diseases.22 Pers et al60 applied DEPICT to 109 genome-wide significant loci outside of the xMHC region from the same PGC data set and found enrichment among genes expressed in the brain as well as 15 canonical pathways, but no enrichment of immune-related gene sets.
With these contrasting observations, it remains unclear whether genetic data support a role for immune cell subsets or immune signaling pathways in the pathogenesis schizophrenia. Differences in gene sets and enrichment methods across studies as well as insufficient statistical power may contribute to the discrepant findings to date, particularly if there is true enrichment of immune-related pathways within specific cell subsets (eg, B-lymphocytes) or signaling pathways (eg, complement cascade). The availability of increasingly massive genetic data sets for schizophrenia presents an important opportunity for follow-up studies to investigate specific immune cell subsets and functional pathways, in order to further clarify their potential role in schizophrenia.
Genetic Correlation Between Schizophrenia and Immune Diseases
Beyond pathway analysis, GWAS of schizophrenia allow comparison to other human diseases with available GWAS data (cross-trait analysis). Methods to detect genome-wide sharing of genetic risk across disorders (genetic correlation) include polygenic risk scoring (PRS),65 restricted maximum likelihood (REML),66 and cross-trait LDSC.67 Similar to pathway analysis methods, each has their own advantages and limitations.68 Already cross-trait analyses have illustrated strong genetic correlation between schizophrenia and other adult onset psychiatric disorders such as bipolar disorder and major depressive disorder,51,52,67 suggesting that these diseases are not as distinct at a pathophysiological level as current diagnostic criteria suggest.
Of interest with respect to the immunogenetic architecture of schizophrenia are cross-trait analyses investigating genetic overlap between schizophrenia and immune-mediated disorders. At least 10 published studies have reported such cross-trait analyses (table 2), although in some earlier studies immune diseases were analyzed as a negative control.51,70 Despite available GWAS data for more than 19 immune diseases in ImmunoBase (www.immunobase.org, accessed March 16, 2018), published studies have only investigated pleiotropy between schizophrenia and nine immune diseases (celiac disease, Crohn’s disease, multiple sclerosis, psoriasis, rheumatoid arthritis, systemic lupus erythematosus, systemic sclerosis, type 1 diabetes, and ulcerative colitis).
Table 2.
Previous Investigations of Genetic Overlap Between Schizophrenia and Immune-Mediated Disease
| Immune Disease | Study | GWAS Samples | N Cases (% Male)a | Method | Resultb |
|---|---|---|---|---|---|
| CEL | Daniel Tylee (personal communication)95 | Schizophrenia PGC228 | 35476 (60%) | LDSC | n.s. |
| CEL69 | 4533 | ||||
| CRO | Purcell et al70 | Schizophrenia ISC70training | 3322 (66%) | PRS | n.s. |
| CRO71target | 1748 (39%) | ||||
| Cross-Disorder Group of the PGC51 | Schizophrenia PGC1–shared samples51 | 9074 (66%) | REML | n.s. | |
| CRO–shared samples72 | 4793 | ||||
| Stringer et al73 | Schizophrenia PGC1–caws28training | 8922 (66%) | PRS | +* | |
| CRO71target | 1664 (39%) | ||||
| Bulik-Sullivan et al67 | Schizophrenia PGC2–Asian28 | 33332 (61%) | LDSC | n.s. | |
| CRO74 | 5956 (45%) | ||||
| Wang et al75 | Schizophrenia PGC Cross-Disorder51 | 9379 (66%) | GPA | +* | |
| CRO72 | 6333 | ||||
| Daniel Tylee (personal communication)95 | Schizophrenia PGC228 | 35476 (60%) | LDSC | + | |
| CRO72 | 6333 | ||||
| MS | Andreassen et al76 | MS77training | 9772 (30%) | CFDR | +* |
| Schizophrenia PGC178target | 9394 (66%) | ||||
| Wang et al75 | Schizophrenia PGC Cross-Disorder52 | 9379 (66%) | GPA | n.s. | |
| MS79 | 931 trios (25%) | ||||
| Daniel Tylee (personal communication)95 | Schizophrenia PGC228 | 35476 (60%) | LDSC | n.s. | |
| MS77 | 9772 (30%) | ||||
| PBC | Daniel Tylee (personal communication)95 | Schizophrenia PGC228 | 35476 (60%) | LDSC | + |
| PBC80 | 2764 | ||||
| PSO | Wang et al75 | Schizophrenia PGC Cross-Disorder52 | 9379 (66%) | GPA | n.s. |
| PSO81 | 1359 (48%) | ||||
| Daniel Tylee (personal communication)95 | Schizophrenia PGC228 | 35476 (60%) | LDSC | + | |
| PSO82 | 2178 | ||||
| RA | Purcell et al70 | Schizophrenia ISC70training | 3322 (66%) | PRS | n.s. |
| RA71target | 1860 (25%) | ||||
| Stringer et al73 | Schizophrenia PGC1–caws28training | 8922 (66%) | PRS | +* | |
| RA71target | 1835 (25%) | ||||
| Lee et al83 | Schizophrenia PGC1+Swe59,78 | 12793 (64%) | REML | −* | |
| RA84,85 | 8064 (32%) | ||||
| Euesden et al86 | Schizophrenia PGC1.559 | 5001 (60%) | PRS | n.s. | |
| RA71 | 1989 (25%) | ||||
| Bulik-Sullivan et al67 | Schizophrenia PGC2–Asian28 | 33332 (61%) | LDSC | n.s. | |
| RA84 | 5539 (37%) | ||||
| Wang et al75 | Schizophrenia PGC Cross-Disorder52 | 9379 (66%) | GPA | + | |
| RA84 | 5539 (37%) | ||||
| Daniel Tylee (personal communication)95 | Schizophrenia PGC228 | 35476 (60%) | LDSC | n.s. | |
| RA85 | 14361 (23%) | ||||
| SLE | Wang et al75 | Schizophrenia PGC Cross-Disorder52 | 9379 (66%) | GPA | + |
| SLE87 | 720 (0%) | ||||
| Daniel Tylee (personal communication) | Schizophrenia PGC228 | 35476 (60%) | LDSC | + | |
| SLE88 | 4036 | ||||
| SSC | Daniel Tylee (personal communication) | Schizophrenia PGC228 | 35476 (60%) | LDSC | n.s. |
| SSC89 | 2296 (11%) | ||||
| T1D | Purcell et al70 | Schizophrenia ISC70training | 3322 (66%) | PRS | n.s. |
| T1D71 target | 1963 (51%) | ||||
| Stringer et al73 | Schizophrenia PGC1–caws28training | 8922 (66%) | PRS | +* | |
| T1D71target | 1894 (51%) | ||||
| Wang et al75 | Schizophrenia PGC Cross-Disorder52 | 9379 (66%) | GPA | n.s. | |
| T1D90 | 7514 | ||||
| Daniel Tylee (personal communication)95 | Schizophrenia PGC228 | 35476 (60%) | LDSC | n.s. | |
| T1D91 | 9934 | ||||
| UC | Bulik-Sullivan et al67 | Schizophrenia PGC2–Asian28 | 33332 (61%) | LDSC | n.s. |
| UC74 | 6968 (52%) | ||||
| Wang et al75 | Schizophrenia PGC Cross-Disorder52 | 9379 (66%) | GPA | +* | |
| UC92 | 6687 | ||||
| Daniel Tylee (personal communication)95 | Schizophrenia PGC228 | 35476 (60%) | LDSC | + | |
| UC92 | 6687 |
Note: n.s., not significant; +, positive genetic correlation between schizophrenia and the immune disease of interest; −, negative genetic correlation between schizophrenia and the immune disease of interest; *, finding survives multiple testing correction; CEL, celiac disease; CFDR, conditional false discovery rate93; CRO, Crohn’s disease; GPA, Genetic analysis incorporating pleiotropy and annotation94; ISC, International Schizophrenia Consortium; LDSC, cross-trait LD Score regression67; MS, multiple sclerosis; PBC, primary biliary cirrhosis; PRS, polygenic risk scoring65; PSO, psoriasis; RA, rheumatoid arthritis; REML, restricted maximum likelihood66; SLE, systemic lupus erythematosus; SSC, systemic sclerosis; T1D, type 1 diabetes; UC, ulcerative colitis.
aPercent of male cases in the GWAS is provided for all studies where it was reported.
bOnly those results that were robust to exclusion of the major histocompatibility complex (MHC) region are presented as significant in this table.
Studies to date have reported evidence of pleiotropy between schizophrenia and Crohn’s disease,73,75,95 multiple sclerosis,76 primary biliary cirrhosis,95 psoriasis,95 rheumatoid arthritis,73,75 systemic lupus erythematosus,75,95 type 1 diabetes73, and ulcerative colitis.75,95 For rheumatoid arthritis negative genetic correlation (whereby genetic risk for rheumatoid arthritis confers protection against developing schizophrenia) has also been reported,83 a result that is more in keeping with the epidemiological observation that schizophrenia patients have lower rates of rheumatoid arthritis than the general population.86 To further complicate the picture, some studies have reported no evidence of shared genetic risk between schizophrenia and these immune diseases (table 2).
There are a number of potential reasons for these apparently conflicting results. False negative findings may be driven by differences in GWAS sample sizes and cross-trait analysis methodologies (table 2), which influence statistical power to detect true genetic correlation. Alternatively, false positive findings may be driven by confounders that are inadequately adjusted for such as population stratification, LD, or nonindependence of samples in the schizophrenia and immune GWAS. With respect to LD, the xMHC is the strongest region of association in schizophrenia GWAS as well as many of the immune diseases. Given the extensive LD in this region, its inclusion in cross-trait analyses can lead to spurious findings. Regarding nonindependence of samples, Wellcome Trust Case Control Consortium (WTCCC) samples were used in the control group for the majority of the immune disease and schizophrenia GWAS, creating the potential for false positives if not accounted for. Another consideration is that, due to the significant sex bias of autoimmune diseases (which have a greater overall prevalence in women96), there may be sex-specific genetic correlations which have not been formally evaluated in existing studies; given the different sex proportions across GWAS samples analyzed to date (% male cases ranging from 0% to 66%; table 2), sex-specific effects could have either diluted or increased true signal leading to different results across studies.
Additional studies are needed to reconcile the inconsistencies in existing cross-trait analyses, with a focus on including all immune diseases with available GWAS data, comparing results across methodological approaches, applying rigorous control for population stratification and overlapping samples, and evaluating sex-specific effects. To this end, our group has undertaken a more comprehensive cross-trait analysis of all immune diseases curated in ImmunoBase, using the largest available GWAS for both schizophrenia and immune diseases, and comparing results from PRS and cross-trait LDSC approaches.97
Overall, there is accumulating evidence of shared genetic risk for schizophrenia and at least a subset of immune-mediated diseases. While it remains unknown what is driving this genetic overlap, it may represent a pathological process shared by immune cells and neurons (eg, a disturbance in calcium channel function or synaptic signaling). Future studies focused on identifying shared biological pathways between schizophrenia and genetically correlated immune diseases are needed to evaluate this hypothesis. Interestingly, genetic overlap with immune diseases has also been reported in Parkinson’s disease98 and Alzheimer’s disease99 using the conditional false discovery rate approach.93 If these findings are replicated, they raise the possibility that a genetic relationship with immune diseases may not be a phenomenon specific to schizophrenia but may instead represent an important etiological pathway for neuropsychiatric disorders in general.
Conclusion
The genomics era provides a new opportunity for evaluation of the immune hypothesis of schizophrenia using large-scale genetic data. Already GWAS have identified several immune candidates in association with schizophrenia. Within the xMHC region, strongest evidence has emerged for C4. Outside of the xMHC, more than 30 immune candidates have been identified including CD14, CLU, DPP4, EGR1, and HSPD1. Proposed immune candidates are expressed in both immune cells and the brain, suggesting dual roles in immunity and CNS function. The best current evidence for this is C4, which encodes a protein responsible for pathogen clearance in the immune system and synaptic pruning in the CNS. Additional fine-mapping and functional studies are needed to validate and understand the role of other immune candidate genes in schizophrenia.
Studies investigating immune pathway enrichment in schizophrenia GWAS have been less conclusive. While some studies have implicated B lymphocytes, others have not identified any immune pathway involvement. Results from cross-trait analyses of schizophrenia and immune-mediated diseases have also been somewhat inconsistent. Genetic correlation with a number of immune diseases has been reported, including Crohn’s disease, multiple sclerosis, rheumatoid arthritis, systemic lupus erythematosus, and type 1 diabetes. However, negative genetic correlation and non-significant findings have also been reported.
Overall, there is strong evidence supporting the involvement of certain immune variants in schizophrenia. However, there is conflicting evidence for enrichment of specific immune cell types or pathways, and for pleiotropy with specific immune diseases. Differences in sample size and methodological approach, clinical heterogeneity, and the strong xMHC association in schizophrenia are all potential factors contributing to these discrepancies across studies. Nevertheless, current data suggest that certain biological pathways with dual roles in immunity and CNS—such as cell death regulation or calcium channel signaling—are involved in both immune diseases and schizophrenia. To further support this hypothesis and identify the specific immune genes/cells/pathways involved in schizophrenia, there is a need for future studies to integrate methods, analyze more clinically homogenous patient samples or use less “noisy” phenotypes (eg, cognitive symptoms, negative symptoms), perform analyses both including and excluding the xMHC region, and follow-up on robust findings with functional studies.
Funding
This work was supported by the Canadian Institutes of Health Research (CIHR) MD/PhD Studentship, Fulbright Canada, the Weston Foundation, and Brain Canada through the Canada Brain Research Fund - a public-private partnership established by the Government of Canada.
Supplementary Material
Supplementary data are available at Schizophrenia Bulletin online.
References
- 1. Wise PM, Babcock W. Is insanity due to a microbe?Sci Am. 1896;75:303. [Google Scholar]
- 2. Mednick SA, Machon RA, Huttunen MO, Bonett D. Adult schizophrenia following prenatal exposure to an influenza epidemic. Arch Gen Psychiatry. 1988;45:189–192. [DOI] [PubMed] [Google Scholar]
- 3. Miller BJ, Buckley P, Seabolt W, Mellor A, Kirkpatrick B. Meta-analysis of cytokine alterations in schizophrenia: clinical status and antipsychotic effects. Biol Psychiatry. 2011;70:663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Upthegrove R, Manzanares-Teson N, Barnes NM. Cytokine function in medication-naive first episode psychosis: a systematic review and meta-analysis. Schizophr Res. 2014;155:101–108. [DOI] [PubMed] [Google Scholar]
- 5. de Witte L, Tomasik J, Schwarz E, et al. Cytokine alterations in first-episode schizophrenia patients before and after antipsychotic treatment. Schizophr Res. 2014;154:23–29. [DOI] [PubMed] [Google Scholar]
- 6. Fillman SG, Cloonan N, Catts VS, et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2013;18:206–214. [DOI] [PubMed] [Google Scholar]
- 7. Khandaker GM, Cousins L, Deakin J, Lennox BR, Yolken R, Jones PB. Inflammation and immunity in schizophrenia: implications for pathophysiology and treatment. Lancet Psychiatry. 2015;2:258–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. [DOI] [PubMed] [Google Scholar]
- 9. Derecki NC, Cardani AN, Yang CH, et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J Exp Med. 2010;207:1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paolicelli RC, Bolasco G, Pagani F, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. [DOI] [PubMed] [Google Scholar]
- 11. Schafer DP, Lehrman EK, Kautzman AG, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Masdeu JC, Dalmau J, Berman KF. NMDA receptor internalization by autoantibodies: a reversible mechanism underlying psychosis?Trends Neurosci. 2016;39:300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Howes OD, McCutcheon R. Inflammation and the neural diathesis-stress hypothesis of schizophrenia: a reconceptualization. Transl Psychiatry. 2017;7:e1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Debnath M, Berk M. Th17 pathway-mediated immunopathogenesis of schizophrenia: mechanisms and implications. Schizophr Bull. 2014;40:1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang K, Dickson SP, Stolle CA, Krantz ID, Goldstein DB, Hakonarson H. Interpretation of association signals and identification of causal variants from genome-wide association studies. Am J Hum Genet. 2010;86:730–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu Rev Genet. 2011;45:203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sekar A, Bialas AR, de Rivera H, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horton R, Wilming L, Rand V, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. [DOI] [PubMed] [Google Scholar]
- 19. Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11:843–854. [DOI] [PubMed] [Google Scholar]
- 20. Wright P, Nimgaonkar VL, Donaldson PT, Murray RM. Schizophrenia and HLA: a review. Schizophr Res. 2001;47:1–12. [DOI] [PubMed] [Google Scholar]
- 21. Kodavali CV, Watson AM, Prasad KM, et al. HLA associations in schizophrenia: are we re-discovering the wheel?Am J Med Genet B Neuropsychiatr Genet. 2014;165B:19–27. [DOI] [PubMed] [Google Scholar]
- 22. Pouget JG, Gonçalves VF, Spain SL, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genome-wide association studies suggest limited immune gene enrichment in schizophrenia compared to 5 autoimmune diseases. Schizophr Bull. 2016;42:1176–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Auton A, Brooks LD, Durbin RM, et al. ; 1000 GPC. A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takeuchi O, Fisher J, Suh H, Harada H, Malynn BA, Korsmeyer SJ. Essential role of BAX,BAK in B cell homeostasis and prevention of autoimmune disease. Proc Natl Acad Sci U S A. 2005;102:11272–11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O’Roak BJ, Stessman HA, Boyle EA, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamdan FF, Gauthier J, Spiegelman D, et al. ; Synapse to Disease Group. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009;360:599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magen D, Georgopoulos C, Bross P, et al. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am J Hum Genet. 2008;83:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones MW, Errington ML, French PJ, et al. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–296. [DOI] [PubMed] [Google Scholar]
- 32. Nasdala I, Wolburg-Buchholz K, Wolburg H, et al. A transmembrane tight junction protein selectively expressed on endothelial cells and platelets. J Biol Chem. 2002;277:16294–16303. [DOI] [PubMed] [Google Scholar]
- 33. Gómez-Sintes R, Lucas JJ. NFAT/Fas signaling mediates the neuronal apoptosis and motor side effects of GSK-3 inhibition in a mouse model of lithium therapy. J Clin Invest. 2010;120:2432–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han BH, DeMattos RB, Dugan LL, et al. Clusterin contributes to caspase-3-independent brain injury following neonatal hypoxia-ischemia. Nat Med. 2001;7:338–343. [DOI] [PubMed] [Google Scholar]
- 35. Lin JR, Cai Y, Zhang Q, Zhang W, Nogales-Cadenas R, Zhang ZD. Integrated post-GWAS analysis sheds new light on the disease mechanisms of schizophrenia. Genetics. 2016;204:1587–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lencz T, Malhotra AK. Targeting the schizophrenia genome: a fast track strategy from GWAS to clinic. Mol Psychiatry. 2015;20:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 38. MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74:656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rietveld CA, Esko T, Davies G, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci U S A. 2014;111:13790–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Siedlinski M, Cho MH, Bakke P, et al. ; COPDGene Investigators; ECLIPSE Investigators. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66:894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steffens M, Leu C, Ruppert AK, et al. ; EPICURE Consortium; EMINet Consortium. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum Mol Genet. 2012;21:5359–5372. [DOI] [PubMed] [Google Scholar]
- 43. Lam M, Trampush JW, Yu J, et al. Large-scale cognitive GWAS meta-analysis reveals tissue-specific neural expression and potential nootropic drug targets. Cell Rep. 2017;21:2597–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bis JC, DeCarli C, Smith AV, et al. ; Enhancing Neuro Imaging Genetics through Meta-Analysis Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Athanasiu L, Smorr LL, Tesli M, et al. Genome-wide association study identifies common variants associated with pharmacokinetics of psychotropic drugs. J Psychopharmacol. 2015;29:884–891. [DOI] [PubMed] [Google Scholar]
- 47. Xie T, Deng L, Mei P, et al. Genome-wide association study combining pathway analysis for typical sporadic amyotrophic lateral sclerosis in Chinese Han populations. Neurobiol Aging. 2014;35:1778.e9–1778.e23. [DOI] [PubMed] [Google Scholar]
- 48. Otowa T, Yoshida E, Sugaya N, et al. Genome-wide association study of panic disorder in the Japanese population. J Hum Genet. 2009;54:122–126. [DOI] [PubMed] [Google Scholar]
- 49. Jun G, Ibrahim-Verbaas CA, Vronskaya M, et al. ; IGAP Consortium. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luciano M, Hagenaars SP, Davies G, et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet. 2018;50:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gale CR, Hagenaars SP, Davies G, et al. ; International Consortium for Blood Pressure GWAS, CHARGE Consortium Aging and Longevity Group. Pleiotropy between neuroticism and physical and mental health: findings from 108 038 men and women in UK Biobank. Transl Psychiatry. 2016;6:e791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lencz T, Knowles E, Davies G, et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consorTium (COGENT). Mol Psychiatry. 2014;19:168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McLaughlin RL, Schijven D, van Rheenen W, et al. ; Project MinE GWAS Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat Commun. 2017;8:14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Y, Hu J, Höfer K, et al. DHHC5 interacts with PDZ domain 3 of post-synaptic density-95 (PSD-95) protein and plays a role in learning and memory. J Biol Chem. 2010;285:13022–13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gupta S, Ellis SE, Ashar FN, et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ripke S, O’Dushlaine C, Chambert K, et al. ; Multicenter Genetic Studies of Schizophrenia Consortium; Psychosis Endophenotypes International Consortium; Wellcome Trust Case Control Consortium 2. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pers TH, Timshel P, Ripke S, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Comprehensive analysis of schizophrenia-associated loci highlights ion channel pathways and biologically plausible candidate causal genes. Hum Mol Genet. 2016;25:1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jia P, Wang L, Meltzer HY, Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr Res. 2010;122:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aberg KA, Liu Y, Bukszár J, et al. A comprehensive family-based replication study of schizophrenia genes. JAMA Psychiatry. 2013;70:573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Finucane HK, Bulik-Sullivan B, Gusev A, et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bulik-Sullivan B, Finucane HK, Anttila V, et al. ; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dubois PC, Trynka G, Franke L, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stringer S, Kahn RS, de Witte LD, Ophoff RA, Derks EM. Genetic liability for schizophrenia predicts risk of immune disorders. Schizophr Res. 2014;159:347–352. [DOI] [PubMed] [Google Scholar]
- 74. Liu JZ, van Sommeren S, Huang H, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang Q, Yang C, Gelernter J, Zhao H. Pervasive pleiotropy between psychiatric disorders and immune disorders revealed by integrative analysis of multiple GWAS. Hum Genet. 2015;134:1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Andreassen OA, Harbo HF, Wang Y, et al. ; Psychiatric Genomics Consortium (PGC) Bipolar Disorder and Schizophrenia Work Groups; International Multiple Sclerosis Genetics Consortium (IMSGC). Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry. 2015;20:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ripke S, Sanders AR, Kendler KS, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hafler DA, Compston A, Sawcer S, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. [DOI] [PubMed] [Google Scholar]
- 80. Cordell HJ, Han Y, Mells GF, et al. ; Canadian-US PBC Consortium; Italian PBC Genetics Study Group; UK-PBC Consortium. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun. 2015;6:8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Feng BJ, Sun LD, Soltani-Arabshahi R, et al. Multiple loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet. 2009;5:e1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsoi LC, Spain SL, Knight J, et al. ; Collaborative Association Study of Psoriasis (CASP); Genetic Analysis of Psoriasis Consortium; Psoriasis Association Genetics Extension; Wellcome Trust Case Control Consortium 2. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee SH, Byrne EM, Hultman CM, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium and Rheumatoid Arthritis Consortium International; Schizophrenia Working Group of the Psychiatric Genomics Consortium Authors; Schizophrenia Working Group of the Psychiatric Genomics Consortium Collaborators; Rheumatoid Arthritis Consortium International Authors; Rheumatoid Arthritis Consortium International Collaborators. New data and an old puzzle: the negative association between schizophrenia and rheumatoid arthritis. Int J Epidemiol. 2015;44:1706–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stahl EA, Raychaudhuri S, Remmers EF, et al. ; BIRAC Consortium; YEAR Consortium. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Okada Y, Wu D, Trynka G, et al. ; RACI Consortium; GARNET Consortium. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Euesden J, Breen G, Farmer A, McGuffin P, Lewis CM. The relationship between schizophrenia and rheumatoid arthritis revisited: genetic and epidemiological analyses. Am J Med Genet B Neuropsychiatr Genet. 2015;168B:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bentham J, Morris DL, Graham DSC, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Radstake TR, Gorlova O, Rueda B, et al. ; Spanish Scleroderma Group. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42:426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Barrett JC, Clayton DG, Concannon P, et al. ; Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bradfield JP, Qu HQ, Wang K, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 2011;7:e1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schork AJ, Thompson WK, Pham P, et al. ; Tobacco and Genetics Consortium; Bipolar Disorder Psychiatric Genomics Consortium; Schizophrenia Psychiatric Genomics Consortium. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10:e1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tylee DS, Sun J, Hess JL, et al. Genetic correlations among psychiatric and immune-related phenotypes based on genome-wide association data. BioRxiv. 2018. doi:10.1101/070730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Fairweather D, Frisancho-Kiss S, Rose NR. Sex differences in autoimmune disease from a pathological perspective. Am J Pathol. 2008;173:600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pouget JG, Han B, Mignot E, et al. Cross-disorder analysis of schizophrenia and 19 immune diseases reveals genetic correlation. BioRxiv. 2018. doi: 10.1101/068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Witoelar A, Jansen IE, Wang Y, et al. ; International Parkinson’s Disease Genomics Consortium (IPDGC), North American Brain Expression Consortium (NABEC), and United Kingdom Brain Expression Consortium (UKBEC) Investigators. Genome-wide pleiotropy between Parkinson disease and autoimmune diseases. JAMA Neurol. 2017;74:780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yokoyama JS, Wang Y, Schork AJ, et al. ; Alzheimer’s Disease Neuroimaging Initiative. Association between genetic traits for immune-mediated diseases and Alzheimer disease. JAMA Neurol. 2016;73:691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.