Abstract
While elevated blood cholesterol has been associated with an increased risk of colorectal cancer (CRC) in observational studies, causality is uncertain. Here we apply a Mendelian randomisation (MR) analysis to examine the potential causal relationship between lipid traits and CRC risk. We used single nucleotide polymorphisms (SNPs) associated with blood levels of total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL), and high-density lipoprotein (HDL) as instrumental variables (IV). We calculated MR estimates for each risk factor with CRC using SNP-CRC associations from 9,254 cases and 18,386 controls. Genetically predicted higher TC was associated with an elevated risk of CRC (odds ratios (OR) per unit SD increase = 1.46, 95% confidence interval [CI]: 1.20-1.79, P=1.68x10-4). The pooled ORs for LDL, HDL, and TG were 1.05 (95% CI: 0.92-1.18, P=0.49), 0.94 (95% CI: 0.84-1.05, P= 0.27), and 0.98 (95% CI: 0.85-1.12, P=0.75) respectively. A genetic risk score for 3-hydoxy-3-methylglutaryl-coenzyme A reductase (HMGCR) to mimic the effects of statin therapy was associated with a reduced CRC risk (OR=0.69, 95% CI: 0.49-0.99, P=0.046). This study supports a causal relationship between higher levels of TC with CRC risk, and a further rationale for implementing public health strategies to reduce the prevalence of hyperlipidaemia.
Keywords: Mendelian randomisation, hyperlipidaemia, cholesterol, colorectal cancer, risk
Introduction
Colorectal cancer (CRC) is the third most common cancer diagnosed in economically developed countries1. The mortality rate from CRC has been declining over the last twenty years as a consequence of improved medical care and probably through the introduction of population screening programs for the early detection of tumours2–4. Despite this improvement in patient outcome, it is still important to understand the risk factors for CRC in order to inform public health policy.
A number of factors influenced by lifestyle have been reported to be associated with the development of CRC in epidemiological observational studies, including a positive correlation with circulating levels of plasma cholesterol and other components of the lipid profile5, 6. It is, however, unclear from these studies if findings reflect a causal relationship or are simply a consequence of confounding by factors common to the aetiology of both CRC and hyperlipidaemia (e.g. common dietary factors) or reverse causality. Because lipid levels can be modified by lifestyle and treatment with statins, deciphering the basis for the association should be informative in formulating and optimizing prevention programs for CRC.
Evidence that statin use will effect a reduction in CRC is highly controversial7, 8. Although an analysis of The Health Improvement Network (THIN) database found that statin usage was associated with reduced CRC (long term usage: odds ratio [OR] = 0.95, 95% confidence interval [CI]: 0.91-0.99; short term usage: OR= 0.92, 95% CI: 0.85-0.99); no difference was shown between continued versus discontinued therapy, suggesting indication bias8. Moreover a recent meta-analysis of data from eight randomized controlled trials (RCTs) failed to demonstrate a beneficial effect which was statistically significant (relative risk = 0.89, 95% CI: 0.74-1.07)9. Each of these RCTs, however have the same limitations of short follow-up time, few CRC cases, and ascertainment of CRC as a secondary outcome.
Mendelian randomisation (MR) provides a useful complement to the traditional epidemiological study10. This strategy makes use of genetic variants that are robustly associated with traits of interest, in this case lipid traits - total cholesterol (TC), low-density lipoprotein (LDL), high-density lipoprotein (HDL), and triglyceride (TG) - as instrumental variables (IV) to infer whether associations between exposure and disease are causal. The use of genetic variants as IV to proxy modifiable exposure therefore avoids confounding by environmental factors, can be reflective of life-long exposure (propensity), and is not be subject to reverse causality. The strength of the IV in MR is important for power, but weak instruments can also lead to inconsistent instrumental variables estimators. Hence using a genetic score derived from a combination of single nucleotide polymorphisms (SNPs), which collectively explains more of the variance in the risk factor, mitigates against weak instrument bias thereby increasing study power.
Genetics scores derived from multiple SNPs for lipid traits have been used in MR studies to investigate associations between blood lipids and coronary heart disease11, and most recently prostate cancer12. Here we have employed MR to examine the impact of lipid traits on the risk of developing CRC.
Methods
Colorectal cancer datasets
We investigated the relationship between genetic risk scores for lipid traits and CRC risk using data from seven previously reported genome-wide association studies (GWAS) of CRC13 (Table 1). Briefly, these GWAS were all based on individuals with European ancestry and comprise: CCFR1, CCFR2, COIN, FINLAND, UK1, Scotland1 and VQ58. All studies were approved by their respective institutional review boards and conducted with appropriate ethical criteria in each country and in accordance with the Declaration of Helsinki. Comprehensive details on the cases and controls are available in previously published work13–16.
Table 1. Summary of the seven genome-wide association studies of colorectal cancer (9,254 cases and 18,386).
| Series | Study setting | Study centre | Sampling | No. cases | No. controls |
|---|---|---|---|---|---|
| CCFR1 | Colon Cancer Family Registry | University of Southern California | Recently diagnosed cases reported to population-based cancer registries in the USA (Seattle Familial Colorectal Cancer Registry). Canada (Ontario Familial Cancer Registry) and Australia (Australasian Colorectal Cancer Family Study). Population-based controls. | 1,290 | 1,055 |
| CCFR2 | Colon Cancer Family Registry | University of Southern California | Recently diagnosed cases reported to population-based cancer registries in the USA (Seattle Familial Colorectal Cancer Registry, Mayo Clinic Cooperative Family Registry for Colon Cancer Studies, USC Consortium Colorectal Cancer Family Registry, University of Hawaii Colorectal Cancer Family Registry). Canada (Ontario Familial Cancer Registry) Australia (Australasian Colorectal Cancer Family Study). Unaffected family controls. | 796 | 2,236 |
| COIN | COIN trial: Multicentre study of cetuximab and other therapies in metastatic CRC. Controls were unselected blood donors | Cardiff University | Cases recruited as a clinical-based series and controls as population-based series. | 2,244 | 2,162 |
| FINLAND | Finnish Colorectal Cancer Predisposition Study | Helsinki University | Cases requited through Finnish Hospitals and Finnish Cancer Registry. Population-based controls from FINRISK, Health 2000, Finnish Twin Cohort and Helsinki Birth Cohort Studies. | 1,172 | 8,266 |
| UK1 | CORGI (colorectal Tumour Gene Identification Consortium) | Oxford University | Cases enriched for family history of CRC, ascertained through UK clinical genetics clinics. Spouse controls with no personal history or family history of CRC. | 940 | 965 |
| Scotland1 | COGS (Colorectal Cancer Susceptibility Study) | Edinburgh University | Scottish population-based incidence cases aged <55 at diagnosis. Population-based controls frequency matched by area of residence. Scotland | 1,012 | 1,012 |
| VQ58 | Cases: VICTOR, post treatment stager of a phase III, randomised trial of rofecoxib (VIOXX) in patients after potentially curative therapy. QUASAR2, multi-centre study of capectibine±bevacizumb as adjuvant treatment. 1958 Birth Cohort controls |
Oxford University | Cases recruited as a clinical-based series and controls as population-based series. | 1,800 | 2,690 |
Genotyping data
Details of the genotyping and quality control of the seven CRC GWAS have been previously published13. Briefly, we excluded SNPs with a minor allele frequency of <1%, low call rate <95%, SNPs violating Hardy-Weinberg equilibrium, and individuals with non-European ancestry as assessed using HapMap v2 reference data17. Imputation of untyped SNP genotypes was performed using IMPUTEv2 software18 using a merged reference panel consisting of Sequencing Initiative Suomi (for the FINLAND data) or UK10K (for the remaining data) in addition to 1000 Genomes Project data. Poorly imputed SNPs (i.e. INFO score of <0.8) were excluded. Summary statistics from the seven GWAS were used to calculate the ORs for lipid-related SNPs.
Gene variants used to construct genetic risk scores
Genetic risk scores as IVs for circulating lipid fractions were developed from SNPs previously identified by the Global Lipids Genetics Consortium (GLGC)19. Median and range of standard deviations of lipid trait measurements in European cohorts of the Global Lipids Genetics Consortium are shown in Supplementary Table 1. We considered only SNPs associated at genome-wide significance (i.e. P ≤ 5.0x10-8) and restricted to individuals with European Ancestry. To avoid co-linearity between SNPs, we excluded SNPs that were correlated (i.e. r2 value ≥ 0.01), only considering the SNP with the strongest effect on the lipid trait for inclusion in genetic risk scores. Pairwise r2 values were calculated using PLINK v1.90 utilising samples of European ancestry from the 1000 Genomes and UK10K sequencing projects (Supplementary Data). This resulted in 58 SNPs for HDL, 29 SNPs for LDL, 26 SNPs for TG, and 38 SNPs for TC (Supplementary Table 2). Because lipid traits share common genetic variants, in addition to calculating an ‘unrestricted allele score’ that included all SNPs associated with the lipid trait, we also calculated a ‘restricted allele score’ as per Holmes et al 11 based on SNPs exclusively associated with HDL (n=43), LDL (n=9), or TG (n=14) to make them as specific as possible (Supplementary Table 3). Risk alleles were those that were positively associated with TC, LDL and TG or negatively associated with HDL levels. For all identified SNPs, we recovered the chromosome positions, the risk alleles, association estimates and standard errors.
Statistical analysis
We performed MR analysis to assess the association between TC, LDL, HDL, TG and CRC using summary statistics as described Burgess et al. (2015) 20. The combined ratio estimate () of all SNPs associated with each lipid trait on CRC was calculated under a fixed-effects model:
Xk corresponds to the association between SNP k with the lipid trait and Yk is the association between SNP k and CRC risk with standard error σYk. The standard error of the combined ratio estimate is given by:
With the statistics generated by following these calculations on the seven different cohorts in the CRC data, we performed a meta-analysis under a fixed-effects model to derive the final ORs and confidence intervals.
A key assumption for this MR analysis is there is no pleiotropism (i.e. a gene influencing multiple traits) between the genes influencing CRC and the lipid traits under study. Therefore, before performing the MR analysis, we performed LD regression to test for global evidence of pleiotropy as per Bulik-Sullivan et al. (2015) 21, 22, and subsequently implemented an MR-Egger regression to examine for violation of the standard IV assumptions in our analysis 23.
For each statistical test we considered a global significance level of P≤0.05 as being satisfactory to derive conclusions. To assess the robustness of our conclusions, we imposed a conservative Bonferroni-corrected significance threshold of 0.0125 (i.e. 0.05/4 lipid traits). We deemed a P-value > 0.05 as non-significant (i.e. no association), a P-value ≤0.05 as evidence for a potential causal association, and a P-value ≤0.0125 as significant evidence for a causal association. All statistical analyses were undertaken using R software (Version 2.14.1).
The power of a MR investigation depends greatly on the proportion of variance in the risk factor that is explained by the IV. We estimated study power using the methodology of Burgess (2014) 24, utilizing published estimates of the heritability of lipid trait associated IV SNPs 19 and the reported effect of each trait on CRC risk in epidemiological studies 8.
In a subsidiary analysis we constructed a genetic risk score for 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) using rs12916, rs17238484, rs5909, rs2303152, rs10066707 and rs2006760. These specific SNPs have previously been used to mimic statin intervention to estimate a causal association of statin use and coronary heart disease and diabetes 25.
Results
Using LD regression, we found no evidence for global pleiotropism (i.e. shared genetic components) between CRC and any of the lipid traits under investigation (Table 2). Following on from these observations we performed MR-Egger regression tests to explicitly examine for infringement of the standard instrumental variable assumptions in our MR analysis. We did not find evidence of any violation in respect to TC, LDL, HDL or TG (Table 2, Supplementary Figure 1). In view of the totality of these findings we were reassured of the validity of our MR-based analysis to infer whether the relation between exposures and CRC were likely to be causal.
Table 2. Testing for global and instrumental-specific pleiotropism.
Point estimates, confidence intervals, and P-values from linkage disequilibrium (LD) regression analysis, and MR-Egger methods. For MR-Egger, the intercept represents the average pleiotropic effect; an intercept significantly different from zero implies directional pleiotropy.
| LD regression results | ||||
|---|---|---|---|---|
| Trait | Heritability estimate | Genetic correlation | Standard error | P-value |
| TC | 0.2408 | 0.049 | 0.0635 | 0.4402 |
| TG | 0.2939 | 0.0322 | 0.0639 | 0.6143 |
| LDL | 0.2122 | 0.0729 | 0.066 | 0.2696 |
| HDL | 0.2499 | -0.0603 | 0.563 | 0.2834 |
| MR-Egger regression results | ||||||
|---|---|---|---|---|---|---|
| Trait | Estimate | Corrected standard error | CI lower | CI upper | P-value | |
| TC | intercept | 1.11x10-2 | 1.25x10-2 | -1.42x10-2 | 3.64x10-2 | 0.38 |
| slope | 0.16 | 0.33 | -0.51 | 0.83 | 0.64 | |
| TG | intercept | -1.13x10-2 | 1.10x10-2 | -3.38x10-2 | 1.12x10-2 | 0.31 |
| slope | 5.65x10-2 | 0.17 | -0.30 | 0.42 | 0.75 | |
| LDL | intercept | -3.41x10-3 | 7.67x10-3 | -1.91x10-2 | 1.23x10-2 | 0.66 |
| slope | 0.10 | 0.11 | -0.11 | 0.32 | 0.34 | |
| HDL | intercept | 2.23x10-3 | 5.58x10-3 | -8.94x10-3 | 1.34x10-2 | 0.69 |
| slope | -0.11 | 0.11 | -0.32 | 0.11 | 0.31 | |
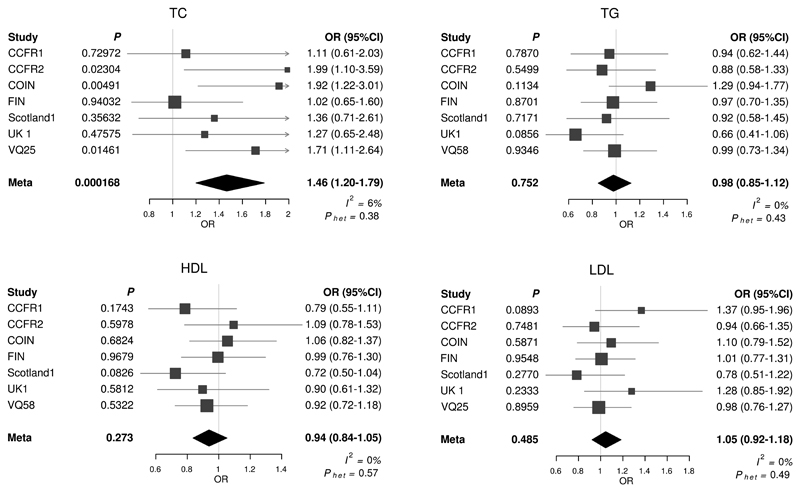
The associations of each unrestricted allele score for respective target lipid traits are shown in Figure 1. A positive correlation between variants associated with higher risk levels of TC and CRC was observed. The pooled OR meta-analysis for CRC by TC, estimated in IV analysis using the allele score was 1.46 per genetically instrumented SD increase in TC (95% CI: 1.20-1.79, P = 1.68 x 10-4, test for heterogeneity between studies I2 = 6%, Phet = 0.38).
Figure 1. Meta-analysis odds ratios (OR) for colorectal cancer per unit increase in genetic risk score (SD trait) for each lipid trait.
TC: Total cholesterol, TG: Triglyceride, LDL: low density lipoprotein, HDL: high density lipoprotein; Horizontal lines: 95% Confidence Intervals (95% CI). Phet: P-value for heterogeneity; I2: proportion of the total variation due to heterogeneity. Box: OR point estimate; its area is proportional to the weight of the study. Diamond: overall summary estimate, with confidence interval given by its width. Vertical line: null value (OR = 1.0).
The strongest reported SNP association for TC levels was provided by rs10401969 (CILP2) and rs12916 (HMGCR)19. To examine if the correlation between TC and CRC risk was primarily driven by these variants, we performed a sensitivity analysis excluding rs10401969 and rs12916. Omission of these two SNPs from the MR analysis did not appreciably affect our MR findings with results remaining significant (OR = 1.69, 95% CI: 1.25-2.28, P = 6.76 x 10-4). Albeit not significant, there was some support for a positive association with LDL (OR = 1.05, 95% CI: 0.92-1.18, P = 0.49) and CRC risk, and a negative association between HDL (OR = 0.94, 95% CI: 0.84-1.05, P = 0.27) and CRC risk.
Following on from these analyses, we performed a MR based analysis of LDL, HDL and TG using genetic scores derived from restricted sets of SNPs. As with the unrestricted analysis, no significant causal effect for each of these lipid traits was observed (Supplementary Figure 2).
Finally, genetically predicted lowered TC using the HMGCR genetic risk score was associated with 43% reduction in CRC (OR=0.69, 95% CI: 0.49-0.99, P=0.046, Phet= I2=56%).
Discussion
The present study strengthens a causal inference between circulating levels of TC and risk of developing CRC that is independent of known confounding effects. The positive correlation between the IV for TC and CRC risk, remained significant even after imposing a Bonferroni-correction to account for multiple testing. It is noteworthy that none of the IV SNPs for TC also represent IVs for obesity26, supporting an independent relationship between TC and CRC. As illustrated here and in previously studies of obesity and CRC 27, 28, insulin levels and uterine cancer 29, and lipid levels and coronary heart disease 30 MR provides an attractive means of establishing causal associations. In addition to demonstrating an association between TC and CRC risk we found that genetic variants that mimic the effect of HMGCR inhibition were associated with a reduced CRC risk, supporting findings from observational epidemiological studies that statins have beneficial effect on the population burden of CRC.
Studies in mice have shown that knocking out the cell surface cholesterol-sensing receptor gene NPC1L1, which plays a critical role in the absorption of intestinal cholesterol, reduces CRC risk31. However, the biological mechanism by which cholesterol may affect CRC risk remains to be established. Cholesterol is thought to have multiple carcinogenic/cancer promoting effects at the cellular level and several mechanisms have been variously suggested, including the cholesterol-mediated activation of the NLRP3 inflammasome32. Since statins are largely retained by hepatocytes, their effect on CRC will be indirect, via HMGCR inhibition. Intriguingly, recent data suggests that any impact of statin therapy on CRC is by prevention of progression of adenomas to frank cancers rather than their development per se 33. Further research on the biological relationship between cholesterol and CRC is needed to address such a proposition.
A major strength of our MR analysis is that it does not suffer from the influence of recall bias and confounding that affects traditional observational studies. Nevertheless, a primary assumption in MR is that the variants used to generate genetic scores are indeed associated with the exposure being examined. To ensure this was the case, we only made use of variants associated with each lipid trait at genome-wide significance from hypothesis-free GWAS. A second assumption is that variants are associated with CRC only through the exposure and are not confounded by shared genetic (i.e. pleiotropy). This would be revealed as an increasing linear relationship between SNPs and their effect size for any lipid trait and CRC risk; we did not observe such a relationship. Although it is not possible to exclude confounding by unknown confounders, the use of multiple independent variants acting through different pathways reduces the likelihood of confounded IV-associations. Moreover by using LD regression, we have been able to exclude pleiotropism on a global basis21. Finally, we only made use of data from individuals of European descent in the GWAS SNPs to limit potential bias from population stratification influencing study findings.
As with any MR analysis, there are potential limitations to our findings, including the limited trait variance explained by genetic variants, restricting statistical power. This is especially relevant for null findings, since wide confidence intervals leave uncertainty over the presence of a causal effect. It is estimated that the SNPs from the Global Lipids Genetics Consortium GWAS explain approximately 8-11% of the total variation in each lipid trait19. Recent analyses of observational studies found higher impact on CRC for TC than LDL or TG; respective ORs and 95% CIs – 1.49 (1.32-1.69), 1.37 (1.11-1.69), and 1.16 (1.06-1.27) 8. Based on these data our MR study was well-powered to demonstrate a causal relation for TC (≈80%, stipulating a P-value of 0.05), but we had limited power to identify associations for other lipid traits, particularly TG and HDL (respective power estimates for TG, LDL and HDL being 13%, 68% and 31%). Hence while the ORs for CRC with LDL and TG are congruous with observational studies 34 larger studies are required to formally establish a relationship using MR.
There are differences in the genomic landscapes of colonic and rectal cancers which presumably may reflect differences in aetiology. Unfortunately, these data were not uniformly collected across datasets, and we therefore did not investigate the possibility of differential effects of cholesterol on risk by anatomical location within the colorectum35.
In conclusion, this study provides evidence for a causal role of higher TC levels in the aetiology of CRC. Hence our findings encouragingly support the overall findings of past observational studies. Our limited power to further refine the relationship between lipid profile and CRC provides a motivational for larger MR studies, which will benefit from enhanced statistical power to demonstrate relationships for the spectrum of colorectal neoplasia. Irrespective of the exact functional basis of the association between TC and CRC risk, reducing hyperlipidaemia is an important target for primary prevention of CRC in the population. Our analysis therefore supports the hypothesis that the increasing use of statins in the population for prevention of cardiovascular disease will have the added bonus of reducing the burden of CRC.
Supplementary Material
Novelty and Impact.
While observational studies have suggested an association between blood cholesterol levels and colorectal cancer (CRC), they do not establish causality and may be influenced by confounding factors. Here we use Mendelian randomisation using genetic instrumental variables to provide evidence for a causal link between blood cholesterol levels and colorectal cancer. Thus, reducing hyperlipidaemia is an important target for primary prevention of CRC in the population.
Acknowledgements
We are grateful to all individuals who participated in the various studies. This study made use of genotyping data from the 1958 Birth Cohort, kindly made available by the Wellcome Trust Case Control Consortium 2. A full list of the investigators who contributed to the generation of the data is available at http://www.wtccc.org.uk/.
Funding
H.R-B was supported by a summer student grant from the Wellcome Trust (202639/Z/16/Z). At the Institute of Cancer Research, this work was supported by Cancer Research UK (C1298/A8362 - Bobby Moore Fund for Cancer Research UK). Additional support was provided by the National Cancer Research Network. A.S. is supported by a clinical fellowship from Cancer Research UK. In Edinburgh the work was supported by Programme Grant funding from Cancer Research UK (C348/A12076). In Oxford additional funding was provided by the Oxford Comprehensive Biomedical Research Centre and the EU FP7 CHIBCHA grant. Core infrastructure support to the Wellcome Trust Centre for Human Genetics, Oxford was provided by grant (090532/Z/09/Z). We are grateful to many colleagues within UK Clinical Genetics Departments (for CORGI) and to many collaborators who participated in the VICTOR and QUASAR2 trials. We also thank colleagues from the UK National Cancer Research Network (for NSCCG). Support from the European Union (FP7/207-2013, grant 258236) and FP7 collaborative project SYSCOL and COST Action in the UK is also acknowledged (BM1206). The COIN and COIN-B trials were funded by Cancer Research UK and the Medical Research Council and were conducted with the support of the National Institute of Health Research Cancer Research Network. COIN and COIN-B translational studies were supported by the Bobby Moore Fund from Cancer Research UK, Tenovus, the Kidani Trust, Cancer Research Wales and the National Institute for Social Care and Health Research Cancer Genetics Biomedical Research Unit (2011-2014). N.A.A., B.F.M. and S.M.W. were funded and supported by KFSHRC. In Finland, this work was supported by grants from the Academy of Finland (Finnish Center of Excellence Program 2012–2017, 250345), the Jane and Aatos Erkko Foundation, the Finnish Cancer Society (K.P.), the European Research Council [ERC; 268648], the Sigrid Juselius Foundation, SYSCOL, the Nordic Information for Action eScience Center (NIASC), the Nordic Center of Excellence financed by NordForsk (project 62721, K.P.) and State Research Funding of Kuopio University Hospital (B1401). We acknowledge the computational resources provided by the ELIXIR node, hosted at the CSC–IT Center for Science, Finland, and funded by the Academy of Finland (grants 271642 and 263164), the Ministry of Education and Culture, Finland. V.S. was supported by the Finnish Academy (grant number 139635). Sample collection and genotyping in the Finnish Twin Cohort has been supported by the Wellcome Trust Sanger Institute, ENGAGE – European Network for Genetic and Genomic Epidemiology, FP7-HEALTH-F4-2007 (201413), the National Institute of Alcohol Abuse and Alcoholism [grants AA-12502 and AA-00145 to Richard J Rose and K02AA018755 to Danielle M Dick] and the Academy of Finland (100499, 205585, 265240 and 263278 to J.K.). The work of the Colon Cancer Family Registry (CCFR) was supported by from the National Cancer Institute (UM1 CA167551), National Institutes of Health and through cooperative agreements with the following CCFR centers: Australasian Colorectal Cancer Family Registry (U01 CA074778, U01/U24 CA097735), USC Consortium Colorectal Cancer Family Registry (U01/U24 CA074799), Mayo Clinic Cooperative Familial Registry for Colon Cancer Studies (U01/U24 CA074800), Ontario Familial Colorectal Cancer Registry (U01/U24 CA074783), Seattle Colorectal Cancer Family Registry (U01/U24 CA074794), and University of Hawaii Colorectal Cancer Family Registry (U01/U24 CA074806). The CCFR Illumina GWAS was supported by funding from the National Cancer Institute, National Institutes of Health (U01 CA122839 and R01 CA143237 to G.C.). Additional support was provided from the Surveillance, Epidemiology and End Results (SEER) Program of the National Cancer Institute to Fred Hutchinson Cancer Research Center (Control Nos. N01-CN-67009, N01-PC-35142 and Contract No. HHSN2612013000121), the Hawai‘i Department of Health (Control Nos. N01-PC-67001, N01-PC-35137, and Contract No. HHSN26120100037C), and the California Department of Public Health (contract HHSN261201000035C) awarded to the University of Southern California. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute, any SEER program or any of the collaborating centers in the CCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, SEER or the CCFR.
Abbreviations
- CI
confidence interval
- CRC
colorectal cancer
- HDL
high-density lipoprotein
- IV
instrumental variables
- LDL
low-density lipoprotein
- MR
Mendelian randomisation
- OR
odds ratios
- SNP
single nucleotide polymorphisms
- TC
total cholesterol
- TG
triglyceride
Footnotes
Conflict of interest statement: None declared.
References
- 1.Forman D, Bray F, Brewster DH, Gombe Mbalawa C, Kohler B, Piñeros M, Steliarova-Foucher E, Swaminathan R, Ferlay J. Cancer incidence in five continents. X. IARC Scientific publications; 2014. p. 917. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Research UK. vol. 2016, 2016.
- 3.Hewitson P, Glasziou P, Watson E, Towler B, Irwig L. Cochrane systematic review of colorectal cancer screening using the fecal occult blood test (hemoccult): an update. Am J Gastroenterol. 2008;103:1541–9. doi: 10.1111/j.1572-0241.2008.01875.x. [DOI] [PubMed] [Google Scholar]
- 4.Shaukat A, Mongin SJ, Geisser MS, Lederle FA, Bond JH, Mandel JS, Church TR. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369:1106–14. doi: 10.1056/NEJMoa1300720. [DOI] [PubMed] [Google Scholar]
- 5.Yao X, Tian Z. Dyslipidemia and colorectal cancer risk: a meta-analysis of prospective studies. Cancer causes & control : CCC. 2015;26:257–68. doi: 10.1007/s10552-014-0507-y. [DOI] [PubMed] [Google Scholar]
- 6.Tian Y, Wang K, Li J, Wang J, Wang Z, Fan Y, Ye Y, Ji G, Li Y. The association between serum lipids and colorectal neoplasm: a systemic review and meta-analysis. Public health nutrition. 2015;18:3355–70. doi: 10.1017/S1368980015000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardou M, Barkun A, Martel M. Effect of statin therapy on colorectal cancer. Gut. 2010;59:1572–85. doi: 10.1136/gut.2009.190900. [DOI] [PubMed] [Google Scholar]
- 8.Mamtani R, Lewis JD, Scott FI, Ahmad T, Goldberg DS, Datta J, Yang YX, Boursi B. Disentangling the Association between Statins, Cholesterol, and Colorectal Cancer: A Nested Case-Control Study. PLoS medicine. 2016;13:e1002007. doi: 10.1371/journal.pmed.1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lytras T, Nikolopoulos G, Bonovas S. Statins and the risk of colorectal cancer: an updated systematic review and meta-analysis of 40 studies. World journal of gastroenterology. 2014;20:1858–70. doi: 10.3748/wjg.v20.i7.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? International journal of epidemiology. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 11.Holmes MV, Asselbergs FW, Palmer TM, Drenos F, Lanktree MB, Nelson CP, Dale CE, Padmanabhan S, Finan C, Swerdlow DI, Tragante V, et al. Mendelian randomization of blood lipids for coronary heart disease. European heart journal. 2015;36:539–50. doi: 10.1093/eurheartj/eht571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bull CJ, Bonilla C, Holly JM, Perks CM, Davies N, Haycock P, Yu OH, Richards JB, Eeles R, Easton D, Kote-Jarai Z, et al. Blood lipids and prostate cancer: a Mendelian randomization analysis. Cancer medicine. 2016;5:1125–36. doi: 10.1002/cam4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlando G, Law PJ, Palin K, Tuupanen S, Gylfe A, Hänninen U, Cajuso T, Tanskanen T, Kondelin J, Kaasinen E, Sarin A-P, et al. Variation at 2q35 (PNKD and TMBIM1) influences colorectal cancer risk and identifies a pleiotropic effect with inflammatory bowel disease. Human Molecular Genetics. 2016 doi: 10.1093/hmg/ddw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houlston RS. members of C. COGENT (COlorectal cancer GENeTics) revisited. Mutagenesis. 2012;27:143–51. doi: 10.1093/mutage/ger059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiffin N, Hosking FJ, Farrington SM, Palles C, Dobbins SE, Zgaga L, Lloyd A, Kinnersley B, Gorman M, Tenesa A, Broderick P, et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Human molecular genetics. 2014;23:4729–37. doi: 10.1093/hmg/ddu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Tassan NA, Whiffin N, Hosking FJ, Palles C, Farrington SM, Dobbins SE, Harris R, Gorman M, Tenesa A, Meyer BF, Wakil SM, et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep. 2015;5:10442. doi: 10.1038/srep10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International HapMap C. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 18.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, Consortium E-I Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. European journal of epidemiology. 2015;30:543–52. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. International Journal of Epidemiology. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. International journal of epidemiology. 2014;43:922–9. doi: 10.1093/ije/dyu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ference BA, Robinson JG, Brook RD, Catapano AL, Chapman MJ, Neff DR, Voros S, Giugliano RP, Davey Smith G, Fazio S, Sabatine MS. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. New England Journal of Medicine. 2016;375:2144–53. doi: 10.1056/NEJMoa1604304. [DOI] [PubMed] [Google Scholar]
- 26.Berndt SI, Gustafsson S, Magi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, Esko T, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvis D, Mitchell JS, Law PJ, Palin K, Tuupanen S, Gylfe A, Hanninen UA, Cajuso T, Tanskanen T, Kondelin J, Kaasinen E, et al. Mendelian randomisation analysis strongly implicates adiposity with risk of developing colorectal cancer. Br J Cancer. 2016;115:266–72. doi: 10.1038/bjc.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thrift AP, Gong J, Peters U, Chang-Claude J, Rudolph A, Slattery ML, Chan AT, Locke AE, Kahali B, Justice AE, Pers TH, et al. Mendelian Randomization Study of Body Mass Index and Colorectal Cancer Risk. Cancer Epidemiol Biomarkers Prev. 2015;24:1024–31. doi: 10.1158/1055-9965.EPI-14-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nead KT, Sharp SJ, Thompson DJ, Painter JN, Savage DB, Semple RK, Barker A, Australian National Endometrial Cancer Study G. Perry JR, Attia J, Dunning AM, et al. Evidence of a Causal Association Between Insulinemia and Endometrial Cancer: A Mendelian Randomization Analysis. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess S, Harshfield E. Mendelian randomization to assess causal effects of blood lipids on coronary heart disease: lessons from the past and applications to the future. Curr Opin Endocrinol Diabetes Obes. 2016;23:124–30. doi: 10.1097/MED.0000000000000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J, Shin H, Wei X, Kadegowda AK, Chen R, Xie SK. NPC1L1 knockout protects against colitis-associated tumorigenesis in mice. BMC Cancer. 2015;15:189. doi: 10.1186/s12885-015-1230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Q, Wang Q, Fan H, Wang J, Liu X, Wang H, Wang Y, Hu R. Dietary cholesterol promotes AOM-induced colorectal cancer through activating the NLRP3 inflammasome. Biochem Pharmacol. 2016;105:42–54. doi: 10.1016/j.bcp.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Jung YS, Park CH, Eun CS, Park DI, Han DS. Statin Use and the Risk of Colorectal Adenoma: A Meta-Analysis. Journal of gastroenterology and hepatology. 2016 doi: 10.1111/jgh.13393. [DOI] [PubMed] [Google Scholar]
- 34.Murai T. Cholesterol lowering: role in cancer prevention and treatment. Biol Chem. 2015;396:1–11. doi: 10.1515/hsz-2014-0194. [DOI] [PubMed] [Google Scholar]
- 35.Frattini M, Balestra D, Suardi S, Oggionni M, Alberici P, Radice P, Costa A, Daidone MG, Leo E, Pilotti S, Bertario L. Different genetic features associated with colon and rectal carcinogenesis. Clin Cancer Res. 2004;10:4015–21. doi: 10.1158/1078-0432.CCR-04-0031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.