Abstract
Background
The usefulness of ribonucleotide reductase catalytic subunit M1 (RRM1) for predicting the therapeutic effects of gemcitabine-containing chemotherapy in patients with non-small cell lung cancer (NSCLC) remains controversial. RRM1-positive patients show unique clinicopathological features.
Methods
Here, we performed a meta-analysis to systematically evaluate the relationship between RRM1 expression and the clinicopathological characteristics of NSCLC patients treated with gemcitabine-containing regimens. A comprehensive electronic and manual search was performed to identify relevant articles. The pooled relative risk (RR) and 95% CI were used to estimate the relation between the clinicopathological characteristics of NSCLC patients and RRM1 expression.
Results
The study included 31 observational studies and 3,667 patients. The analysis showed no significant association between RRM1 expression and pathological type, stage, and smoking status; however, RRM1 positivity was significantly lower in women than in men (43.0% vs 51.7%, RR=0.84, 95% CI: 0.74–0.94, P=0.004).
Conclusion
The present pooled analyses demonstrated that RRM1 positivity in women with advanced NSCLC was associated with a higher rate of response to gemcitabine-containing regimens. Immunohistochemistry may be valuable to prescreen for RRM1 expression in clinical practice, whereas PCR can be routinely used as a verification method. These findings will help design suitable molecular-targeted therapies for NSCLC.
Keywords: RRM1, gemcitabine, meta-analysis, clinicopathological features, NSCLC
Introduction
Lung cancer is a common malignancy and the main cause of cancer-related mortality in the world. Non-small cell lung cancer (NSCLC) accounts for 80% of all lung cancer patients worldwide.1–3 Lung carcinoma is divided into two subtypes according to the response to conventional treatments and the histological features of tumors: small cell lung carcinoma and NSCLC. Despite advances in the therapeutic approaches to the treatment of lung cancer, including immunotherapy, chemotherapy, radiotherapy, and noninvasive surgery, the 5-year relative survival rate of patients with lung cancer iŝ18% in the USA.1
First-line chemotherapy for NSCLC usually consists of a platinum-containing doublet regimen. Patients with advanced NSCLC are generally treated with chemotherapy drugs, including cisplatin and gemcitabine.4 Gemcitabine, a pyrimidine nucleoside antimetabolite, is active in advanced NSCLC, especially in combination with a platinum derivative or a next-generation anticancer agent.5,6 Pharmacoeconomic evaluation suggests that a gemcitabine-based regimen is the least costly regimen for the treatment of advanced NSCLC.7 However, patients with advanced NSCLC may develop resistance to gemcitabine, which is associated with a poor prognosis. Therefore, identification of the markers for predicting clinical outcomes and treatment sensitivity in patients receiving gemcitabine chemotherapy would be of great value.
Ribonucleotide reductase catalyzes the reduction of ribonucleotide 5-diphosphate to deoxyribonucleotide 5-diphosphate for DNA synthesis and damage repair. The ribonucleotide reductase catalytic subunit M1 (RRM1) gene is located on chromosome segment 11p15.5 and encodes a key enzyme involved in DNA synthesis that catalyzes the biosynthesis of deoxyribonucleotides.8,9 Preclinical studies show that RRM1 is involved in gemcitabine sensitivity in NSCLC.10,11 High levels of RRM1 are associated with a decreased response to gemcitabine-containing regimens, whereas RRM1 downregulation is associated with a high rate of response to gemcitabine-containing regimens.12,13 Therefore, RRM1 may be a predictive biomarker for gemcitabine chemotherapy in NSCLC.
Reverse transcriptase PCR (RT-PCR) and immunohistochemistry (IHC) are currently used to detect RRM1. However, an effective algorithm for RRM1 gene screening in the clinical lung cancer population remains undetermined because these two approaches have both advantages and disadvantages. Thus, to increase the detection efficiency of the two methodologies, we studied whether combining the clinicopathological features of NSCLC with RRM1 detection would yield valuable information for the effective prescreening of patients in clinical practice. Although a large number of patients carrying the RRM1 gene show unique clinicopathological characteristics,13–17 the detailed clinicopathological profiles remain unclear because of the small number of cases identified. Here, we performed a pooled analysis of a series of studies to assess the relationship between RRM1 gene expression in NSCLC and the clinicopathological characteristics of patients treated with gemcitabine-containing regimens. To the best of our knowledge, the present study is the first comprehensive and systematic analysis of the clinicopathological characteristics of NSCLC patients harboring the RRM1 gene.
Materials and methods
Literature search
EMBASE, PubMed, China National Knowledge Internet (CNKI), the WanFang database, and the Cochrane Library were searched for relevant studies up to September 2017. Electronic searches were performed using the terms “non-small cell lung cancer or NSCLC,” “gemcitabine,” and “ribonucleotide reductase M1 or RRM1.” The detailed search strategies are showed in Table S1. This meta-analysis followed the PRISMA guidelines.
Eligibility criteria
Articles were included based on the following criteria: 1) included NSCLC patients, regardless of the pathological phenotypes; 2) provided information about the RRM1 gene detection method and the clinicopathological characteristics of lung carcinoma patients harboring the RRM1 gene, including gender, clinical stage, smoking habits, and pathological type; and 3) were published as full-text articles. Review articles, conference abstracts, case reports, and those lacking sufficient data (because of the limited data) to evaluate the relative risk (RR) and 95% CI were excluded.
Data collection
Data extraction was independently performed by two authors (Chao Wu and Qiu-Ping Leng) using a standardized form. Data such as first author, year of publication, gender, number of patients, pathological type of tumors, clinical stage, detection method, and RRM1 expression were collected. Discrepancies were settled by discussion, with disagreements resolved by consensus.
Statistical analysis
The pooled RR and 95% CI were used to estimate the strength of the clinicopathological characteristics of NSCLC patients harboring the RRM1 gene. The Cochran’s Q-test and I2 statistics were used to measure the statistical heterogeneity. The random-effects model was implemented if the heterogeneity was significant (P<0.05 or I2>50%)18; otherwise, the fixed-effects model was used.19 Sensitivity analysis was performed by sequentially excluding studies from the current analysis to evaluate the stability of the pooled results. Begg’s test was performed to evaluate the possible potential publication bias of the studies (P<0.05 was considered significant).20 Meta-analyses were performed using Review Manager 5.3 (RevMan 5.3®; Nordic Cochrane Center, Copenhagen, Denmark). Categorical variables were analyzed by the χ2 test using SPSS 24.0 (SPSS Inc., Chicago, IL, USA), and P<0.05 was considered statistically significant.
Results
Characteristics of included studies
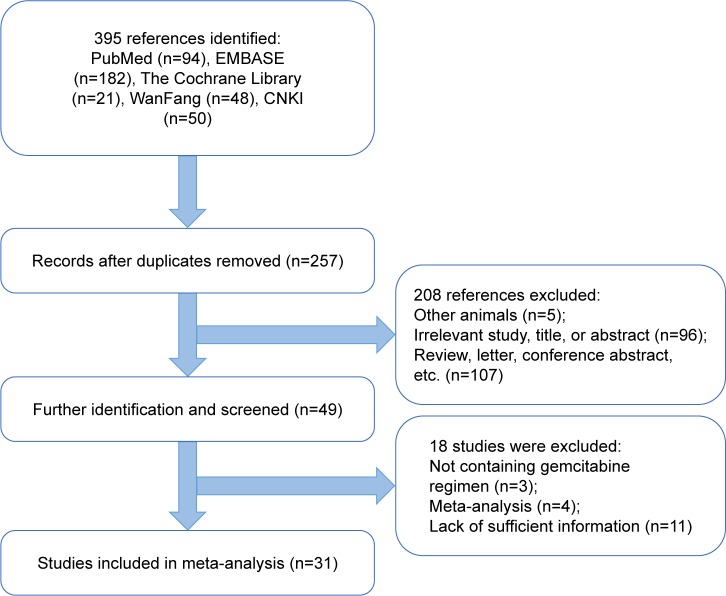
As shown in the flow diagram for the study selection process (Figure 1), 395 potentially relevant studies were initially obtained from PubMed, EMBASE, CNKI, the WanFang database, and the Cochrane Central Library. After screening the abstracts, titles, and contents, 31 observational studies including 3,667 patients were identified as eligible for the present study. Of the 31 studies, 16 (1,425 patients) reported on the relationship between RRM1 expression and gender in NSCLC patients treated with gemcitabine-containing chemotherapy, whereas the remaining 15 studies (1,242 patients) showed the relationship between RRM1 expression and pathological type. Three studies (288 patients) focused on the relationship between RRM1 expression and clinical stage, whereas eight studies (907 patients) focused on the association of RRM1 expression in NSCLC with smoking status. The baseline features of the selected studies are listed in Table 1.
Figure 1.
Flow diagram of study selection.
Table 1.
Baseline characteristics of the included studies
| Author | Year | Total | Gender
|
Smoking
|
Pathological type
|
TNM (stage)
|
RRM1 overall response
|
RRM1 detection | ||
|---|---|---|---|---|---|---|---|---|---|---|
| M (F) | Y (N) | AD (NAD) | I–II | III–IV | +(H) | −(L) | ||||
| Bepler et al12 | 2006 | 35 | 18 (17) | 33 (2) | 11 (24) | 1 | 34 | 4/16 | 11/19 | PCR |
| Rosell et al13 | 2004 | 75 | 62 (13) | NR | 33 (42) | NR | 75 | 1/4 | 6/12 | PCR |
| Rosell et al14 | 2004 | 67 | NR | NR | NR | NR | NR | 11/15 | 16/17 | PCR |
| Souglakos et al15 | 2008 | 53 | 45 (8) | NR | NR | NR | 53 | 6/25 | 7/17 | PCR |
| Guo et al29 | 2015 | 52 | 38 (14) | 33 (19) | 28 (24) | 15 | 28 | 4/25 | 3/13 | IHC |
| Xing et al30 | 2014 | 138 | 82 (56) | 72 (66) | 40 (98) | NR | 138 | 7/32 | 105/209 | IHC |
| Zeng and Shan31 | 2009 | 51 | 23 (28) | 21 (30) | 33 (18) | NR | 51 | 3/17 | 16/34 | IHC |
| Zhao et al32 | 2014 | 158 | 115 (43) | 36 (45) | 38 (43) | NR | 158 | 19/86 | 14/72 | PCR |
| Xian-Jun et al33 | 2014 | 208 | 158 (50) | 69 (139) | 92 (115) | NR | 208 | 40/104 | 49/104 | PCR |
| Wang et al34 | 2014 | 418 | 316 (102) | 154 (264) | 181 (237) | NR | 418 | 68/209 | 105/209 | PCR |
| Liang et al35 | 2014 | 377 | 269 (108) | 130 (247) | 118 (259) | NR | 377 | 114/236 | 50/114 | PCR |
| Dong et al36 | 2014 | 229 | 45 (36) | 36 (45) | 38 (43) | NR | 81 | 7/29 | 19/52 | IHC |
| Jian-Wei et al37 | 2013 | 294 | 202 (92) | 97 (197) | 101 (193) | NR | 294 | 101/185 | 51/109 | PCR |
| Su et al38 | 2011 | 85 | 67 (18) | 22 (63) | 44 (41) | NR | 85 | 0/4 | 8/18 | PCR |
| Wu et al39 | 2014 | 50 | 23 (27) | NR | NR | 18 | 32 | NR | NR | PCR |
| Wang et al40 | 2010 | 124 | 80 (44) | NR | 83 (41) | NR | 124 | 18/50 | 40/74 | IHC |
| Guo et al41 | 2010 | 142 | 82 (60) | NR | 75 (67) | NR | 142 | 22/71 | 27/71 | PCR |
| Lee et al42 | 2010 | 40 | 31 (9) | NR | 20 (20) | NR | 40 | 1/13 | 6/25 | IHC |
| Ma et al43 | 2014 | 68 | 47 (21) | 41 (27) | 31 (37) | 44 | 24 | NR | NR | IHC |
| Jiang et al44 | 2013 | 60 | 44 (16) | NR | 30 (30) | NR | 60 | 10/34 | 15/26 | IHC |
| Lin et al45 | 2016 | 40 | 24 (16) | 21 (19) | 30 (10) | NR | NR | 11/19 | 14/21 | IHC |
| Vilmar et al46 | 2013 | 140 | 83 (57) | NR | 61 (79) | NR | 140 | NR | NR | IHC |
| Xu et al47 | 2015 | 257 | 138 (119) | 99 (158) | NR | NR | NR | NR | NR | PCR |
| Liu et al48 | 2017 | 66 | 46 (20) | NR | 49 (17) | 36 | 30 | NR | NR | IHC |
| Liu et al49 | 2009 | 61 | 43 (18) | NR | 29 (32) | NR | 61 | 5/26 | 16/35 | IHC |
| Li and Liu50 | 2010 | 71 | 48 (23) | NR | 46 (25) | NR | 71 | 3/40 | 11/31 | IHC |
| Yang et al51 | 2009 | 30 | 24 (6) | NR | 5 (25) | NR | 30 | 2/16 | 11/14 | IHC |
| Zhang et al52 | 2012 | 49 | 33 (16) | 18 (31) | 35 (14) | NR | 49 | 4/25 | 12/24 | PCR |
| Bepler et al53 | 2008 | 52 | 26 (26) | 50 (2) | NR | 34 | 18 | 2/17 | 10/18 | PCR |
| Boukovinas et al54 | 2008 | 102 | 87 (9) | NR | NR | NR | 96 | 8/31 | 21/64 | PCR |
| Gao et al55 | 2011 | 75 | 50 (25) | 47 (28) | 49 (26) | NR | 75 | 9/29 | 19/46 | IHC |
Abbreviations: AD, adenocarcinoma; F, female; H, high; IHC, immunohistochemistry; L, low; M, male; NAD, non-adenocarcinoma; NR, no report; RRM1, ribonucleotide reductase catalytic subunit M1; RT-PCR, real-time reverse transcriptase PCR.
Meta-analysis results
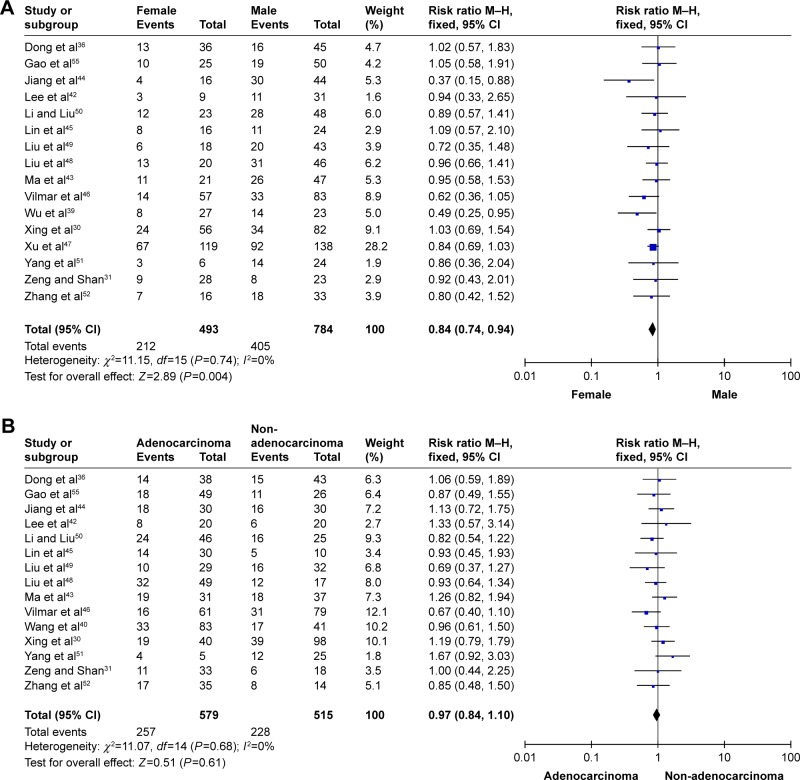
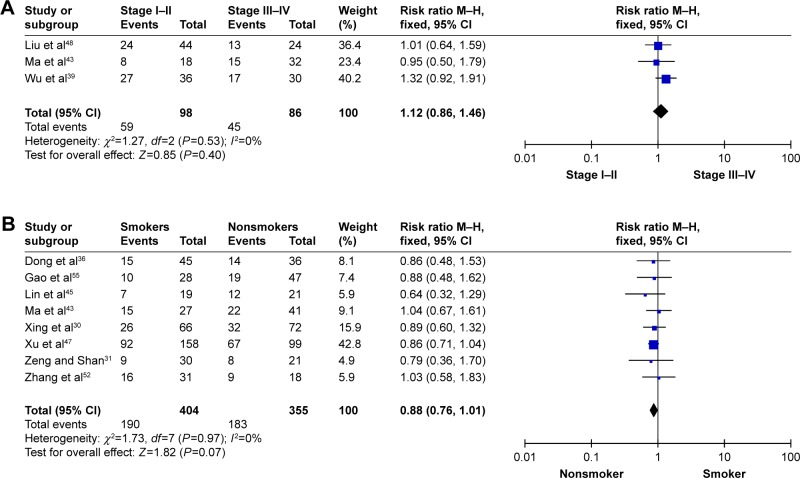
In total, 3,667 participants from 31 observational studies were included in the present meta-analysis; 1,772 patients (49.1%) harbored the RRM1 gene. Of the 31 studies, 16 studies demonstrated an association between RRM1 expression in NSCLC and gender. There was no significant heterogeneity among these studies (Pheterogeneity=0.74, I2=0%); therefore, a fixed-effects model was used to analyze the data. The pooled findings are shown in Figure 2A. Women with positive RRM1 expression had a statistically significantly lower risk of NSCLC than men (43.0% vs 51.7%, RR=0.84, 95% CI: 0.74–0.94, P=0.004). Fifteen studies assessed the role of RRM1 expression in the adenocarcinoma and non-adenocarcinoma groups. There was no significant heterogeneity among the studies (Pheterogeneity=0.68, I2=0%). The results of the study showed no significant differences between the adenocarcinoma and non-adenocarcinoma groups (44.4% vs 44.3%, RR=0.97, 95% CI: 0.84–1.10, P=0.61) (Figure 2B). Three studies investigated the association between RRM1 expression and tumor staging. There was no significant heterogeneity among these studies (Pheterogeneity=0.53, I2=0%); therefore, the data were analyzed using a fixed-effects model. The present meta-analysis demonstrated that there was no significant difference between stage I–II and stage III–IV (RR=0.88, P=0.07) (Figure 3A). In addition, eight studies evaluated the correlation between RRM1 gene expression and smoking history. There was no significant heterogeneity among these studies (Pheterogeneity=0.97, I2=0%), and hence the data were analyzed using a fixed-effects model. The results demonstrated that there was no significant difference between smokers and nonsmokers (RR=0.88, P=0.07) (Figure 3B).
Figure 2.
Meta-analysis of data for RRM1.
Notes: (A) Female vs male. (B) Adenocarcinomas vs non-adenocarcinomas.
Abbreviations: RRM1, ribonucleotide reductase catalytic subunit M1c; M–H, Mantel–Haenszel.
Figure 3.
Meta-analysis of data for RRM1.
Notes: (A) Stage I–II vs stage III–IV. (B) Nonsmokers vs smokers.
Abbreviations: M–H, Mantel–Haenszel; RRM1, ribonucleotide reductase catalytic subunit M1.
The research was extended to the analysis of two detection methods (RT-PCR and IHC) for direct comparison of sensitivity and specificity. Of 1,231 patients analyzed by IHC in 15 studies, 539 (43.8%) had high-level expression of RRM1; 2,265 patients had successful RT-PCR detection of RRM1 expression in 16 studies, of which 1,256 (55.5%) expressed the RRM1 gene (Table 2).
Table 2.
Methods for the detection of RRM1 expression
| Method | RRM1 expression
|
No of studies (cases) | |
|---|---|---|---|
| High | Low | ||
| IHC | 539 (43.8%) | 692 (56.2%) | 15 (1,231) |
| PCR | 1,256 (55.5%) | 1,009 (44.5%) | 16 (2,265) |
| Total | 1,795 (51.3%) | 1,701 (48.7%) | 31 (3,496) |
Note: χ2 test indicated that there was significant difference between the two methods in the detection rate of RRM1 expression (χ2=43.46, P=0.000).
Abbreviations: IHC, immu nohistochemistry; RRM1, ribonucleotide reductase catalytic subunit M1.
Sensitivity analysis and publication bias
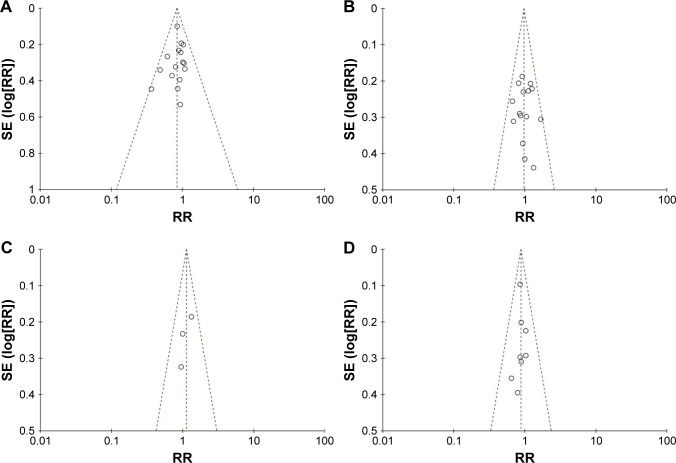
Sensitivity analysis was performed by sequentially excluding individual studies to examine the impact of each study on the summarized findings. This revealed that the findings were statistically robust and credible (data not shown). Begg’s test was performed to evaluate the potential publication bias of the studies.20 The shape of the funnel plot was symmetrical, suggesting there was no obvious publication bias (Figure 4).
Figure 4.
Funnel plot of the outcome of clinicopathological feature and the RRM1 gene.
Notes: (A) Female vs male; (B) adenocarcinomas vs non-adenocarcinomas; (C) stage I–II vs stage III–IV; (D) nonsmokers vs smokers.
Abbreviation: RRM1, ribonucleotide reductase catalytic subunit M1.
Discussion
Gemcitabine (2′,2′-difluorodeoxycytidine), which is active against NSCLC, is a deoxycytidine analog that is incorporated into DNA and competitively inhibits DNA synthesis.21 Resistance to gemcitabine is associated with RRM1 overexpression.22–24 RRM1 catalyzes the biosynthesis of deoxyribonucleotide and participates in DNA synthesis and damage repair. Preclinical studies show that RRM1 expression is associated with chemosensitivity to gemcitabine-containing therapies and that a low level of RRM1 expression is associated with a better response to gemcitabine-containing chemotherapy.10,11
RRM1, which is located on chromosome segment 11p15.5, is involved in the regulation of tumor proliferation, invasiveness, and metastasis. This region, also known as LOH11A, shows frequent loss of heterozygosity in NSCLC.8,9 Previous clinical studies demonstrated a close relationship between RRM1 expression and the response to gemcitabine-containing regimens in different types of cancer, including pancreatic cancer, nasopharyngeal carcinoma, and NSCLC.8,12,15,17,25–56
Here, we reviewed almost all available published articles and conducted the present meta-analysis to examine the correlation between RRM1 expression and the clinicopathological features of NSCLC. Analysis of 3,667 participants from 31 studies indicated a low rate (49.1%) of RRM1 expression in NSCLC patients. Therefore, we examined the clinicopathological characteristics of RRM1-positive lung cancer patients to improve the screening efficacy. The present analysis demonstrated that RRM1 positivity in lung cancer patients was more prevalent in men than in women (43.0% vs 51.7%, RR=0.84, 95% CI: 0.74–0.94, P=0.004). The proportion of RRM1-positive patients in adenocarcinoma was slightly lower than that in non-adenocarcinoma, although the relation was not statistically significant (RR=0.97, 95% CI: 0.84–1.10, P=0.61). The proportion of RRM1-positive patients was slightly lower in nonsmokers than in smokers with NSCLC (47.0% vs 51.5%, RR=0.88, 95% CI: 0.76–1.01, P=0.07). Similarly, RRM1 gene positivity was not strongly correlated with clinical stage in NSCLC patients (RR=1.12, 95% CI: 0.86–1.46, P=0.40).
The present study provided evidence to guide the pre-screening of NSCLC patients for selecting a population likely to harbor this specific gene. Dong et al36 reported that RRM1 expression in tumor tissues could be a predictive biomarker in patients with advanced NSCLC receiving gemcitabine-containing chemotherapy. These authors showed that 83 of 229 (36.20%) NSCLC tumors were RRM1-positive, and 13 RRM1-positive tumors were detected in women (36.11% [13/36]). Vilmar et al46 reported that 47 of 140 (33.57%) NSCLC patients were RRM1-positive, and 14 RRM1-positive tumors were found in women (24.56% [14/57]). These results are consistent with those of the present meta-analysis showing a prevalence of 43.00% in women. As the incidence of RRM1 is low in NSCLC patients, investigating the clinicopathological characteristics of RRM1-positive lung cancer patients is necessary to improve the screening efficacy and reduce medical costs. Evaluation of the clinicopathological characteristics of NSCLC patients was the first prescreening step.
RT-PCR and IHC are the most reliable methodologies to detect the RRM1 gene. Reynolds et al used a fluorescence-based IHC method combined with automated quantitative analysis, but failed to observe important differences in survival in NSCLC patients with different levels of RRM1 gene expression who received gemcitabine-containing chemotherapy.57,58 Zheng et al59 used these two methods simultaneously and found that the mRNA expression levels were closely related to protein expression levels. In the present study, the positive rates of IHC and RT-PCR were 43.8% and 55.5%, respectively, indicating that these methods are reliable for detecting RRM1 expression. IHC, which is a well-established method for detecting RRM1 expression, is a convenient and cost-effective technique for detecting RRM1 expression in patients with NSCLC.60 Currently, RT-PCR is the most sensitive method for detecting and quantifying mRNA. Large-sample prospective studies are necessary to further evaluate the efficacy of RT-PCR detection for the prediction of chemosensitivity to gemcitabine associated with RRM1 expression.
This study had several limitations. First, the conclusions are mainly based on observational studies. A meta-analysis of well-designed nonrandomized studies can be as accurate as that of randomized-controlled trials.61 Second, only studies written in English or Chinese were included in the meta-analysis. This means that eligible studies published in other languages may have been overlooked, which may have introduced selection bias. Third, most of the participants of the current pooled analysis were Asian. Therefore, future studies should include subjects from diverse ethnicities. The present survey also lacked sufficient individual patient data. Therefore, additional details and subgroup data such as sex, smoking status, pathologic classification, and clinical stages are needed for further analysis.
Conclusion
Our meta-analysis suggests that RRM1 positivity is related to a higher response rate to gemcitabine-based regimens in women with advanced NSCLC. RRM1 may be used as a biomarker to predict the response rate to gemcitabine-based chemotherapy in NSCLC. IHC could be used to prescreen for RRM1 expression in the clinic, and RT-PCR could be used for confirmation. Further large-scale, well-designed prospective studies are necessary to determine the potential correlation between RRM1 expression in NSCLC and the clinicopathological characteristics of patients treated with gemcitabine-containing chemotherapy.
Supplementary material
Table S1.
Search strategy
| PubMed via the NCBI Entrez system from inception to 1 September 2017 | ||
| #1 | “Carcinoma, Non-Small-Cell Lung” [Mesh] | 43491 |
| #2 | NSCLC [Title/Abstract] | 31187 |
| #3 | “Lung cancer*” [Title/Abstract] | 123251 |
| #4 | “Lung carcinoma*” [Title/Abstract] | 16074 |
| #5 | “Lung neoplasm*” [Title/Abstract] | 370 |
| #6 | “Lung tumor*” [Title/Abstract] | 5242 |
| #7 | “Lung tumour*” [Title/Abstract] | 833 |
| #8 | “Non-small-cell*” [Title/Abstract] | 48315 |
| #9 | “Non-small cell*” [Title/Abstract] | 48315 |
| #10 | “Non small-cell*” [Title/Abstract] | 48315 |
| #11 | “Non small cell*” [Title/Abstract] | 48315 |
| #12 | (#3 OR #4 OR #5 OR #6 OR #7) AND (#8 OR #9 OR #10 OR #11) | 46076 |
| #13 | #1 OR #2 OR #12 | 58288 |
| #14 | “Gemcitabine” [Supplementary Concept] OR “gemcitabine” [All Fields] | 14018 |
| #15 | “RRM1” [All Fields] OR “ribonucleotide reductase M1” [All Fields] OR “ribonucleotide reductase subunit M1” [All Fields] OR “ribonucleotide reductase large subunit” [All Fields] | 675 |
| #16 | #13 AND #14 AND #15 | 94 |
| EMBASE (via Elsevier) Search Strategy from 1980 to 1 September 2017 | ||
| #1 | “Lung cancer”/exp OR “lung cancer” | 345530 |
| #2 | “Non small cell lung cancer”/exp | 119859 |
| #3 | “Nonsmall cell”:tn,lnk,ab,ti | 4485 |
| #4 | “Lung tumor”/exp | 370444 |
| #5 | “Lung carcinoma”/exp | 171987 |
| #6 | “Lung neoplasm”:tn,lnk,ab,ti | 379 |
| #7 | “Lung tumour”/exp | 370444 |
| #8 | “Thoracic cancer”:tn,lnk,ab,ti | 297 |
| #9 | “Nsclc”:tn,lnk,ab,ti | 60146 |
| #10 | #1 OR #2 OR #3 OR #4 OR #5 OR #6 OR #7 OR #8 OR #9 | 393353 |
| #11 | “Gemcitabine”:tn,lnk,ab,ti | 23161 |
| #12 | “RRM1”:tn,lnk,ab,ti | 974 |
| #13 | “Ribonucleotide reductase m1”/exp | 113 |
| #14 | “Ribonucleotide reductase subunit m1”/exp | 30 |
| #15 | “Ribonucleotide reductase large subunit”/exp | 57 |
| #16 | #12 OR #13 OR #14 OR #15 | 1060 |
| #17 | #10 AND #11 AND #16 | 182 |
| Cochrane Library (from inception to 01 September 2017) Search Strategy | ||
| #1 | MeSH descriptor: [Lung Neoplasms] explode all trees | 6056 |
| #2 | MeSH descriptor: [Carcinoma, Non-Small-Cell Lung] explode all trees | 3099 |
| #3 | “Lung cancer*”:ti,ab,kw | 11984 |
| #4 | “Non-small cell*”:ti,ab,kw | 7740 |
| #5 | “Non small cell*”:ti,ab,kw | 7740 |
| #6 | “Nonsmall cell*”:ti,ab,kw | 277 |
| #7 | “Nsclc”:ti,ab,kw | 5108 |
| #8 | #1 or #2 or #3 or #4 or #5 or #6 or #7 | 13862 |
| #9 | Gemcitabine* | 3393 |
| #10 | “Ribonucleotide reductase subunit m1” | 7 |
| #11 | “Ribonucleotide reductase large subunit” | 1 |
| #12 | “Ribonucleotide reductase m1” | 9 |
| #13 | “RRM1” | 38 |
| #14 | #10 or #11 or #12 or #13 | 40 |
| #15 | #8 or #9 or #14 | 21 |
| CNKI Search Strategy from inception to 1 September 2017 | ||
| #1 | 肺癌 AND RRM1 AND 吉西他滨 | 50 |
| WanFang Database Search Strategy from inception to 1 September 2017 | ||
| #1 | 肺癌 AND RRM1 AND 吉西他滨 | 48 |
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A, Statistics C. CA Cancer J Clin. 2017;2017(67):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40(4):558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132(5):1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 4.Paz-Ares L, de Marinis F, Dediu M, et al. Maintenance therapy with pemetrexed plus best supportive care versus placebo plus best supportive care after induction therapy with pemetrexed plus cisplatin for advanced non-squamous non-small-cell lung cancer (PARAMOUNT): a double-blind, phase 3, randomised controlled trial. Lancet Oncol. 2012;13(3):247–255. doi: 10.1016/S1470-2045(12)70063-3. [DOI] [PubMed] [Google Scholar]
- 5.Georgoulias V, Androulakis N, Kotsakis A, et al. Docetaxel versus docetaxel plus gemcitabine as front-line treatment of patients with advanced non-small cell lung cancer: a randomized, multicenter phase III trial. Lung Cancer. 2008;59(1):57–63. doi: 10.1016/j.lungcan.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 6.Toschi L, Cappuzzo F. Gemcitabine for the treatment of advanced nonsmall cell lung cancer. Onco Targets Ther. 2009;2:209–217. doi: 10.2147/ott.s4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reaume MN, Leighl NB, Mittmann N, et al. Economic analysis of a randomized phase III trial of gemcitabine plus vinorelbine compared with cisplatin plus vinorelbine or cisplatin plus gemcitabine for advanced non-small-cell lung cancer (Italian GEMVIN3/NCIC CTG BR14 trial) Lung Cancer. 2013;82(1):115–120. doi: 10.1016/j.lungcan.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Pitterle DM, Kim YC, Jolicoeur EM, Cao Y, O’Briant KC, Bepler G. Lung cancer and the human gene for ribonucleotide reductase subunit M1 (RRM1) Mamm Genome. 1999;10(9):916–922. doi: 10.1007/s003359901114. [DOI] [PubMed] [Google Scholar]
- 9.Bepler G, Gautam A, Mcintyre LM, et al. Prognostic significance of molecular genetic aberrations on chromosome segment 11p15.5 in non-small-cell lung cancer. J Clin Oncol. 2002;20(5):1353–1360. doi: 10.1200/JCO.2002.20.5.1353. [DOI] [PubMed] [Google Scholar]
- 10.Bergman AM, Eijk PP, Ruiz van Haperen VW, et al. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res. 2005;65(20):9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- 11.Oguri T, Achiwa H, Sato S, et al. The determinants of sensitivity and acquired resistance to gemcitabine differ in non-small cell lung cancer: a role of ABCC5 in gemcitabine sensitivity. Mol Cancer Ther. 2006;5(7):1800–1806. doi: 10.1158/1535-7163.MCT-06-0025. [DOI] [PubMed] [Google Scholar]
- 12.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24(29):4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 13.Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide Reductase Messenger RNA Expression and Survival in Gemcitabine/Cisplatin-Treated Advanced Non-Small Cell Lung Cancer Patients. Clinical Cancer Research. 2004;10(4):1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 14.Rosell R, Felip E, Taron M, et al. Gene expression as a predictive marker of outcome in stage IIB-IIIA-IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clin Cancer Res. 2004;10(12 Pt 2):4215S–4219S. doi: 10.1158/1078-0432.CCR-040006. [DOI] [PubMed] [Google Scholar]
- 15.Souglakos J, Boukovinas I, Taron M, et al. Ribonucleotide reductase subunits M1 and M2 mRNA expression levels and clinical outcome of lung adenocarcinoma patients treated with docetaxel/gemcitabine. Br J Cancer. 2008;98(10):1710–1715. doi: 10.1038/sj.bjc.6604344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordheim LP, Sève P, Trédan O, Dumontet C. The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol. 2011;12(7):693–702. doi: 10.1016/S1470-2045(10)70244-8. [DOI] [PubMed] [Google Scholar]
- 17.Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17(12):1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 18.Dersimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 19.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 20.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 21.Chabner BA, Longo DL. Cancer Chemotherapy and Biotherapy: Principles and Practice. Philadelphia, PA: Lippincott Williams & Wilkins; 2011. [Google Scholar]
- 22.Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22(14):2135–2142. doi: 10.1038/sj.onc.1206232. [DOI] [PubMed] [Google Scholar]
- 23.Goan YG, Zhou B, Hu E, Mi S, Yen Y. Overexpression of ribonucleotide reductase as a mechanism of resistance to 2,2-difluorodeoxycytidine in the human KB cancer cell line. Cancer Res. 1999;59(17):4204–4207. [PubMed] [Google Scholar]
- 24.Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64(11):3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- 25.Aoyama T, Miyagi Y, Murakawa M, et al. Clinical implications of ribonucleotide reductase subunit M1 in patients with pancreatic cancer who undergo curative resection followed by adjuvant chemotherapy with gemcitabine. Oncol Lett. 2017;13(5):3423–3430. doi: 10.3892/ol.2017.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mlak R, Krawczyk P, Ciesielka M, et al. The relationship between RRM1 gene polymorphisms and effectiveness of gemcitabine-based first-line chemotherapy in advanced NSCLC patient. Clin Transl Oncol. 2016;18(9):915–924. doi: 10.1007/s12094-015-1461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maus MK, Mack PC, Astrow SH, et al. Histology-related associations of ERCC1, RRM1, and TS biomarkers in patients with non-small-cell lung cancer: implications for therapy. J Thorac Oncol. 2013;8(5):582–586. doi: 10.1097/JTO.0b013e318287c3c5. [DOI] [PubMed] [Google Scholar]
- 28.Zhao LP, Xue C, Zhang JW, et al. Expression of RRM1 and its association with resistancy to gemcitabine-based chemotherapy in advanced nasopharyngeal carcinoma. Chin J Cancer. 2012;31(10):476–483. doi: 10.5732/cjc.012.10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo N, Zhang W, Zhang B, et al. EGFR and K-RAS mutations and ERCC1, TUBB3, TYMS, RRM1 and EGFR mRNA expression in non-small cell lung cancer: Correlation with clinical response to gefitinib or chemotherapy. Mol Clin Oncol. 2015;3(5):1123–1128. doi: 10.3892/mco.2015.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing X, Wang L, Xiao X, et al. Analysis of RRM1 expression in advanced NSCLC and its relationship with chemotherapy. Chin J Cancer Prev Treat. 2014;21:1893–1896. [Google Scholar]
- 31.Zeng F, Shan L. Investigation about expression of RRM1 (ribonucleotide reductase M1) and the correlation with Gemcitabine-based chemotherapy sensitivity in the advanced non-small cell lung cancer. Journal of Taishan Medical College. 2009;30:570–573. [Google Scholar]
- 32.Zhao H, Zhang H, du Y, Gu X. Prognostic significance of BRCA1, ERCC1, RRM1, and RRM2 in patients with advanced non-small cell lung cancer receiving chemotherapy. Tumour Biol. 2014;35(12):12679–12688. doi: 10.1007/s13277-014-2592-7. [DOI] [PubMed] [Google Scholar]
- 33.Xian-Jun F, Xiu-Guang Q, Li Z, et al. ERCC1 and BRCA1 mRNA expression predicts the clinical outcome of non-small cell lung cancer receiving platinum-based chemotherapy. Pak J Med Sci. 2014;30(3):488–492. doi: 10.12669/pjms.303.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Meng L, Wang XW, Ma GY, Chen JH. Expression of RRM1 and RRM2 as a novel prognostic marker in advanced non-small cell lung cancer receiving chemotherapy. Tumour Biol. 2014;35(3):1899–1906. doi: 10.1007/s13277-013-1255-4. [DOI] [PubMed] [Google Scholar]
- 35.Liang JG, Jin ZY, Gao XD, et al. Predictive role of RRM1 and BRCA1 mRNA expression on the clinical outcome of advanced non-small cell lung cancer. Genet Mol Res. 2014;13(3):5292–5298. doi: 10.4238/2014.July.24.8. [DOI] [PubMed] [Google Scholar]
- 36.Dong X, Hao Y, Wei Y, Yin Q, du J, Zhao X. Response to first-line chemotherapy in patients with non-small cell lung cancer according to RRM1 expression. PLoS One. 2014;9(3):e92320. doi: 10.1371/journal.pone.0092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jian-Wei B, Yi-Min M, Yu-Xia S, Shi-Qing L. Expression levels of ERCC1 and RRM1 mRNA and clinical outcome of advanced non-small cell lung cancer. Pak J Med Sci. 2013;29(5):1158–1161. doi: 10.12669/pjms.295.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su C, Zhou S, Zhang L, et al. ERCC1, RRM1 and BRCA1 mRNA expression levels and clinical outcome of advanced non-small cell lung cancer. Med Oncol. 2011;28(4):1411–1417. doi: 10.1007/s12032-010-9553-9. [DOI] [PubMed] [Google Scholar]
- 39.Wu GQ, Liu NN, Xue XL, et al. Multiplex real-time PCR for RRM1, XRCC1, TUBB3 and TS mRNA for prediction of response of non-small cell lung cancer to chemoradiotherapy. Asian Pac J Cancer Prev. 2014;15(10):4153–4158. doi: 10.7314/apjcp.2014.15.10.4153. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Zhao J, Yang L, et al. Positive expression of ERCC1 predicts a poorer platinum-based treatment outcome in Chinese patients with advanced non-small-cell lung cancer. Med Oncol. 2010;27(2):484–490. doi: 10.1007/s12032-009-9239-3. [DOI] [PubMed] [Google Scholar]
- 41.Guo QZ, Wang J, Bai H, et al. High expression of ERCC1 is a poor prognostic factor in Chinese patients with non-small cell lung cancer receiving cisplatin-based therapy. Chin J Cancer Res. 2010;22(4):296–302. [Google Scholar]
- 42.Lee JJ, Maeng CH, Baek SK, et al. The immunohistochemical overex-pression of ribonucleotide reductase regulatory subunit M1 (RRM1) protein is a predictor of shorter survival to gemcitabine-based chemotherapy in advanced non-small cell lung cancer (NSCLC) Lung Cancer. 2010;70(2):205–210. doi: 10.1016/j.lungcan.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Ma K, Li E, Guo Y, Wang X, Sun H, Shao G. Relationship between the efficacy of gemcitabine/cisplatin adjuvant chemotherapy and RRM1 protein expression in postoperative NSCLC patients. Zhonghua Zhong Liu Za Zhi. 2014;36(7):505–510. [PubMed] [Google Scholar]
- 44.Jiang B, Tu C, He W, Yang C, Zhao J. Analysis of RRM1 expression and efficacy of gemcitabine in advanced non-small cell lung cancer. Cancer Research on Prevention and Treatment. 2013;40:68–71. [Google Scholar]
- 45.Lin L, Zhang X, Hu J, Huang F, Li W. Analysis of the relationship between RM1 mRNA expression level and efficacy of gemcitabine maintenance therapy for advanced non-small cell lung cancer. The Practical Journal of Cancer. 2016;31:22–25. [Google Scholar]
- 46.Vilmar AC, Santoni-Rugiu E, Sorensen JB. Predictive impact of RRM1 protein expression on vinorelbine efficacy in NSCLC patients randomly assigned in a chemotherapy phase III trial. Ann Oncol. 2013;24(2):309–314. doi: 10.1093/annonc/mds335. [DOI] [PubMed] [Google Scholar]
- 47.Xu CW, Wang G, Wang WL, et al. Association between epidermal growth factor receptor mutations and the expression of excision repair cross-complementing protein 1 and ribonucleotide reductase subunit M1 mRNA in patients with non-small cell lung cancer. Exp Ther Med. 2015;9(3):880–884. doi: 10.3892/etm.2015.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu D, Nakashima N, Nakano J, et al. Customized Adjuvant Chemotherapy Based on Biomarker Examination May Improve Survival of Patients Completely Resected for Non-small-cell Lung Cancer. Anticancer Res. 2017;37(5):2501–2507. doi: 10.21873/anticanres.11591. [DOI] [PubMed] [Google Scholar]
- 49.Liu H, Wang H, Yang J. RRM1 Expression and Efficacy Analysis of Gemcitabine in Advanced Non-small Cell Lung Cancer. Pract Prevent Med. 2009;16:11–14. [Google Scholar]
- 50.Li L, Liu X. Correlation of expression of ERCC1/RRM1 with cisplatin combined with gemcitabine chemotherapy sensitivity and prognosis in non-small cell lung cancer. Bull Acad Mil Med Sci. 2010;34:265–268. [Google Scholar]
- 51.Yang Z, Liu D, Chen F, Chen J, Tan S, Lin S. The effect of RRM1 expression in lung cancer tissues for predict chemotherapeutic effect of GP. Chin J Oncol Prev Treat. 2009;1:124–126. [Google Scholar]
- 52.Zhang GB, Chen J, Wang LR, et al. RRM1 and ERCC1 expression in peripheral blood versus tumor tissue in gemcitabine/carboplatin-treated advanced non-small cell lung cancer. Cancer Chemother Pharmacol. 2012;69(5):1277–1287. doi: 10.1007/s00280-012-1834-x. [DOI] [PubMed] [Google Scholar]
- 53.Bepler G, Sommers KE, Cantor A, et al. Clinical efficacy and predictive molecular markers of neoadjuvant gemcitabine and pemetrexed in resectable non-small cell lung cancer. J Thorac Oncol. 2008;3(10):1112–1118. doi: 10.1097/JTO.0b013e3181874936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boukovinas I, Papadaki C, Mendez P, et al. Tumor BRCA1, RRM1 and RRM2 mRNA expression levels and clinical response to first-line gemcitabine plus docetaxel in non-small-cell lung cancer patients. PLoS One. 2008;3(11):e3695. doi: 10.1371/journal.pone.0003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Z, Han B, Shen J, Gu A, Zhong H. Relations between RRM1 protein expression levels and effects of gemcitabine and cisplatin chemotherapy in advanced non-small cell lung cancer patients. Zhongguo Fei Ai Za Zhi. 2011;14(4):340–344. doi: 10.3779/j.issn.1009-3419.2011.04.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosell R, Scagliotti G, Danenberg KD, et al. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene. 2003;22(23):3548–3553. doi: 10.1038/sj.onc.1206419. [DOI] [PubMed] [Google Scholar]
- 57.Reynolds C, Obasaju C, Schell MJ, et al. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J Clin Oncol. 2009;27(34):5808–5815. doi: 10.1200/JCO.2009.21.9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–1328. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 59.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356(8):800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 60.Nalbandian A, Yan BS, Pichugin A, Bronson RT, Kramnik I. Lung carcinogenesis induced by chronic tuberculosis infection: the experimental model and genetic control. Oncogene. 2009;28(17):1928–1938. doi: 10.1038/onc.2009.32. [DOI] [PubMed] [Google Scholar]
- 61.Abraham NS, Byrne CJ, Young JM, Solomon MJ. Meta-analysis of well-designed nonrandomized comparative studies of surgical procedures is as good as randomized controlled trials. J Clin Epidemiol. 2010;63(3):238–245. doi: 10.1016/j.jclinepi.2009.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Search strategy
| PubMed via the NCBI Entrez system from inception to 1 September 2017 | ||
| #1 | “Carcinoma, Non-Small-Cell Lung” [Mesh] | 43491 |
| #2 | NSCLC [Title/Abstract] | 31187 |
| #3 | “Lung cancer*” [Title/Abstract] | 123251 |
| #4 | “Lung carcinoma*” [Title/Abstract] | 16074 |
| #5 | “Lung neoplasm*” [Title/Abstract] | 370 |
| #6 | “Lung tumor*” [Title/Abstract] | 5242 |
| #7 | “Lung tumour*” [Title/Abstract] | 833 |
| #8 | “Non-small-cell*” [Title/Abstract] | 48315 |
| #9 | “Non-small cell*” [Title/Abstract] | 48315 |
| #10 | “Non small-cell*” [Title/Abstract] | 48315 |
| #11 | “Non small cell*” [Title/Abstract] | 48315 |
| #12 | (#3 OR #4 OR #5 OR #6 OR #7) AND (#8 OR #9 OR #10 OR #11) | 46076 |
| #13 | #1 OR #2 OR #12 | 58288 |
| #14 | “Gemcitabine” [Supplementary Concept] OR “gemcitabine” [All Fields] | 14018 |
| #15 | “RRM1” [All Fields] OR “ribonucleotide reductase M1” [All Fields] OR “ribonucleotide reductase subunit M1” [All Fields] OR “ribonucleotide reductase large subunit” [All Fields] | 675 |
| #16 | #13 AND #14 AND #15 | 94 |
| EMBASE (via Elsevier) Search Strategy from 1980 to 1 September 2017 | ||
| #1 | “Lung cancer”/exp OR “lung cancer” | 345530 |
| #2 | “Non small cell lung cancer”/exp | 119859 |
| #3 | “Nonsmall cell”:tn,lnk,ab,ti | 4485 |
| #4 | “Lung tumor”/exp | 370444 |
| #5 | “Lung carcinoma”/exp | 171987 |
| #6 | “Lung neoplasm”:tn,lnk,ab,ti | 379 |
| #7 | “Lung tumour”/exp | 370444 |
| #8 | “Thoracic cancer”:tn,lnk,ab,ti | 297 |
| #9 | “Nsclc”:tn,lnk,ab,ti | 60146 |
| #10 | #1 OR #2 OR #3 OR #4 OR #5 OR #6 OR #7 OR #8 OR #9 | 393353 |
| #11 | “Gemcitabine”:tn,lnk,ab,ti | 23161 |
| #12 | “RRM1”:tn,lnk,ab,ti | 974 |
| #13 | “Ribonucleotide reductase m1”/exp | 113 |
| #14 | “Ribonucleotide reductase subunit m1”/exp | 30 |
| #15 | “Ribonucleotide reductase large subunit”/exp | 57 |
| #16 | #12 OR #13 OR #14 OR #15 | 1060 |
| #17 | #10 AND #11 AND #16 | 182 |
| Cochrane Library (from inception to 01 September 2017) Search Strategy | ||
| #1 | MeSH descriptor: [Lung Neoplasms] explode all trees | 6056 |
| #2 | MeSH descriptor: [Carcinoma, Non-Small-Cell Lung] explode all trees | 3099 |
| #3 | “Lung cancer*”:ti,ab,kw | 11984 |
| #4 | “Non-small cell*”:ti,ab,kw | 7740 |
| #5 | “Non small cell*”:ti,ab,kw | 7740 |
| #6 | “Nonsmall cell*”:ti,ab,kw | 277 |
| #7 | “Nsclc”:ti,ab,kw | 5108 |
| #8 | #1 or #2 or #3 or #4 or #5 or #6 or #7 | 13862 |
| #9 | Gemcitabine* | 3393 |
| #10 | “Ribonucleotide reductase subunit m1” | 7 |
| #11 | “Ribonucleotide reductase large subunit” | 1 |
| #12 | “Ribonucleotide reductase m1” | 9 |
| #13 | “RRM1” | 38 |
| #14 | #10 or #11 or #12 or #13 | 40 |
| #15 | #8 or #9 or #14 | 21 |
| CNKI Search Strategy from inception to 1 September 2017 | ||
| #1 | 肺癌 AND RRM1 AND 吉西他滨 | 50 |
| WanFang Database Search Strategy from inception to 1 September 2017 | ||
| #1 | 肺癌 AND RRM1 AND 吉西他滨 | 48 |