Abstract
Systems medicine holds many promises, but has so far provided only a limited number of proofs of principle. To address this road block, possible barriers and challenges of translating systems medicine into clinical practice need to be identified and addressed. The members of the European Cooperation in Science and Technology (COST) Action CA15120 Open Multiscale Systems Medicine (OpenMultiMed) wish to engage the scientific community of systems medicine and multiscale modelling, data science and computing, to provide their feedback in a structured manner. This will result in follow-up white papers and open access resources to accelerate the clinical translation of systems medicine.
Keywords: systems medicine, modelling, data science, computing
Introduction
Human health and disease are characterized by a complex interplay of multiple factors, from the genome to the exposome. For many complex diseases, a sufficiently detailed understanding of the underlying mechanisms has remained elusive, and therefore, the development of effective cures continues to be major challenge. The socioeconomic burden (morbidity, mortality, financial cost) of complex diseases thus remains high and is likely to grow within Europe’s aging population. Systems medicine is an emerging interdisciplinary framework that aims to improve our understanding, prevention and treatment of complex diseases by integrating knowledge and data across multiple levels of biomedical organization [1] (see also Supplementary Figure S1 and Supplementary Table S1 for further references). It represents the implementation of systems biology approaches in medical concepts, research and practice, where the outcome is measurable improvement of patient health. The clinical and societal drivers of systems medicine include (i) mechanism-based drugs combined with patient stratification approaches; (ii) biomarkers and their multidimensional combination; (iii) rational design of therapies; and (iv) reduction of discovery, development and healthcare costs [2, 3].
The ultimate challenge and vision of multiscale systems medicine is a radical paradigm shift, from a scale-specific reductionistic to multiscale systems medicine. To facilitate this process, the European Cooperation in Science and Technology (COST) Action CA15120 Open Multiscale Systems Medicine (OpenMultiMed) has been initiated [4], in synergy with other European systems medicine/biology research and infrastructure efforts, e.g. Coordinating Systems Medicine across Europe (CASyM), European Association of Systems Medicine (EASyM) and Infrastructure for Systems Biology in Europe (ISBE). The two elements of CA15120 represent the main specific S&T challenge:
To develop novel multiscale systems medicine concepts, methods and technologies that provide effective, efficient and economical solutions for emerging and future approaches to multiscale systems medicine.
To develop a transdisciplinary multiscale systems medicine framework that integrates systems medicine, multiscale modelling, multiscale data science and multiscale computing at the level of research, education and training.
It is agreed that a major challenge of today’s medicine is coping with the technological revolution, specifically with the application of the big data approach to massive information stored in multiple formats, at different clinical sites, and with sensitive ethical issues. The goal is to integrate this within the everyday clinical practice in the benefit of the patient. This integration will likely be a long process, partially because of drawbacks in the current medical and other higher education programmes that do not cover adequately the aspects of data-driven and multimodal next-generation human medicine.
This last aspect is worth highlighting: to tackle the challenges inherent in the future healthcare and systems medicine, researchers and clinicians will have to join efforts, using a shared conceptual framework and highly interdisciplinary approaches. This is nevertheless far from trivial, as different disciplines seldom share the same language in tackling problems. If having a single review of the state of the art might prima facie seem as a good starting point to synchronize and homogenize all knowledge in the field, a previous step is essential: clinical researchers should be able to pose questions and demands to the next generation of scientists, and vice versa. In this opinion article, we present OpenMultiMed’s effort to achieve such communication. We discuss four questionnaires, aimed at gathering feedbacks from the community, and review their main themes. We further invite all readers to participate in this collective effort, which we hope will help materialize the large number of systems medicine’s promises into concrete achievements.
Exploring the barriers and challenges of multiscale systems medicine
The dialogue we here aim at achieving requires the inputs from different scientific disciplines: systems medicine, including clinical expertise, multi scale modelling, multiscale data science and multiscale computing. For the sake of simplicity, four different questionnaires have been created (see Table 1 for access links), whose content is reviewed in this section. Note that, beside strictly scientific questions, we also gather aggregated background and geographical information of researchers, to ensure a correct stratification and the absence of biases in the results. To verify the quality and accessibility of the questionnaires, we conducted two trials, first involving few members of the OpenMultiMed action for then opening to the full consortium, whose feedbacks have been used to add new questions and improve the global user experience. Participants are going to be rewarded by listing their names as part of the consortium of scientific collaborators, which will be one of the co-authors of all manuscripts based on the use of the collected data.
Table 1.
Access to the four questionnaires—also available through the OpenMultiMed Web portal [3]
| Topic | Questionnaire URL |
|---|---|
| Systems medicine | https://goo.gl/forms/uyjONFlVfeSZzc5y1 |
| Multiscale modelling | https://goo.gl/forms/wipuMU8Af1d8QE6y2 |
| Multiscale data science | https://goo.gl/forms/Nbme7OxFFeH3fDB32 |
| Multiscale computing | https://goo.gl/forms/Etb9r2iZe92i5UmF2 |
Systems medicine
We are facing a technological revolution, where we can screen individual human genomes, measure numerous biochemical parameters of the blood and tissues and search organs by imaging techniques, resulting in enormous amounts of data from humans with multifactorial diseases. However, the technological and information revolution has so far only barely influenced the clinics. A key fact is that most patients with common diseases are still treated in a rather one-dimensional or standardized manner not taking the individual complexity of multifactorial diseases into account, which is a challenge to modern healthcare, causing both suffering and enormous costs [5]. Only a global (systems) view on physiology and pathophysiology, where data sets from distinct fields of science, at different scales and set-ups are combined, from heterogenous populations of patients, model organisms, in different time frames, modes of sample analyses, etc., can bring a major step forward. This has recently been exemplified, e.g. in novel drug target identifications and treatment options [6–8].
Another difficult aspect is the quality and content of the databases used in clinical and basic research [9]. Their content is subject to constant torrent of novel data sets, whereas past data sets are being discarded through curation and quality control. Although complete data sets encompassing sequence and metadata information at the levels of genomes are progressively being deposited to public databases, there are multiple ethical restrictions for human data. Additionally, proteomic, post-translational and metabolomic levels are frequently missing; hence, in most cases, preventing the assembly of data sets amenable for complex multiscale analyses required for improved medicine. In essence, we are talking about data limitations, despite an increasing quantity of data, and the problem of reproducibility. Hence, we call for a more concerted way of organizing multiscale systems medicine research, as it is initiated already in certain subfields like cancer research, with The Cancer Genome Atlas [10] serving as platform for molecular and clinical data sets.
In cancer, no longer organ-based disease definitions are used. In contrast, all major non-communicable and chronic disease definitions rely on an apparent symptom (hypertension; asthma; depression), the affected organ (heart failure, retinopathy, nephropathy, etc.) or the name of a doctor (Parkinson; Alzheimer). Drug discovery often relies on correcting symptoms or normalizing risk factors (e.g. blood cholesterol; glucose; blood pressure) assuming that by modulating a surrogate parameter or risk factor, the relevant outcome (e.g. prevention of a heart attack, stroke or death) will also be achieved. But neither symptoms nor risk factors are mechanistic definitions of a disease. In fact, in most cases, we do not understand what exactly causes a disease phenotype. Exceptions are rare diseases, where a precise (often single and severe) mutation is known and sometimes a specific therapy is available. If all diseases will be mechanistically defined, it is predicted that all diseases, also those that are currently labelled as ‘common’ diseases, will become rare diseases. This implies that we need to achieve entirely different disease definitions as today. The use of such 18th/19th century disease definitions explains why 20th/21st century techniques such as multi-omics have still not achieved major breakthroughs and applications in clinical medicine.
The systems medicine concept constitutes thus a paradigm shift that should initially be dominated by integrative types of analyses (meta-analyses of existing fragmentary data) to generate predictive and hypothesis-generating mathematical models, as recently exemplified for liver pathologies [11] and chronic immune disorders [12] and eventually their clinical application in new drugs and diagnostics. The final aim is to understand human disease at the systems level, and predict the progression and treatment options for each individual patient. This concept of integrative meta-analyses should be deemed as the new frontiers of a systems approach.
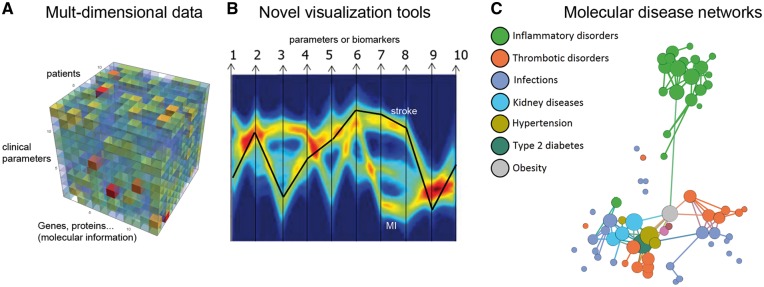
The human genome contains about 19 000 genes [13] and a proteomic potential of 106 proteins with innumerable post-translational modifications creating a vast diversity of functions and regulatory processes. Another level of complexity is caused by the dynamic processes of metabolism with about 4000 metabolites in human serum and the complex interactions with commensal microbes. All these different data, comprising genes, proteins, regulatory RNAs, as well as metabolites have to be combined with clinical data for sufficiently large groups of individuals to enable a more systematic view of human physiology. To achieve this, novel tools of analysis and visualization are needed to elucidate the underlying molecular disease networks and to identify crucial disease hubs suitable for therapeutic interventions (Figure 1). To further catalyse an advancement of systems medicine and to obtain an overview of the current state, expectations, needs and visions, and to achieve coherence and cooperativity in the community, an online questionnaire has been set up to collect feedback from scientists in the field (Table 1).
Figure 1.
Illustration of a systems medicine workflow: (A) multidimensional data comprising molecular information (gene expression, proteins, lipids, metabolites, etc.) for a large group of individuals as well as clinical parameters or biomarkers (plotted in a three-dimensional format with Mathematica); (B) example of visualization of multiple factors or biomarkers by a multi-parallel coordinate plot—adapted from [14] with permission from Wiley Online (e.g. patients with thrombotic diseases). Each arrow from 1 to 10 represents a biomarker, a clinical or a molecular parameter and is plotted in arbitrary units on a separate y-axis. Each patient represents a line linking the values of all the parameters (1–10). Data from numerous patients result in a high-density bundle of lines, which are represented in a pseudocolor mode to visualize their frequencies. In this example, the parameters of a single patient are shown as black line implying that the individual falls within the subgroup of stroke patients, although the person did not present with clear symptoms of stroke. The second major subgroup represents myocardial infarction, as indicated. (C) Molecular data from comorbidities can be used to calculate disease networks to identify nodes and hubs as promising targets for drug combination strategies and precision medicine.
Multiscale modelling
Mathematical and computational models allow formalizing our mechanistic knowledge of biological systems through sets of rules and equations, which can be simulated or analysed taking advantage of the computational power nowadays available [15] (see also ‘Multiscale computing’ section). Modelling efforts in systems biology originally started in the lower part of the complexity spectrum, focusing on isolated regulatory, signalling or metabolic pathways. The development of systems biology into systems medicine brought us to a state where much more complex systems has to be analysed and understood in the context of pathogenesis. Some diseases will require to consider the whole human body as the right scale to understand and fight them.
Avoiding complexity is not a viable option, as processes at a molecular level have to be linked with the behaviour of tissues, entire organs or even the whole organism. This is for instance the rationale for the interplay between individual cells and organism-level pharmacokinetic and pharmacodynamics models [16]. Different types of molecular and cellular processes (metabolism, signalling, gene regulation networks, cell-to-cell communication, etc.) require the use of distinctive modelling approaches (e.g. stoichiometric, kinetic or boolean models). However, considering a biomedical problem from different organizational (molecular interactions, cells, tissues, etc.) and time (from femtoseconds to minutes, to years) scales and interlink them will require constructing complex, multiscale and potential hybrid models. In other words, having a modular system in which the monolithic problem is split into smaller, interconnected fragments [17].
Such change in strategy entails several challenges, which are dealt with in the second questionnaire. The questionnaire is about analysing the features of multiscale models currently used in systems medicine, and compare them with those developed in other fields like astrophysics, process engineering or geophysics. The underlying idea is to extract the common and distinctive key features of multiscale modelling across scientific disciplines. Further, the modelling of biological events spanning different organizational levels requires the use of different modelling frameworks because of the nature of the processes investigated and the features of the experimental data available. Thus, one may need to develop hybrid models combining different modelling frameworks at different scales. The questionnaire deals with the analysis of multiscale hybrid modelling strategies used in systems medicine now or in a next future. Specifically, we foresee that a community effort is needed to create standards, procedures and tools for interlinking models at different scales, dynamics, locations and other dimensions. This would enable re-using the existing models in larger constructs, facilitating the coordinated development of interlinkable and specialized Lego-like modelling building blocks.
Multiscale data science
The concept of data analysis is an old one, stemming from the classical fields of statistical learning [18], Bayesian statistics and machine learning [19]; yet, the application of analysis techniques to large sets of real data has only recently emerged, thanks to the advances in computation and storage capabilities. Data science holds the prospect of generating new knowledge about diseases and their cures, thus enabling an evolving and learning medicine [20]. Yet, as any new paradigm, it also faces several important challenges and barriers, slowing down its adoption by the community. First, data science has no well-defined boundaries: it leverages on and integrates several disciplines, from computer science and user interaction, to applied mathematics. The practitioner should thus be aware of multiple techniques, many of them outside his or her core expertise. Secondly, these techniques have initially been developed for tackle other problems, and as such their adaptation gives birth to multiple problems. For instance, topics like ethics [21] and data confidentiality [22], secondary in other fields, are of utmost importance in medicine. The aim of the third questionnaire is 2-fold: (i) to review what are the data analysis techniques expected by the community to be key in the next years, and (ii) to identify the problems commonly encountered by practitioners. Detailed information on modelling approaches is gathered.
Multiscale computing
Multiscale computing encompasses all computational challenges regarding multiscale modelling and multiscale data science. The challenges involve the technical coupling of different simulation models, integrative computational algorithms, multiscale data processing and analytics at scale, and doing all this using large-scale computer infrastructures. A multiscale simulation consists of two or more submodels, each operating on its unique temporal and spatial scale [23]. Multiscale computing has met its suitable implementation infrastructure in the Cloud computing paradigm, which contains shared computing resources, providing high processing power [24]. The resources required by the deployed services, such as CPU power, internal memory and network load, are allocated on demand, providing an autonomous and highly dynamic scalable environment. This paradigm however is suitable only if the sub-model components are only loosely interconnected in terms of dynamical evolution, otherwise other ‘classical’ high-performance computing solutions are to be preferred.
Multiscale computing in systems medicine has grown steadily in the past decades [25], and researchers now recognize that many of the challenges in multiscale computing transcend disciplinary boundaries. Although there are a range of discipline-specific toolkits (e.g. VPH Hypermodelling Framework [26]), in recent years, portable and generic tools to facilitate multiscale computing are becoming more prevalent. For example, the MUSCLE2 coupling toolkit [27] has been applied to climate, fusion, astrophysics and biomedicine models. Likewise, workflow engines such as Kepler [28], formalisms such as the multiscale modelling and simulation framework [23] and generic paradigms such as hierarchical multiscale modelling [29] and multiscale computing patterns [30] are finding uptake across different scientific domains. Future challenges in multiscale computing include the effective mapping of these applications at high-end computational resources, as well as quantifying the error of such models [31]. In addition, the complexity involved in deploying and using these applications in computational research has led to the development of new automation approaches such as FabSim [32] and MultiGrain/MAPPER [33], the latter of which was used to automate the inference of gene regulatory networks.
The questionnaire on multiscale computing is intended to help gathering the consensus on the meaning of multiscale in these contexts, to analyse the extent to which these techniques have been adopted and to determine the areas where major challenges reside.
Conclusions
In a first effort to promote a structured exchange of opinions and knowledge between the four communities that will participate in the future systems medicine playground (medicine, modelling, data science and computing), we here presented four questionnaires, which we share with all readers, and whose results will be made public in the future. It is our belief that such collective effort will help outlining the needs and challenges that will surely emerge from this new endeavour, ultimately leading to concrete life-improving achievements. We further invite all readers to interact with the authors of this contribution and with OpenMultiMed, by sharing any idea these questionnaires may have generated.
Supplementary Material
Massimiliano Zanin received his PhD in electrotecnical engineering by the Universidade Nove de Lisboa. His research interests include data analysis and complex network theory.
Ivan Chorbev completed his PhD studies at the Faculty of Electrical Engineering and Information Technologies in Skopje in 2009. He is currently the Dean of the Faculty for Computer Science and Engineering, University Ss. Cyril and Methodius in Skopje.
Blaz Stres is an Associate Professor of Microbiology at University of Ljubljana, Slovenia. His research interests include human intestinal tract microbiome multi-omics data analyses, bioinformatics and systems medicine.
Egils Stalidzans has received doctoral degree in information technology. He is active in systems biology field since 2006 developing models and modeling software. He is member of Virtual Physiological Human Institute.
Julio Vera is professor in Systems Tumor Immunology at the FAU Erlangen-Nuremberg, Germany. He works in data-driven mathematical modeling of biochemical pathways and networks relevant to diseases.
Paolo Tieri is a physicist with expertise in immunology. He is mainly involved in network biology, systems medicine and theoretical immunology.
Filippo Castiglione is a computer scientist interested in complex systems and mathematical biology, in particular in the immune system and related pathologies.
Derek Groen is a Lecturer in Simulation and Modelling at Brunel University London. Derek specializes in multiscale computing and high-performance computing, has published about 25 journal papers and is one of the main developers of the HemeLB bloodflow solver.
Huiru Zheng is a Professor of Computer Science at Ulster University, United Kingdom. Her research interests include integrative data analytics in the field of systems biology and systems medicine.
Jan Baumbach is Professor at the University of Southern Denmark where he is heading the Computational Biomedicine unit. He is known for his research on unsupervised machine learning in bioinformatics.
Johannes Schmid is coordinating a research network of 10 groups investigating links between inflammation, cardiovascular diseases and cancer, at molecular, cellular and clinical levels.
José Basilio is bioinformatician at the Medical University of Vienna with a biology background, mainly working on omics- and pathway analyses.
Nataša Debeljak, is an Associate Professor of University of Ljubljana, Faculty of Medicine, and a Board member of the COST CA15120, specialized in wet-lab part of systems medicine of multifactorial human conditions like erythrocytosis.
Peter Klimek is an Associate Professor at the Section for Science of Complex Systems at the Medical University of Vienna. His research focuses on novel data analysis and modelling approaches in medicine and healthcare.
Damjana Rozman is a Full Professor of University of Ljubljana, Faculty of Medicine, covering systems medicine of multifactorial metabolic disorders, deputy member of the COST CA15120 Board and member of the EASyM Board.
Harald Schmidt is a physician scientist in target discovery and drug development, chairing the COST action OpenMultiMed and the in silico drug repositioning H2020 programme, REPO-TRIAL.
Key Points
Most diseases are characterized by a complex interplay of multiple factors, which is the focus of the new systems medicine approach.
Most disease definitions are 18th/19th century and organ- or symptom-based and may mechanistically combine entirely different phenotypes.
A necessary step is the identification of barriers and challenges of multiscale systems medicine through a collective effort.
We here present four questionnaires through which we aim at gathering feedbacks from the community.
Funding
J.A.S. and J.B. were supported by the Austrian Science Fund: Special Research Program SFB-F54. The European Cooperation in Science and Technology (COST) Action CA15120 OpenMultiMed (http://openmultimed.net).
References
- 1. Auffray C, Chen Z, Hood L.. Systems medicine: the future of medical genomics and healthcare. Genome Med 2009;1(1):2 10.1186/gm2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hood L, Flores M.. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. New Biotechnol 2012;29(6):613–24. 10.1016/j.nbt.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 3. Castellani GC, Menichetti G, Garagnani P, et al. Systems medicine of inflammaging. Brief Bioinform 2016;17(3):527–40. 10.1093/bib/bbv062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.OpenMultiMed’s. http://openmultimed.net/.
- 5. Benson M. Clinical implications of omics and systems medicine: focus on predictive and individualized treatment. J Int Med 2016;279(3):229–40. 10.1111/joim.12412 [DOI] [PubMed] [Google Scholar]
- 6. Singh N, Freiesleben S, Gupta SK, et al. Identification of antineoplastic targets with systems approaches, using resveratrol as an in-depth case study. Curr Pharm Des 2017; doi: 10.2174/1381612823666170710152918. [DOI] [PubMed] [Google Scholar]
- 7. Gupta SK, Jaitly T, Schmitz U, et al. Personalized cancer immunotherapy using systems medicine approaches. Brief Bioinform 2016;17(3):453–67. 10.1093/bib/bbv046 [DOI] [PubMed] [Google Scholar]
- 8. Ballesta A, Innominato PF, Dallmann R, et al. Systems chronotherapeutics. Pharmacol Rev 2017;69(2):161–99. 10.1124/pr.116.013441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McMurry J, Juty N, Blomberg N, et al. Identifiers for the 21st century: how to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. PLoS Biol 2017;15(6):e2001414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Cancer Genome Atlas’s. https://cancergenome.nih.gov/.
- 11. Cvitanović T, Reichert MC, Moškon M, et al. Large‐scale computational models of liver metabolism: how far from the clinics? Hepatology 2017;66(4):1323–34. [DOI] [PubMed] [Google Scholar]
- 12. Te Velde AA, Bezema T, van Kampen AH, et al. Embracing complexity beyond systems medicine: a new approach to chronic immune disorders. Front Immunol 2016;7:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ezkurdia I, Juan D, Rodriguez JM, et al. Multiple evidence strands suggest that there may be as few as 19, 000 human protein-coding genes. Hum Mol Genet 2014;23(22):5866–78. 10.1093/hmg/ddu309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Streit M, Ecker RC, Österreicher K, et al. 3D parallel coordinate systems—a new data visualization method in the context of microscopy‐based multicolor tissue cytometry. Cytometry A 2006;69A(7):601–11. [DOI] [PubMed] [Google Scholar]
- 15. Kitano H. Computational systems biology. Nature 2002;420(6912):206–10. 10.1038/nature01254 [DOI] [PubMed] [Google Scholar]
- 16. Wolkenhauer O, Auffray C, Brass O, et al. Enabling multiscale modeling in systems medicine. Genome Med 2014;6(3):21 10.1186/gm538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ayton GS, Noid WG, Voth GA.. Multiscale modeling of biomolecular systems: in serial and in parallel. Curr Opin Struct Biol 2007;17(2):192–8. 10.1016/j.sbi.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 18. Lee PM. Bayesian Statistics: An Introduction. New York: John Wiley & Sons, 2012. [Google Scholar]
- 19. Vapnik VN, Vapnik V.. Statistical Learning Theory, Vol. 1 New York, NY: Wiley, 1998. [Google Scholar]
- 20. Krumholz HM. Big data and new knowledge in medicine: the thinking, training, and tools needed for a learning health system. Health Aff 2014;33(7):1163–70. 10.1377/hlthaff.2014.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mittelstadt BD, Floridi L.. The ethics of big data: current and foreseeable issues in biomedical contexts In: The Ethics of Biomedical Big Data. Berlin, Germany: Springer International Publishing, 2016. [DOI] [PubMed] [Google Scholar]
- 22. Roski J, Bo-Linn GW, Andrews TA.. Creating value in health care through big data: opportunities and policy implications. Health Aff 2014;33(7):1115–22. 10.1377/hlthaff.2014.0147 [DOI] [PubMed] [Google Scholar]
- 23. Chopard B, Borgdorff J, Hoekstra AG.. A framework for multi-scale modelling. Philos Trans A Math Phys Eng Sci 2014;372(2021):20130378 10.1098/rsta.2013.0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoefer CN, Karagiannis G.. Taxonomy of cloud computing services In: GLOBECOM Workshops (GC Wkshps), 2010. IEEE, Miami, USA. [Google Scholar]
- 25. Groen D, Zasada SJ, Coveney PV.. Survey of multiscale and multiphysics applications and communities. IEEE Comput Sci Eng 2014;16(2):34–43. 10.1109/MCSE.2013.47 [DOI] [Google Scholar]
- 26. Tartarini D, Duan K, Gruel N, et al. The VPH Hypermodelling framework for cancer multiscale models in the clinical practice. In: Proceedings of the 6th International Advanced Research Workshop on In Silico Oncology and Cancer Investigation (IARWISOCI), 2014, pp. 1–4. IEEE, Athens, Greece.
- 27. Borgdorff J, Mamonski M, Bosak B, et al. Distributed multiscale computing with MUSCLE 2, the multiscale coupling library and environment. J Comput Sci 2014;5(5):719–31. 10.1016/j.jocs.2014.04.004 [DOI] [Google Scholar]
- 28. Ludäscher B, Altintas I, Berkley C, et al. Scientific workflow management and the Kepler system. Concurr Comput 2006;18(10):1039–65. [Google Scholar]
- 29. Weinan E, Engquist B, Li X, et al. Heterogeneous multiscale methods: a review. Commun Comput Phys 2007;2(3):367–450. [Google Scholar]
- 30. Alowayyed S, Groen D, Coveney PV, et al. Multiscale computing in the exascale era. J Comput Sci 2017;22:15–25. 10.1016/j.jocs.2017.07.004 [DOI] [Google Scholar]
- 31. Hoekstra A, Chopard B, Coveney P.. Multiscale modelling and simulation: a position paper. Philos Trans R Soc Lond A 2014;372(2021):20130377 10.1098/rsta.2013.0377 [DOI] [PubMed] [Google Scholar]
- 32. Groen D, Bhati AP, Suter J, et al. FabSim: facilitating computational research through automation on large-scale and distributed e-infrastructures. Comput Phys Commun 2016;207:375–85. 10.1016/j.cpc.2016.05.020 [DOI] [Google Scholar]
- 33. Mizeranschi A, Kennedy N, Thompson P, et al. The influence of network topology on reverse-engineering of gene-regulatory networks. Procedia Comput Sci 2014;29:410–21. 10.1016/j.procs.2014.05.037 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.