ABSTRACT
Autophagy is essential for appressorium-mediated plant infection by Magnaporthe oryzae, the causal agent of rice blast disease and a major threat to global food security. The regulatory mechanism of pathogenicity-associated autophagy, however, remains largely unknown. Here, we report the identification and functional characterization of a plausible ortholog of yeast SNT2 in M. oryzae, which we term MoSNT2. Deletion mutants of MoSNT2 are compromised in autophagy homeostasis and display severe defects in autophagy-dependent fungal cell death and pathogenicity. These mutants are also impaired in infection structure development, conidiation, oxidative stress tolerance and cell wall integrity. MoSnt2 recognizes histone H3 acetylation through its PHD1 domain and thereby recruits the histone deacetylase complex, resulting in deacetylation of H3. MoSnt2 binds to promoters of autophagy genes MoATG6, 15, 16, and 22 to regulate their expression. In addition, MoTor controls MoSNT2 expression to regulate MoTor signaling which leads to autophagy and rice infection. Our study provides evidence of a direct link between MoSnt2 and MoTor signaling and defines a novel epigenetic mechanism by which MoSNT2 regulates infection-associated autophagy and plant infection by the rice blast fungus.
Abbreviations: M. oryzae: Magnaporthe oryzae; S. cerevisiae: Saccharomyces cerevisiae; F. oxysporum: Fusarium oxysporum; U. maydis: Ustilago maydis; Compl.: complemented strains of ΔMosnt2 expressing MoSNT2-GFP; ATG: autophagy-related; HDAC: histone deacetylase complex; Tor: target of rapamycin kinase; MTOR: mechanistic target of rapamycin kinase in mammals; MoSnt2: DNA binding SaNT domain protein in M. oryzae; MoTor: target of rapamycin kinase in M. oryzae; MoAtg8: autophagy-related protein 8 in M. oryzae; MoHos2: hda one similar protein in M. oryzae; MoeIf4G: eukaryotic translation initiation factor 4 G in M. oryzae; MoRs2: ribosomal protein S2 in M. oryzae; MoRs3: ribosomal protein S3 in M. oryzae; MoIcl1: isocitrate lyase in M. oryzae; MoSet1: histone H3K4 methyltransferase in M. oryzae; Asd4: ascus development 4; Abl1: AMP-activated protein kinase β subunit-like protein; Tig1: TBL1-like gene required for invasive growth; Rpd3: reduced potassium dependency; KAT8: lysine (K) acetyltransferase 8; PHD: plant homeodomain; ELM2: Egl-27 and MTA1 homology 2; GFP: green fluorescent protein; YFP: yellow fluorescent protein; YFPCTF: C-terminal fragment of YFP; YFPNTF: N-terminal fragment of YFP; GST: glutathione S-transferase; bp: base pairs; DEGs: differentially expressed genes; CM: complete medium; MM-N: minimum medium minus nitrogen; CFW: calcofluor white; CR: congo red; DAPI: 4ʹ, 6-diamidino-2-phenylindole; BiFC: bimolecular fluorescence complementation; RT: reverse transcription; PCR: polymerase chain reaction; qPCR: quantitative polymerase chain reaction; RNAi: RNA interference; ChIP: chromatin immunoprecipitation
KEYWORDS: Autophagy, Magnaporthe oryzae, MoSnt2, MoTor signaling, pathogenicity
Introduction
Macroautophagy/autophagy is an evolutionarily conserved catabolic process whereby intracellular materials, such as proteins and organelles, are delivered into the lysosome in animals, or the vacuole in plants and fungi, for degradation and recycling [1]. In eukaryotes, autophagy plays pivotal roles in a wide range of physiological and pathophysiological processes, such as cellular differentiation and tissue remodeling, development, metabolic adaptation, stress and immune responses [2]. It is also becoming increasingly apparent that autophagy is crucial for pathogenicity of plant pathogenic fungi [3–5], but how infection-associated autophagy is regulated remains largely unknown. In this study, we set out to characterize the regulation of autophagy in the blast fungus Magnaporthe oryzae (M. oryzae), which causes severe yield losses to the global rice harvest and, more recently, to wheat production in South America and Bangladesh [6,7].
The rice blast disease cycle involves attachment of a conidium to the leaf surface, conidial germination, appressorium development and maturation, penetration peg emergence, invasive hyphal proliferation, necrotic lesion development and sporulation [6]. The appressorium, a dome-shaped, melanized infection structure, enables M. oryzae to penetrate the cuticle of host plants by generating enormous turgor [8]. Development of a functional appressorium requires spatiotemporally-regulated autophagy, which occurs in both the conidium and appressorium [9]. Conidial autophagy leads to degradation of conidial components and programmed cell death, while appressorial autophagy exploits storage products and cellular components recycled from conidial cells to aid in appressorium maturation and penetration peg deployment. The autophagy-related genes MoATG1, MoATG4, MoATG5, MoATG8, MoATG9 and MoATG18 regulate conidial cell death, appressorium maturation and penetration peg formation, supporting a critical role for autophagy in these processes [3,4]. Autophagy is also important for conidiation of M. oryzae through coordinating glycogen breakdown to fuel asexual development [10,11]. However, it is not yet clear how infection-associated autophagy in M. oryzae is genetically regulated.
The Tor (target of rapamycin) kinase (MTOR [mechanistic target of rapamycin kinase] in mammals) is an evolutionarily conserved protein kinase central to cell growth control and the regulation of autophagy [12]. Post-translational covalent modification of histone proteins – an epigenetic control mechanism – also plays an important role in regulation of autophagy [13]. Tor signaling is associated with the chromatin-remodeling complex Rsc1 in the regulation of autophagy in yeast (Saccharomyces cerevisiae) [14]. The mammalian MTOR signaling modulates transcription of histone acetyltransferase KAT8/hMOF/MYST1 to control the expression of autophagy-related genes and the outcome of autophagy [15]. In M. oryzae, Tor signaling is suggested to be coordinated with the pre-mRNA processing protein Rbp35 (RNA-binding protein), glutaminolysis regulator Asd4 (ascus development 4), glucose-metabolizing enzyme Tkl1 (transketolase) and the carbon-responsive regulator Abl1 (AMP-activated protein kinase β subunit-like protein) to regulate host infection [16–19]. Asd4 and Abl1 also regulate autophagy via Tor signaling [17,19]. However, our knowledge of Tor signaling with regard to the regulation of autophagy is still very limited in M. oryzae.
A growing body of evidence suggests that epigenetic regulators of histone proteins play important roles in the development and pathogenicity of both animal and plant fungal pathogens. In the human pathogen Candida albicans, for instance, acetylation of histone H3 at position K56 (H3K56) by histone acetyltransferase Rtt109 (regulation of Ty1 transposition) is critical for DNA damage response and fungal pathogenesis [20]. The histone deacetylase Hos2 (hda one similar) affects extracellular depolymerase expression, conidiation and pathogenicity of the maize pathogen Cochliobolus carbonum [21]. Hos2 also regulates mating-type gene expression to control the dimorphic switch and virulence of the maize pathogen Ustilago maydis [22]. In M. oryzae, infection structure morphogenesis and physiological transitions during host infection are accompanied by global transcriptional changes which require epigenetic histone modifications. For example, both histone deacetylase complex (HDAC) Tig1 (TBL1-like gene required for invasive growth) and NAD-dependent histone deacetylase Sir2 (silent information regulator, sirtuin) regulate infectious growth within host cells [23,24]. The histone epigenetic regulator MoSet1 (histone H3K4 methyltransferase in M. oryzae) also determines appressorium formation and conidiation by coordinating transcription of infection-associated genes [25]. Recently, the histone acetyltransferase Gcn5 (general control nonderepressible) of M. oryzae has been shown to negatively regulate light-induced autophagy and conidiation by acetylating the autophagy protein Atg7 [26]. It is not known how epigenetic modifications are associated with autophagy-dependent plant infection by M. oryzae.
In eukaryotic cells, histone modifications modulate DNA-related processes such as gene expression, DNA replication, and repair, by recruiting chromatin remodeling complexes to target chromatin regions [27]. The Snt2 protein, named for the presence of the DNA-binding domain SaNT [28,29], is widely distributed in fungal species and contains chromatin recognizing- and remodeling-associated domains, such as PHD (Plant Homeodomain), ELM2 (Egl-27 and MTA1 homology 2) and BAH (Bromo Adjacent Homology) [30]. In yeast, Snt2 forms a complex with the histone deacetylase Rpd3 (reduced potassium dependency) and is enriched at promoters of genes associated with oxidative and starvation stress responses to control their expression, in a process mediated by Tor [31]. Yeast Snt2 also functions as a histone E3 ligase to regulate intracellular levels of histone proteins H3 and H4 [29]. In the phytopathogenic fungus Fusarium oxysporum, Snt2 furthermore regulates oxidative stress response and respiration [32]. Interestingly, F. oxysporum Snt2 regulates autophagy and pathogenicity by affecting expression of genes in the Tor signaling pathway [30]. Despite evidence supporting the importance of Snt2 in fungal development and pathogenicity, its regulatory mechanism has not been explored.
In this report, we demonstrate that MoSnt2 regulates infection-associated autophagy by deacetylation of histone protein H3. MoSnt2 regulates H3 acetylation status by recruiting HDAC. Disruption of MoSNT2 impairs autophagic homeostasis and pathogenicity, and results in hypersensitivity to oxidative stress and impairment in cell wall integrity. We also report that MoTor regulates MoSNT2 expression and that MoSnt2 mediates MoTor signaling.
Results
MoSNT2 regulates autophagy in M. oryzae
M. oryzae genome contains a single SNT2 plausible ortholog, which we named MoSNT2 (MGG_04421). We cloned the full-length MoSNT2 cDNA, which is 6669 base pairs (bp) in length and putatively encodes a 1794-amino acid protein. MoSnt2 displays high similarities in both sequence (approximately 48 to 63%) and domain architecture with Snt2 orthologs from different fungi (Figures S1 and S2). To investigate biological functions of MoSNT2, we generated 5 independent targeted deletion mutants (ΔMosnt2.1 to ΔMosnt2.5) in the wild-type rice-pathogenic Guy11 strain of M. oryzae (Figure S3). As all deletion mutants showed similar phenotypes, we selected 2 deletion mutants, ΔMosnt2.1 and ΔMosnt2.2, for further analysis. We also complemented ΔMosnt2.1 with a MoSNT2-GFP gene fusion, and used one of the complemented strains (ΔMosnt2 expressing MoSNT2-GFP, abbreviated as Compl.) and Guy11 as control strains. After 10 days of growth on CM agar medium, the ΔMosnt2 deletion mutants formed much smaller colonies (40.7 ± 1.9 and 37.7 ± 2.1 mm in diameter) than the control strains (61.0 ± 1.0 and 62.7 ± 1.2 mm) (Figure 1(A,B)). The ΔMosnt2 mutants formed thin and flat colonies due to reduced aerial hyphal growth. By contrast, the control strains formed thick and fluffy colonies with extensive aerial hyphae (Figure 1(A)). The mutants also produced reduced numbers of conidia (Figure 1(C,D)). While the control strains produced pyriform conidia, the ΔMosnt2 mutants generated mostly abnormal, spindle-shaped or round conidia (Figure 1(E)). When assayed for sexual reproduction, crosses of control strains (MAT1-2) with a standard opposite strain, TH3 (MAT1-1), formed numerous perithecia at mycelial junctions, whereas the ΔMosnt2 × TH3 cross formed no perithecia (Figure 1(F)). These results suggest that MoSNT2 plays an important role in hyphal growth, asexual and sexual reproduction of M. oryzae.
Figure 1.
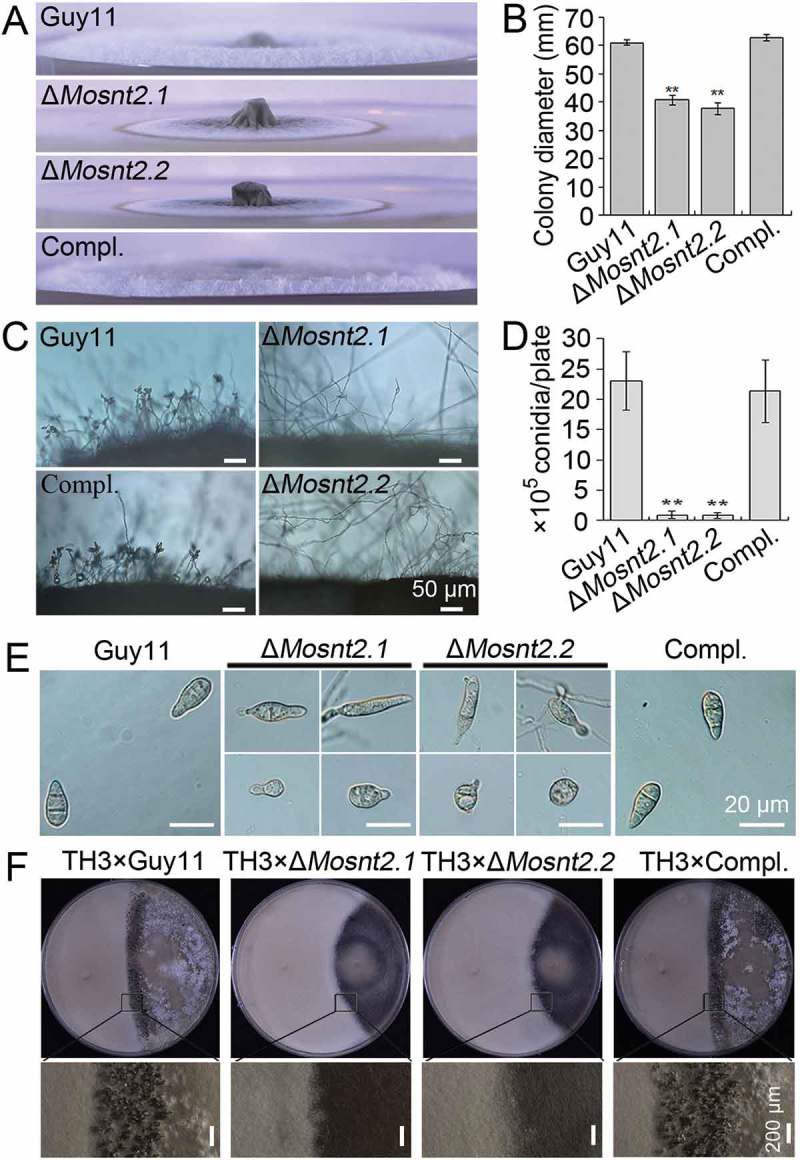
MoSNT2 is critical for growth and reproduction of M. oryzae. (A) Hyphal growth of plate colonies on CM agar medium. (B) Quantified diameters of colonies. Error bars represent standard deviations. Asterisk indicates significant difference (** P < 0.01). (C) Microscopy observation of conidiphore development. Scale bar: 50 μm. (D) Quantified conidial production. (E) Microscopic observation of conidial morphology. Scale bar: 20 μm. (F) Fertility assay. The standard testing strain TH3 was crossed with each indicated strain on oatmeal medium in an inductive condition. Scale bar: 200 μm.
To test whether MoSNT2 regulates autophagy, we introduced an autophagy marker fusion gene GFP-MoATG8 into the ΔMosnt2 mutant for autophagic flux analysis by epifluorescence microscopy [9]. When grown in CM rich medium, the wild-type Guy11 strain contained very few autophagosomes and little GFP-MoAtg8 fluorescence was observed within hyphal cytoplasm (Figure 2(A-C)). In contrast, the ΔMosnt2 mutant accumulated more autophagosomes and exhibited strong GFP-MoAtg8 fluorescence inside cytoplasm. When starved for nitrogen in minimum medium minus nitrogen (MM-N), the wild-type accumulated autophagosomes and displayed GFP-MoAtg8 fluorescence; the ΔMosnt2 mutant showed similar GFP-MoAtg8 fluorescence as in CM (Figure 2(A-C)). These results suggest that the ΔMosnt2 mutant is compromised in maintaining homeostatic regulation of autophagy.
Figure 2.
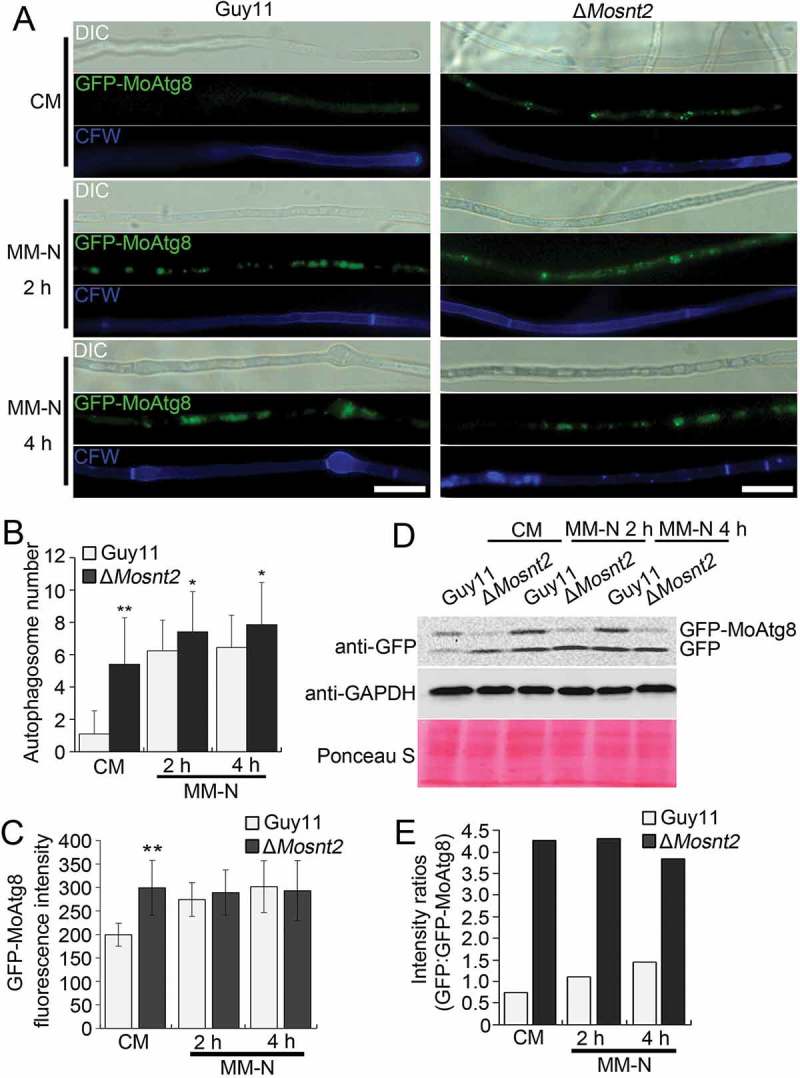
MoSNT2 plays critical roles in autophagy of M. oryzae. (A) Epifluorescence micrographs of autophagosomes. Transformants expressing the GFP-MoATG8 fusion gene were grown in CM liquid medium for 48 h, then transferred into MM-N for the indicated time. Mycelium was stained with 10 μg/ml CFW before photographing. Scale bar: 20 μm. (B) Autophagosome number within hyphae. The mean autophagosome number was calculated from at least 25 hyphal segments, each of which was defined as a hyphal region separated by 2 neighboring CFW-stained septa. (C) Fluorescence intensity of GFP-MoAtg8. The mean value of GFP fluorescence intensity was calculated from at least 25 hyphal segments with a length of 50 μm. (D) Immunoblot analysis of GFP-MoAtg8 proteolysis. (E) Quantified intensity of GFP:GFP-MoAtg8 ratios. The GFP-MoAtg8 band in the Guy11 strain was defined as reference with an intensity of 1.0.
We further assessed autophagic flux by analyzing vacuolar delivery and subsequent breakdown of GFP-MoAtg8 [33]. In both the wild-type and the ΔMosnt2 mutant expressing GFP-MoATG8, a full length GFP-MoAtg8 fusion protein (42 kDa) and free GFP (27 kDa) could be readily detected in immunoblots with an anti-GFP antibody. When grown in CM, the Guy11 strain contained lower amounts of free GFP than GFP-MoAtg8 (Figure 2(D)), suggesting that rich nutrients repressed delivery of GFP-MoAtg8 into vacuoles for cleavage, maintaining relatively low levels of autophagic flux. By contrast, the ΔMosnt2 mutant accumulated higher amounts of free GFP than GFP-MoAtg8 in CM (Figure 2(D)), indicating an increase in autophagic flux as a consequence of MoSNT2 deletion. When grown in MM-N medium, the wild-type exhibited increases in amounts of both GFP-MoAtg8 and GFP, and the ratio of GFP:GFP-MoAtg8 (Figure 2(D,E)). However, the ΔMosnt2 mutant grown in MM-N displayed a GFP:GFP-MoAtg8 ratio similar as that observed following growth in CM. Collectively, these results indicate that MoSNT2 plays a significant role in repressing autophagy in rich growth conditions.
MoSNT2 is essential for autophagy-dependent plant infection by M. oryzae
We examined the virulence of ΔMosnt2 deletion mutants by inoculation of seedlings of the susceptible rice variety CO39. Because ΔMosnt2 mutants were severely compromised in conidial production, we failed to harvest sufficient conidia for spray assays. Therefore, we inoculated detached leaves using mycelial plugs to evaluate fungal pathogenicity, as described previously [34]. Six days postinoculation, the wild-type produced large chlorotic blast lesions on rice leaves (Figure 3(A)). The ΔMosnt2 mutants, however, produced only small necrotic-like, dark brown spots at the inoculation sites that likely represent a hypersensitive response resulting from an incompatible Magnaporthe-rice interaction. Determination of fungal DNA in infected plant tissue by qPCR confirmed that ΔMosnt2 mutants were severely compromised in invasive growth within rice tissue (Figure 3(B)). A rice-root infection assay also revealed that the ΔMosnt2 mutants failed to colonize rice roots (Figure 3(C,D)). These results suggest that MoSNT2 is essential for plant infection by M. oryzae.
Figure 3.
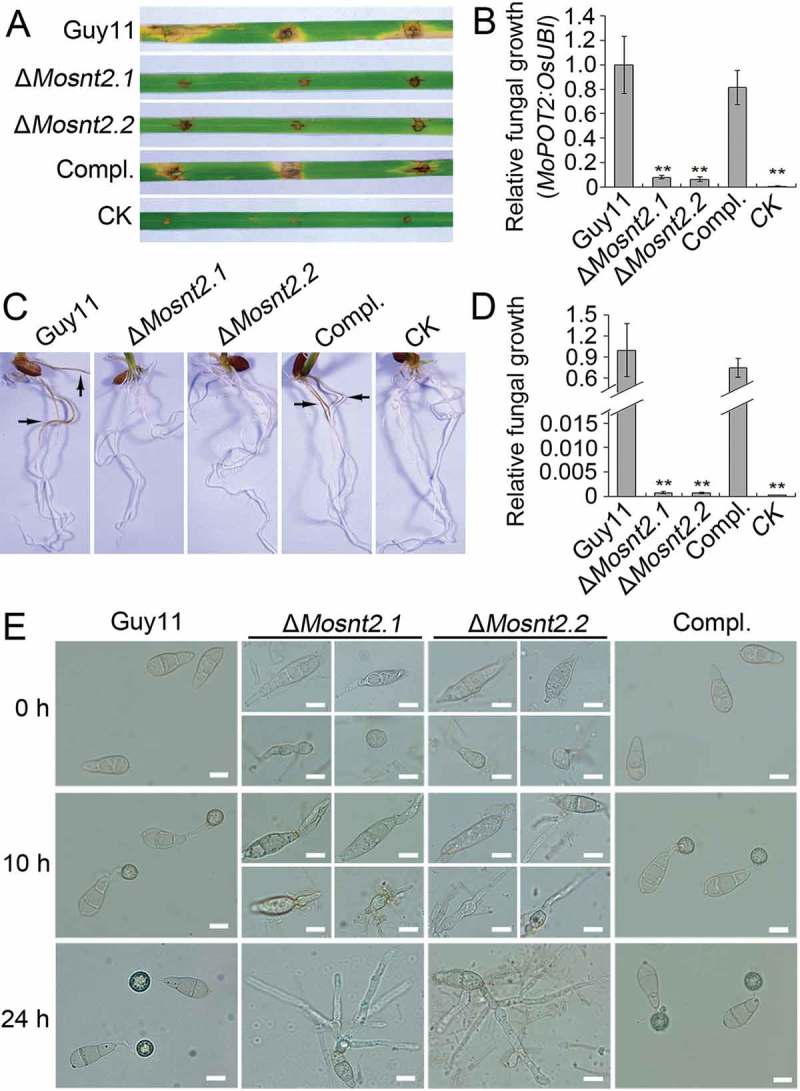
MoSNT2 is essential for plant infection by M. oryzae. (A) Rice leaf segments infected with fungal mycelium. (B) Measurement of fungal biomass in infected leaves based on qPCR analysis of the MoPOT2 repetitive element. CK, rice leaf segments inoculated with agar plugs without fungal mycelium. (C) Rice root infection assay. Arrows indicate typical necrotic lesions. (D) qPCR analysis of fungal biomass in inoculated rice roots. (E) Appressorium development on hydrophobic coverslip. Scale bar: 10 μm.
To understand the reasons for the compromised pathogenicity of the ΔMosnt2 mutants, we examined the development of infection structure. At 10 h postinoculation (hpi) on hydrophobic coverslip, conidia germinated and produced melanized appressoria at the tips of germ tubes in the control strains. Meanwhile, the abnormally shaped conidia of ΔMosnt2 mutants failed to form appressoria, even though germ tubes were produced (Figure 3(E)). At 24 hpi, conidial cells of the control strains collapsed leading to autophagic cell death. However, conidia of ΔMosnt2 mutants produced hyphal branches without formation of appressoria and failed to collapse (Figure 3(E)). We further examined infection structure development on onion epidermis, and observed that ΔMosnt2 mutants produced dramatically less appressoria than control strains (Figure 4(A,B)). The ΔMosnt2 mutants produced neither penetration pegs nor invasive hyphae inside onion epidermis (Figure 4(B)). By contrast, 70% of appressoria in the control strains produced penetration pegs and infectious hyphae. The ΔMosnt2 mutants were also severely compromised in collapse of conidial cells compared to the control strains (Figure 4(A,C)). In rice leaf sheath inoculation assays, we obtained similar results showing that the ΔMosnt2 mutants have defects in autophagic cell death and penetration peg formation (Figure 4(D)).
Figure 4.
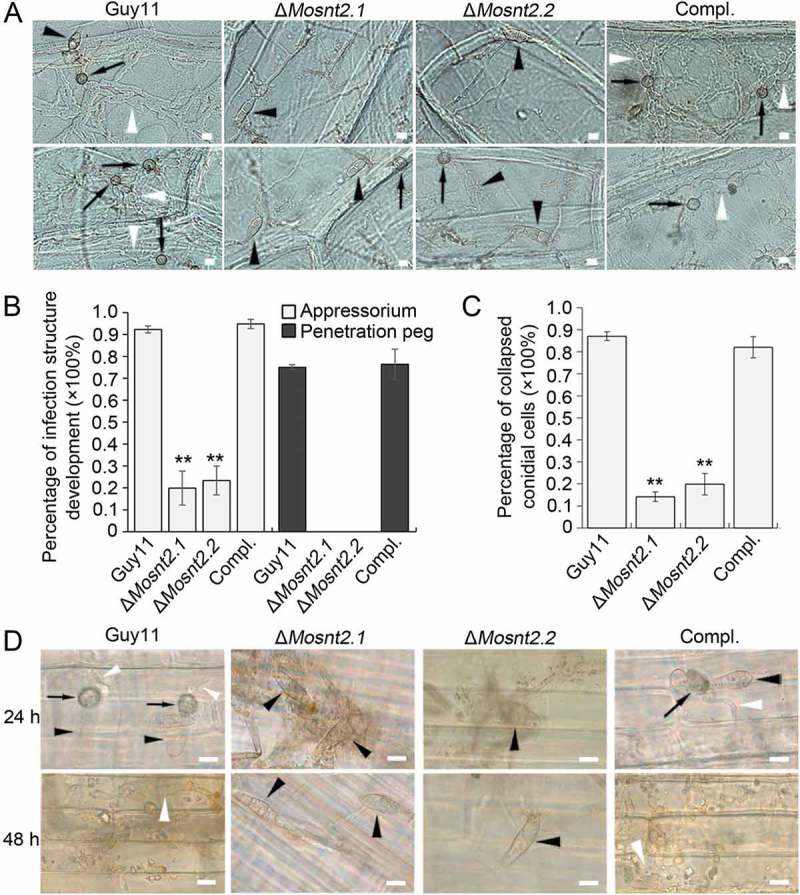
MoSNT2 is crucial for infection structure development and autophagic cell death. (A) Infection structure development on onion epidermis at 36 hpi. The black arrowhead and arrow indicates conidium and appressorium respectively, while the white arrowhead indicates invasive hypha. (B) Percentage of appressorium and penetration peg formation on onion epidermis (n > 50, triple replications, ** P < 0.01). (C) Percentage of spores containing 3 totally-collapsed conidial cells on onion epidermis (n > 50, triple replications). (D) Infection structure development on rice leaf sheath. Scale bar: 10 μm.
M. oryzae is able to infect rice host by developing appressoria from the tips of vegetative hyphae [35]. On hydrophobic surface, hyphae of all examined strains produced appressoria at hyphal tips (Figure S4(A)). However, unlike the control strains which developed normal penetration pegs and invasive hyphae, the ΔMosnt2 mutants failed to differentiate penetration pegs or penetrate host tissue (Figure S4(A)). Hyphal appressoria of control strains penetrated rice leaf epidermis, eliciting modest ROS production, observed by diaminobenzidine (DAB) staining at the penetration site (Figure S4(B,C)). In contrast, the ΔMosnt2 mutants produced no obvious DAB staining around hyphal tip appressoria, consistent with the lack of penetration peg development. In a hyphal immersion inoculation assay, rice leaves developed blast lesions after inoculation with control strains, whereas leaves inoculated with ΔMosnt2 mutants did not form typical blast lesions and contained very few fungal DNA (Figure S4(D,E)). These results indicate that MoSNT2 is essential for repolarisation of appressoria formed at hyphal tips and is essential for all forms of plant infection by M. oryzae.
Disruption of MoSNT2 impairs cell wall integrity and oxidative stress response
Previous studies have shown the importance of cell wall integrity in hyphal development and pathogenicity [36]. Because ΔMosnt2 mutants display defects in both hyphal growth and host penetration, we examined growth of ΔMosnt2 mutants under cell wall stress conditions triggered by exposure to Calcofluor White (CFW) and Congo Red (CR), which inhibit assembly of chitin and β-1,4-glucans in the cell wall, respectively [37]. The ΔMosnt2 mutants showed increased sensitivity to both CFW and CR compared to control strains, suggesting alteration of cell wall integrity as a consequence of MoSNT2 deletion (Figure 5(A)). Peroxidases catalyze the degradation of CR [38]. Consistent with this, peroxidase activity and expression of peroxidase-encoding genes were greatly reduced in the ΔMosnt2 mutants (Figure S5(A,B)). Oxidizing activity of laccase enzymes and expression of laccase-encoding genes were also reduced in the ΔMosnt2 mutants (Figure S5(C-E)).
Figure 5.
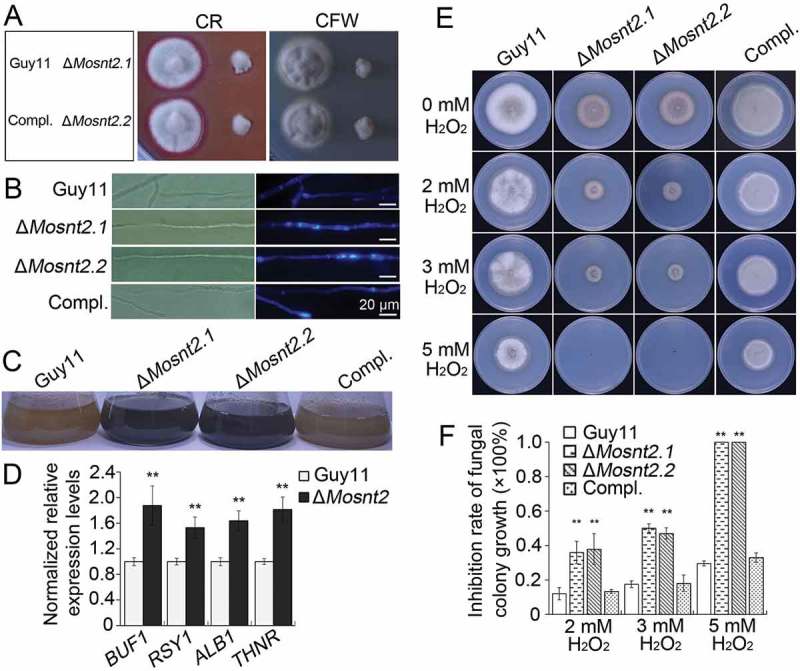
MoSNT2 regulates cell wall integrity and oxidative stress response. (A) Growth of M. oryzae on CM agar medium containing 200 μg/ml CFW or 200 μg/ml CR for 5 days. (B) CFW staining and epifluorescence microscopy of cell wall chitin of mycelium grown in liquid CM. Scale bar: 20 μm. (C) Increased hyphal melanization as a consequence of MoSNT2 deletion. (D) qRT-PCR analysis on the expression levels of melanin biosynthesis genes in mycelium grown in liquid CM. (E) Mycelial growth on CM agar medium in the presence of different concentrations of H2O2. (F) Statistical analysis of the inhibition rate under H2O2-induced oxidative stress on mycelial growth. Asterisks represent significant differences (** P < 0.01).
We next tested whether cell wall properties in the mutants were altered by using CFW to stain chitin. In control strains, newly synthesized chitin, revealed by CFW fluorescence, was mainly concentrated at hyphal septa and tips (Figure 5(B)). In ΔMosnt2 mutants, CFW-stained chitin was not only present at septa and hyphal tips, but also distributed on the lateral cell wall along hyphal axes. After growth in liquid CM for 2 days, hyphae of ΔMosnt2 mutants became noticeably darker than the control strains (Figure 5(C)), suggesting excess melanin accumulation in the ΔMosnt2 mutants. Consistent with this, expression levels of melanin biosynthesis genes were significantly higher in the ΔMosnt2 mutants (Figure 5(D)). Fungal growth was inhibited by increasing concentrations of H2O2; noticeably, ΔMosnt2 mutants showed hypersensitivity (Figure 5(E,F)). Growth of the ΔMosnt2 mutants was completely (100%) inhibited at 5 mM H2O2 while growth of the control strains was only reduced by 35%. Together, these results indicate that MoSNT2 is required for cell wall integrity and oxidative stress tolerance.
MoSnt2 binds the acetylated H3 histone and recruits the MoHos2 histone deacetylase
We next investigated the mechanism by which MoSNT2 regulates autophagy and pathogenicity. Yeast Snt2 is known to associate with histone H3 and epigenetically regulate expression of genes involved in response to both oxidative and starvation stress [29,31]. We therefore hypothesized that MoSnt2 might mediate epigenetic control of gene expression. Our results showed that MoSnt2-GFP colocalized with 4ʹ, 6-diamidino-2-phenylindole (DAPI) staining of nuclei in mycelia, conidia, appressoria and invasive hyphae of M. oryzae (Figure S6), suggesting that MoSnt2 is a nuclear protein. We then examined the potential in vivo interaction between MoSnt2 and H3 using a bimolecular fluorescence complementation (BiFC) assay [39]. MoSnt2 was fused to the C-terminal fragment of YFP (YFPCTF) and H3 fused to the N-terminal fragment of YFP (YFPNTF). The 2 constructs expressing MoSnt2-YFPCTF and YFPNTF-H3 were cotransformed into the Guy11 strain; as a negative control, the constructs MoSnt2-YFPCTF and YFPNTF were cotransformed into the Guy11 strain. YFP signals were only detected in transformants coexpressing MoSnt2-YFPCTF and YFPNTF-H3, but not in the negative control (Figure 6(A)), suggesting that MoSnt2 interacts with H3.
Figure 6.
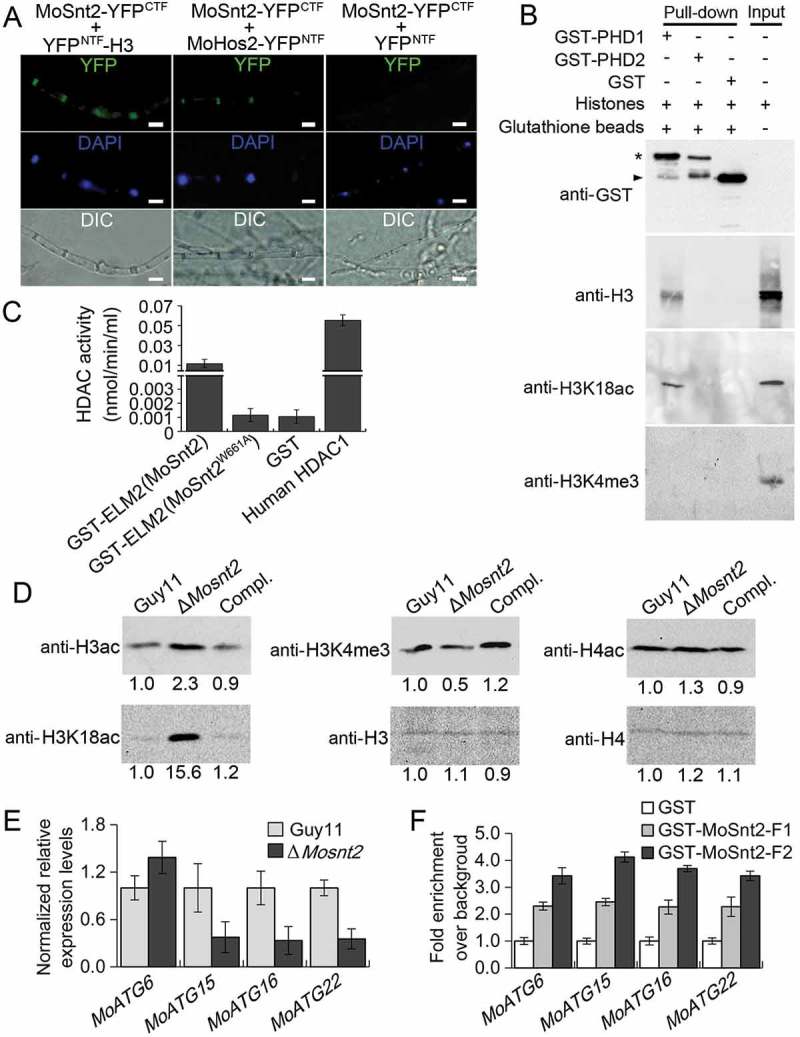
MoSnt2 mediates H3 deacetylation and regulates expression of autophagy genes. (A) Visualization of the interaction between proteins as shown in the BiFC assay. Vegetative hyphae were stained with DAPI and then analyzed by epifluorescence microscopy. Scale bar: 10 μm. (B) GST-PHD1 coimmunoprecipitates H3 histones. E. coli-expressed fusion proteins were used for affinity isolation of histones of calf thymus and immunoblot analysis conducted with the antibodies indicated. The star indicates GST-PHD1 and GST-PHD2, while arrowhead indicates GST. (C) Histone deacetylase activity in affinity isolation complexes. (D) Immunoblot analysis of histone proteins in M. oryzae with the indicated primary antibodies. (E) qRT-PCR analysis on the expression levels of autophagy genes. (F) In vitro affinity isolation of autophagy gene DNA by MoSnt2. GST-MoSnt2-F1, GST-MoSnt2-F2 or GST alone were incubated with sheared chromatin, affinity isolated, washed and subjected to qPCR for autophagy genes. Similar results were obtained from 3 independent biological experiments.
We further assessed the nature of the protein interaction. The PHD finger is a small zinc-binding motif (C4HC3) mediating protein-protein interaction in chromatin-dependent transcriptional regulation commonly found in nuclear proteins, and independent lines of evidence have shown that PHD domains possess the ability to bind the N terminus of histone H3 [40]. The SMART program [41] predicted that MoSnt2 contains 3 PHD finger domains (Figure S1), whereas, ClustalOmega predicted typical zinc-binding signature C4HC3 in the first (PHD1) and second PHD (PHD2) domains, but not in the third putative PHD domain (PHD3) (Figure S2). We reasoned that the MoSnt2 PHD domains are capable of binding histone H3. To test this idea, we performed an affinity isolation assay by incubating each of the bacterial-expressed glutathione S-transferase (GST) fusion proteins GST-PHD1 and GST-PHD2 with calf thymus total histones. Our results revealed that PHD1 can bind histone H3, but PHD2 cannot (Figure 6(B)). Interestingly, PHD1 interacts with acetylated H3, but not with methylated H3 (Figure 6(B)). These results suggest that MoSnt2 binds histone H3 through its PHD1 domain.
MoSnt2 contains a conserved ELM2 domain which is well characterized in recruiting histone deacetylase for transcriptional regulation [42]. To address whether the MoSnt2 ELM2 domain has a similar role, we assayed the Guy11 strain protein complexes affinity-isolated by GST-ELM2(MoSnt2) and GST individually, for histone deacetylase activity. The results showed that with affinity isolation assays, the GST-ELM2(MoSnt2) complex contained 10-fold higher histone deacetylase activity than the GST control (Figure 6(C)). Consistent with previous reports [42], replacement of a conserved, critical tryptophan by alanine within the ELM2 domain (GST-ELM2[MoSnt2W661A], according to the MoSnt2 aa sequence) completely abolishes the specific interaction, reducing histone deacetylase activity to that of the GST-only control (Figure 6(C)). The ELM2 domain of MoSnt2 therefore functions to recruit histone deacetylase. We further examined whether MoSnt2 interacts with the histone deacetylase MoHos2, which belongs to the Tig1 HDAC and also regulates pathogenicity and conidiation in M. oryzae, C. carbonum and U. maydis [21–23]. BiFC analysis showed that MoSnt2 is associated with MoHos2 in vivo (Figure 6(A)). Together, these results indicate that MoSnt2 recruits histone deacetylase with its ELM2 domain.
Deletion of MoSNT2 elevates acetylation of histone H3 and changes global gene expression
We then explored whether MoSnt2 directly regulates deacetylation of histone H3. Protein extracts of M. oryzae grown on CM agar medium were subjected to immunoblot analysis. Total acetylation levels of H3, detected with an anti-H3ac antibody, were increased in the ΔMosnt2 mutant, whereas acetylation levels of histone H4 detected by anti-H4ac were similar among the ΔMosnt2 mutant and control strains (Figure 6(D)). A previous study has revealed that H3K18 acetylation (H3K18ac) regulated by the Tig1 HDAC is critical for pathogenicity of M. oryzae [23]. Using an anti-H3K18ac antibody, we observed that H3K18ac levels were highly elevated in the ΔMosnt2 mutant (Figure 6(D)). Histone lysine methyltransferase MoSet1 is known to epigenetically regulate global gene expression during infection-associated development by modifying H3K4 trimethylation (H3K4me3) [25]. When analyzed with an anti-H3K4me3 antibody, H3K4me3 levels were slightly reduced in the ΔMosnt2 mutant (Figure 6(D)). Similar results were obtained when analyses were performed on the hyphae grown in liquid CM (Figure S7(A)). Immunoblot analysis of 5 independent ΔMosnt2 mutants further confirmed the role of MoSnt2 in regulating H3 acetylation levels (Figure S7(B-F)). Collectively, these results demonstrate that deletion of MoSNT2 elevates acetylation levels of H3, in particular at H3K18, indicating that MoSnt2 functions to promote H3 deacetylation in M. oryzae.
To gain insight into which genes are regulated by MoSNT2, we utilized RNA-seq to compare global gene expression profiles between a ΔMosnt2 mutant and the wild-type. We identified many differentially expressed genes (DEGs), with 1554 genes that were upregulated and 1323 downregulated in the ΔMosnt2 mutant (based on the log2Ratio [ΔMosnt2/Guy11 strain] > 1 or < −1, P < 0.05). These DEGs contain many functional groups associated with conidiation, pathogenicity, autophagy, chitin and glucan biogenesis, xylanase, cell wall integrity, amino acids metabolism, lipase, permease and transporter, multidrug resistance, histone modification and cell-cycle regulation (Table S1, P < 0.05). We utilized qRT-PCR to validate 41 downregulated or upregulated genes that were functionally characterized as associated with infection and development (Figure S8). Our qRT-PCR results validated the RNA-seq data. The differential expression of these genes is consistent with the roles of MoSnt2 in regulating conidiation, pathogenicity, cell wall integrity and autophagy, and moreover suggests a role for MoSnt2 in adaptation to nutrition and environmental stresses. Interestingly, several genes encoding histone acetyltransferases are upregulated, in line with MoSnt2 regulation of H3 deacetylation. Together, these results indicate that MoSnt2 coordinates global gene expression by regulating H3 deacetylation.
Several autophagy-related genes were among the DEGs (Table S1, P < 0.05), including the autophagy core genes MoATG6, MoATG15, MoATG16 and MoATG22 that are important for plant infection by M. oryzae [9]. qRT-PCR analysis confirmed the upregulation of MoATG6, and downregulation of MoATG15, MoATG16 and MoATG22 in the ΔMosnt2 mutant (Figure 6(E)). We further assessed whether MoSnt2 physically associates with these autophagy genes to regulate their expression. Due to the large size of MoSnt2 (196.2 kDa) or its instability, we failed to detect the MoSnt2-GFP fusion protein in immunoblots and were thus unable to conduct in vivo chromatin immunoprecipitation (ChIP)-qPCR. We therefore adopted an in vitro affinity isolation assay to determine binding of MoSnt2 to the promoters of autophagy genes. MoSnt2 containing 5 domains was expressed and purified as 2 overlapping GST fusion fragments GST-MoSnt2-F1 (covering the BAH, PHD1 and ELM2 domains) and GST-MoSnt2-F2 (covering ELM2, PHD2 and PHD3 domains) (Figure S9). These 2 fragments and GST control were each used to pull down Guy11 strain chromatin DNA. qPCR analysis on MoATG6, MoATG15, MoATG16 and MoATG22 showed that their promoters were 2- to 4-fold enriched by GST-MoSnt2-F1 or GST-MoSnt2-F2 compared to that by the GST control (Figure 6(F)). These results indicate that MoSnt2 binds to promoters of autophagy genes to regulate their expression and thereby control autophagic homeostasis.
MoSNT2 mediates MoTor signaling and its expression is dependent on MoTor
Tor is an evolutionarily conserved protein kinase central to growth control and autophagy regulation [43]. We assessed the relationship between MoSnt2 and M. oryzae Tor kinase (MoTor, MGG_15156). When assayed for inhibition by rapamycin, a specific inhibitor of Tor kinase, hyphal growth of the ΔMosnt2 mutants was more severely inhibited (~ 50% reduction) compared to the wild-type (~ 35% reduction) (Figure 7(A,B)). In analysis of expression profiles, we observed that MoSNT2 and MoTOR were ubiquitously expressed, but both peaked during conidiation (Figure 7(C)). Both were repressed by oxidative stress. Moreover, a highly positive linear correlation coefficient existed between their expression levels (R2 = 0.8159, Figure 7(D)). Inhibition of MoTor by rapamycin reduced MoSNT2 expression levels by about 70% in the wild-type (Figure 7(E)), suggesting a requirement of MoTor kinase activities for MoSNT2 expression. We attempted to delete MoTOR, but did not recover any ΔMotor deletion mutants, suggesting that MoTOR is likely an essential gene. We therefore employed a conditional RNA interference (RNAi) strategy [44] to knock down expression of MoTOR. In 4 independent MoTOR RNAi transformants (pSilent1-TorFRB.7, pSilent1-TorFRB.28, pSilent1-TorFAT.39 and pSilent1-TorFAT.54), MoTOR expression levels in vegetative hyphae grown in CM remained similar to that in the wild-type, but were reduced to ~ 50% of wild-type level when the RNAi was induced in MM+ NaAc medium (Figure S10(A)). More importantly, MoSNT2 expression levels were greatly reduced to about ~ 10% of wild-type level when MoTOR was knocked down (Figure S10(B)). Together, these results suggest that MoSNT2 expression is positively regulated by MoTor and likely functions downstream of MoTor signaling.
Figure 7.
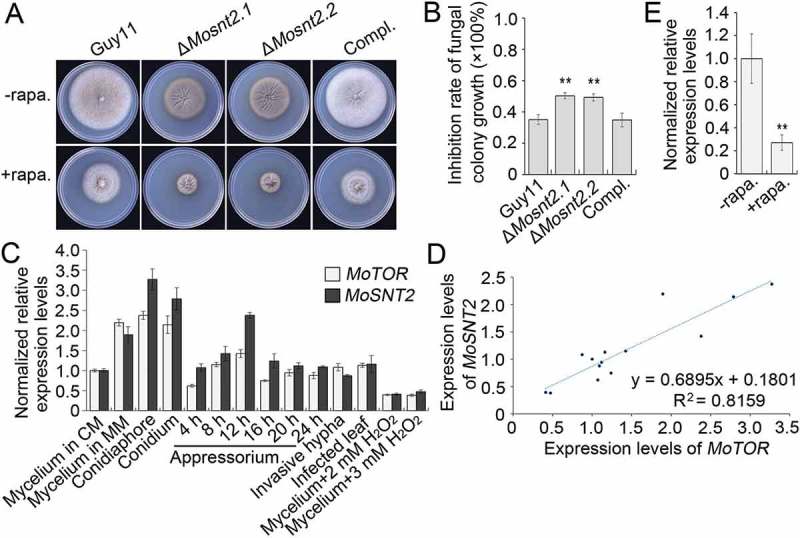
MoSNT2 is associated with the MoTor signaling pathway. (A) Vegetative growth of M. oryzae on CM agar medium supplemented with or without 1 μg/ml rapamycin (rapa.). (B) Inhibition rate of rapamycin on the mycelial growth. (C) Expression profiles of MoSNT2 and MoTOR in the wild-type Guy11 strain at different developmental processes. (D) Linear correlation between qRT-PCR-measured expression levels of MoSNT2 and MoTOR. (E) qRT-PCR analysis of MoSNT2 expression levels in the Guy11 strain in response to rapamycin. The Guy11 strain grown in liquid CM for 48 h was transferred into fresh liquid CM in the presence or absence of 1 μg/ml rapamycin for 6 h before total RNA extraction.
We further investigated the role of MoSNT2 in MoTor signaling by comparing the expression of genes regulated by MoTor between the wild-type and the ΔMosnt2 mutant. MoRS2 and MoRS3 (encoding ribosomal proteins), MoeIF4G (encoding translation initiation factor 4 G) and MoATG8 represent genes associated with MoTor-regulated ribosomal biogenesis, translation initiation and autophagy respectively [18]. In the wild-type, MoRS2, MoRS3 and MoeIF4G expression levels were lower in MM-N than in CM (Figure S10(C-E)), indicating downregulation of MoTor signaling by nutrient depletion. However, in the ΔMosnt2 mutant, MoRS2, MoRS3 and MoeIF4G expression levels in MM-N remained similar to those in CM (Figure S10(C-E)), suggesting that the ΔMosnt2 mutant is unable to properly regulate expression of genes in the MoTor signaling pathway upon nutrient deprivation. In MM-N medium, MoATG8, which is upregulated when Tor is inactivated, is induced higher in ΔMosnt2 than in wild-type (Figure S10(F)). Together, these results indicate that MoSNT2 mediates MoTor-regulated cellular processes in M. oryzae, including ribosomal protein biogenesis, translation initiation and autophagy.
We also determined the role of MoTOR in growth, autophagy and pathogenicity. Rapamycin exposure reduced the diameter of hyphal colonies of the Guy11 strain grown in CM (Figure 8(A,B)), suggesting a requirement of MoTor for vegetative growth. In nutrient-rich CM medium, rapamycin induced the generation of autophagosomes, to a similar extent as caused by starvation stress (Figure S11), indicating a role for MoTor in maintaining autophagic homeostasis. We also observed that during appressorium development, inhibition of MoTor by rapamycin strongly compromised autophagic conidial cell death. At 24 hpi, ~ 90% of conidia receiving no treatment or the solvent-only DMSO control fully collapsed due to autophagic cell death, whereas less than 35% of conidia collapsed in the presence of rapamycin (Figure 8(C,D)). Consistently, glycogen and lipid degradation was inhibited by exposure to rapamycin (Figure 8(E-G)), confirming the effects of rapamycin on autophagic cell death. Inoculation of rice leaves with conidial suspensions of the Guy11 strain, showed that virulence of M. oryzae was obviously reduced upon exposure to increasing concentrations of rapamycin (Figure 8(H-J)). In the MoTOR RNAi transformants, downregulation of MoTOR also impaired vegetative growth, autophagic cell death and pathogenicity (Figure S12). Collectively, these results demonstrate that MoTOR plays a role similar to MoSNT2 in regulating levels of developmentally conditioned autophagy that is necessary for M. oryzae to infect rice.
Figure 8.
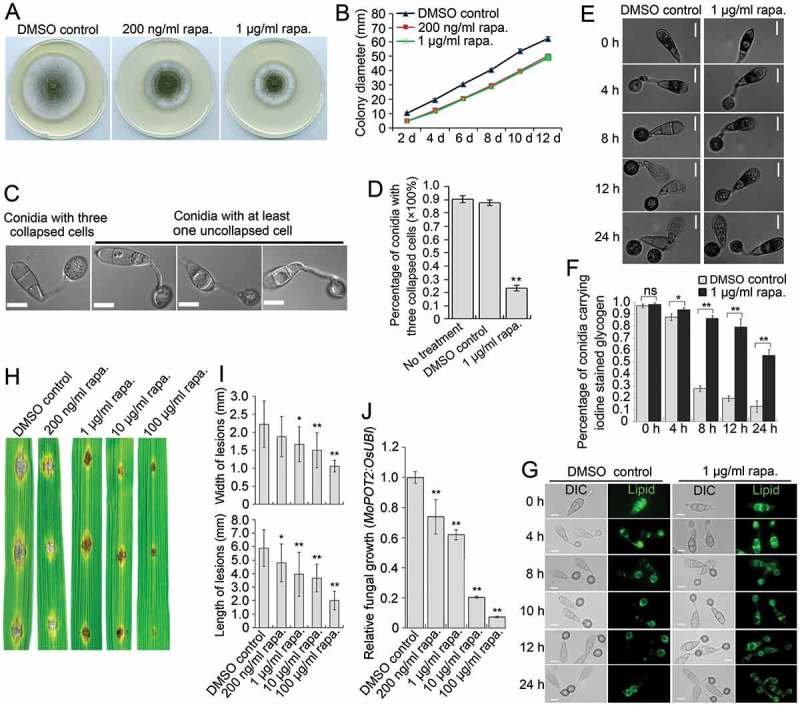
Effects of rapamycin on vegetative growth, autophagic cell death and pathogenicity of M. oryzae. (A) Plate colonies of the Guy11 strain in CM agar medium. The Guy11 strain was grown on CM medium supplemented with rapamycin (rapa.) at the indicated concentration. Solvent DMSO was seperately added into medium as a control. (B) Diameters of plate colonies recorded every 2 days. (C) Autophagic conidial cell death of the Guy11 strain at 24 hpi of appressorium development on hydrophobic coverslip in the presence of rapamycin. Scale bar: 10 μm. (D) Percentage of Guy11 strain spores containing 3 totally-collapsed conidial cells at 24 hpi (n > 100, triple replications, ** P < 0.01). (E) Glycogen distribution during appressorium development on hydrophobic surface. Glycogen was stained by iodine solution and microscopically visualized as yellowish-brown deposits. (F) Percentage of spores containing glycogen content in conidial cells (n > 100, triple replications, * P < 0.05, ** P < 0.01). (G) Lipid body translocation and degradation during appressorium morphogenesis as revealed by Bodipy staining. (H) Blast lesions on rice leaf segments infected with Guy11 strain. (I) Mean width and length of lesions calculated from at least 15 independent blast lesions. (J) qPCR analysis of fungal biomass in the infected rice leaf segments.
Discussion
In this study, we report a critical role for the MoSnt2 protein as an epigenetic factor required for regulation of infection-associated autophagy in M. oryzae (Figure 9). Autophagy is crucial for virulence of many pathogenic fungi, such as M. oryzae, Colletotrichum lindemuthianum, C. orbiculare, F. graminearum, U. maydis and Cryptococcus neoformans [3,4]. In M. oryzae, targeted deletion of any of the genes regulating autophagic cell death prevents plant infection [9]. We have demonstrated that MoSNT2 plays a critical role in maintaining autophagic homeostasis in M. oryzae and that its expression is positively regulated by the MoTor kinase. MoSnt2 is nuclear-localized and recognizes acetylated histone H3 through its PHD1 domain and recruits HDAC by interaction with MoHos2 via its ELM2 domain to target chromatin regions. MoSnt2-mediated recruitment of HDAC thereby decreases acetylation of H3, altering expression of a large set of genes associated with developmental processes including autophagy.
Figure 9.
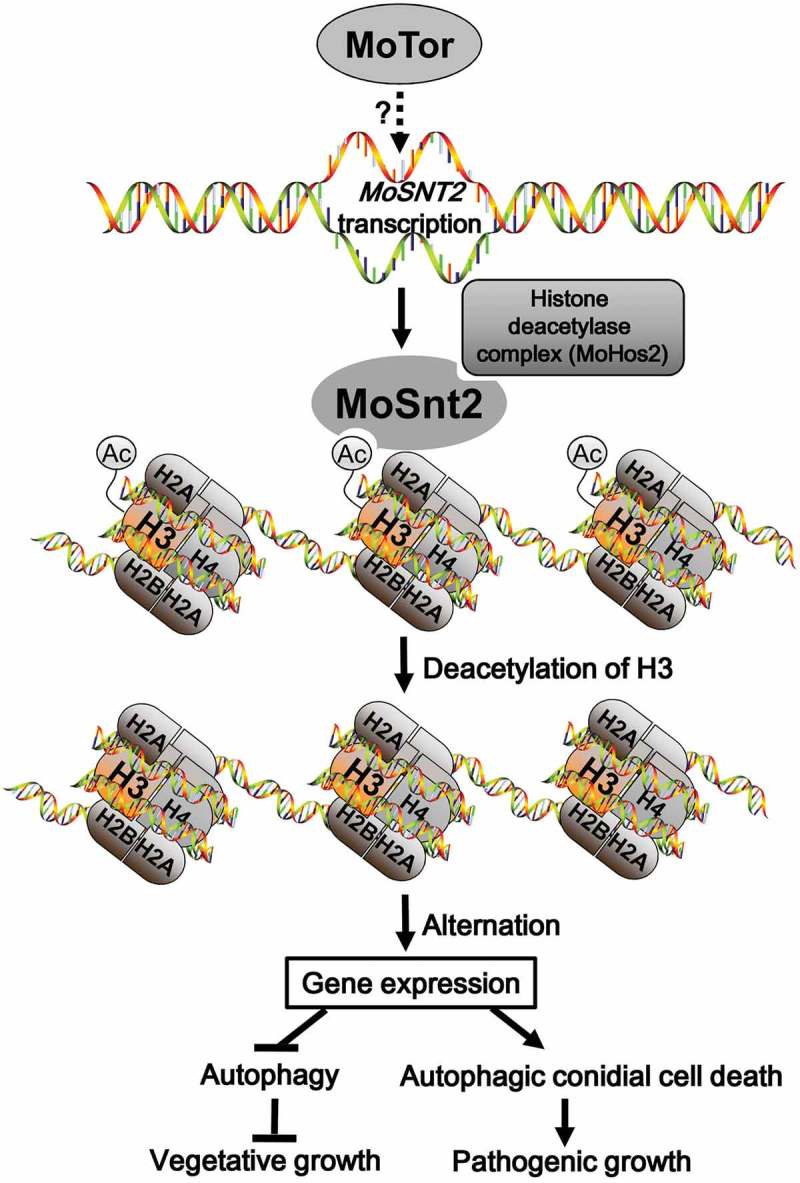
Model for MoSnt2-mediated epigenetic control of pathogenicity in M. oryzae. MoTor promotes the expression of MoSNT2 through unidentified effector(s) or transcription factor(s). Nucleus-localized MoSnt2 recognizes acetylated histone H3 and recruits the histone HDAC deacetylase complex to targeted chromatin regions. The MoSnt2-recruited HDAC then deacetylates H3 and alters expression of genes. MoSnt2-regulated gene expression functions to repress autophagy to promote hyphal proliferation during vegetative growth in nutrient rich conditions, while promoting autophagic conidial cell death and assisting pathogenic growth on host rice.
MoSnt2 determines autophagy-dependent plant infection by M. oryzae
Although SNT2 is widely distributed among fungal pathogens, its role has only been described previously in F. oxysporum [30,32]. Deletion mutants of SNT2 in F. oxysporum show defects in autophagy, vegetative growth, conidiation and oxidative stress sensitivity [30,32], but the molecular mechanism underlying the regulatory roles of Snt2 remains unknown. We have provided evidence that MoSnt2 functions as an epigenetic regulator by deacetylation of histone H3, thereby determining autophagy-related growth and pathogenicity in M. oryzae. We observed that acetylated-H3 levels were increased in ΔMosnt2 mutants (Figure 6), which displayed similar defects to the F. oxysporum Δsnt2 mutant, including disruption of autophagic homeostasis (Figure 2). These defects mirror those of M. oryzae mutants lacking autophagy genes including MoATG1, MoATG4, MoATG5, MoATG8, MoATG9 and MoATG18. Autophagy-associated defects of ΔMosnt2 deletion mutants are therefore consistent with MoSnt2 being necessary for regulation of these processes during appressorium-mediated rice infection, as shown in the model presented in Figure 9.
The importance of MoSnt2 in autophagy is highlighted by its role in transcriptional regulation of autophagy genes, such as MoATG6, MoATG15, MoATG16 and MoATG22 through association with their promoters (Figure 6). Atg6 and Atg16 are both important for autophagosome formation, while Atg15 and Atg22 determine degradation of autophagic bodies within vacuoles [3]. It is therefore likely that MoSnt2 affects autophagic homeostasis through modulating the expression of autophagy genes involved in different stages of autophagic progression. Infection-associated autophagy in M. oryzae is regulated by various cytoplasmic events. The Rab GTPase MoYpt7, for instance, mediates fusion of autophagosomes with vacuoles and is thus required for autophagy-dependent plant infection [33]. MoVps35, a component of the endosomal protein sorting retromer complex, regulates biogenesis of autophagosomes and is essential for pathogenicity of M. oryzae [45]. Our study suggests that autophagy in fungal pathogens is further regulated by nuclear events, including modification of histone by deacetylation.
The ΔMosnt2 mutant shows aberrant cell wall integrity, which is apparent from its hypersensitivity to cell wall perturbing agents, disordered cell wall melanization, and chitin deposition (Figure 4). It has been shown that cell wall integrity regulators, such as the mitogen-activated protein kinase Mps1 [46] and NADPH oxidases Nox1 and Nox2 [36], are essential for penetration of host by M. oryzae. MoYpt7, an important regulator of autophagy and cell wall integrity, is also critical for pathogenicity of M. oryzae [33]. MoSnt2 may therefore be necessary for pathogenicity through its regulatory role in both autophagy and cell wall integrity. Signaling pathways other than the MAPK Mps1, Pkc/protein kinase C and Ca2+ have already been shown to determine cell wall integrity and plant penetration by M. oryzae [44,47,48]. We observed that MoSnt2 is transcriptionally regulated by MoTor and mediates MoTor-dependent cellular processes. These results suggest that a signal cascade involving MoTor and MoSnt2 is also important for the cell wall integrity-dependent plant infection by M. oryzae.
MoSnt2 modulates autophagy through coordination with MoTor kinase signaling
Tor kinase functions as a key regulator of autophagy in a broad range of eukaryotes from yeast to mammals [43]. It has been recently reported that MoTor signaling is negatively regulated by a carbon-responsive regulator Abl1 and plays an important role in infection-associated autophagy in M. oryzae [19]. This previous study [19] does not, however, report the inhibitory effects of rapamycin on autophagic conidial cell death observed in our present report. A much lower concentration of rapamycin has been previously used [19], and it may have been inadequate to completely inhibit MoTor, particularly given the melanization of conidia and appressoria. Our study reveals that inhibition or knockdown of MoTor can cause defects similar to those observed in ΔMosnt2. These findings not only provide extra evidence that MoTor regulates infection-associated autophagy in M. oryzae, but also highlight how coordinated regulation of autophagy by MoSnt2 and MoTor is likely to be essential for rice blast disease. Expression of MoSNT2 is positively correlated with that of MoTOR (Figure 7). In yeast, Snt2 has been shown to mediate Tor signaling, because it is necessary for the transcriptional response to starvation stress triggered by rapamycin [31]. Deletion of SNT2 in F. oxysporum elevates expression of Tor-regulated genes including IDI4, PDC and eEF1G [30]. This suggests that Snt2 orthologs may play conserved functions among fungi in determining autophagy through a common pathway involving Tor.
Our results indicate that MoTor functions in concert with MoSnt2 to negatively regulate autophagy during vegetative growth, while also being required for autophagic conidial cell death during pathogenic growth. During vegetative growth in CM, inhibition of MoTor induced the occurrence of autophagosomes (Figure S11), in a similar manner to the deletion of MoSnt2 (Figure 2). Because autophagy is active when Tor kinase is inactive, MoTor is likely to normally repress autophagy during vegetative growth of M. oryzae under nutrient rich conditions. During infection, however, MoTor and MoSnt2 are both required for autophagic cell death of conidia, which is necessary for appressorium-mediated plant infection (Figures 3(E), 4A-C and Figure 8(C-G)). There are 2 possible explanations for this observation. First, it is possible that MoTor signaling during host infection is distinct from that during vegetative growth. It is known, for instance, that Atg13, a core component of an Atg1/ULK1-Atg13 complex essential for autophagy regulation in various species [49–51], is dispensable for infection-related autophagy and pathogenicity in M. oryzae [9,52]. M. oryzae can dispense with Atg13 function during its infection process, and so may have evolved an alternative molecular machinery to transduce Tor signaling during infection-associated autophagy. Appressorium morphogenesis of M. oryzae is, for example, tightly coupled with cell cycle progression, which can also lead to compromised autophagic cell death [5,53]. Since MoTor signaling is important for maintaining proper cell cycle progression [18,19], it may also be essential for the onset of conidial cell death.
Secondly, it is possible that a critical threshold of autophagy is essential for conidial cell death to occur, such that overstimulation of autophagy – resulting from MoTor inhibition, rapamycin treatment, or in MoSnt2 mutants – prevents the prodeath signals that are essential for completion of conidial collapse, thereby preventing trafficking of the contents to the incipient appressorium and impairing appressorium function. This would suggest that the precise level of autophagy in conidia is essential for plant infection by M. oryzae, such that loss of autophagy, due to mutation of autophagy genes will prevent conidial cell death, while overstimulation of autophagy, by loss of MoTor activity would also prevent completion of autophagic cell death. Future experiments will address which of these scenarios is the most likely.
Emerging evidence suggests that histone modifications mediate Tor kinase-regulated autophagy in yeast and mammals. The yeast chromatin-remodeling complex Rsc1, for example, regulates ATG8 transcription and is required for proper downregulation of Tor kinase complex 1 activity through stimulation of the Rho-Kog1 interaction under starvation stress conditions [14]. Mammalian histone acetyltransferase KAT8 is transcriptionally regulated by Tor kinase activity to determine acetylation of H4K16 on autophagy-related genes and the occurrence of autophagy [15]. In mammals, the methyltransferase EZH2 (enhancer of zeste homolog 2, a subunit of polycomb repressive complex 2) controls autophagic progression by modifying H3K27me3 of several genes encoding negative regulators of MTOR [54]. With regard to fungal pathogens, it remains largely unknown how autophagy is regulated at the molecular level. In M. oryzae, MoTor signaling plays a role in regulation of mitosis and autophagy through coordination with Rbp35, Asd4, Tkl1 and Abl1 [16–19]. Our study shows that the epigenetic factor MoSnt2 functions downstream of MoTor signaling to maintain proper autophagic homeostasis by recruiting the HDAC MoHos2 and determining H3 acetylation levels, regulating downstream gene expression. These results provide new insight into genetic regulation of infection-associated autophagy in fungal pathogens, and furthermore advance the autophagic histone code concept in eukaryotes [55].
MoSnt2 recognizes acetylated H3 and recruits HDAC to epigenetically regulate gene expression
Previous studies have shown that development of M. oryzae may be subject to epigenetic regulation [23,25]. Our study identifies MoSnt2 as a key epigenetic regulator in M. oryzae because disruption of MoSNT2 significantly increases H3 acetylation levels (Figure 6) and compromises expression of large sets of genes associated with conidiation, autophagy, virulence, cell wall remodeling and integrity, amino acid metabolism, membrane transport (permease and transporter), multidrug resistance, histone modification and cell cycle control (Table S1). In M. oryzae, the HDAC component Tig1 and Hos2 [23], and histone methyltransferase MoSet1 [25] are epigenetic regulators with similar, potentially overlapping functions. Several defects, such as reduced vegetative growth and conidiation, increased sensitivity to oxidative stress, defective infection structure development and loss of pathogenicity, which were observed in Δtig1, Δhos2 and ΔMoset1 deletion mutants, are also found in the ΔMosnt2 deletion mutants. Together, our observations suggest that MoSnt2 plays a crucial role in histone modification, likely involving the Tig1 complex and MoSet1. Characterization of histone acetylation- and methylation-related enzymes reveals the importance of histone epigenetic modifications for the virulence of fungal pathogens [56,57]. However, to the best of our knowledge, it has not been shown how histone marks are recognized and chromatin remodeling complexes recruited onto chromatin in these species. Our study reveals that MoSnt2 lacks typical catalytic domains for histone acetylation or methylation, but instead executes an epigenetic regulatory function that requires its PHD1 domain to recognize acetylated-H3 and the ELM2 domain for recruitment of histone deacetylase. Thus, the role of MoSnt2 in histone modification represents a novel mode of epigenetic regulation for fungal pathogenesis.
PHD domains have been previously characterized as versatile epigenetic readers for recognizing H3K4me3, H3K4me or H3K14ac [40], but these functions of PHD domains are mainly known in mammalian studies. ELM2 domains have been reported in several nuclear proteins for binding histone deacetylases to assist in transcriptional regulation [42,58]. The yeast Snt2 protein is reported to bind and recruit histone deacetylase Rpd3 to regulate transcription of genes related to oxidative and starvation stress [31]. Our study shows that the MoSnt2 PHD1 domain and ELM2 domain bind acetylated H3 and recruits HDAC, respectively. Snt2 may therefore share a similar function among fungi in epigenetic recruitment of HADC. Furthermore, Snt2 seems to have functionally diverged between yeast and M. oryzae in its regulation of histone levels. Yeast Snt2 mediates histone degradation and loss of function of SNT2 results in accumulation of excess histone [29]. However, we did not find any differences in protein levels of either histone H3 or H4 between wild-type and ΔMosnt2 mutants in M. oryzae (Figure 6(D)).
Certain histone modifications, such as H3K56ac, H3K4me3, H3K9me2, H4K16ac and H4K20me3, contribute to control of autophagic flux in yeast and mammals [13,59]. The modifying enzymes responsible for the autophagy-associated changes in these histone marks, for example, include KAT8 (histone acetyltransferase), deacetylase SIRT1 (sirtuin 1), EHMT2/G9a (euchromatic histone lysine N-methyltransferase 2) and EZH2 (enhancer of zeste homolog 2) [13,54]. We observed that deletion of MoSNT2 elevates H3K18ac level and subsequently increases autophagic flux. This suggests that H3K18ac modification constitutes another important autophagic histone code in eukaryotes [55]. Autophagy is subject to regulation by HDAC at both the transcriptional and translational levels, including epigenetic control over expression of autophagy genes and modification of acetylation of autophagy proteins [13,59]. For instance, the HDAC Ume6-Sin3-Rpd3 serves as a negative regulator of transcription of yeast ATG8 and the genes encoding Atg8-family orthologs in mammalian cells [60]. Atg3, an E2-like protein necessary for autophagosome assembly, is acetylated at K19 and K48 by coordinated action of histone acetyltransferase Esa1 and deacetylase Rpd3 to control its interaction with Atg8, regulating starvation-induced autophagy [61]. In M. oryzae, the histone acetyltransferase Gcn5 is identified to repress autophagy via acetylation on Atg7 [26]. Our study reveals that MoSnt2 regulates transcription of autophagy genes MoATG6, MoATG15, MoATG16 and MoATG22. However, considering the role of MoSnt2 in recruiting HDAC and fine-tuning homeostatic distribution of histone deacetylases, we do not exclude the possibility that MoSnt2 may further regulate autophagy through modulating deacetylation of potential autophagy proteins in M. oryzae.
In various mammalian cell types, HDAC inhibition generally results in elevation of histone acetylation leading to induction of autophagy [59,62]. However, at present, very few nuclear factors have been characterized that link HDAC inhibition with autophagy induction. Among them, the transcription factor FOXO1, for example, mediates the effects of HDAC inhibition on autophagy induction [63]. Its expression and nuclear import increases upon HDAC inhibition, which then enhances expression of ATG genes and suppresses MTOR via transcription of SESN3 (Sestrin 3). Our results indicate that MoSnt2 controls autophagy in a manner associated with MoTor signaling. The increased histone H3 acetylation and autophagy induction in ΔMosnt2 mutant resembles those observed in other species encountering HDAC inhibition. This demonstrates that MoSnt2 represents another new epigenetic regulator that links MoTor signaling, histone deacetylation and autophagy.
In conclusion, this study uncovers a critical link between MoTor signaling, H3 deacetylation, and autophagy induction in M. oryzae and reveals a novel histone-based molecular switch for autophagy controlled by a signaling cascade regulated by MoTor and MoSnt2. The identification of epigenetic histone modifications in the regulation of infection-associated autophagy process offers an attractive conceptual framework to further understand the molecular pathogenesis of fungal pathogens.
Materials and methods
Molecular manipulations with RNA and DNA
The information for all primers, PCR templates and amplicons is listed in Table S2. Total RNA of M. oryzae was prepared using an RNeasy Plant Mini Kit (Qiagen, 74904) and RNase-Free DNase (Qiagen, 79254) was used for on-column digestion of DNA. For analyzing the expression pattern of MoSNT2 and MoTOR, total RNA was extracted from Guy11 strain grown under following conditions: (1) submerged mycelium grown in liquid CM; (2) conidiophore grown on CM agar; (3) conidium produced on CM agar; (4) appressorium formed on hydrophobic surface of GelBond film (Lonza, 53734) for 4 h, 8 h, 12 h, 16 h, 20 h and 24 h respectively; (5) invasive hypha colonizing rice sheath for 48 h; (6) invasive hypha infecting rice leaf for 6 days, (7) submerged mycelium under oxidative stress caused by 2 mM or 3 mM H2O2 (Sigma, 323381). For qRT-PCR analysis, 1st strand cDNA was generated by PrimeScrip RT reagent Kit with gDNA Eraser (Takara, RR047A) from 500 ng of total RNA. Real-time PCR was performed on 5 ng of 1st cDNA template with QuantiNova SYBR Green PCR kit (Qiagen, 208056) on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, 1855196). Each qRT-PCR reaction was repeated in at least 2 biological experiments independently with 3 technical replicates per sample. Fold changes in gene expression were calculated by 2−ΔΔCT method [64] with the Tubulin/β-tubulin gene (Tub, MGG_00604) as an internal control for normalization.
For gene cloning, first strand cDNA was produced using SuperScript III Reverse Transcriptase (Invitrogen, 18080093). Phusion high fidelity DNA polymerase (Thermo Scientific, F530S) was used for PCR amplification. MoSNT2 double-stranded cDNA sequences with a length of 6669 base pairs (bp) were divided into 4 overlapping fragments (F1, F2, F3, F4) that were respectively amplified with the primer pairs MoSNT2-F1-For and MoSNT2-F1-Rev, MoSNT2-F2-For and MoSNT2-F2-Rev, MoSNT2-F3-For and MoSNT2-F3-Rev, as well as MoSNT2-F4-For and MoSNT2-F4-Rev. The 4 PCR amplicons with lengths of 2026 bp, 2195 bp, 1512 bp and 2047 bp were cloned into the pEASY-blunt cloning vector (TransGen, CB101) and sequenced.
Fungal strains and phenotypic analyses
Growth and maintenance of M. oryzae, medium composition and fungal transformation are described previously [65]. For plate growth assays, fungal strains were grown on CM agar medium at 25°C and colony diameters measured from 3 replicates. The CM agar medium is prepared with following composition [65]: 10 g/L glucose, 2 g/L peptone, 1 g/L yeast extract, 1 g/L casamino acids, 0.1% (v/v) trace elements, 0.1% (v/v) vitamin supplement, 6 g/L NaNO3, 0.5 g/L KCI, 0.5 g/L MgSO4, 1.5 g/L KH2PO4, 15 g/L agar, pH 6.5. Quantification of the inhibitory effects of chemicals including CFW (Sigma, F3543) and CR (Sigma, C6277) on vegetative growth is calculated using the following formula: inhibition rate = (the diameter of untreated strain – the diameter of strain treated with chemicals)/(the diameter of untreated strain). Determination of fertility by genetic crosses was conducted as described previously [39], using the oatmeal agar medium (5% oatmeal [w:v] and 2.5% agar).
CFW and DAPI (Sigma, D9542) staining, measurement of extracellular peroxidase and laccase activities using colorimetric substrate 2, 2′-azino-bis-(3-ethylbenzothiazoline-6 sulfonic acid) (ABTS) (Sigma, A1888) were performed, as previously described [34]. Conidiation ability, infection structure morphogenesis and virulence of M. oryzae were determined according to the methods described previously [66]. Rice root inoculation assays were conducted following a previously described protocol [16]. Relative fungal invasive growth within rice tissue was quantified by qPCR with a previously described 2−ΔΔCT method [67]. A rapamycin (Selleckchem, S1039) stock solution was prepared in DMSO (Amresco, 0231) at a concentration of 10 mg/ml. When observing infection-related morphogenesis, the rapamycin stock solution was diluted 10 fold to a final concentration of 1 mg/ml and the diluted solution added to the Guy11 strain spore suspension (1 × 105 spores/ml) at a concentration of 1 μg/ml. In the control experiment spore suspensions was supplemented only with DMSO. All phenotypic analyses were performed using at least 2 biological replications. Images of conidial germination and appressorium development were recorded using an Olympus IX81 motorized inverted microscope (Olympus Corporation, Japan) or an Axio imager A2 microscope (Carl Zeiss, Germany).
Sequence analyses of MoSnt2
Snt2 sequences from different fungi were obtained by searching against the reference proteins database in GenBank (www.ncbi.nlm.nih.gov), and their similarities determined using the BlastP algorithm. ClustalOmega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and BoxShade (http://www.ch.embnet.org/software/BOX_form.html) programs were used for amino acid sequence alignment. Phylogenetic tree was generated using the Mega 6.05 program with 1000 bootstrap replications [68]. Domains of Snt2 orthologs were identified using the SMART software (http://smart.embl-heidelberg.de/).
Generation of MoSNT2 gene deletion and complementation mutants
A PCR-based, split-marker deletion method was used for targeted gene deletion [9]. A MoSNT2 partial coding region with a length of 2.5 kbp was targeted for replacement by selection marker gene HPH conferring resistance to hygromycin B (Roche Applied Science, 10843555001). HY and YG, 2 overlapping fragments derived from HPH, were respectively fused by PCR with the left flanking (LF) and right flanking (RF) fragment of MoSNT2 targeted replacement sequences. The 2 fusion amplicons, LF-HY and YG-RF, were cotransformed into protoplasts of the Guy11 strain. Selection and confirmation of targeted gene deletion mutants were performed, as previously described [9].
The plasmid pNEB-MoSnt2-GFP-BAR was constructed with PCR-directed recombination in S. cerevisiae [66], for expression of the MoSnt2-GFP fusion protein and complementation of the ΔMosnt2 deletion mutant. In brief, 4 overlapping fragments of MoSNT2, a fragment containing the GFP-coding sequences and terminator, a fragment of bialaphos resistance selection marker gene BAR, were respectively amplified with the primer pairs MoSNT2-For1 and MoSNT2-Rev1, MoSNT2-For2 and MoSNT2-Rev2, MoSNT2-For3 and MoSNT2-Rev3, MoSNT2-For4 and MoSNT2-Rev4, GFP-MoSNT2-For and GFP-MoSNT2-Rev, as well as BAR-For and BAR-Rev. PCR amplicons were gel-purified and cotransformed into the relevant yeast strain together with the yeast plasmid pNEB-Nat-Yeast which had been linearized with HindIII and SacI. The desired yeast transformants harboring the pNEB-MoSnt2-GFP-BAR plasmid were selected on yeast synthetic drop-out medium lacking uracil. Plasmids were extracted from yeast cells, reintroduced into Escherichia coli strain XL1-Blue competent cells for propagation followed by plasmid extraction. pNEB-MoSnt2-GFP-BAR was transformed into a ΔMosnt2 mutant and transformants were screened for their resistance to herbicide glufosinate-ammonium (Sigma, 45520).
Generation of MoTOR gene silencing mutants
Approximately 500 bp cDNA sequences encoding FRB or FAT domains of MoTor were chosen as 2 independent RNAi target sites, respectively. A fungal RNAi plasmid pSilent-1 [69], capable of transcribing single self-complementary hairpin RNA, was used to generate the MoTOR RNAi plasmids. The promoter of the MoICL1 (isocitrate lyase) gene, which is induced in expression by exposure to acetate as sole carbon source, and repressed in glucose-rich medium [70], was used to drive the transcription of self-complementary hairpin RNA. The pSilent1 vector was first modified by replacing the constitutive TrpC promoter with the promoter of MoICL1 in order to achieve regulation by RNAi. The MoICL1 promoter was amplified with the primer pair MoICL1P-SpeI-For and MoICL1P-SnaBI-Rev, then cloned into the SpeI and SnaBI sites within the pSilent-1 vector. To construct the RNAi plasmid pSilent1-TorFRB, 2 FRB cDNA sequences that are reverse-complementary to each other were respectively amplified with the primer pairs TorFRB50.1 and TorFRB30.1, as well as TorFRB50.2 and TorFRB30.2, and then cloned step by step into the SnaBI and HindIII, ApaI and BglII sites of the modified pSilent1. The RNAi plasmid pSilent1-TorFAT was generated with a similar approach, with the exception that the 2 FAT sequences were respectively amplified with the primer pairs TorFAT50.1 and TorFAT30.1, as well as TorFAT50.2 and TorFAT30.2. pSilent1-TorFRB and pSilent1-TorFAT were individually transformed into the Guy11 strain.
Generation of GFP-MoAtg8 fusion protein expressing transformant
To monitor the autophagic process in M. oryzae, a GFP-MoATG8 fusion gene expression plasmid pNEB-GFP-MoAtg8-BAR was constructed using PCR-directed recombination in S. cerevisiae as described above. The native promoter of the MoATG8 gene, GFP-coding fragment, a fragment containing the MoATG8 coding region and terminator, and the selection marker gene BAR, were respectively amplified with the primer pairs MoATG8Pro-SmaI-For and MoATG8Pro-AgeI-Rev, GFP-MoATG8-For and GFP-MoATG8-Rev, MoATG8-XhoI-For and MoATG8ter-BglII-Rev, as well as BAR-For and BAR-Rev. The pNEB-GFP-MoAtg8-BAR plasmid was transformed into the wild-type strain Guy11 and a ΔMosnt2 mutant.
Transcriptional profile analyses by RNA-seq
Total RNA was extracted from wild-type Guy11 strain and a ΔMosnt2 mutant grown on CM agar medium for 10 days. Two biological replicate samples were prepared for each strain. RNA concentration was measured using Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, Q32855). RNA-seq libraries were prepared using 3 μg of total RNA with NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, E7530S). Clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina Inc., PE-401–3001). After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform (Illumina Inc., USA) and 150 bp paired-end reads were generated. Raw reads of fastq format were filtered with in-house perl scripts to obtain clean data. Paired-end clean reads were aligned to the reference genome of the M. oryzae strain 70–15 [71] using TopHat v2.0.12 [72]. HTSeq v0.6.1 was used to count the reads numbers mapped to each gene [73]. Differential expression analysis was performed using the DESeq R package [74]. Genes with an adjusted P-value < 0.05 found by DESeq were assigned as differentially expressed.
BiFC assays
BiFC analysis of protein-protein interactions in vivo of M. oryzae was performed, as previously described [39]. A C-terminal fragment spanning approximately amino acids 156 to 239 of YFP (YFPCTF) was fused at the carboxyl terminus of MoSnt2 to form a plasmid pNEB-MoSnt2-YFPCTF-BAR. To construct this plasmid, the native MoSNT2 promoter in the vector pNEB-MoSnt2-GFP-BAR was firstly replaced by the constitutive promoter of RP27 gene. The RP27 gene promoter was amplified with the primer pair RP27pro-MoSNT2-For and RP27pro-MoSNT2-Rev, then used to replace the fragment between SpeI and EcoRI sites in pNEB-MoSnt2-GFP-BAR, yielding the pNEB-RP27pro-MoSnt2-GFP-BAR vector. The pNEB-RP27pro-MoSnt2-GFP-BAR was depleted of the GFP-TrpCter cassette by digestion with XmaI, then ligated with the YFPCTF fragment amplified with the primer pair MoSNT2-YCxmaI-For and MoSNT2-YCxmaI-Rev, resulting in the final pNEB-MoSnt2-YFPCTF-BAR plasmid.
An N-terminal fragment spanning approximately amino acids 1 to 155 of YFP (YFPNTF) was fused at the N terminus of H3 to form a pEASY-simple-YFPNTF-H3-HYG plasmid. The selection marker gene HPH was amplified with the primer pair TrpC-Pro-For and HygORF-Rev, and cloned into the pEASY-simple vector (TransGen, CB111) to produce a vector pEASY-simple-HYG. Meanwhile, the RP27 gene promoter was amplified with the primer pair RP27-SpeXho-For and RP27-XmaKpnSac-Rev, and used to replace the fragment between SpeI and XmaI sites in the pNEB-RP27pro-MoSnt2-GFP-BAR plasmid, yielding a pNEB-RP27pro-TrpCter-BAR vector. An RP27pro-TrpCter cassette was amplified with the primer pair RP27-SpeXho-For and TrpCter-XhoI-Rev from plasmid pNEB-RP27pro-TrpCter-BAR, digested with XhoI and then cloned into the SalI site of pEASY-simple-HYG to form a pEASY-simple-RP27pro-TrpCter-HYG plasmid. The primer pairs YFPNTF-SacI-For and YFPNTF-nostop-Rev, as well as H3-For and H3-BamHI-Rev were respectively used to amplify 2 cDNA fragments YFPNTF and H3, which were joined together by fusion PCR with primers YFPNTF-SacI-For and H3-BamHI-Rev. The joint fragment YFPNTF-H3 was finally ligated into the SacI and BamHI sites in pEASY-simple-RP27pro-TrpCter-HYG to form the final pEASY-simple-YFPNTF-H3-HYG vector.
YFPNTF was fused at the C terminus of MoHos2 to form a pEASY-simple-MoHos2-YFPNTF-HYG plasmid. The cDNA sequences of MoHOS2 and YFPNTF were respectively amplified with the primer pairs MoHOS2-01633-For1 and MoHOS2-01633-Rev1, as well as MoHOS2-YFPNTF-For and MoHOS2-YFPNTF-Rev, and then cloned into the SacI and BamHI sites of pEASY-simple-RP27pro-TrpCter-HYG using the ClonExpress II One Step Cloning Kit (Vazyme, C112). For BiFC analysis, the pNEB-MoSnt2-YFPCTF-BAR plasmid was transformed into the Guy11 strain in combination with pEASY-simple-YFPNTF-H3-HYG or pEASY-simple-MoHos2-YFPNTF-HYG. Transformants were screened on CM containing hygromycin and glufosinate-ammonium, and at least 3 independent transformants were examined by epifluorescence microscopy on Axio imager A2 microscope (Carl Zeiss, Germany).
GST-affinity isolation and immunoblot analyses
For expression and purification of fusion proteins GST-PHD1, GST-PHD2, GST-ELM2(MoSnt2), GST-MoSnt2-F1 and GST-MoSnt2-F2, cDNA sequences of MoSNT2 were respectively amplified with the primer pairs PHD1-For and PHD1-Rev, PHD2-For and PHD2-Rev, ELM2(MoSnt2)-EcoRI-For and ELM2(MoSnt2)-NotI-Rev, MoSNT2ex1-For and MoSNT2ex1-Rev, as well as MoSNT2ex2-For and MoSNT2ex2-Rev. PCR amplicon was cloned into the EcoRI and NotI sites of vector pGEX6p-1 (Amersham Biosciences, 28-9546-48) using the ClonExpress II One Step Cloning Kit. The ELM2 domain of MoSnt2 containing the W661A [ELM2(MoSnt2W661A)] point mutation was produced by site-directed mutagenesis. Two overlapping fragments, which were respectively amplified by the primer pairs ELM2(MoSnt2)-EcoRI-For and ELM2(MoSnt2W661A)-Rev, as well as ELM2(MoSnt2W661A)-For and ELM2(MoSnt2)-NotI-Rev, were fused by PCR. The fusion fragment was cloned into the EcoRI and NotI sites of pGEX6P-1 to form a vector expressing GST-ELM2(MoSnt2W661A).
Expression and affinity-purification of GST fusion proteins were conducted as previously described using Glutathione Sepharose 4B (GE Healthcare, 17075601) [54]. The bicinchoninic acid method was employed for determining concentration of purified proteins using a BCA Protein Assay Kit (Solarbio, PC0020). GST-affinity isolation assay was performed following the method as described previously [75]. The affinity isolation mixtures were subjected to immunoblot analysis with anti-GST (Abcam, ab19256), anti-H3 (Active Motif, 39163), anti-H3K18ac (Abcam, ab1191) or anti-H3K4me3 (Active Motif, 39160) antibodies. Proteins reacting with the primary antibodies were visualized by appropriate peroxidase HRP-conjugated secondary antibodies and ECL substrate detection reagents (Bio-Rad, 1705060).
In the GFP-MoAtg8 proteolysis assay, mycelium expressing GFP-MoAtg8 was ground into powder with liquid nitrogen. Protein extraction and immunoblot analysis were conducted as described previously [25] with an anti-GFP antibody (Thermo Scientific, Pierce, MA1-952) and an internal control anti-GAPDH antibody (Sangon Biotech, D110016). For immunoblot analysis of histone, protein extracts were probed with primary antibodies including anti-H3, anti-H4 (Active Motif, 39270), anti-H3ac pan-acetyl (Active Motif, 61638), anti-H4ac pan-acetyl (Active Motif, 39926), anti-H3K18ac and anti-H3K4me3, respectively.
ChIP-qPCR assays
Chromatin of the Guy11 strain and purified GST fusion protein were used for in vitro ChIP assays adapted from a previous report [24]. GST fusion proteins GST-MoSnt2-F1 or GST-MoSnt2-F2 were affinity purified as mentioned above. Mycelium grown in liquid CM for 2 days was collected for cross-linking by formaldehyde as previously described [24]. After grinding the cross-linked mycelium into powder with liquid nitrogen, the cross-linked DNA was extracted using the CelLytic™ PN Plant Nuclear Isolation Kit (Sigma, CELLYTPN1) and sheared into 200- to 1000-bp fragments by ultrasonication. The sheared nuclear lysates were precleared with Sepharose beads (Sigma, 4B200) at 4°C for 4 h, and then equal amounts of lysates used to respectively incubate with GST-MoSnt2-F1, GST-MoSnt2-F2 or GST overnight at 4°C in the presence of Glutathione Sepharose 4B beads. An aliquot of twenty percent of DNA was used as input chromatin. After washing the beads 4 times, affinity isolation products were eluted with 200 μl elution buffer (1% SDS, 0.1 M NaHCO3, 0.25 mg/ml proteinase K [Invitrogen, 25530049], 1 mM DTT). For reverse cross-linking, the eluate was supplemented with NaCl at a final concentration of 0.2 M and incubated at 65°C overnight. After treatment with RNase A (Roche Applied Science, 10109169001), DNA fragments were extracted using the phenol-chloroform methods. qPCR quantification on DNA fragments of autophagy genes MoATG6, 15, 16, 22 was performed with the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, 1855196) in triplicate using specific primer pairs listed in Table S2. Fold enrichment of DNA was determined by calculating the values of affinity isolation DNA relative to the levels of DNA in the input controls, then comparing the signal-to-input ratio derived from GST-MoSnt2-F1 or GST-MoSnt2-F2 against that of GST control.
Histone deacetylase activity assays
The GST-ELM2(MoSnt2) and GST-ELM2(MoSnt2W661A) fusion proteins, as well as GST were used to affinity isolate nuclear proteins in extracts of the Guy11 strain. Preparation of M. oryzae nuclear protein extracts was performed as previously described [23]. Nuclear extracts were incubated with 20 μg of GST-ELM2(MoSnt2), GST-ELM2(MoSnt2W661A) or GST proteins plus 50 μl of glutathione beads. After incubation at 4°C for 2 h with gentle mixing, the beads were collected by centrifugation and washed for a total of 6 times. The affinity isolation products were finally examined for HDAC activities using a HDAC fluorometric activity assay kit (Cayman Chemical, 10011563). Human HDAC provided in the kit was used as a positive control when testing HDAC activities.
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: MoSNT2, MGG_04421; MoTOR, MGG_15156.
Statistical analyses
In immunoblot analysis, the intensity of the protein band was determined by the Bio-Rad Image Lab 3.0 software and a band of the Guy11 strain was defined as a reference with intensity of 1.0. For analyzing statistical significance of difference between the wild-type Guy11 strain and ΔMosnt2 mutant, the probability value (P-value) was calculated by the two-sample Student’s t-test using the Microsoft Excel version 15.0. The Pearson correlation coefficient on gene expression levels of MoSNT2 and MoTOR was calculated by the correlation test using the Microsoft Excel version 15.0.
Funding Statement
This work was supported by the National Natural Science Foundation of China (NSFC) grant 31301626, International Cooperation and Exchange Program of Sichuan Science and Technology Department 2014HH0066, Science foundation of Education Department of Sichuan Province 17ZA0313 to M. He, grants from the Transgenic Projects from the Chinese Ministry of Agriculture 2014ZX0800903B, and National Key Research and Development Plan of China 2016YFD0100601 to X. W. Chen, the NSFC grant 31501627 to W. T. Li, and the Transgenic Projects from the Chinese Ministry of Agriculture grant 2016ZX08001002 to B. T. Ma.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mizushima N, Komatsu M.. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. [DOI] [PubMed] [Google Scholar]
- [3].Talbot NJ, Kershaw MJ. The emerging role of autophagy in plant pathogen attack and host defence. Curr Opin Plant Biol. 2009;12:444–450. [DOI] [PubMed] [Google Scholar]
- [4].Liu XH, Gao HM, Xu F, et al. Autophagy vitalizes the pathogenicity of pathogenic fungi. Autophagy. 2012;8:1415–1425. [DOI] [PubMed] [Google Scholar]
- [5].Veneault-Fourrey C, Barooah M, Egan M, et al. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–583. [DOI] [PubMed] [Google Scholar]
- [6].Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7:185–195. [DOI] [PubMed] [Google Scholar]
- [7].Callaway E. Devastating wheat fungus appears in Asia for first time. Nature. 2016;532:421–422. [DOI] [PubMed] [Google Scholar]
- [8].De Jong JC, McCormack BJ, Smirnoff N, et al. Glycerol generates turgor in rice blast. Nature. 1997;389:244. [Google Scholar]
- [9].Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci USA. 2009;106:15967–15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Deng YZ, Ramos-Pamplona M, Naqvi NI. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy. 2009;5:33–43. [DOI] [PubMed] [Google Scholar]
- [11].Deng YZ, Naqvi NI. A vacuolar glucoamylase, Sga1, participates in glycogen autophagy for proper asexual differentiation in Magnaporthe oryzae. Autophagy. 2010;6:455–461. [DOI] [PubMed] [Google Scholar]
- [12].González A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017;36:397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. [DOI] [PubMed] [Google Scholar]
- [14].Yu F, Imamura Y, Ueno M, et al. The yeast chromatin remodeler Rsc1-RSC complex is required for transcriptional activation of autophagy-related genes and inhibition of the TORC1 pathway in response to nitrogen starvation. Biochem Biophys Res Commun. 2015;464:1248–1253. [DOI] [PubMed] [Google Scholar]
- [15].Fullgrabe J, Lynch-Day MA, Heldring N, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Franceschetti M, Bueno E, Wilson RA, et al. Fungal virulence and development is regulated by alternative pre-mRNA 3′end processing in Magnaporthe oryzae. PLoS Pathog. 2011;7:e1002441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Marroquin-Guzman M, Wilson RA. GATA-dependent glutaminolysis drives appressorium formation in Magnaporthe oryzae by suppressing TOR inhibition of cAMP/PKA signaling. PLoS Pathog. 2015;11:e1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fernandez J, Marroquin-Guzman M, Wilson RA. Evidence for a transketolase-mediated metabolic checkpoint governing biotrophic growth in rice cells by the blast fungus Magnaporthe oryzae. PLoS Pathog. 2014;10:e1004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marroquin-Guzman M, Sun G, Wilson RA. Glucose-ABL1-TOR signaling modulates cell cycle tuning to control terminal appressorial cell differentiation. PLoS Genet. 2017;13:e1006557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lopes Da Rosa J, Boyartchuk VL, Zhu LJ, et al. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc Natl Acad Sci USA. 2010;107:1594–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Baidyaroy D, Brosch G, Ahn JH, et al. A gene related to yeast HOS2 histone deacetylase affects extracellular depolymerase expression and virulence in a plant pathogenic fungus. Plant Cell. 2001;13:1609–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Elias-Villalobos A, Fernandez-Alvarez A, Moreno-Sanchez I, et al. The Hos2 histone deacetylase controls Ustilago maydis virulence through direct regulation of mating-type genes. PLoS Pathog. 2015;11:e1005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ding SL, Liu W, Iliuk A, et al. The tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell. 2010;22:2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fernandez J, Marroquin-Guzman M, Nandakumar R, et al. Plant defence suppression is mediated by a fungal sirtuin during rice infection by Magnaporthe oryzae. Mol Microbiol. 2014;94:70–88. [DOI] [PubMed] [Google Scholar]
- [25].Pham KT, Inoue Y, Vu BV, et al. MoSET1 (histone H3K4 methyltransferase in Magnaporthe oryzae) regulates global gene expression during infection-related morphogenesis. PLoS Genet. 2015;11:e1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang S, Liang M, In N, et al. Phototrophy and starvation-based induction of autophagy upon removal of Gcn5-catalyzed acetylation of Atg7 in Magnaporthe oryzae. Autophagy. 2017;13:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. [DOI] [PubMed] [Google Scholar]
- [28].Yang Y, Zhang Z, Li Y, et al. Identifying cooperative transcription factors by combining ChIP-chip data and knockout data. Cell Res. 2010;20:1276–1278. [DOI] [PubMed] [Google Scholar]
- [29].Singh RK, Gonzalez M, Kabbaj M-HM, et al. Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS One. 2012;7:e36295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Denisov Y, Freeman S, Yarden O. Inactivation of Snt2, a BAH/PHD-containing transcription factor, impairs pathogenicity and increases autophagosome abundance in Fusarium oxysporum. Mol Plant Pathol. 2011;12:449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Baker LA, Ueberheide BM, Dewell S, et al. The yeast Snt2 protein coordinates the transcriptional response to hydrogen peroxide-mediated oxidative stress. Mol Cell Biol. 2013;33:3735–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Denisov Y, Yarden O, Freeman S. The transcription factor SNT2 is involved in fungal respiration and reactive oxidative stress in Fusarium oxysporum and Neurospora crassa. Physiol Mol Plant Pathol. 2011;76:137–143. [Google Scholar]
- [33].Liu XH, Chen SM, Gao HM, et al. The small GTPase MoYpt7 is required for membrane fusion in autophagy and pathogenicity of Magnaporthe oryzae. Environ Microbiol. 2015;17:4495–4510. [DOI] [PubMed] [Google Scholar]
- [34].Guo M, Chen Y, Du Y, et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011;7:e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kong LA, Li GT, Liu Y, et al. Differences between appressoria formed by germ tubes and appressorium-like structures developed by hyphal tips in Magnaporthe oryzae. Fungal Genet Biol. 2013;56:33–41. [DOI] [PubMed] [Google Scholar]
- [36].Egan MJ, Wang ZY, Jones MA, et al. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc Natl Acad Sci USA. 2007;104:11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wood PJ, Fulcher RG. Dye interactions. A basis for specific detection and histochemistry of polysaccharides. J Histochem Cytochem. 1983;31:823–826. [DOI] [PubMed] [Google Scholar]
- [38].Chi MH, Park SY, Kim S, et al. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 2009;5:e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liu TB, Liu XH, Lu JP, et al. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy. 2010;6:74–85. [DOI] [PubMed] [Google Scholar]
- [40].Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36:364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ding Z, Gillespie LL, Paterno GD. Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol Cell Biol. 2003;23:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Soulard A, Cohen A, Hall MN. TOR signaling in invertebrates. Curr Opin Cell Biol. 2009;21:825–836. [DOI] [PubMed] [Google Scholar]
- [44].Penn TJ, Wood ME, Soanes DM, et al. Protein kinase C is essential for viability of the rice blast fungus Magnaporthe oryzae. Mol Microbiol. 2015;98:403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zheng W, Zhou J, He Y, et al. Retromer is essential for autophagy-dependent plant infection by the rice blast fungus. PLoS Genet. 2015;11:e1005704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xu JR, Staiger CJ, Hamer JE. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci USA. 1998;95:12713–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu XH, Ning GA, Huang LY, et al. Calpains are involved in asexual and sexual development, cell wall integrity and pathogenicity of the rice blast fungus. Sci Rep. 2016;6:31204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Qi Z, Wang QI, Dou X, et al. MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae. Mol Plant Pathol. 2012;13:677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dong B, Liu XH, Lu JP, et al. MgAtg9 trafficking in Magnaporthe oryzae. Autophagy. 2009;5:946–953. [DOI] [PubMed] [Google Scholar]
- [53].Saunders DG, Aves SJ, Talbot NJ. Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell. 2010;22:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wei FZ, Cao Z, Wang X, et al. Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy. 2015;11:2309–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fullgrabe J, Heldring N, Hermanson O, et al. Cracking the survival code: autophagy-related histone modifications. Autophagy. 2014;10:556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Brosch G, Loidl P, Graessle S. Histone modifications and chromatin dynamics: a focus on filamentous fungi. FEMS Microbiol Rev. 2008;32:409–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dubey A, Jeon J. Epigenetic regulation of development and pathogenesis in fungal plant pathogens. Mol Plant Pathol. 2017;18:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Barrios ÁP, Gómez AV, Sáez JE, et al. Differential properties of transcriptional complexes formed by the CoREST family. Mol Cell Biol. 2014;34:2760–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Banreti A, Sass M, Graba Y. The emerging role of acetylation in the regulation of autophagy. Autophagy. 2013;9:819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bartholomew CR, Suzuki T, Du Z, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci USA. 2012;109:11206–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yi C, Ma M, Ran L, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. [DOI] [PubMed] [Google Scholar]
- [62].Truee O, Matthias P. Interplay between histone deacetylases and autophagy - from cancer therapy to neurodegeneration. Immunol Cell Biol. 2012;90:78–84. [DOI] [PubMed] [Google Scholar]
- [63].Zhang J, Ng S, Wang J, et al. Histone deacetylase inhibitors induce autophagy through FOXO1-dependent pathways. Autophagy. 2015;11:629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- [65].Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].He M, Kershaw MJ, Soanes DM, et al. Infection-associated nuclear degeneration in the rice blast fungus Magnaporthe oryzae requires non-selective macro-autophagy. PLoS One. 2012;7:e33270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Park CH, Chen S, Shirsekar G, et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell. 2012;24:4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nakayashiki H, Hanada S, Quoc NB, et al. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol. 2005;42:275–283. [DOI] [PubMed] [Google Scholar]
- [70].Wang ZY, Thornton CR, Kershaw MJ, et al. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol Microbiol. 2003;47:1601–1612. [DOI] [PubMed] [Google Scholar]
- [71].Dean RA, Talbot NJ, Ebbole DJ, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. [DOI] [PubMed] [Google Scholar]
- [72].Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Huyen Y, Zgheib O, Ditullio RA Jr., et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


