Abstract
A better understanding of cancer biology has led to the development of targeted anticancer drugs. However, the full potential of these agents has not been realised due to the presence of de novo resistance, often resulting from compensatory signalling pathways or the development of acquired resistance in cancer cells that undergo clonal evolution under selective pressures of treatment. Combinations of targeted treatments can circumvent some mechanisms of resistance yielding clinical benefit. We explore the challenges of how to identify the best drug combinations, the best combination strategies, and the complexities of delivering these to patients. Recognizing treatment-induced toxicity and the inability to use continuous pharmacodynamically effective doses of many targeted treatments needs creative intermittent scheduling. Serial tumour profiling and the use of parallel co-clinical trials contribute to understanding mechanisms of resistance, and will guide the development of adaptive clinical trial designs that can accommodate hypothesis testing, to enable the full potential of combination therapies.
Introduction
A better understanding and characterisation of cancer cells, their microenvironment, and their interaction with the host immune system has resulted in a paradigm shift in the way new cancer therapies are developed.[1] Over the past 20 years, the focus of new drug development has been on the known oncogenic drivers. Notable success examples have included the high rate of cytogenetic response rates and long-term remissions seen patients with BCR–ABL driven chronic myeloid leukaemia (CML) treated with imatinib [2]. While imatinib is regarded the poster child of the ‘one genetic abnormality - one drug’ drug development paradigm, in reality the efficacy seen was the exception rather than a rule and reflects the biological complexity that governs the vast majority of cancers. As our arsenal of targeted anticancer agents grows, despite the promise observed in preclinical experiments and initial high response rates, a large number of targeted drugs (such as PI3K, AKT, MET and IGFR inhibitors) have not been successful in providing reproducible improvements in survival in patients with cancer when used as single agents [3–5]. The good intentions but unrealistic expectation set by the early success of imatinib in CML have been sobering, and some might posit they have led to premature termination of the development of classes of drugs such as IGF/IGFR inhibitors because they did not show single agent activity [6].
Efforts focussed on identifying small subsets of tumours hypersensitive to these drugs in ‘basket’ studies have yielded some benefit,[7, 8] but these approaches may not be constitute commercially viable strategies to help patients with cancer. This is, in part, because intrinsic or de novo resistance is a major reason for treatment failure and, in some instances, has been successfully overcome by a combination therapy approach, either with chemotherapy (such as rituximab with chemotherapy) [9, 10] or with other targeted treatments (such as everolimus with letrozole).[11] In addition, a large number of mechanisms of acquired resistance have emerged that exist in clones of tumour cells that have evolved and proliferated under the selective pressure of effective treatment.[12–15] While some mechanisms of acquired resistance are due to the presence of events that make the drug ineffective against its target (for example, the development of T790M mutations in cancer cells that cause resistance to first-generation EGFR inhibitors, such as erlotinib and gefitinib) [16, 17], some of these mechanisms of resistance still enable a drug to be effective against its target but might make the tumour become resistant in time owing to compensatory signalling (such as MEK signalling in BRAF-driven melanoma treated with BRAF inhibitors).[18–21] It is possible to make a difference in this latter category, and use a ‘window of opportunity’ to prevent development of acquired resistance by combining targeted agents, such as the combination of BRAF and MEK inhibitors, to circumvent compensatory pathway activation, which has shown improved overall survival in melanoma BRAF mutant melanoma. [22–24]
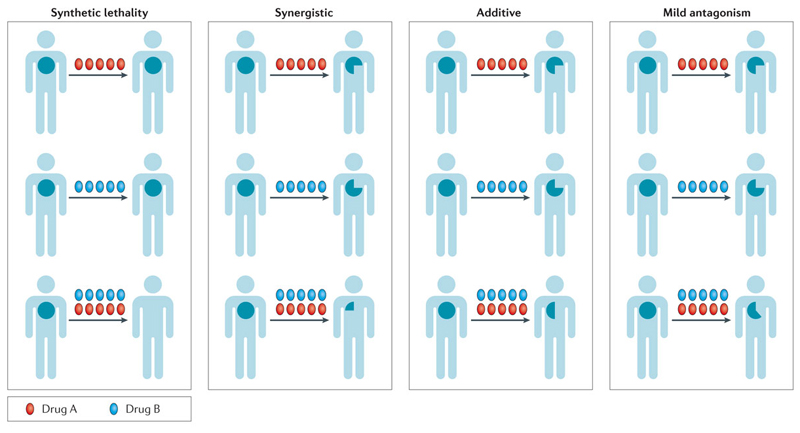
Hence, a focus on understanding mechanisms of de novo and acquired resistance is required to translate the preclinical promise of these targeted anticancer drugs when used in combination into strategies that can be registered in trials. In devising combination studies, the strategies should be judged by the ultimate impact made on the patient’s tumour. This can range from and number of approaches. First, synthetic lethality where drugs as single agents have minimal effects, to significant antitumour activity when a genomic defect or compensatory pathway is targeted therapeutically in combination. Second, the concept of synergy, where one or all partners in the combination have some clinical activity, but that the sum of clinical activity is greater than the effect of each individual drug (the whole if greater than the sum of its parts). Third, additivity, where the effect on the tumour is equal to the sum of activities of both drugs. Finally, mild antagonism, where the sum of activity of all drugs in the combination is less than the sum of activity of individual agents, but is more than each individual drug in the combination (Fig 1). Most targeted drug combinations in the clinic operate in the realms of additivity and mild antagonism; however, efforts are now focussed on identifying synthetically lethal or synergistic drug combinations. In the setting of metastatic (and, currently in most instances, incurable) disease, the clinical outcomes should not be based entirely on the effects of the combination on the tumour, but should also consider the associated direct and indirect costs of toxicity to patients.
Figure 1. Clinical impact of drug combinations on the tumour.
The size of the patient tumour is demarcated within the illustrated human cartoon. Drugs A and B are colour-coded as shown. While drugs in a given combination will have common effects on a proportion of clones of tumour cells, they can have differential effects on other clones thus influencing clonal evolution owing to selective pressure. The ultimate common readout is the size of the tumour and its rate of growth that eventually translates to progression-free survival and overall survival. The effects of clinically used combinations can be broadly classified as synthetically lethal, synergistic, additive, or mildly antagonistic.
We have deliberately included discussion of only combinations of targeted anticancer agents with other anticancer agents, but not with chemotherapy to ensure the focus is retained. Researchers face complex challenges in designing and executing these drug combinations. We aim to define these challenges and propose creative strategies to implement them, with the eventual aim of improving outcomes in cancer patients.
Challenges for targeted combinations
There are two major challenges in developing drug combination strategies for treating cancer. The first challenge is to find the best drug combinations to explore. While relatively small hypothesis-testing combination experiments have resulted in successful combination strategies, for example the clinical co-targeting of MEK and AKT signalling in KRAS mutant tumours [25] the scale of the biological complexity of the mechanisms of resistance to targeted treatment is too large to be tackled by multiple single-hypothesis-testing experiments and instead requires novel approaches. Furthermore, there are hundreds of targeted treatments in development and in clinical evaluation and the number of combination trials outweigh the number of patients who can be entered into trials to evaluate them [26]. Thus, experiments to discover and prioritize the optimal combinations for the treatment of patients with cancer are needed. The second challenge is the difficulties involved in combining targeted anticancer agents in a clinical setting. Issues related to realising the full potential of combination therapy in the clinic using targeted anticancer agents include toxicity, pharmacokinetic interactions and finding the correct timing and context of using these combinations. We have focussed on addressing the two challenges above related to combination therapy using targeted anticancer agents.
Challenge 1: the best combination
Unbiased chemical screens
Novel and unexpected drug combinations can be identified from unbiased high-throughput systems-based approaches.[26] Combination high-throughput screening of all FDA licensed drugs has been carried out to discover unexpected synergistic interactions, as exemplified by the combination of the anti-parasitic drug pentamidine with antipsychotic chlorpromazine that that has synergistic anti-mitotic activity in vitro [27, 28]. In some instances, the use of unbiased modelling on the basis of reported clinical adverse effects has been used to predict the tolerability of a drug combination [29, 30] but it remains to be seen if clinically efficacious combinations can be identified by such an approach. One limitation of this strategy is its lack of mechanistic insight, for example, an anti-helminthic that is designed to remain extracellular is unlikely to have in vivo activity in humans. An alternative to chemical screening would be the systematic genome-wide loss-of-function screens that are utilized to assess the role of specific target genes in tumour cells and rapidly identify novel drugs for combination [31].
Synthetic lethality and siRNA screens
The concept of synthetic lethality originates from studies in Drosophila model systems in which a combination of mutations in two or more separate genes leads to cell death and exploits inherent differences between cancer cells and normal cells.[32] Genome-wide short hairpin RNA (shRNA) and small interfering RNA (siRNA)–mediated synthetic lethal drug-sensitization screens are uncovering novel therapeutic combinations that can be targeted to oncogenic drivers, including those previously thought to be undruggable, such as KRAS.[33, 34] Investigators screened human non-small-cell lung cancer (NSCLC) cell lines carrying either wildtype or mutant KRAS with an siRNA library developed against 7,000 human genes and uncovered a novel synthetic lethal interaction between the DNA transcription factor GATA2 and oncogenic KRAS.[35] It was not possible to directly drug GATA2, which is a transcription factor, but combined gene-expression analysis and chromatin-occupancy analysis revealed a broad network of pathways controlled by GATA2 in RAS-pathway mutant NSCLC cells, which included NF-κB and RHO-related signalling pathways. The combined inhibition of the proteasome and Rho/ROCK signalling with the clinical compounds bortezomib (a proteasome inhibitor) and the Rho-kinase inhibitor fasudil led to almost complete regression of well-established lung tumours in a KRAS-driven mouse model[35]. There are currently clinically licensed proteasome inhibitors [36], and although ROCK inhibitors are being developed for use in cancer and have not yet been tested in the clinic, this combination could be exemplar of this approach of finding combinations to treat KRAS mutant cancers—an area of unmet need.
A more-common strategy for identifying potential drug combinations uses RNA interference (RNAi) to simulate models of pharmacological inhibition. In this method, wild-type cell lines or cell lines with induced loss of function for a particular target are screened against RNAi libraries with or without exposure to a particular drug to look for synergism or drug sensitization.[37, 38] An example is the large-scale unbiased combinatorial drug screen to identify cocktails active against melanomas with activating BRAF or RAS mutations, including those with intrinsic or acquired resistance to the approved BRAF inhibitor vemurafenib. This identified a triple combination, involving BRAF, EGFR and AKT inhibition, which was highly effective in BRAF-mutant melanomas and overcame vemurafenib resistance in vitro. Moreover, it revealed that combinations of statins and pan-CDK inhibitors were effective in NRAS-mutant melanomas.[39] Of note, the effective combinations were not predicted by single-agent screening, which highlights the potential for this approach to identify novel feedback signalling circuits and nodes that can be targeted by synergistic drug combinations.
SiRNA screens can also be used to understand and solve mechanisms of resistance in different tumour contexts. For example, BRAF inhibitors are successful in treating BRAFV600E mutant melanoma [40], but not BRAFV600E mutant colorectal cancer.[41] A focused kinase siRNA screen showed that knockdown of EGFR was synthetically lethal with vemurafenib treatment in BRAFV600E mutant colorectal cancer cells suggesting that the combination therapy of BRAF and EGFR inhibitors may be clinically beneficial.[42]. Subsequently, dual RAF and EGFR inhibition led to improved in vivo efficacy in BRAF mutant CRC xenografts and multiple clinical groups are exploring this combination in hypothesis-testing clinical trials (NCT01719380, NCT01791309, NCT01750918) and preliminary results suggest clinical activity [43, 44, 45].
Although siRNA screens are powerful to identify and validate novel targets, there remain significant limitations.[46] Firstly, siRNA inhibition can have off-target effects, or only partially knock-down the protein of interest. This might be overcome by the advancement of CRISPR-Cas9 genome-editing technologies that can complement and verify targets.[47, 48] Second, silencing of gene expression, and the loss of all functions of the protein, may not recapitulate the effects of targeting a single function of the protein, such as a kinase activity.
Systems biology
Systems-biology approaches involve analysis of a plethora of data, which could be genetic, transcriptomic, proteomic or factors that affect post-translational modification.[49] Different computational models can be created, iteratively refined and implemented. Examples of models include Bayesian, logic based or mass action models to analyse phosphoproteomic data and to provide a testable hypothesis for combinations to overcome resistance to targeted anticancer agents.[26, 50, 51]. Some approaches study the importance of proteins in intracellular networks and are used to inform on combinations of drugs outside of traditional portfolio of anticancer drugs based on differences these drug targets make on the intracellular interactome.[52] One recent study benchmarked the ‘ideal’ state of genetic extinction of mutant NRAS in a mouse model, and identified the drug combination of MEK and CDK4 inhibitors as most closely approximating the transcriptomic landscape of NRAS extinction, thereby proposing a workable hypothesis using this combination of targeted anticancer drugs to develop a clinically effective combination.[53] This benchmarking approach also enables the flexibility to adapt the design of combinations to address issues of tumour heterogeneity.[54] While, thus far, these data sets have involved the interpretation of biological data using machine-learning algorithms, future applications could involve the use of artificial intelligence.
Systems biology approaches have also directed researchers towards drug combinations of targeted agents and agents that target epigenetic mechanisms. Examples include combinations targeting HER2 and BET bromodomain inhibitors or BRAF and BET bromodomain inhibitors in breast cancer and melanoma, respectively.[55, 56] This strategy of targeting a node in the network, such as MEK or RAF in combination with a second agent that has broader epigenetic or post-translational effects (for example, BET bromodomain, HDAC or HSP90 inhibitors) is an emerging theme in preclinical experiments.
Translating systems biology approaches has significant challenges. The results from such studies need to be validated in preclinical experiments and then distilled into clinically usable combinations in subsets of patients defined by biomarkers. However, if refined and distilled into clinically usable combinations, these approaches are likely to open up the possibilities of combinations of entire new sets of drugs and biological targets, currently overlooked by current approaches.
Human tissue and primary cell-based assays
Looking forward, disease models based on human samples that accurately reflect the complexity of human cancers will be critical to the success of the development of new drugs and combinations. Large repositories of ‘established’ cell lines exist; however, there are significant differences in their genetic landscape when compared to tumour samples from patients [57]. Investigators are developing strategies to establish patient-derived cell culture models, for example, from biopsy samples of patients with lung cancer whose disease had progressed on EGFR or ALK inhibitors, and then testing them on a pharmacogenomic platform that facilitated the rapid discovery of multiple, effective drug combinations such as the combination of ALK and SRC inhibitors. [58] While these models reflect the biological complexities of resistant human tumours better than established cell lines models, the results of randomized clinical trials remain to be seen to prove that drug combinations suggested by such experiments can change clinical outcomes.
Patient-derived xenografts (PDXs) are also emerging as a powerful tool for investigating tumour biology and resistance mechanisms because they more faithfully recapitulate the molecular diversity, cellular heterogeneity, and histology seen in patient tumours.[59–61] The power of PDXs in studying preclinical drug combination efficacy is illustrated by recent work studying a PDX model of EGFR mutant lung cancer that developed ERK1 and ERK2 signalling reactivation days following continuous treatment with an EGFR inhibitor [62] [57]. The combination of EGFR inhibition with MEK inhibition not only delayed the onset of resistance, but achieved cure some mouse xenograft models. Furthermore, tumours that were resistant to the dual combination were shown to have developed downstream mTOR activation, offering an avenue for further combination strategies.[62] The limitation currently is that the development and propagation of mouse models, PDXs and drug testing can take up to 6 months. With the development of PDX encyclopaedias [63] and improvements in efficiency, speed and cost, this technology is likely to yield exciting results that will directly impact upon patient care.
Challenge 2: Executing the combination
Once a rationally designed combination is selected, its early clinical development is a complex process that requires attention to detail. Many issues influence implementation of combination strategies that are indicated by well-conducted scientific experiments. These include toxicity, pharmacokinetic and pharmacodynamic interactions, the timing of the development of resistance and, finally, finding a robust biomarker to predict response. In our mind, there are three major challenges facing the clinical community when testing a drug combination: first, how do we implement the combination; second, when do we implement the combination; and third, in whom do we implement the combination.
How to implement a drug combination
The biggest challenge in developing combination therapies of targeted anticancer drugs is the narrow therapeutic index of each drug owing to overlapping toxicities.[64] In some instances it has been possible to combine the full dose of each targeted agent at the original schedule; for example, in the case of combining dabrafenib and trametinib, and vemurafenib and cobimetinib in the setting of malignant melanoma.[23, 65–67] In addition to expected overlapping toxicities, unexpected toxicities relatively specific to the combination, such as fever have been observed.[66, 67] However, being able to deliver the recommended phase 2 dose of single agents within a combination is rare, and not infrequently, supra-additive toxicities are seen. [68] (Table 1).
Table 1. Examples of toxicities seen with combinations of novel anticancer agents.
| Overlapping mechanisms | Example drug combination | Toxicities (grade 3–4) | Comment | Refs |
|---|---|---|---|---|
| Toxicity of the combination is predominantly attributable to one drug in the combination | Everolimus + exemestane | Stomatitis, rash, fatigue, and diarrhoea | Toxicity attributable to everolimus makes the combination treatment more toxic than aromatase-inhibitor therapy alone | 117 |
| Overlapping toxicity of the combination is attributable to both drugs | MEK inhibitor + AKT inhibitor | Skin rash and diarrhoea | Overlapping toxicity of each drug | 25 |
| Significant non-overlapping toxicity in addition to overlapping toxicity | lpilimumab + nivolumab | Transaminitis | Unusual with either drug, but significantly more common with the combination therapy | 118 |
| Diarrhoea, fatigue, rash | Seen with both drugs but additive or greater than additive with the combination treatment | |||
| Significant non-overlapping toxicity and overlapping toxicity, and also reduction of certain toxicities | BRAF inhibitor + MEK inhibitor | Pyrexia | Unusual with either drug, but significantly more common with the combination therapy | 119 |
| Fatigue and diarrhoea | Expected overlapping toxicities | |||
| Keratoaca nthoma/squamous carcinoma of skin | Incidence lower with the combination therapy compared with BRAF-inhibitor treatment alone |
Toxicities could be attributable to one or both drugs. Toxicities could be additive or supra-additive (rare with either drug but significant in the combination). Rarely the combination can reduce the incidence of toxicity seen with one of the drugs.
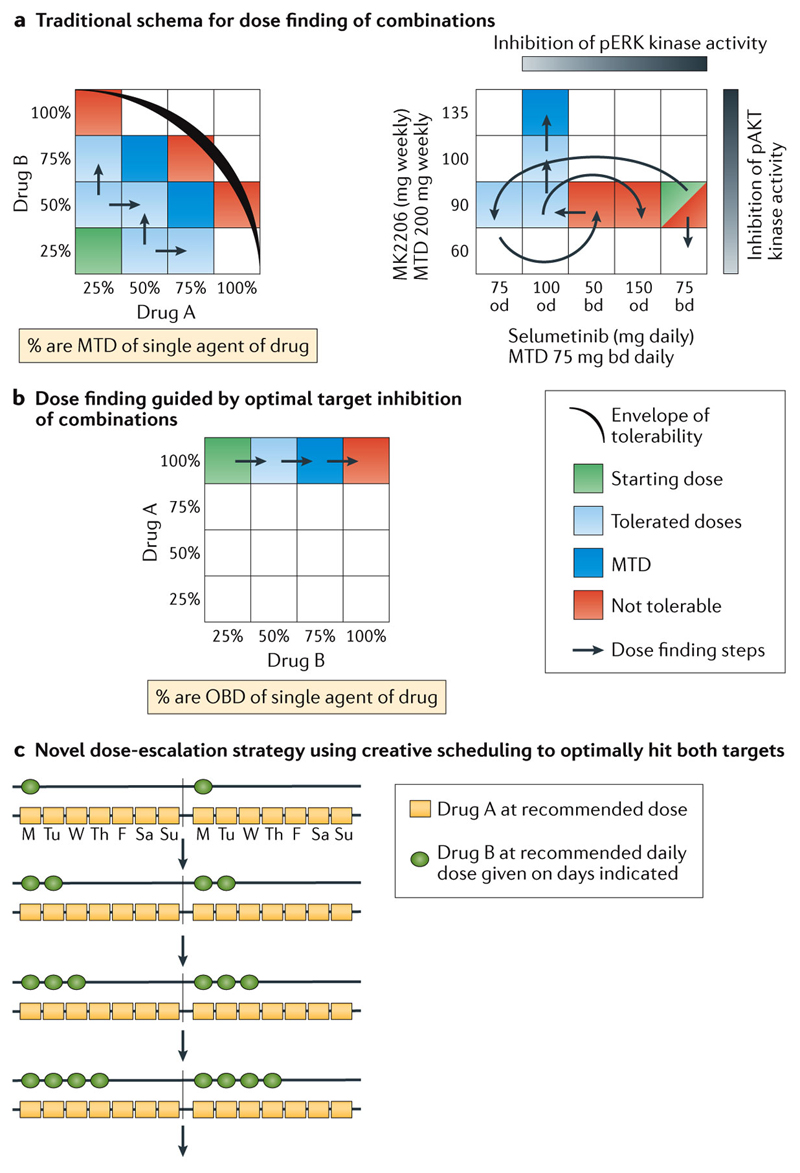
At present, preclinical models are not yet able to reliably predict toxicity [69], which adds to the challenge of optimising the toxicity-efficacy balance of drugs given in combination. For example, combining drugs that inhibit the PI3K/AKT/mTOR and the oncogenic RAS/RAF/MEK/ERK networks are exciting because resistance to inhibition of one pathway is attributed to signalling crosstalk.[4] However, it has been exceedingly difficult to deliver full does of combinations of MEK and PI3K pathway inhibitors in the clinic because of overlapping toxicities caused by drugs that act on these targets.[70, 71] For example, the adverse effects associated with PI3K inhibitors, such as hyperglycaemia, nausea, fatigue, rash and gastrointestinal toxicities, as well as rash, diarrhoea and dermatitis associated with MEK inhibition, illustrates the challenge in ongoing early phase dose-finding trials that have not been able to define the doses of drugs which, as single agents, caused maximal pharmacodynamics effects because of dose-limiting toxicities. Rather, the trials to date have required multiple dose de-escalation steps to determine a tolerable dose (Figure 2A right panel).[25] Furthermore, continuous exposure to both PI3K and MEK inhibitors is not feasible in the clinical setting, with most patients requiring treatment interruptions and dose modifications with chronic dosing.[72]
Figure 2. The challenge of optimising drugs dosed in combinations.
A) Left panel shows the traditional rule-based dose-escalation strategy for combinations based on toxicity. A starting dose is selected utilising a combination of a fraction of the maximum tolerated dose (MTD) of Drug A with a fraction of the MTD of Drug B. Each drug is in turn escalated in the dose-escalation approach depending on tolerability. Green shaded cubes mark the starting dose for the combination with proposed dose-escalation steps marked with red arrows. Cubes are coded for tolerability as marked in the key, with the black curve indicating a hypothetical envelope of tolerability. Right panel illustrates a real-life example of the complicated intricacies of dose-finding [25]. The MTD of selumetinib and MK-2206 are as indicated, with the grey bars indicating doses where target inhibition is seen. The green box mark the starting dose for the combination, with the red arrows marking the multiple de-escalation and re-escalation steps required to find a tolerable dose. Abbreviations: od = once daily; bd = twice daily
B) Combination dose-finding schema based on attaining adequate target inhibition. The optimal biological doses (OBD) are pre-defined based on pharmacological parameters, for example, target saturation of the drug, or optimal target modification of downstream pathways. Drug A is given at its recommended dose, with escalating doses of Drug B with the aim of reaching its OBD. The arrows indicate proposed steps for escalation with the cubes coded for tolerability as shown. Cubes are coded as in A) above.
C) Novel combination dose-finding schema [Au: Title too long, so I’ve added to the legend] In this novel combination scheme, the fixed OBDs of each drug are used but and are given in an increasing schedule to ensure drug tolerability. Drug A is given at its recommended dose and schedule, while Drug B is given at its recommended dose but for only one day a week. If tolerable, the number of dosing days is increased with each cohort until the recommended schedule is attained. This strategy aims to hit both targets hard, but in a tolerable schedule aiming to avoid the problems of sub-optimal target blockade.
Preclinical studies in a panel of cell lines quantifying the signalling transduction output of combined MEK and AKT inhibition in the background of specific driver mutations showed that sub-maximal inhibition of MEK and AKT pathways did not cause a significantly greater growth inhibition compared with growth inhibition caused by maximal MEK or AKT inhibition alone[73]. This finding indicates that a more biological meaningful strategy to develop drug combinations would start with 100% of the optimal biological dose (OBD) of drug A and escalate drug B (Figure 2B) or use the daily OBDs of both drugs but escalate their schedule (Figure 2C).
Other issues that can influence how to combine targeted anticancer agents include pharmacokinetic (PK) interactions. For example, lapatinib is both a substrate and a moderate inhibitor of CYP450-3A4, and can significantly reduce the clearance of other drugs that are CYP450-3A4 substrates and vice versa.[74] A phase I trial of lapatinib and pazopanib compared PK parameters of the combination with historical data of the drugs and it was decided that combining them did not alter exposure of either drug.[75] However, more detailed subsequent PK analyses in a glioma patient population at the phase II recommended dose level indicated that there was a significant drug–drug interaction leading to subtherapeutic lapatinib dosing[76] [72]. In addition, the frequent use of anti-epileptics in this population also decreased exposure to pazopanib leading to the poor outcomes seen.[76] This highlights the importance of incorporating detailed PK assessments into the evaluation of combinations of targeted anticancer agents.
When to implement a drug combination
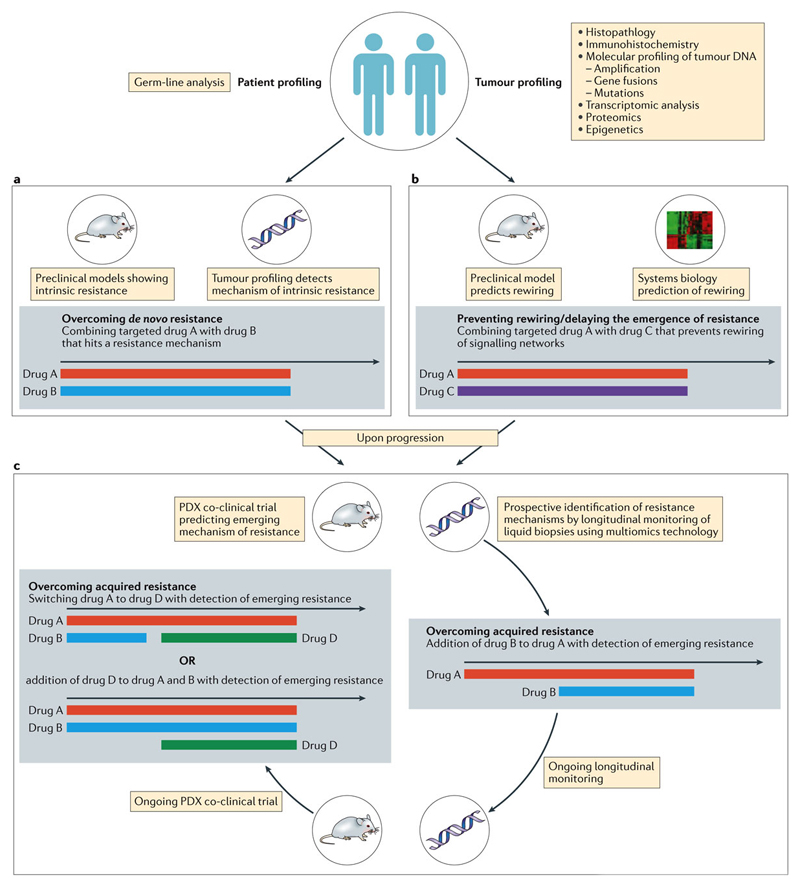
Initial studies recommending the dose and schedule of drugs in a drug combination are often conducted, and the dose determined, in the setting of advanced-stage disease. However, it is critical to define the setting for evaluating the efficacy of the combination in later phase clinical trials. The settings where combinations can be evaluated should be guided by the hypothesis being tested (Figure 3).
Figure 3. The future: proposed clinical trial designs to overcome resistance.
Patients are their tumours are profiled using -omics technology as shown and various strategies proposed for each hypothetical scenario A) Should profiling indicate a degree of de-novo resistance to the proposed Drug A, in which case, could it be reversed by addition of Drug B, and the combination used up-front. B) Should profiling indicate sensitivity to Drug A, but preclinical models and system biology predict the eventual rewiring of signalling networks causing resistance, Drug A can be combined with Drug C that has a broader epigenetic role. C) Co-clinical strategies employing patient-derived xenografts alongside prospective longitudinal monitoring of circulating cell-free DNA (cfDNA) in patients treated with Drug A. At the earliest sign of the development of resistant clones, Drug B is added in combination with continuation of the PDXs in a co-clinical trial and cfDNA monitoring. With emergence of an alternative mechanism of acquired resistance, Drug D can be added to the cocktail of Drug A & B, or the patient could be switched to a combination of Drug A & D. The black arrows are timelines, with the coloured bars indicating the various drugs described as colour coded.
[Au: I have removed Box 1 as this is just repeating the subheadings in the text. However, I think a Box of glossary items could be useful. Suggested terms include synthetic lethality, Synergistic Additive etc.
Firstly, is the combination being instituted to delay the onset of resistance rather than increase the degree of tumour shrinkage, that is, to increase the length rather than the depth of the response? For example, preclinical arguments have been put forward to pre-emptively combine HSP90 inhibitors with hormonal therapy or BRAF inhibitors to prevent the emergence of resistance.[77, 78] The HSP90 chaperone machinery ‘buffers’ disadvantageous metabolic and environmental stress within cancers cells caused by secondary mutations, which can lead to acquired resistance [79]. Inhibiting HSP90 prevents the evolution of such resistance clones and, therefore, these combinations are likely best evaluated in the frontline adjuvant or first-line metastatic setting.
Secondly, is the combination being tested because there is a degree of de novo resistance to one of the drugs that is being reversed by the addition of the second drug? Again, in this case the efficacy of the combination is best conducted in the first-line or neoadjuvant treatment setting, as shown with the addition of pertuzumab to trastuzumab and chemotherapy regimens.[80, 81] It could be argued that once patients have developed resistance to chemotherapy or trastuzumab this combination is unlikely to succeed, as the addition of pertuzumab does not realistically reverse multiple mechanisms of acquired resistance to trastuzumab or chemotherapy. While a drug holiday can on occasion enable patients to regain sensitivity to the same drug, this is usually due to reversal of compensatory feedback loops and expansion of sensitive tumours clones.[82, 83] Another example is the lack of single-agent activity of MEK inhibitors in multiple settings, which is attributed to activation of signal transduction pathways, such as the PI3/AKT/mTOR pathways in light of de novo resistance.[84, 85] As this hypothesis involves reversal of de novo resistance, efficacy of these combinations is best conducted when MEK inhibitors are first used rather than in a setting when resistance has already occurred. Both these examples of using combinations to reverse de novo resistance predominantly increase the depth of the response, but as a consequence they improve progression-free survival rather than uniquely inhibiting the development of resistance.[25]
Lastly, is the combination being developed to reverse acquired resistance? The development of secondary mutations, gene amplifications and late activation of signal transduction pathways in tumour cells is a common feature in the development of acquired resistance [Ref]. Adding a second drug as part of the combination in this setting takes the dynamic nature of clonal evolution into consideration and assumes that the tumour consists of clones that are still sensitive to the first drug and that addition of the second drug to the therapy combination will target clones resistant to the first drug. An example of this type involves the combination of HER2-targeting drugs in combination with mTOR inhibitors,[86–88] where secondary mutations in PIK3CA or increased signalling though PI3K has been shown to be a mechanism of acquired resistance in women with HER2-positive advanced-satge breast cancer.[89] In the future, innovative clinical trial design in both these settings can involve adding the second drug when resistance has occurred following initial response to the first drug. It is, however, important that the second drug is added when there are still clones within the tumour that are sensitive to the first drug. This is greatly facilitated by the analysis of circulating tumour DNA (ctDNA).[90, 91] Such molecular studies can be used as cues to initiate a combination before resistance occurs. However, profiling of mutations in tumours is unable to guide combination strategies that target epigenetic alterations that rewire signal transduction as a mechanism of resistance.
Implementing combinations and biomarker
Biomarkers for positive selection of patients likely to respond to targeted treatment can include mutated genes (BRAF, EGFR), amplification (HER2) or translocations (ALK) or alterations in protein products (ER) [92–94] and there are a few examples of the use of such biomarkers for negative selection of patients for a particular therapy, such as the use of KRAS mutations to predict resistance to cetuximab in colorectal cancer. [95–97] However, such markers might not be useful to guide drug combinations. For example, although BRAF mutation is now established as a biomarker for MEK inhibition,[98] and PI3K mutation for AKT inhibition,[4] recent work suggests that KRAS mutations are a biomarker for the successful use of a drug combination of MEK and PI3K/AKT inhibitors.[99] Furthermore, the experience with MEK and AKT inhibitor combination regimens has revealed a potential lineage context of therapeutic importance; these combinations have shown preliminary antitumour activity in KRAS-mutant NSCLC and ovarian cancer, but not in colorectal cancer.[25, 99] Thus, there is a real unmet need to understand and develop robust biomarkers in the context of an evolving complex signalling network.
The increased use of reliable genetically-engineered mouse models and PDX models that reflect mutations for the study of drug combinations might also facilitate the more-rapid identification of potential biomarkers.[100, 101] For example, dual MEK and mTORC1/2 blockade results in synergistic anti-proliferative effects in PDX models of colorectal cancer bearing alterations in KRAS/BRAF and PIK3CA/PTEN [102] [Ref]. However, this marked pro-apoptotic response to combination therapy was observed exclusively in wild-type P53 models, [102] suggesting that the combination of concomitant KRAS and PI3K mutations in a P53 wild-type setting could be predictive of clinical activity for the combination, and this can easily be tested in early phase trials.
The immune system and microenvironment
Another potential strategy that has garnered much attention is the use of combination cancer immunotherapy. The hypothesis that tumour cell death triggered by targeted therapies could result in antigen release and immunomodulation that could potentiate immune responses and enable eradication of tumour cells is exciting.[103–105]
Studies focusing on the specific mechanisms of tumour immune evasion are still at a relatively early stage. Although expression of both PD-1 and CTLA-4 dampens T-cell activation, we have yet to identify a robust signature of immune evasion and adaptive immune resistance.[106–108] For example, if there is a sufficient density of T cells in tumours that are turned off by adaptive immune resistance, PD-1–PD-L1 combinations might be effective. If T cells have not made it into the tumour, or are unable to recognise tumour antigens, combination strategies that activate an immune response or manipulate the network of cellular interactions in the tumour microenvironment that converge to establish immune tolerance would be required. [109, 110]
Exciting recent work has uncovered the effect of oncogenic signalling pathways on the modulation of tumour–immune interactions, and indicates that the combination of targeted treatments with immunotherapy may be an effective approach for cancer treatment. For example, preclinical models of melanoma have shown that PTEN loss in tumour cells increases the expression of immunosuppressive cytokines resulting in decreased T-cell infiltration into tumours. [111]. Treatment with a PI3K inhibitor improved the efficacy of checkpoint inhibition in this model and this could be studied in hypothesis-testing clinical trials. Other murine models of resistance to targeted treatments, such as imatinib have shown that resistance critically depends on indirect effects from the immune system and that concurrent administration of imatinib with checkpoint blockade can be synergistic,[112]. This possibility is now being pursued in an early phase trial (NCT01738139).[113]
While the use of PDX models based on patient samples harvested at diagnosis to study mechanisms of resistance are well known, their use in studying immune mechanisms of resistance are limited by the intrinsic murine immune system; thus, very complex mouse models will be required to exploit the use of PDXs in the setting of immunotherapy.
Despite concerns that immune checkpoint combinations can be overtly toxic, early studies of the combinations of the BRAF inhibitor vemurafenib with anti-PD-1 or anti-CTLA-4 antibodies showed they were reasonably tolerated, albeit with a significant incidence of unanticipated adverse events, such as hepatotoxicity, cutaneous and neurologic toxicities. [114] [115]. This situation might be due to the paradoxical ability of BRAF inhibitors to activate T-cells via ERK signalling, highlighting the need for careful assessment of the therapeutic indices of combinations of targeted treatments with immunotherapy.
Efforts are ongoing to identify biomarkers that can predict patients that are likely to respond to immunotherapy. Retrospective analyses suggest that the maximum benefits of dual anti-CTLA-4 and anti-PD-1 therapy was seen in patients with tumours that were PD-L1 negative, which if confirmed in future trials, suggests biomarkers are critical in choosing combinations with immune checkpoint regulators.
Conclusions
Thinking surrounding the development of anticancer strategies is evolving. The initial euphoria of early breakthroughs exploiting targeted treatments was followed by disappointment related to the observation of de novo resistance to large numbers of these agents and, later, acquired resistance in patients who had an initial response. While combinations of targeted therapies have made some inroads to address de novo and acquired resistance, we have faced considerable challenges and the full potential of combination approaches has not been realised.
We feel that there is reason to be optimistic. Newer technologies, such as the widespread use of gene-silencing tools, including shRNA and CRISPR to find synthetically lethal drug combinations, and phosphoproteomic technologies to understand and predict complex compensatory signalling mechanisms, will greatly enable researchers to propose and predict effective combinations that would not have come to light or would have been much slower if conventional small hypothesis testing experiments were done. Furthermore, the promise of the widespread ability to serially profile genomic, transcriptomic and epigenetic events in cancer cells and in the blood circulation of patients has enabled oncologists to help decide when combinations should or can be instituted. Finally, clinical trial designs that reflect the realities of toxicity and the use of intermittent dosing, and adaptive trial designs to allow the dynamic institution of combinations based on emergence of resistance, will greatly accelerate more-effective therapy combinations to improve patient care.
Glossary.
Basket trials are a new and evolving form of clinical trial design and are predicated on the hypothesis that the presence of a molecular marker predicts response to a targeted therapy independent of tumor histology.
Synthetic lethality originates from studies in Drosophila model systems in which a combination of mutations in two or more separate genes leads to cell death and exploits inherent differences between cancer cells and normal cells. In the drug development scene, this has expanded to included scenarios where drugs as single agents have minimal effects, but have significant anti-tumor activity in combination.
Synergy refers to the scenario where one or all partners in a combination have some clinical activity, but the sum of the clinical activity of the combination is greater than the effect of each drug
Additivity refers to the scenario where each partner in a combination has some clinical activity, and the effect on the tumour is equal to the sum of activities of both drugs.
Mild antagonism refers to the scenario where one or all partners in a combination have some clinical activity, but the sum of the clinical activity of the combination is less than the sum of activity of each individual drug
Systems biology is the computational and mathematical modelling of complex biological systems. In the context of drug development, these approaches aim to advance the prediction of effective drug combinations and the most common strategies include computational modelling, gene signature analysis, functional genomics, and high-throughput drug combination screening.[119]
Computational modellling of signaling networks is an important tool for increasing understanding of pathological signalling networks and prioritizing drug targets to test experimentally. Through model simulations, one can predict the relative importance of various proteins in the network, the presence of signal amplification, and the role of feedback and cross-talk.
Footnotes
Declaration of competing financial interests
The authors declare no competing interests.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Westin JR, Kurzrock R. It's about time: lessons for solid tumors from chronic myelogenous leukemia therapy. Molecular cancer therapeutics. 2012;11(12):2549–55. doi: 10.1158/1535-7163.MCT-12-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molife LR, Dean EJ, Blanco-Codesido M, et al. A phase I, dose-escalation study of the multitargeted receptor tyrosine kinase inhibitor, golvatinib, in patients with advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(24):6284–94. doi: 10.1158/1078-0432.CCR-14-0409. [DOI] [PubMed] [Google Scholar]
- 4.Yap TA, Bjerke L, Clarke PA, et al. Drugging PI3K in cancer: refining targets and therapeutic strategies. Current opinion in pharmacology. 2015;23:98–107. doi: 10.1016/j.coph.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis LM, Hicklin DJ. Resistance to Targeted Therapies: Refining Anticancer Therapy in the Era of Molecular Oncology. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(24):7471–7478. doi: 10.1158/1078-0432.CCR-09-1070. [DOI] [PubMed] [Google Scholar]
- 6.Basu B, Olmos D, de Bono JS. Targeting IGF-1R: throwing out the baby with the bathwater? British journal of cancer. 2011;104(1):1–3. doi: 10.1038/sj.bjc.6606023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grisham RN, Sylvester BE, Won H, et al. Extreme Outlier Analysis Identifies Occult Mitogen-Activated Protein Kinase Pathway Mutations in Patients With Low-Grade Serous Ovarian Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015 doi: 10.1200/JCO.2015.62.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Ahmadie H, Iyer G, Hohl M, et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer discovery. 2014;4(9):1014–21. doi: 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mounier N, Briere J, Gisselbrecht C, et al. Rituximab plus CHOP (R-CHOP) overcomes bcl-2--associated resistance to chemotherapy in elderly patients with diffuse large B-cell lymphoma (DLBCL) Blood. 2003;101(11):4279–84. doi: 10.1182/blood-2002-11-3442. [DOI] [PubMed] [Google Scholar]
- 10.Coiffier B, Lepage E, Briere J, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. The New England journal of medicine. 2002;346(4):235–42. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
- 11.Johnston SR. Enhancing Endocrine Therapy for Hormone Receptor-Positive Advanced Breast Cancer: Cotargeting Signaling Pathways. Journal of the National Cancer Institute. 2015;107(10) doi: 10.1093/jnci/djv212. [DOI] [PubMed] [Google Scholar]
- 12.Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational implications of tumor heterogeneity. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(6):1258–66. doi: 10.1158/1078-0432.CCR-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt MW, Loeb LA, Salk JJ. The influence of subclonal resistance mutations on targeted cancer therapy. Nature reviews. Clinical oncology. 2015 doi: 10.1038/nrclinonc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holohan C, Van Schaeybroeck S, Longley DB, et al. Cancer drug resistance: an evolving paradigm. Nature reviews. Cancer. 2013;13(10):714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 15.Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nature reviews. Cancer. 2012;12(7):487–93. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS medicine. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart EL, Tan SZ, Liu G, et al. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Translational lung cancer research. 2015;4(1):67–81. doi: 10.3978/j.issn.2218-6751.2014.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nature medicine. 2013;19(11):1401–9. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 19.Van Allen EM, Wagle N, Sucker A, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer discovery. 2014;4(1):94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, Hugo W, Kong X, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer discovery. 2014;4(1):80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romano E, Pradervand S, Paillusson A, et al. Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(20):5749–57. doi: 10.1158/1078-0432.CCR-13-0661. [DOI] [PubMed] [Google Scholar]
- 22.Long GV, Stroyakovskiy D, Gogas H, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. The New England journal of medicine. 2014;371(20):1877–88. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 23.Long GV, Stroyakovskiy D, Gogas H, et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet. 2015 doi: 10.1016/S0140-6736(15)60898-4. [DOI] [PubMed] [Google Scholar]
- 24.Robert C, Karaszewska B, Schachter J, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. The New England journal of medicine. 2015;372(1):30–9. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 25.Tolcher AW, Khan K, Ong M, et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(4):739–48. doi: 10.1158/1078-0432.CCR-14-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nature biotechnology. 2012;30(7):679–92. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 27.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nature reviews. Drug discovery. 2005;4(1):71–8. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 28.Lee MS, Johansen L, Zhang Y, et al. The novel combination of chlorpromazine and pentamidine exerts synergistic antiproliferative effects through dual mitotic action. Cancer research. 2007;67(23):11359–67. doi: 10.1158/0008-5472.CAN-07-2235. [DOI] [PubMed] [Google Scholar]
- 29.Zhao S, Nishimura T, Chen Y, et al. Systems pharmacology of adverse event mitigation by drug combinations. Science translational medicine. 2013;5(206):206ra140. doi: 10.1126/scitranslmed.3006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JA, Shinn P, Jaken S, et al. Novel Phenotypic Outcomes Identified for a Public Collection of Approved Drugs from a Publicly Accessible Panel of Assays. PloS one. 2015;10(7):e0130796. doi: 10.1371/journal.pone.0130796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrera-Abreu MT, Pearson A, Campbell J, et al. Parallel RNA interference screens identify EGFR activation as an escape mechanism in FGFR3-mutant cancer. Cancer discovery. 2013;3(9):1058–71. doi: 10.1158/2159-8290.CD-12-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLornan DP, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. The New England journal of medicine. 2014;371(18):1725–35. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
- 33.Steckel M, Molina-Arcas M, Weigelt B, et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell research. 2012;22(8):1227–45. doi: 10.1038/cr.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pourdehnad M, Truitt ML, Siddiqi IN, et al. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(29):11988–93. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar MS, Hancock DC, Molina-Arcas M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149(3):642–55. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 36.Richardson PG, Mitsiades C, Hideshima T, et al. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annual review of medicine. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 37.Whitehurst AW, Bodemann BO, Cardenas J, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446(7137):815–9. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 38.O'Grady M, Raha D, Hanson BJ, et al. Combining RNA interference and kinase inhibitors against cell signalling components involved in cancer. BMC cancer. 2005;5:125. doi: 10.1186/1471-2407-5-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Held MA, Langdon CG, Platt JT, et al. Genotype-selective combination therapies for melanoma identified by high-throughput drug screening. Cancer discovery. 2013;3(1):52–67. doi: 10.1158/2159-8290.CD-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopetz S, D J, Chan E, Hecht JR, O'Dwyer PJ, Lee RJ, Nolop KB, Saltz L. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J Clin Oncol. 2010;28:15s. (suppl; abstr 3534), 2010. [Google Scholar]
- 42.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 43.Van Geel Robin, E E, Bendell Johanna C, Faris Jason Edward, Lolkema Martijn PJK, Eskens Ferry, Spreafico Anna, Kavan Petr, Delord Jean-Pierre, Schuler Martin H, Wainberg Zev A, et al. Phase I study of the selective BRAFV600 inhibitor encorafenib (LGX818) combined with cetuximab and with or without the α-specific PI3K inhibitor BYL719 in patients with advanced BRAF-mutant colorectal cancer. J Clin Oncol. 2014;32(5s) (suppl; abstr 3514) [Google Scholar]
- 44.Atreya Chloe Evelyn, VC E, Bendell Johanna C, Andre Thierry, Schellens Jan H M, Gordon Michael S, McRee Autumn Jackson, O'Dwyer Peter J, Muro Kei, Tabernero Josep, van Geel Robin, et al. Updated efficacy of the MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in patients (pts) with BRAF V600E mutated (BRAFm) metastatic colorectal cancer (mCRC) J Clin Oncol. 2015;33(2015) (suppl; abstr 103) [Google Scholar]
- 45.Yaeger Rona D, C A, O'Reilly Eileen Mary, Reidy Diane Lauren, Kemeny Nancy E, Wolinsky Tamar, Gollub Marc J, Lacouture Mario E, Rosen Neal, Vakiani Efsevia, Saltz Leonard. Pilot study of vemurafenib and panitumumab combination therapy in patients with BRAF V600E mutated metastatic colorectal cancer. J Clin Oncol. 2015;33(suppl 3) abstr 611. [Google Scholar]
- 46.Mohr SE, Smith JA, Shamu CE, et al. RNAi screening comes of age: improved techniques and complementary approaches. Nature reviews. Molecular cell biology. 2014;15(9):591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor J, Woodcock S. A Perspective on the Future of High-Throughput RNAi Screening: Will CRISPR Cut Out the Competition or Can RNAi Help Guide the Way? Journal of biomolecular screening. 2015;20(8):1040–51. doi: 10.1177/1087057115590069. [DOI] [PubMed] [Google Scholar]
- 48.Shi J, Wang E, Milazzo JP, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nature biotechnology. 2015;33(6):661–7. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pathway and network analysis of cancer genomes. Nature methods. 2015;12(7):615–21. doi: 10.1038/nmeth.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelander S, Wang W, Nilsson B, et al. Models from experiments: combinatorial drug perturbations of cancer cells. Molecular systems biology. 2008;4:216. doi: 10.1038/msb.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iadevaia S, Lu Y, Morales FC, et al. Identification of optimal drug combinations targeting cellular networks: integrating phospho-proteomics and computational network analysis. Cancer research. 2010;70(17):6704–14. doi: 10.1158/0008-5472.CAN-10-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitsopoulos C, Schierz AC, Workman P, et al. Distinctive Behaviors of Druggable Proteins in Cellular Networks. PLoS computational biology. 2015;11(12):e1004597. doi: 10.1371/journal.pcbi.1004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwong LN, Costello JC, Liu H, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nature medicine. 2012;18(10):1503–10. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwong LN, Heffernan TP, Chin L. A systems biology approach to personalizing therapeutic combinations. Cancer discovery. 2013;3(12):1339–44. doi: 10.1158/2159-8290.CD-13-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korkut A, Wang W, Demir E, et al. Perturbation biology nominates upstream-downstream drug combinations in RAF inhibitor resistant melanoma cells. eLife. 2015;4 doi: 10.7554/eLife.04640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuhlmiller TJ, Miller SM, Zawistowski JS, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell reports. 2015;11(3):390–404. doi: 10.1016/j.celrep.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domcke S, Sinha R, Levine DA, et al. Evaluating cell lines as tumour models by comparison of genomic profiles. Nature communications. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480–6. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cassidy JW, Caldas C, Bruna A. Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer research. 2015;75(15):2963–8. doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tentler JJ, Tan AC, Weekes CD, et al. Patient-derived tumour xenografts as models for oncology drug development. Nature reviews. Clinical oncology. 2012;9(6):338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gandara DR, Lara PN, Jr, Mack PC. Patient-Derived Xenografts for Investigation of Acquired Resistance in Oncogene-Driven Cancers: Building a Better Mousetrap. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(26):2839–40. doi: 10.1200/JCO.2015.61.9692. [DOI] [PubMed] [Google Scholar]
- 62.Tricker EM, Xu C, Uddin S, et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer discovery. 2015;5(9):960–71. doi: 10.1158/2159-8290.CD-15-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao H, Korn JM, Ferretti S, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nature medicine. 2015;21(11):1318–25. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 64.Park SR, Davis M, Doroshow JH, et al. Safety and feasibility of targeted agent combinations in solid tumours. Nature reviews. Clinical oncology. 2013;10(3):154–68. doi: 10.1038/nrclinonc.2012.245. [DOI] [PubMed] [Google Scholar]
- 65.Grob JJ, Amonkar MM, Karaszewska B, et al. Comparison of dabrafenib and trametinib combination therapy with vemurafenib monotherapy on health-related quality of life in patients with unresectable or metastatic cutaneous BRAF Val600-mutation-positive melanoma (COMBI-v): results of a phase 3, open-label, randomised trial. The Lancet. Oncology. 2015;16(13):1389–98. doi: 10.1016/S1470-2045(15)00087-X. [DOI] [PubMed] [Google Scholar]
- 66.Ribas A, Gonzalez R, Pavlick A, et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: a phase 1b study. The Lancet. Oncology. 2014;15(9):954–65. doi: 10.1016/S1470-2045(14)70301-8. [DOI] [PubMed] [Google Scholar]
- 67.Larkin J, Ascierto PA, Dreno B, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. The New England journal of medicine. 2014;371(20):1867–76. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 68.Soria JC, Massard C, Izzedine H. From theoretical synergy to clinical supra-additive toxicity. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(9):1359–61. doi: 10.1200/JCO.2008.20.8595. [DOI] [PubMed] [Google Scholar]
- 69.Blomme EA, Will Y. Toxicology Strategies for Drug Discovery: Present and Future. Chemical research in toxicology. 2015 doi: 10.1021/acs.chemrestox.5b00407. [DOI] [PubMed] [Google Scholar]
- 70.Shimizu T, Tolcher AW, Papadopoulos KP, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(8):2316–25. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 71.Britten CD. PI3K and MEK inhibitor combinations: examining the evidence in selected tumor types. Cancer chemotherapy and pharmacology. 2013;71(6):1395–409. doi: 10.1007/s00280-013-2121-1. [DOI] [PubMed] [Google Scholar]
- 72.Bedard PL, Tabernero J, Janku F, et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(4):730–8. doi: 10.1158/1078-0432.CCR-14-1814. [DOI] [PubMed] [Google Scholar]
- 73.Stewart A, Thavasu P, de Bono JS, et al. Titration of signalling output: insights into clinical combinations of MEK and AKT inhibitors. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2015;26(7):1504–10. doi: 10.1093/annonc/mdv188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ho HK, Chan JC, Hardy KD, et al. Mechanism-based inactivation of CYP450 enzymes: a case study of lapatinib. Drug metabolism reviews. 2015;47(1):21–8. doi: 10.3109/03602532.2014.1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Jonge MJ, Hamberg P, Verweij J, et al. Phase I and pharmacokinetic study of pazopanib and lapatinib combination therapy in patients with advanced solid tumors. Investigational new drugs. 2013;31(3):751–9. doi: 10.1007/s10637-012-9885-8. [DOI] [PubMed] [Google Scholar]
- 76.Reardon DA, Groves MD, Wen PY, et al. A phase I/II trial of pazopanib in combination with lapatinib in adult patients with relapsed malignant glioma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(4):900–8. doi: 10.1158/1078-0432.CCR-12-1707. [DOI] [PubMed] [Google Scholar]
- 77.Whitesell L, Santagata S, Mendillo ML, et al. HSP90 empowers evolution of resistance to hormonal therapy in human breast cancer models. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(51):18297–302. doi: 10.1073/pnas.1421323111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smyth T, Paraiso KH, Hearn K, et al. Inhibition of HSP90 by AT13387 delays the emergence of resistance to BRAF inhibitors and overcomes resistance to dual BRAF and MEK inhibition in melanoma models. Molecular cancer therapeutics. 2014;13(12):2793–804. doi: 10.1158/1535-7163.MCT-14-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lindquist S. Protein folding sculpting evolutionary change. Cold Spring Harbor symposia on quantitative biology. 2009;74:103–8. doi: 10.1101/sqb.2009.74.043. [DOI] [PubMed] [Google Scholar]
- 80.Swain SM, Baselga J, Kim SB, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. The New England journal of medicine. 2015;372(8):724–34. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. The Lancet Oncology. 2012;13(1):25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 82.Sun C, Wang L, Huang S, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508(7494):118–22. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 83.Das Thakur M, Salangsang F, Landman AS, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494(7436):251–5. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deuker MM, Marsh Durban V, Phillips WA, et al. PI3'-kinase inhibition forestalls the onset of MEK1/2 inhibitor resistance in BRAF-mutated melanoma. Cancer discovery. 2015;5(2):143–53. doi: 10.1158/2159-8290.CD-14-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nature reviews Clinical oncology. 2014;11(7):385–400. doi: 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- 86.Gandhi L, Bahleda R, Tolaney SM, et al. Phase I study of neratinib in combination with temsirolimus in patients with human epidermal growth factor receptor 2-dependent and other solid tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32(2):68–75. doi: 10.1200/JCO.2012.47.2787. [DOI] [PubMed] [Google Scholar]
- 87.Gadgeel SM, Lew DL, Synold TW, et al. Phase I study evaluating the combination of lapatinib (a Her2/Neu and EGFR inhibitor) and everolimus (an mTOR inhibitor) in patients with advanced cancers: South West Oncology Group (SWOG) Study S0528. Cancer chemotherapy and pharmacology. 2013;72(5):1089–96. doi: 10.1007/s00280-013-2297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andre F, O'Regan R, Ozguroglu M, et al. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): a randomised, double-blind, placebo-controlled phase 3 trial. The Lancet Oncology. 2014;15(6):580–91. doi: 10.1016/S1470-2045(14)70138-X. [DOI] [PubMed] [Google Scholar]
- 89.O'Brien NA, McDonald K, Tong L, et al. Targeting PI3K/mTOR overcomes resistance to HER2-targeted therapy independent of feedback activation of AKT. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(13):3507–20. doi: 10.1158/1078-0432.CCR-13-2769. [DOI] [PubMed] [Google Scholar]
- 90.Yates LR, Campbell PJ. Evolution of the cancer genome. Nature reviews Genetics. 2012;13(11):795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burrell RA, McGranahan N, Bartek J, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–45. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 92.Fisher R, Larkin J. Vemurafenib: a new treatment for BRAF-V600 mutated advanced melanoma. Cancer management and research. 2012;4:243–52. doi: 10.2147/CMAR.S25284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382(9893):720–31. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 94.Advani PP, Crozier JA, Perez EA. HER2 testing and its predictive utility in anti-HER2 breast cancer therapy. Biomarkers in medicine. 2015;9(1):35–49. doi: 10.2217/bmm.14.95. [DOI] [PubMed] [Google Scholar]
- 95.Ashraf N, Kothari N, Kim R. Predictive biomarkers for anti-epidermal growth factor receptor therapy: beyond KRAS testing. Journal of the National Comprehensive Cancer Network : JNCCN. 2014;12(10):1433–42. doi: 10.6004/jnccn.2014.0140. [DOI] [PubMed] [Google Scholar]
- 96.Perkins G, Pilati C, Blons H, et al. Beyond KRAS status and response to anti-EGFR therapy in metastatic colorectal cancer. Pharmacogenomics. 2014;15(7):1043–52. doi: 10.2217/pgs.14.66. [DOI] [PubMed] [Google Scholar]
- 97.Therkildsen C, Bergmann TK, Henrichsen-Schnack T, et al. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta oncologica. 2014;53(7):852–64. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
- 98.Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer cell. 2011;19(1):11–5. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 99.Tolcher AW, Patnaik A, Papadopoulos KP, et al. Phase I study of the MEK inhibitor trametinib in combination with the AKT inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer chemotherapy and pharmacology. 2015;75(1):183–9. doi: 10.1007/s00280-014-2615-5. [DOI] [PubMed] [Google Scholar]
- 100.Hidalgo M, Amant F, Biankin AV, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer discovery. 2014;4(9):998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Robles AI, Varticovski L. Harnessing genetically engineered mouse models for preclinical testing. Chemico-biological interactions. 2008;171(2):159–64. doi: 10.1016/j.cbi.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 102.Celina GG, Rivas MA, Ibrahim YH, et al. MEK plus PI3K/mTORC1/2 therapeutic efficacy is impacted by TP53 mutation in preclinical models of colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-14-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nature reviews. Cancer. 2012;12(4):237–51. doi: 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Apetoh L, Ladoire S, Coukos G, et al. Combining immunotherapy and anticancer agents: the right path to achieve cancer cure? Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2015;26(9):1813–23. doi: 10.1093/annonc/mdv209. [DOI] [PubMed] [Google Scholar]
- 105.Zamarin D, Postow MA. Immune checkpoint modulation: rational design of combination strategies. Pharmacology & therapeutics. 2015;150:23–32. doi: 10.1016/j.pharmthera.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 106.Vinay DS, Ryan EP, Pawelec G, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Seminars in cancer biology. 2015 doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 107.Ribas A. Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer discovery. 2015;5(9):915–9. doi: 10.1158/2159-8290.CD-15-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Teng MW, Ngiow SF, Ribas A, et al. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer research. 2015;75(11):2139–45. doi: 10.1158/0008-5472.CAN-15-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Das R, Verma R, Sznol M, et al. Combination therapy with anti-CTLA-4 and anti-PD-1 leads to distinct immunologic changes in vivo. Journal of immunology. 2015;194(3):950–9. doi: 10.4049/jimmunol.1401686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fridman WH, Pages F, Sautes-Fridman C. The immune contexture in human tumours: impact on clinical outcome. Nature reviews. Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 111.Peng W, Chen JQ, Liu C, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer discovery. 2015 doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Balachandran VP, Cavnar MJ, Zeng S, et al. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nature medicine. 2011;17(9):1094–100. doi: 10.1038/nm.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ipilimumab and Imatinib Mesylate in Advanced Cancer. ClinicalTrials.gov Identifier: NCT01738139.
- 114.Ribas A, Hodi FS, Callahan M, et al. Hepatotoxicity with combination of vemurafenib and ipilimumab. The New England journal of medicine. 2013;368(14):1365–6. doi: 10.1056/NEJMc1302338. [DOI] [PubMed] [Google Scholar]
- 115.Johnson DB, Wallender EK, Cohen DN, et al. Severe cutaneous and neurologic toxicity in melanoma patients during vemurafenib administration following anti-PD-1 therapy. Cancer immunology research. 2013;1(6):373–7. doi: 10.1158/2326-6066.CIR-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. The New England journal of medicine. 2012;366(6):520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. The New England journal of medicine. 2015;373(1):23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. The New England journal of medicine. 2012;367(18):1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ryall KA, Tan AC. Systems biology approaches for advancing the discovery of effective drug combinations. Journal of cheminformatics. 2015;7:7. doi: 10.1186/s13321-015-0055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]