Abstract
Earlier we created a chemical hazard database via natural language processing of dossiers submitted to the European Chemical Agency with approximately 10 000 chemicals. We identified repeat OECD guideline tests to establish reproducibility of acute oral and dermal toxicity, eye and skin irritation, mutagenicity and skin sensitization. Based on 350–700+ chemicals each, the probability that an OECD guideline animal test would output the same result in a repeat test was 78%–96% (sensitivity 50%–87%). An expanded database with more than 866 000 chemical properties/hazards was used as training data and to model health hazards and chemical properties. The constructed models automate and extend the read-across method of chemical classification. The novel models called RASARs (read-across structure activity relationship) use binary fingerprints and Jaccard distance to define chemical similarity. A large chemical similarity adjacency matrix is constructed from this similarity metric and is used to derive feature vectors for supervised learning. We show results on 9 health hazards from 2 kinds of RASARs—“Simple” and “Data Fusion”. The “Simple” RASAR seeks to duplicate the traditional read-across method, predicting hazard from chemical analogs with known hazard data. The “Data Fusion” RASAR extends this concept by creating large feature vectors from all available property data rather than only the modeled hazard. Simple RASAR models tested in cross-validation achieve 70%–80% balanced accuracies with constraints on tested compounds. Cross validation of data fusion RASARs show balanced accuracies in the 80%–95% range across 9 health hazards with no constraints on tested compounds.
Chemical structure determines the biological properties of substances, though the connection is typically too complex to derive rules for larger parts of the chemical universe, whether by computational means or human understanding (Hartung and Hoffmann, 2009; Patlewicz and Fitzpatrick, 2016). Practical use of structure activity relationships has therefore been largely limited to so-called read-across, ie, the pragmatic comparison to 1 or few similar chemicals, with case-by-case reasoning about the validity of the approach (Patlewicz et al., 2014). This subjective expert-driven approach cannot be quickly applied to large numbers of chemicals. Read-across dependence on human opinion makes evaluation of the technique difficult and prevents reliable estimates of method reproducibility.
Read-across approaches have become the predominant nonanimal-testing source of data (https://echa.europa.eu/documents/10162/13639/alternatives_test_animals_2017_en.pdf/075c690d-054c-693a-c921-f8cd8acbe9c3; last accessed June 30, 2018) in the European REACH (Registration, Evaluation, Authorization and Restriction of Chemicals) registration process (Aulmann and Pechacek, 2014; Hartung, 2010; Williams et al., 2009), the largest investment into consumer safety ever, requesting data equivalent to multibillion Euro of animal testing for tens of thousands of chemicals (Hartung and Rovida, 2009; Rovida and Hartung, 2009). Increasing experience with read-across (Patlewicz et al., 2013) allowed the development of the first read-across assessment framework by the European Chemical’s Agency (https://echa.europa.eu/documents/10162/13628/raaf_en.pdf; last accessed June 30, 2018) and the development of Good Read-Across Practices (Ball et al., 2016), but the utility of the approach is limited by data access and the unknown uncertainty of these predictions.
A large machine-readable database of toxicological information makes automation of read-across approaches viable by allowing computational modeling of chemicals and chemical analogs. Machine readable databases of chemical testing also allow assessment of the quality of testing data by analysis of repeatedly tested substances (Hartung, 2016a). Knowledge of testing data reliability also provides a baseline against which to compare computational models.
A major advance of the European chemicals legislation is, that key information of registrations on tens of thousands of chemicals are made public, though not in a standardized manner to allow computation. Previously, we used natural language pattern matching to make the public information from the REACH registration process machine-readable (Luechtefeld et al., 2016a). Interestingly, many chemicals have been tested more than once, some shockingly often: For example, one of the often-challenged animal tests is the Draize rabbit eye test, where for more than 70 years now, test chemicals are administered into rabbit eyes. Two chemicals were tested more than 90 times, 69 chemicals were tested more than 45 times, shows the database (Luechtefeld et al., 2016c). This excessive and redundant animal testing facilitated an assessment of the reproducibility of OECD guideline tests, based on hundreds of chemicals for each endpoint, presented here in a comprehensive manner for the first time. Notably, the 9 most frequently done animal tests analyzed here consumed 57% of all animals for toxicological safety testing in Europe 2011 (http://ec.europa.eu/environment/chemicals/lab_animals/reports_en.htm; last accessed June 30, 2018).
Chemical skin sensitization extracted from ECHA dossiers enables assessment of chemical similarity-based hazard models. Using the simple 1-nearest neighbor approach, where a chemical is classified by its closest analog (defined by a binary vector—PubChem 2D and Jaccard distance) balanced accuracies of 80%–92% have been previously demonstrated (Luechtefeld et al., 2016d). These accuracies are made possible by requiring a minimum threshold of similarity. This threshold states that a chemical can be modeled when an analog of similarity greater than or equal to the threshold is present in the available data. Low minimum similarity thresholds allow simple similarity methods to model more chemicals at the cost of lower accuracy. This paper demonstrates that increasing similarity (PubChem 2D + Jaccard method) leads to increased model accuracy and supports the use of chemical similarity methods in chemical modeling.
Success in modeling chemical hazards in earlier work prompted 2 developments to support a Toxicology for the 21st century (Hartung, 2009). First, the demonstration of the predictive power of big data led us to focus on generating larger databases. Our data integration pipeline pulls chemical property data from PubChem, ECHA, and an NTP-curated acute oral toxicity dataset. The combined data continues to grow but at present contains 833 844 chemical property values used for modeling across 80, 908 chemicals for an average of approximately 10 properties per chemical. Second, a simple method of automating read-across was created to model chemical properties. Tentatively named read-across structure activity relationships—RASAR was created and presented here for the first time. RASAR combines conventional chemical similarity with supervised learning. Chemical similarity is done by generating a binary fingerprint for each chemical and using Jaccard distance (similarity = 1–distance) on fingerprints. Supervised learning methods then provide a statistical model of the insights deliverable from chemical similarity. Due to automation, the approach can be applied to large datasets and thus validated according to common statistical methods such as cross-validation. Supervised learning models built on chemical similarity also allow assignment of confidence to individual predictions. Similar approaches using the large datasets from robotized testing within ToxCast have been recently reported (Shah et al., 2016; Zhu et al., 2016).
We demonstrate a “simple” RASAR which trains a logistic regression model to predict chemical hazards from the similarity to the closest chemical tested negative (maxNeg) and similarity to the closest chemical which has tested positive (maxPos). The approach has been applied to 9 of the most frequently used hazard determinations in REACH and toxicology in general (Skin Sensitization, Eye Irritation, Acute Oral Toxicity, Acute Dermal Toxicity, Acute Inhalation Toxicity, Dermal Irritation, Acute/Chronic Aquatic Toxicity and Mutagenicity). “Simple” RASARs obtain cross-validated sensitivities above 80% with specificities of 50%–70%. This is on par with the reproducibility of the respective animal tests.
A further improvement to the simple RASAR trains random forest models from diverse chemical information on analogs. A broad variety of 19 categories of GHS classifications (74 in total) of similar chemicals was considered to inform each endpoint. This “Data Fusion” approach boosts cross-validated balanced accuracies into the 80%–95% range.
The models presented here are part of the Underwriters Laboratories Cheminformatics Suite.
MATERIALS AND METHODS
Pairwise Evaluation of OECD Guideline Test Reproducibility
The generation of the machine-readable REACH registration database has been described earlier (Luechtefeld et al., 2016a) using language pattern matching, database and web manipulation packages (mongodb, htmlunit and beautifulsoup). It contains data for 9801 unique substances, 3609 unique study descriptions and 816 048 study documents.
To evaluate guideline study repeatability, results must be assigned to each study. Every OECD guideline dossier reports a “submitters_conclusions” field, from which a text result was mapped to a controlled term for each related test. To evaluate repeatability each test result was mapped to either “positive” or “negative” and potencies were ignored.
The reproducibility evaluations used all chemicals with multiple results for each of the listed guidelines. A pairwise approach is used, ie, all chemicals that have been tested multiple times for a given hazard are collected. The set of outcomes for a given chemical are then mapped to the set of all pairs of outcomes. The conditional probability of one outcome (T2 in below equation) given another (T1 in below equation) is then calculated using the definition of conditional probability.
Sensitivity and specificity are estimated by the conditional probability that a test is positive given that its paired test was positive (sensitivity) or that a test is negative given that its paired test is negative (specificity).
A no information (NOI) rate was calculated by combining random tests with the same OECD guideline. This NOI rate helps distinguish accuracy due to repeatability from accuracy due to high imbalances in prevalence. For example, a guideline test that always results in the same outcome is no more repeatable for a specific chemical than it is for any random pair of tests.
Demonstration of Network Effects for Chemical Similarity
Analog-based coverage of a set of chemicals can be defined as the proportion of chemicals in the set, for which a labeled analog is defined. Large chemical sets can be covered by a relatively small set of analogs when single labeled compounds can behave as analogs for many elements of the set. In practice this “rapid coverage” of a large set by a small set occurs quite often, primarily due to (1) the method of fingerprinting, (2) the similarity metric, and (3) the physical constraints of possible chemical structures.
A graphical illustration (Figure 7) of the coverage of a set of approximately 33, 000 commercial compounds by a relatively small number of chemicals with data was attempted. In this experiment, 1387 chemicals from REACH Annex VI Table 3.1 (a table of hazard classifications[http://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:02006R1907-20171010&from=EN:#page=491; last accessed June 30, 2018]) were treated as a labeled set of compounds and compared with 33 000 commercial substances in the European INventory of Existing Commercial chemical Substances (EINECS) (https://echa.europa.eu/information-on-chemicals/ec-inventory; last accessed June 30, 2018). Annex 3.1 compounds were considered analogs of EINECS compounds when they were ≥70% similar as measured via the Tanimoto index on PubChem 2D fingerprints. Connections are shown between unlabeled EINECS compounds (blue) and highly similar Annex compounds (red). By using only 1387 labeled chemicals, we can cover 33 000 unknowns providing similar neighbors. This demonstrates that a relatively small number of compounds are needed to find an analog for every compound in a much larger (here >20× larger) set.
Figure 7.
Modeling of sufficiently close neighbor availability with increasing number of chemicals with data. Two substance lists of 33 383 substances (European Inventory of Existing Commercial Chemical Substances [EINECS]), representing here chemicals with no data, and 1387 chemicals (Annex VI of the REACH legislation) are used, representing chemicals with labels. Please note that these are used here only as random lists of chemicals with CAS numbers. EINECS compounds are represented in blue and ANNEX VI Table 3.1 compounds are in red. At start, none of the 33 383 has neighbors with data. Choosing randomly an increasing number from the 1387-chemcial list, more and more chemicals find neighbors indicated by the contraction of dots linked by Jaccard similarities. We use a minimum similarity of 70% in these figures. The number of neighbors is symbolized by the size of red dots. Edges represent similarities between EINECS compounds and Annex compounds. These visualizations are made with the aid of Gephi graph visualization software.
Table 1.
Example Data Fusion Features
| SMILES | H225+ | H225− | H225T | … | H410+ | H410− | H410T |
|---|---|---|---|---|---|---|---|
| C(#CCCCCC)CCCCC | 0.35 | 0.63 | 0 | 0.34 | 0.56 | 0.82 | |
| C(#C[P+](C=1C=CC=CC1) | 0.01 | 0.46 | 1 | 0.67 | 0.32 | 0.50 | |
| C1CCC2CCCC2C1 | 0.12 | 0.03 | 1 | 0.80 | 0.38 | 0.53 |
This mock table illustrates how data fusion features are built during the unsupervised step of model training. Each column represents the similarity to the closest positive (eg, H225+) or closest negative (eg. H225-) or binary target values (eg. H225T).
RASAR Database
A database was created combining the REACH database from prior work with PubChem and an NTP-curated acute oral toxicity dataset. The portion of the database used in this publication contains 80 908 chemicals with sometimes missing information on 74 properties resulting in 833 844 chemical labels.
European Chemical Agency ClassificationandLabeling: UN GHS hazards derived from submissions to the REACH chemical regulatory program. ECHA prohibits the re-publication of these data but has made registration data available for download (https://iuclid6.echa.europa.eu/reach-study-results; last accessed June 30, 2018). In agreement with ECHA, the database is available from the authors for collaborative projects.
PubChem: UN GHS hazards derived from HSDB, ECHA C&L, and other sources. Noteworthy, at the time of this writing, PubChem only reports positive hazard results, thus adding a skew towards positive results to this data. Some balancing of the dataset for training was therefore necessary and is described in the implementation section.
NTP—Predictive Models for Acute Oral Systemic Toxicity: A set of acute oral toxicity LD50 values derived from HSDB (https://toxnet.nlm.nih.gov/cgi-bin/sis/htmlgen?HSDB; last accessed June 30, 2018) and other sources.
For the REACH database chemical hazard labels are derived from the ECHA classification and labeling (C&L) data. ECHA C&L are ultimately regulatory calls on chemical classifications. These calls are made on the basis of OECD guideline studies, read across studies, QSAR studies and other information available in chemical dossiers submitted in service of REACH legislation. Labels generated from PubChem are themselves derived from ECHA, CAMEO chemicals, HSDB, and other sources. Labels generate from the NTP curated acute oral toxicity dataset are available and described at https://ntp.niehs.nih.gov/pubhealth/evalatm/test-method-evaluations/acute-systemic-tox/models/index.html, last accessed June 30, 2018.
All chemicals in the database are (1) entered as INCHI Identifiers, (2) mapped to SMILES identifiers, and (3) mapped to PubChem 2D vectors: The approach uses PubChem 2D chemical fingerprints, ie, binary vectors with 881 features describing atom counts, ring counts and other structural descriptors. A description of each feature is available (ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.txt; last accessed June 30, 2018).
A Supplementary Material is available that describes each of the hazard labels used in this work. The labels used include the UN GHS health hazards H300-H399, the UN GHS chemical properties H200-H300, and an NTP label for acute oral toxicity. The labels also include simple functions of the aforementioned labels termed here “dependency labels”. Dependency labels capture well-defined relationships between UN GHS hazards. For example H300-H305 describes different potencies of acute oral toxicity. The dependency label “acute_oral_binary” is true if any H300-H305 is true and answers the question “is this chemical an acute oral hazard or not?”
Currently discordance in a chemical label is handled by selecting the most hazardous value (aka the precautionary principle). The most common known label with 49 609 known values is “eye_irritation_binary”, which is true if H319 (the UN GHS hazard for serious eye damage) or H318 (eye irritation) is true and false when both hazards are false.
Read-Across Structure Activity Relationship
RASARs are constructed in an unsupervised learning step and a supervised learning step.
Unsupervised Step
In the unsupervised learning step distances between all chemicals in the modeled database are built. Currently we do this exhaustively by comparing every chemical to every other chemical (an O[n2] operation), but it can be improved using locality sensitive hashing methods. After building chemical similarities a local graph can be constructed for each modeled compound. This local graph describes distances to each of the chemicals surrounding the compound of interest. Finally, the unsupervised step applies an aggregation function on the local graph to generate a feature vector. K-nearest neighbors (KNNs) can be treated as an aggregation function on local networks. KNN creates an n-dimensional vector with a count of the number of times each of n labels occurs in the k closest neighbors. In this work the Simple RASAR aggregation function generates 2D vectors describing similarity to the closest positive and negative in the local graph (Figure 1).
Figure 1.
Illustration of aggregation functions on the local network of 1-decene. 1-decene is marked as target. Positive and small dots indicates analogs that are positive for a modeled endpoint. Negative indicates analogs that are negative for the modeled endpoint. The table illustrates well known aggregation functions. Data Fusion aggregation function not given.
Supervised Step
The supervised learning step applies a supervised learning model to the vectors generated by the unsupervised learning step. In this work we use logistic regression in the Simple RASAR and a random forest in the Data Fusion RASAR.
Simple RASAR
The simple RASAR combines an unsupervised aggregation function with logistic regression. The unsupervised aggregation function generates a 2D vector for each chemical. The generating function is specific to the modeled hazard H. In an equation the generated vector for a chemical c can be described as:
is the similarity of the target compound to an analog . The function if the chemical, , is positive for the hazard of interest and -1 if the chemical is negative for the hazard of interest. The first element of the generated vector is the similarity to the analog that maximizes from all analogs that are positive for the hazard of interest [ie, ]. The second element is the same but for chemicals such that .
Data Fusion RASAR
The Data Fusion RASAR creates large feature vectors from the local graph of each chemical and uses these vectors to train a random forest. In the unsupervised step, the data fusion RASAR extends the simple RASAR by building similarity-based features for every catalogued chemical and property. The Data Fusion RASAR also records feature data for the target chemical of interest.
For n hazards Data Fusion RASAR’s generating function can be mathematically described as the concatenation of 3 types of vectors. describes similarities to closest analogs that are positive for each catalogued hazard . describes similarities to analogs that are negative for each catalogd hazard. describes the known hazard values for the compound of interest:
In the supervised step, is used to train a random forest. Unlike the Simple RASAR, the data fusion aggregation vector is the same for all hazard models. Thus, the data fusion RASAR consists of the creation of a large aggregation vector for each compound and then the training of a random forest for each hazard of interest. It should be noted that each label used to build these feature vectors is a binary label.
Data fusion allows for strong predictions even in the absence of data for a modeled hazard. For example, a prediction for eye irritation may falter if there are no nearby chemicals with eye irritation data. If reliable skin irritation data is available for similar chemicals, then a prediction that uses skin irritation data will outperform a prediction that does not.
Feature Hiding
An extra step is necessary for data fusion RASARS to prevent fitting trivial models. Some values in the aggregation vector must be hidden during model training. As an example, one can see that contains the hazard value being modeled. This value must be hidden because the trained model would simply become the identity model. Other values must be hidden as well because of some well-defined dependencies between chemical properties/hazards, for instance when modeling binary acute oral hazard, we must hide all target features pertaining to acute oral toxicity.
The final data fusion vector is composed of called target features of , called positive analog features and called negative analog features. We give a more detailed description of each below.
Target features
Target features are the known labels for the chemical in question. There are 74 labels used as target features in the data fusion approach. These include the UN GHS labels, NTP acute oral toxicity labels and dependency labels, which are simple functions of the former. These hazards fall under 19 different categories:
Acute Toxicity—Dermal/Inhalation/Oral
Hazardous to the aquatic environment - acute/chronic
Skin or Respiratory Sensitization/Corrosion/Irritation
Serious Eye Damage or Irritation
Water contact flammable
Substances and Mixtures corrosive to Metals
Self-heating Substances and Mixtures
Reproductive Toxicity
Pyrophoric Solids/Liquids
Oxidizing Solids/Gases
Organic Peroxides
Hazardous to the ozone layer
Germ Cell Mutagenicity
Gases Under Pressure
Flammable Solids/Liquids/Gases
Explosives
Effects on or via Lactation
Carcinogenicity
Aspiration Hazard
A list of all these features is given in the appendix. Although the Simple RASAR uses only distances to the closest analogs, the Data Fusion Rasar uses both additional information about the target as well as distances to all catalogued labels. When target features are missing they are marked as missing in the generated feature vector. The supervised learning algorithms must be able to manage missing features.
Positive/negative analogs
After compiling target features, the Data Fusion RASAR finds all the analogs for a target compound. The distance—defined by Jaccard distance on PubChem 2D descriptors—to the closest positive analog for each of the 74 labels (see appendix) makes up the 74D positive analog vector. The same vector is made for negatives.
All together each chemical is given 74 target features (ie, values for the substance itself where available), 74 positive analog features (Tanimoto similarity to the closest positive analog) and 74 negative analog features (Tanimoto similarity to the closest negative analog) making a 74*3 = 222D feature vector for each compound. Table 1 gives a mock example of data fusion features.
RASAR Implementation Details—Spark Pipeline
The RASAR algorithms are constructed with Apache Spark (https://spark.apache.org/docs/latest/index.html), an open-source cluster-computing framework, originally developed at the University of California, Berkeley’s AMPLab, the Spark codebase was later donated to the Apache Software Foundation. The entire training process takes place on an Amazon EC2 cluster, which is primarily necessary for the construction of the large adjacency matrix in the unsupervised step of both RASAR methods.
The endpoints evaluated here include 9 binary hazards: acute oral binary, acute dermal binary, acute inhalation binary, acute aquatic binary, skin sensitization binary, skin corrosion binary, eye irritation binary and mutagenic binary, all of which we consider “dependency labels”, because they are dependent on simple functions of other labels (ie, acute oral binary is true whenever any UN GHS hazard for acute oral toxicity is true). The other trained models are enumerated in the Supplementary Table 1.
In the unsupervised step, the Spark pipeline builds descriptive feature vectors. This is the most computationally expensive part of the process and is done across a computing cluster using a custom built spark “User Defined Aggregation Function”.
In the final step of the RASAR algorithm, unsupervised features are used to build supervised learning models for 51 of the 74 labels. This means that the RASAR algorithm is the composition of one unsupervised step and 51 supervised steps. The supervised step consists of data sampling and model training.
Data Sampling
Both RASAR algorithms perform oversampling and undersampling. Many of the tracked UN GHS hazards have an imbalanced ratio of positives and negatives. The sampling method oversamples the low-prevalence class up to 5× and undersamples the high-prevalence class down to one-third. This means that a hazard with 100 positives and 1000 negatives will have positives resampled up to 500 and negatives randomly removed down to 500. Balanced datasets are important for many learning algorithms, particularly in the absence of very large datasets. Care is taken to perform resampling within the cross-validation circuit to prevent model evaluation on chemicals used in model training.
Model Training
The Simple RASAR (2 aggregation features) uses a different supervised learning model than the Data Fusion RASAR (222 features) due to differences in the feature vectors. The Simple RASAR uses the spark.ml Logistic Regression model with a maximum of 300 000 iterations of training, a tolerance for convergence of 1E-12 and a regularization parameter of 1E-4. The Data Fusion RASAR uses the spark.ml.RandomForest model with 20 trees and 10 minimum instances per leaf node and otherwise default features.
Model Evaluation
Unsupervised feature generation is done only once (outside of cross-validation) for both RASAR algorithms due to its computational cost. Once the unsupervised features have been generated for all chemicals, the supervised learning algorithms are trained and tested in 5-fold cross-validation to generate the evaluation statistics (sensitivity, specificity, etc.) reported here (Tables 3 and 4).
Table 3.
Simple RASAR Prediction Accuracy in a Leave-One-Out Cross-Validation
| Hazard | Chemicals With Data | Positive (Toxic) | Negative (Nontoxic) | Threshold Negative % | Threshold Positive % | Sensitivity % | Specificity % | Coverage % | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| Skin sensitization | 4783 | 2886 | 1897 | 43 | 50 | 80 | 50 | 85 | 68% |
| Eye damage | 15 760 | 14 794 | 966 | 47 | 55 | 81 | 51 | 88 | 79% |
| Acute oral | 12 157 | 10 225 | 1932 | 40 | 50 | 80 | 64 | 87 | 77% |
| Acute dermal | 6427 | 4430 | 1997 | 40 | 60 | 80 | 69 | 73 | 77% |
| Skin irritation / corrosion | 15 223 | 13 846 | 1377 | 37 | 52 | 80 | 51 | 75 | 77% |
| Mutagenicity | 3395 | 600 | 2795 | 42 | 50 | 80 | 55 | 83 | 59% |
| Chronic aquatic | 2844 | 2582 | 262 | 40 | 54 | 80 | 50 | 80 | 77% |
| Acute aquatic | 2055 | 1129 | 926 | 40 | 50 | 80 | 52 | 82 | 67% |
| Acute inhalation | 6184 | 4812 | 1372 | 41 | 59 | 74 | 75 | 83 | 74% |
The table shows the number of toxic (positive) and nontoxic (negative) chemicals in the expanded database. The chosen negative (negT) and positive (posT) thresholds of probability resulted in the sensitivities, specificities and coverage of chemicals indicated.
Table 4.
Data Fusion RASAR—5-Fold Cross-Validation Results for 9 Hazard Classifications
| Hazard | Chemicals | Sensitivity | Specificity | BAC % | ACC % |
|---|---|---|---|---|---|
| Acute aquatic binary | 10 541 | 95 | 94 | 95 | 95 |
| Acute dermal binary | 11 252 | 89 | 94 | 92 | 90 |
| Acute inhalation binary | 11 369 | 90 | 91 | 91 | 90 |
| Acute oral binary | 32 411 | 94 | 86 | 90 | 93 |
| Chronic aquatic binary | 17 295 | 98 | 66 | 82 | 98 |
| Eye irritation binary | 48 767 | 99 | 70 | 84 | 98 |
| Mutagenic binary | 3703 | 76 | 92 | 84 | 88 |
| Skin corrosion binary | 46 331 | 98 | 75 | 86 | 97 |
| Skin sensitization binary | 7670 | 80 | 96 | 88 | 84 |
Five-fold cross-validation of the model based on all GHS classifications. In 5 iterations, 20% of randomly picked chemicals were predicted by a model trained on the remaining 80% of chemicals. The resulting average accuracies compared with the actual test data are given. Please note, that all chemicals were predicted, ie, the applicability domain is 100%.
Abbreviations: BAC balanced accuracy; ACC, accuracy.
Visualization of Chemical Universe
The most demanding step of model training is the evaluation of chemical similarity pairs. To visualize this process on large datasets a proprietary force layout algorithm was built at ToxTrack LLC. We applied this algorithm to an adjacency matrix built from approximately 10 million compounds in PubChem (50 trillion comparisons).
In this process, a “cross-join” is performed on a Spark computing cluster to compare all chemicals with each other. Currently this is an O(n2) operation (n = number of compounds). Similarities <70% are dropped. A similarity is calculated by the number of PubChem 2D features shared by 2 chemicals divided by the total number of PubChem 2D features in both compounds (Tanimoto or Jaccard similarity).
The RASAR database contains 81 089 chemicals with label data, which alone require 3.3E9 choose-2-comparisons. The millions of structures in PubChem require approximately 1014 comparisons. This was done on a large computing cluster on the Amazon cloud. Specifically, the task of similarity calculation was divided into many parts across 20+ spot instances running Apache Spark with Yarn resource manager, the method for setting up this cluster is described by Berkeley Amplab at https://github.com/amplab/spark-ec2, last accessed June 30, 2018. Once this large adjacency matrix has been calculated, visualization of the chemical space remains a challenge. ToxTrack LLC’s proprietary graphical layout algorithm applies the force layout algorithm on massive datasets and generates the visualizations shown here (Figure 2).
Figure 2.
Force layout graph of 10 million chemicals. A, where each dot represents a chemical and their distance reflects chemical similarity, calculated by the number of PubChem 2D features shared by 2 chemicals divided by the total number of PubChem 2D features in both compounds (Jaccard similarity). B–D, Step-wise zooming in, where the frame indicates the area shown in the next graph, until in D, individual chemicals are seen with their similarity connections as gray lines, whose length represents % similarity.
Variable Importance Analysis
Variable importance can be assessed for data fusion models by using a feature subset evaluation algorithm. In short, each of the 51 supervised learning models (random forests) is retrained and reevaluated with one feature removed. The importance of the removed feature is evaluated by its impact on the resulting model accuracy. A feature whose removal results in a large reduction in accuracy is considered an “important feature”.
Packages
This work involves the use of dozens of software packages. The packages of paramount importance in chemical and data manipulation are:
org.openscience.cdk cdk-bundle: The chemical development kit is used to manipulate chemical structure and build chemical fingerprints.
org.apache.spark spark-core, spark-mllib: Apache spark libraries provide the means for cluster deployment and statistical model building on this cluster.
RESULTS
Overview
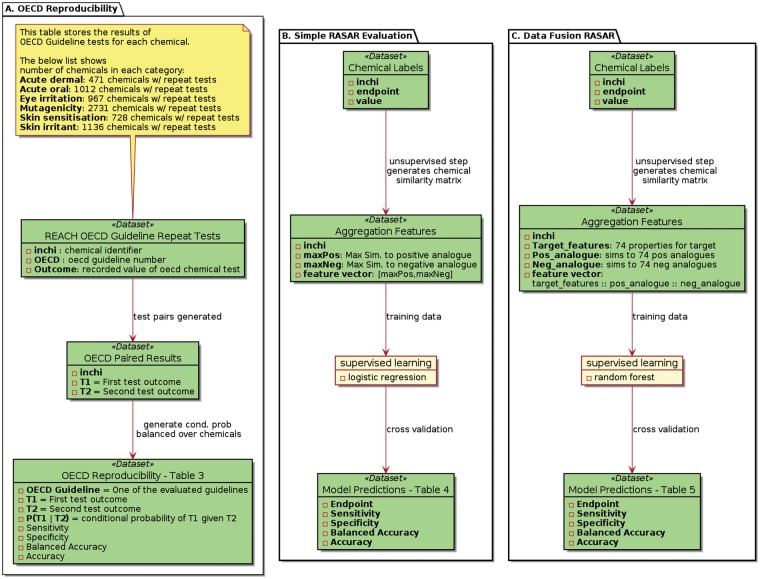
Three main results are presented herein. Due to the number and complexity of the workflow, Figure 3 provides an overview of these 3 results presented in Tables 2–4. The first workflow “A. OECD Reproducibility” is an evaluation of the reproducibility of animal tests performed according to OECD guidelines. This evaluation is done by evaluating how well 1 animal test can predict a repeated animal test. The second workflow “B. Simple RASAR Evaluation” evaluates a simple RASAR. This RASAR is built and evaluated on chemical classification and labeling data detailed in the Database section. It is distinguished from a “Data Fusion RASAR” by the simplicity of the aggregation features generated prior to supervised learning. These features capture the similarity to the closest negative and positive analog for the endpoint of interest. The last workflow “C. Data Fusion Rasar” evaluates a RASAR built from “data fusion” features which include descriptions of (1) off-target properties of the predicted compound, (2) similarities to positive analogs for 74 properties, and (3) similarities to negative analogs for 74 properties.
Figure 3.
Workflow of the presented studies. A, OECD Reproducibility is evaluated via conditional probabilities generated from repeated test pairs found in ECHA dossiers. B, A Simple RASAR built from ECHA C&L data is evaluated in cross-validation. C, A Data Fusion RASAR built from ECHA C&L data is evaluated in cross-validation.
Table 2.
Reproducibility of OECD Animal Test Guideline Studies for Acute and Topical Hazards
| Hazard | OECD TG | Chemicals | Se | Sp | Acc | NOI | BAC |
|---|---|---|---|---|---|---|---|
| Acute oral | 401 | 707 | 87 | 97 | 94 | 72 | 92 |
| Acute dermal | 402 | 384 | 65 | 91 | 88 | 84 | 78 |
| Skin irritation | 404 | 709 | 68 | 83 | 78 | 59 | 75.5 |
| Eye irritation | 405 | 718 | 75 | 92 | 88 | 74 | 83.5 |
| Skin sensitization | 406 | 493 | 70 | 95 | 92 | 87 | 82.5 |
| 429 | 97 | 82 | 89 | 87 | 56 | 85.5 | |
| Mutagenicity | 474 | 207 | 51 | 97 | 94 | 89 | 74 |
| 475 | 154 | 50 | 100 | 99 | 97.5 | 75 |
The database from Luechtefeld et al. (2016a) based on REACH registrations until December 2014 was used to extract multiple guideline studies on the same chemical. Conditional pairwise probabilities were calculated to derive sensitivity, specificity and accuracy of a repeat experiment. Please note that TG 401 has been abandoned, but the newer tiered TG has not relevant numbers of repeated studies. Mutagenicity includes only 8 positive compounds.
Abbreviations: Se, Sensitivity; Sp, Specificity; Acc, Accuracy; NOI, Accuracy on random selected pairs of tests; BAC, balanced accuracy.
Animal OECD Test Reproducibility
The machine-readable REACH database includes many replicate tests (Luechtefeld et al., 2016c). All available testing data must be registered by the consortia called Substance Information Exchange Forum, a mandatory requirement to register jointly (“one substance, one registration” principle of REACH), which leads for the first time to the compilation of essentially all available studies. The extent of repeated tests is often surprising. For example, chemicals tested for acute eye irritation guideline 405 are tested on average 2.91 times. Chemicals tested for acute oral toxicity guideline 401 have on average 3.56 tests. The medians for both guidelines are 1 indicating a skew due to some chemicals with many repeat tests. The average OECD guideline test is repeated 2.21 times. This number is skewed upwards because guidelines with many repeats will have more records (there are 7560 eye irritation TG 405 tests, which have mean repeats = 2.9, and only 108 skin irritant TG 431 tests, which have mean repeats = 1.01). There are 1.57 average test repeats balanced across all OECD guidelines. This high repetition factor is owed to the fact that before the introduction of the OECD mutual acceptance of data, a given substance had to be registered in each country with data generated in this country and that competitors registering the same substance were not aware and/or had no access to data from earlier registrations.
The extracted REACH data allowed a systematic quality assessment of the animal studies to derive a benchmark for the in silico predictions presented here. As reproducibility is substance-dependent (nontoxic and highly toxic substances are more reproducible in their biological effect), modeling of a repeat test was calculated for each chemical, for which multiple OECD guideline tests were available, and then averaged for the study type. Table 2 shows the sensitivities, specificities and respective overall accuracies, based on conditional probabilities if the result is true and asking for the correspondence of the second draw.
For each animal test several hundred retested chemicals were available (361–718). Incidences of low sensitivity (eg, TG 402) indicate that a substance tested positive in the first test, would often test negative in the second. The higher specificity is owed to the fact that overall far more nontoxic than toxic substances were found among the retested ones in the ECHA database. This reflects the general somewhat surprising finding that toxic hazards are less frequent in the database than might be expected, typically below 20% of chemicals for any given health hazard label (Luechtefeld et al., 2016a). This is probably due to the selection bias of nontoxic substances for use in products, ie, the mostly high-production volume chemicals registered in 2010 and 2013. These results would be even worse if the reproducibility of potency classes would be included, but as benchmarks are needed here for a tool for hazard identification, all weak, moderate, strong, etc. effects were combined simply as positive outcomes. Noteworthy, the ECHA database includes potency information (Supplementary Figure 1), which has not yet been exploited for analysis of reproducibility or prediction.
Simple RASAR Models Can Estimate Chemical Hazard
Visualize similarity
To demonstrate that chemicals tend to create rich adjacency matrices due to complex relationships on the possible structural combinations, we built a force layout diagram from approximately 10 million compounds in PubChem (50 trillion similarities). The visualized graph (built with a proprietary graphical package from ToxTrack LLC) shows large numbers of highly similar chemicals and clustering at multiple zoom levels. The figure zooms into a part of the map, which reflects shared functional groups (Jaccard similarity on PubChem 2D vectors) of minimum 70% by connecting lines and the exact similarity by distance of the substances.
Simple rasar feature evaluation
Following the vision toward a read-across-based prediction of hazard and its validation (Hartung, 2016a), an expanded database was curated. The RASAR database contains 80 908 chemicals with hazard labeling data. This database allowed now to establish a probability of hazard based on the proximity (similarity) to toxic substances and similarly a probability of nonhazard based on the proximity to nontoxic ones.
The Simple RASAR generates feature vectors for each chemical by recording similarities to the closest positive analog and closest negative analog. Figure 4 is a local graph for 1-DECENE (“target”) and shows positive analogs (“positive” and small dots) and negative analogs (in negative). The closest analogs (large nodes) are used to build the Simple RASAR feature vector.
Figure 4.

Illustration of the closest positive and negative neighbor approach for 1-DECENE. The graph shows chemicals with similarity > 0.9 according to PubChem 2D Tanimoto. The RASAR uses similarity to the closest Positive (large positive node—1, 7-OCTADIENE) and closest Negative (large negative node—MYRCENE) along with other features to characterize a local similarity space. All small nodes here are positives.
Figure 5 demonstrates the value of the similarity to the closest positive (termed maxPos) and similarity to closest negative (maxNeg). In this work maxPos and maxNeg are cubed, which has the effect of emphasizing larger similarities. Figure 5A demonstrates that negative compounds (in green) tend to have greater similarity to the closest negative than the closest positive and the same is seen for positive compounds. One of the benefits of providing these features to a supervised learner is that activity cliffs can be identified when chemicals are very similar to both positives and negatives (diagonal line from lower left to upper right). The activity cliff region tends to be mixed between positives and negatives. The supervised learner (logistic regression for Simple RASAR) can fit these observations and provides probabilities visualized in Figure 5B. Probabilities of hazard can be seen to fall as one move from the lower right to the upper left of the diagram. Figures 5C and 5D visualize counts of negative and positive chemicals, respectively. Negatives are largely in the upper left and positives largely in the lower right.
Figure 5.
Proximity to negative and positive neighbors and probability of skin sensitization. These graphs show how skin sensitizers and nonsensitizers distribute over features describing the closest negative and positive chemicals. A, shows how proximity to closest negative (MaxNeg3) and positive neighbor (MaxPos3) distribute for actually toxic (red) and non-toxic (green) chemicals. B, The associated probability for a positive classification (color gradient from green as low probability for a toxic to red for high probability of toxic property). (C, D) 2D histograms for negative (C) and positive (D) chemicals. In (C), hexes to the upper left of the red line are correct classifications. In (D), classifications to the lower right of the red line are correct classifications. Brighter hexes indicate more chemicals with the given feature values.
The efficacy of similarity metrics can be evaluated by measuring the probability of chemical hazard as a function of similarity to hazardous/nonhazardous compounds. Stronger metrics identify chemicals as similar when they share the activities of interest. This means that similarity metrics should be defined in the context of the activities of interest. It also means that a similarity metric is flawed when it identifies a compound as simultaneously similar to differently labeled chemicals (so called “activity cliffs”).
The RASAR approach allows us to build a model to evaluate similarity-based predictions on evidence. Rather than posing that 2 chemicals are similar will tend to share properties, we model this effect in a supervised model. No commonly used fingerprint + metrics are globally applicable. To identify chemicals for which a similarity metric is likely to be less applicable, we can measure similarity to analogs with diverging properties. We would expect that chemicals that are both similar to negative and positive chemicals would be less predictable from their closest neighbor. This statement is visually supported by Figure 5.
Simple RASAR evaluation
Figure 6 demonstrates how the Simple RASAR logistic regression model (predictions visualized in Figure 5B) separates sensitizers (green) and nonsensitizers (red). On both ends of the graph, the judgement is easy as the probability of hazard versus nonhazard is very distinct. In the middle, such decisions are difficult, creating a gray zone, where decisions are less trustworthy. By setting thresholds for negative and positive probabilities, this gray zone is defined, which determines the applicability domain (coverage of the chemical universe). Conservative choices lead to very good predictions but for less chemicals. The manual setting of these thresholds (Table 3, columns 5 and 6) aimed to optimize sensitivity, specificity and coverage and in the current implementation the metrics give a sensitivity of 80+ % with specificities of 50+ % and 65+ % coverage.
Figure 6.
Distribution of sensitizers/nonsensitizers over Simple RASAR hazard estimates. The upper figure (A) is a histogram counting the number of ± chemicals receiving different probabilistic estimates (2.5% increments). The lower figure (B) shows the percentage of ± chemicals at each hazard probability estimate.
The database includes between 5 and 15 thousand chemicals with animal test-based classifications (Table 3, column 4). This allowed now making predictions for each of them in a leave-one-out cross-validation. The achieved accuracies are shown in Table 3 (columns 7 and 8). This shows that while choosing 80+ % sensitivity, and maintaining specificities between 51% and 69%, the approach worked for on average 82% of substances (73%–88%). It should be noted that the Simple RASAR approach achieves significantly lower specificities than animal reproducibility shows. This is improved via the more complex Data Fusion RASAR.
Development of a data fusion RASAR
Data fusion integrates multiple data sources to achieve more consistent, accurate, and useful information than the individual datasets. Although the Simple RASAR makes only use of one type of hazard information, the data fusion approach uses all labels of the neighboring chemicals. There were 74 labels considered including the UN GHS labels, NTP acute oral toxicity nontoxic label and dependency labels, which are simple functions of the former. There were 23 features no longer considered due to their extreme label imbalances. A feature vector combining 3 kinds of features (known labels for the chemical in question, Jaccard similarities to the closest positive and negative analog) for every label is created to combine different kinds of hazard data.
The RASAR algorithm builds feature vectors for every compound in the unsupervised step and then fits the resulting vectors with Random Forest models for 51 chemical hazard/property labels. Results are shown here for 9 binary (toxic vs. nontoxic) hazards: acute oral toxicity, acute dermal toxicity, acute inhalation toxicity, acute- and chronic-aquatic toxicity, skin sensitization, skin corrosion, eye irritation and mutagenicity. Model accuracy metrics have been assessed in 5-fold cross-validation given in Table 4.
The evaluation metrics for modeling approaches in Tables 3 and 4 compares well with reproducibility metrics from Table 2. This provides encouraging evidence for the ability of computational models to provide valuable predictions on untested chemicals. There are 2 potential reasons for the strength demonstrated in our Data Fusion RASAR. First, the data fusion approach can handle noise in the data and potential activity cliffs via its integration of information on many analogs across many chemical properties. Second, the use of supervised learning methods on high-dimensional data allows for the capture of complex relationships and the avoidance of pitfalls associated heuristic or simplistic structure/activity-relationships (SAR) and Quantitative SAR (QSAR) models. This latter novel approach is based on analysis on how different hazards predict each other. The algorithm incorporated this relationship.
Figure 7 demonstrates how similarity approaches benefit from the quick generation of many analogs for each chemical in a dataset. This “network” effect allows RASAR models to quickly cover a large number of compounds with a relatively small number of labeled compounds. In each graph, more labeled compounds from REACH Annex VI Table 3.1 are added to 33 000 compounds selected from the European INventory of Existing Commercial chemical Substances (EINECS). Connections are shown between unlabeled EINECS compounds (blue) and highly similar Annex compounds (red). The figure helps to visualize how the increasing number of neighbors makes the database cluster as more and more chemicals find a neighbor from the 1 387-chemical list. The X/Y graph shows the coverage of the chemical space. We can see a small number of ANNEX chemicals cover a very large number of EINECs chemicals. By using only 1 387 labeled chemicals we can cover 33 000 unknowns. The bottom right panel of Figure 7 shows how the number of compounds without labeled analogs (termed “lonely EINECS”) decreases as more labeled compounds are added.
The coverage rate, ie, for how many chemicals sufficiently close neighbors are available to make a call, depends mainly on the number of chemicals with data in the database and is thus improved with any addition of further data-points. Counter-intuitively, the information gain of adding a single chemical increases the more, the larger the database already is; this is owed to the fact that the new data-point can be paired with all already included, being one reason for the power of big data for machine learning. The figure demonstrates that chemical similarity networks appear to obey this also known as Metcalfe’s law (Metcalfe’s law states the effect of a telecommunications network is proportional to the square of the number of connected users of the system (n2). Generalized, a network is more valuable the more nodes it has.). Models that can integrate different kinds of data have a much larger benefit from this effect due to the increase in labeled chemicals.
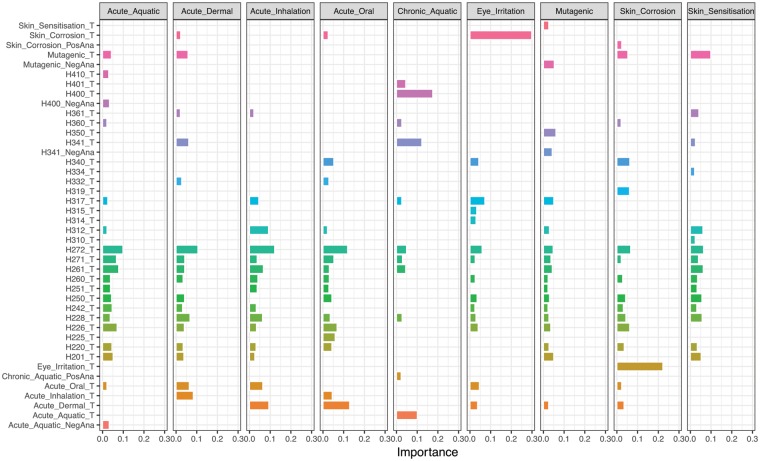
An unfortunate side effect of integrating more data is the loss of a clear explanation for predictions. This can be ameliorated to some degree via analysis of feature importance. Figure 8 shows the variable importance derived by a wrapper approach wherein a model is built with and without a feature and changes in accuracy are treated as importance. The entire results are given as Supplementary Table 2. Notably the H200’s are chemical physical properties and appear to consistently provide value across the 9 shown hazards. This observation is probably due to the intrinsic value of these features but also simply to the high number of chemicals with H200-H299 labeling data. It is satisfying that “skin corrosion binary” (true for a chemical whenever any of the skin corrosion UN GHS hazards is true) appears to be strongly predictive of “eye irritation binary”.
Figure 8.
Select features contributing to the Data Fusion RASAR prediction. The most important information sources in the data fusion approach for 9 hazards are shown. The length of each bar shows the relative importance of the feature (given on the left) towards prediction of the hazard (given at top). Row names take the form <feature>_T for a feature describing the target compound, <feature>_PosAna for a feature describing distance to closest negative analog, and <feature>_NegAna for a feature describing similarity to the closest negative analog. All relative contributions are given as Supplementary Table 1.
DISCUSSION
These results provide evidence that animal tests as described in OECD test guidelines are not strongly reproducible. The reproducibility of an animal test is an important consideration when considering acceptance of associated computational models and other alternative approaches. These results additionally show that computational methods, both simple and complex, can provide predictive capacity similar to that of animal testing models and potentially stronger in some domains.
The measurement of animal test reproducibility results here has potential shortcomings. Test reproducibility is not independent of the chemical being tested. For example, a soluble acid may be more reproducible than an insoluble allergen in eye irritation tests. Thus, the results should not be taken as a global predictor of reproducibility. Additionally, chemicals that have been tested multiple times may be biased to those that are more difficult to evaluate. Notwithstanding these shortcomings, it seems that animal test results are highly variable. Computational models such as those presented here now obtain accuracies in line to the animal tests, on which they are based. It is possible that these models obtain stronger results than single animal tests in cases where they can leverage reliable data on analogs or off-target hazards of the predicted compound. Shortcomings in animal testing have been discussed earlier (Basketter et al., 2012; Hartung, 2008, 2013); a recent publication of ours (Smirnova et al., 2018) summarizes this for the systemic endpoints though the balance between opinion and evidence is difficult in the absence of systematic reviews (Hartung, 2017a). For the acute and topical hazards addressed here, some analyses available are in line with our earlier findings (Luechtefeld et al., 2016c,d) and those reported here: The variability of the LLNA was pointed out by Urbisch et al. (2015): By retesting 22 LLNA performance standards in the standard LLNA protocol, a reproducibility of only 77% was found (Kolle et al., 2011). Recently, Hoffmann (2015) analyzed the variability of the LLNA test, using the NICEATM database. Repeat experiments for more than 60 substances were analyzed in terms of skin sensitization potential, ie, discriminating sensitizer from nonsensitizers: The false positive rate ranged from 14% to 20% (false negative rate 4%–5%). For eye irritation, Adriaens et al. (2014) showed by resampling Draize eye test from more than 2000 studies, analyses an overall probability of at least 11% that chemicals classified as category 1 could be equally identified as category 2 and of about 12% for category 2 chemicals to be equally identified as no category. Hoffmann et al. (2010) reported somewhat better reproducibility of the acute oral toxicity (LD50) in mice and rats, corresponding to the 94% accuracy observed here and similar in Luechtefeld et al. (2016b). Noteworthy, TG 401 for acute oral toxicity has been replaced in the meantime by tiered test guidelines, for which no such reproducibility data are available.
Noteworthy, the animal test reproducibility should be higher in toxicology than other areas of the life sciences as these highly standardized tests are carried out under Good Laboratory Practice by skilled professionals addressing high doses of substances (often so-called maximum tolerated doses) in healthy animals, ie, without further modeling of a disease as in most drug-related studies. Still mere reproducibility ranges only in the 70s–80s percent as shown here for the first time in a comprehensive way for the most commonly used toxicity tests based on a very robust sample of hundreds of studies used for regulatory purposes. The benchmarks established here should be of value for decisions on replacing any of these methods by alternatives far beyond the RASAR tool presented here. This sheds another light on the reproducibility crisis in science (Baker, 2016).
Higher reproductions of such data are either due to better reference datasets or over-fitting, ie, an optimization for the given training set, which will then not hold for the later application. The RASAR development described made use of classifications not the underlying raw animal data, ie, expert judgment has already integrated the available knowledge. It can therefore be somewhat better than the reproducibility of the animal test. Relying on both positive and negative neighbors and other covariates also reduces the impact of misclassified neighbors. Noteworthy, preliminary work using more than 1 positive/negative chemical did not improve predictions to relevant extent, probably reflecting the redundancy of this information.
Supervised models for hazard that output probabilities of hazard allow for setting thresholds to achieve 80+ % sensitivity, ie, to find most toxic substances and thus reflecting regulatory needs. A balance between specificity and coverage was attempted as neither a tool, which has too many false-positives, nor one, which can make predictions only for a small portion of chemicals, is of any use. It should be noted that most toxicological tools are rendered rather unspecific in order to increase sensitivity with specificities often below 10%–20% (Basketter et al., 2012; Hoffmann and Hartung, 2005), ie, a true-positive rate of 1:5–1:10. The sensitivities achieved are thus remarkably high, which is desirable from a company perspective as unnecessary restrictions of use are avoided. Setting thresholds even more conservatively, even higher sensitivities (but then lower coverage) could be obtained depending on the use scenario for these results. Setting model thresholds based on animal reproducibility metrics makes sense in a regulatory context.
With sufficient data on the target compound and its analogs, computational models presented here show accuracy commensurate to the repeat animal test. By data fusion, this predictivity was considerably boosted, even exceeding animal test reproducibility. For the 6 tests often referred to as “toxicological 6-pack” a reproducibility sensitivity of on average 70% was found (Table 2); the Simple RASAR matched this with on average the same 70%; by data fusion, 89% average sensitivity was achieved clearly outperforming the respective animal test. As cited above, these 6 tests consume 55% of all animals for toxicological safety testing in Europe 2011. These methods are constrained by the availability of training data. Careful construction of training data should be considered to optimize future model training and reduce the use of animals.
The data fusion model covers the standard tests for the REACH 2018 registration. In 2009, we predicted the numbers of chemicals to be registered under REACH (Hartung and Rovida, 2009; Rovida and Hartung, 2009). For phases 1–2, we predicted a minimum of 12 007 and 13 328 were received (http://www.cefic.org/Documents/IndustrySupport/REACH-Implementation/Workshops/RIEF-IV-16-6-2015/12%20Reach%20and%20%20non-animal%20testing%20-%20Katy%20Taylor.pdf; last accessed June 30, 2018). For 2018, we predicted a minimum of 56 202 chemicals to be registered and ECHA now talks of expected 60 000 registrations in 2018: “We estimate to process around 60 000 dossiers and to assign them registration numbers so that companies can continue to manufacture, import or sell their substances on the European market.” (https://www.echa.europa.eu/documents/10162/13609/work_programme_2018_in_brief_en.pdf/9412a2bd-64f1-13a8-9c49-177a9f853372; last accessed June 30, 2018).
So, if the estimates from 2009 stand, theoretically stopping REACH after the 2013 deadline (the data we are using) and using the data fusion RASAR, we would have saved 2, 8 million animals and 490 million testing costs and received even more reliable data. This is a very theoretical calculation based on the assumption that the ECHA test guidance to industry is actually being enforced. The estimate takes already into consideration available data, waiving, (Q)SAR and in vitro studies. However, as discussed earlier (for reproductive toxicity, the main challenge in REACH and not covered by the RASAR approach yet) industry did in fact only propose a fraction of these studies (Rovida et al., 2011). It will be crucial now to see how ECHA enforces its testing guidelines and RASAR-like approaches might still help to fill data-gaps.
The critical question is the validity of the approach. The internal validation of both approaches uses an unprecedented number of (tens of) thousands of chemicals for the leave-one-out cross-validation of the RASAR and 5-fold cross-validation of the data fusion RASAR. What the RASAR models lose in mirroring biological complexity, they gain by their automatable nature. We will have to learn, where for each hazard the areas of uncertainty or misclassification lie, to possibly flag or alert for them. While the current approach already aligns with the OECD validity criteria for (Q)SAR (http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?doclanguage=en&cote=env/jm/mono(2004)24) agreed in 2004 (http://www.oecd.org/chemicalsafety/risk-assessment/37849783.pdf; last accessed June 30, 2018), a formal validation of the approach is under way with the U.S. Inter-Agency Coordinating Committee for the Validation of Alternative Methods, which shall identify such shortcomings. The methods are made practically available in a collaboration with Underwriters Laboratories (UL) (as REACHacrosshttps://www.ulreachacross.com; last accessed June 30, 2018; and the UL Cheminformatics Suite, respectively).
The method is agnostic to the endpoint of interest—whenever there is a sufficiently large curated dataset of a given property of substances it can predict and determine the confidence of the prediction. For example, the addition of aquatic toxicity endpoints and inhalation toxicity complemented the array of prediction models. The critical dependence on availability and quality of data is acknowledged, urging also those with access to such data, ie, industry and regulatory agencies to make such data in suitable formats publicly available. Questions of legitimate access to these data for use registration purposes, if used in aggregated manner for predicting other substances, have fortunately been clarified by ECHA: “A registrant would need permission to use protected data to read-across from a single substance to the target substance, … But they would not need this to make a Qsar prediction.” ( Chemical Watch, July 5, 2017: Echa gives clarity on IP issues for Qsar predictions)
The approach also allows addressing the backlog of other untested substances, eg, the almost ten thousand flavors used in e-cigarettes (Hartung, 2016b) or several thousand food additives and contact materials (Hartung, 2018) or help with emergency assessments or frontload toxicity testing in product development. The latter is referred to as Green Toxicology (Crawford et al., 2017; Maertens et al., 2014; Maertens and Hartung, 2018), ie, synthesizing (“benign design”) likely nontoxic substances or sort out toxic ones by earlier informing the product development process. The tool might also be helpful to address substances, for which simply not enough material is available (up to 20 kg required for comprehensive testing in animal studies) such as impurities in drugs and food. Further uses can be seen in the prioritization of testing or risk assessment or the comparison of substances where alternative chemistry shall replace a substance of concern, avoiding simply that the toxic one is only replaced by a less tested but similarly toxic one. RASAR, a QSAR based on read-across, combines the best of these 2 worlds, the robustness of local chemical similarity driving similarity in biological properties and the objective and fast execution of a QSAR, which can be validated and has established reporting and acceptability criteria.
Future Directions
Every year, about 3 billion Euro are spent for animal tests in toxicology (Bottini and Hartung, 2009), mainly the tests addressed here. These results indicate that large parts of this could be carried out by in silico prediction at a fraction of time and costs. In May 2018, several ten thousand additional substances have to be registered for the European REACH legislation to continue marketing them. The data fusion RASAR approach could satisfy the information requirements for the most prominent endpoints for this deadline without using animals and at a fraction of the costs. However, it is coming obviously too late for this process. It might still help to overcome the shortage in laboratory capacities currently experienced in preparation of dossiers for the deadline. Noteworthy, prices for tests have now often tripled because of these shortages (Dr C. Rovida, personal communication). However, there is not only REACH in Europe. Similar programs from new and emerging policies in the United States (the Toxic Substance Control Act reauthorization, known as the Lautenberg Chemical Safety for the 21st Century Act), Canada, Turkey, Korea, Taiwan, China, India, and more are following.
Very important, the method is endpoint-agnostic. Any sufficiently large dataset of organic chemicals with a given property could be subjected to the RASAR, opening up for further hazards such as endocrine disruption (Juberg et al., 2014), but even chemicophysical properties could be predicted, perhaps also forming an interesting dependency label for the prediction. Future optimizations of the approach beside the expansion and curation of the database should address the similarity metrics employed (Luechtefeld and Hartung, 2017) and validate prediction for more difficult chemistries such as inorganic molecules, ions and polymers. Preliminary data show that especially combinations of different metrics are very promising. A key challenge is metabolism of substances. A combination with software to predict metabolites and subjecting them to the same assessment could be envisaged.
The RASAR represents an enabling technology with uses beyond regulatory toxicology (Luechtefeld et al., 2018): Green Chemistry (or better here Green Toxicology), ie, the frontloading of toxicological considerations in the chemical and product development is one, the identification of problematic substances in the supply chain and the search for alternative chemicals is another one. Interestingly, these large databases also impact on the derivation of thresholds of toxicological concern (TTC) (Hartung, 2017b; van Ravenzwaay et al., 2017), which might synergize with the in silico approach of a RASAR: the RASAR would prioritize substances from the hazard properties side, while TTC bring in the relevant exposure for the respective toxicological space. This could be an integral part for the strategic development of a new safety sciences paradigm (Busquet and Hartung, 2017).
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
ACKNOWLEDGMENT
Editing of the article by Sean Doughty is gratefully appreciated. Craig Rowlands is an employee of Underwriters Laboratories (UL). The other authors consult UL on computational toxicology, especially read-across, and have a share of their respective sales. Tom Luechtefeld and Dan Marsh have created ToxTrack LLC to develop such computational tools.
FUNDING
Thomas Luechtefeld was supported by an NIEHS training grant (T32 ES007141). This work was supported by the EU-ToxRisk project (An Integrated European “Flagship” Program Driving Mechanism-Based Toxicity Testing and Risk Assessment for the 21st Century) funded by the European Commission under the Horizon 2020 program (Grant Agreement No. 681002).
Supplementary Material
REFERENCES
- Adriaens E., Barroso J., Eskes C., Hoffmann S., McNamee P., Alepée N., Bessou‐Touya S., De Smedt A., De Wever B., Pfannenbecker U., et al. (2014). Retrospective analysis of the Draize test for serious eye damage/eye irritation: importance of understanding the in vivo endpoints under UN GHS/EU CLP for the development and evaluation of in vitro test methods. Arch. Toxicol. 88, 701–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulmann W., Pechacek N. (2014). Reach (and CLP). Its role in regulatory toxicology In Regulatory Toxicology (Reichl F.-X., Schwenk M., Eds.), pp. 779–795. Springer, Berlin, Heidelberg. [Google Scholar]
- Baker M. (2016). 1, 500 scientists lift the lid on reproducibility. Nature 533, 452–454. [DOI] [PubMed] [Google Scholar]
- Ball N., Cronin M. T. D., Shen J., Blackburn K., Booth E. D., Bouhifd M., Donley E., Egnash L., Hastings C., Juberg D. R., et al. (2016). Toward Good Read-Across Practice (GRAP) guidance. ALTEX 33, 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basketter D. A., Clewell H., Kimber I., Rossi A., Blaauboer B., Burrier R., Daneshian M., Eskes C., Goldberg A., Hasiwa N., et al. (2012). A roadmap for the development of alternative (non-animal) methods for systemic toxicity testing. ALTEX 29, 3–89. [DOI] [PubMed] [Google Scholar]
- Bottini A. A., Hartung T. (2009). Food for thought… on economics of animal testing. ALTEX 26, 3–16. [DOI] [PubMed] [Google Scholar]
- Busquet F., Hartung T. (2017). The need for strategic development of safety sciences. ALTEX 3–21. 34, [DOI] [PubMed] [Google Scholar]
- Crawford S. E., Hartung T., Hollert H., Mathes B., van Ravenzwaay B., Steger-Hartman T., Studer C., Krug H. F. (2017). Green toxicology: a strategy for sustainable chemical and material development. Environ. Sci. Europe 29, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T. (2008). Food for thought … on animal tests. ALTEX 25, 3–9. [DOI] [PubMed] [Google Scholar]
- Hartung T. (2009). Toxicology for the twenty-first century. Nature 460, 208–212. [DOI] [PubMed] [Google Scholar]
- Hartung T., Rovida C. (2009). Chemical regulators have overreached. Nature 460, 1080–1081. [DOI] [PubMed] [Google Scholar]
- Hartung T., Hoffmann S. (2009). Food for thought on…. in silico methods in toxicology. ALTEX 26, 155–166. [DOI] [PubMed] [Google Scholar]
- Hartung T. (2010). Food for thought… on alternative methods for chemical safety testing. ALTEX 27, 3–14. [DOI] [PubMed] [Google Scholar]
- Hartung T. (2013). Look Back in anger – what clinical studies tell us about preclinical work. ALTEX 30, 275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T. (2016a). Making big sense from big data in toxicology by read-across. ALTEX 33, 83–93. [DOI] [PubMed] [Google Scholar]
- Hartung T. (2016b). E-Cigarettes and the need and opportunities for alternatives to animal testing. ALTEX 33, 211–224. [DOI] [PubMed] [Google Scholar]
- Hartung T. (2017a). Opinion versus evidence for the need to move away from animal testing. ALTEX 34, 193–200. [DOI] [PubMed] [Google Scholar]
- Hartung T. (2017b). Thresholds of Toxicological Concern—Setting a threshold for testing where there is little concern. ALTEX 34, 331–351. [DOI] [PubMed] [Google Scholar]
- Hartung T. (2018). Rebooting the Generally Recognized as Safe (GRAS) approach for food additive safety in the US. ALTEX 35, 3–25. [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Hartung T. (2005). Diagnosis: toxic! – Trying to apply approaches of clinical diagnostics and prevalence in toxicology considerations. Toxicol. Sci. 85, 422–428. [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Kinsner-Ovaskainen A., Prieto P., Mangelsdorf I., Bieler C., Cole T. (2010). Acute oral toxicity: variability, reliability, relevance and interspecies comparison of rodent LD50 data from literature surveyed for the ACuteTox project. Regulat. Toxicol. Pharmacol. 58, 395–407. [DOI] [PubMed] [Google Scholar]
- Hoffmann S. (2015). LLNA variability: an essential ingredient for a comprehensive assessment of non-animal skin sensitization test methods and strategies. ALTEX 32, 379–383. [DOI] [PubMed] [Google Scholar]
- Juberg D. R., Borghoff S. J., Becker R. A., Casey W., Hartung T., Holsapple M., Marty S., Mihaich E., Van Der Kraak G., Wade M. G., et al. (2014). Lessons learned, challenges, and opportunities: The U.S. endocrine disruptor screening program. ALTEX 31, 63–78. [DOI] [PubMed] [Google Scholar]
- Kolle S. N., Kandárová H., Wareing B., van Ravenzwaay B., Landsiedel R. (2011). In-house validation of the EpiOcular™ eye irritation test and its combination with the bovine corneal opacity and permeability test for the assessment of ocular irritation. Altern. Lab. Anim. 39, 365–387. [DOI] [PubMed] [Google Scholar]
- Luechtefeld T., Maertens A., Russo D. P., Rovida C., Zhu H., Hartung T. (2016a). Global analysis of publicly available safety data for 9, 801 substances registered under REACH from 2008-2014. ALTEX 33, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T., Maertens A., Russo D. P., Rovida C., Zhu H., Hartung T. (2016b). Analysis of public oral toxicity data from REACH registrations 2008-2014. ALTEX 33, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T., Maertens A., Russo D. P., Rovida C., Zhu H., Hartung T. (2016c). Analysis of Draize eye irritation testing and its prediction by mining publicly available 2008-2014 REACH data. ALTEX 33, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T., Maertens A., Russo D. P., Rovida C., Zhu H., Hartung T. (2016d). Analysis of publically available skin sensitization data from REACH registrations 2008-2014. ALTEX 33, 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T., Hartung T. (2017). Computational approaches to chemical hazard assessment. ALTEX 34, 459–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T., Rowlands C., Hartung T. (2018). Big-data and machine learning to revamp computational toxicology and its use in risk assessment. Toxicol. Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens A., Anastas N., Spencer P. J., Stephens M., Goldberg A., Hartung T. (2014). Green Toxicology. ALTEX 31, 243–249. [DOI] [PubMed] [Google Scholar]
- Maertens A., Hartung T. (2018). Green toxicology – Know early about and avoid toxic product liabilities. Toxicol. Sci. 161, 285–289. [DOI] [PubMed] [Google Scholar]
- Patlewicz G., Ball N., Booth E. D., Hulzebos E., Zvinavashe E., Hennes C. (2013). Use of category approaches, read-across and (Q)SAR: General considerations. Regulat. Toxicol. Pharmacol. 67, 1–12. [DOI] [PubMed] [Google Scholar]
- Patlewicz G., Ball N., Becker R. A., Blackburn K., Booth E., Cronin M., Kroese D., Steup D., van R. B., Hartung T. (2014). Read-across approaches - Misconceptions, promises and challenges ahead. ALTEX 31, 387–396. [DOI] [PubMed] [Google Scholar]
- Patlewicz G., Fitzpatrick J. M. (2016). Current and future perspectives on the development, evaluation, and application of in silico approaches for predicting toxicity. Chem. Res. Toxicol. 29, 438–451. [DOI] [PubMed] [Google Scholar]
- Rovida C., Hartung T. (2009). Re-evaluation of animal numbers and costs for in vivo tests to accomplish REACH legislation requirements. ALTEX 26, 187–208. [PubMed] [Google Scholar]
- Rovida C., Longo F., Rabbit R. R. (2011). How are reproductive toxicity and developmental toxicity addressed in REACH dossiers? ALTEX 28, 273–294. [DOI] [PubMed] [Google Scholar]
- Shah I., Liu J., Judson R. S., Thomas R. S., Patlewicz G. (2016). Systematically evaluating read-across prediction and performance using a local validity approach characterized by chemical structure and bioactivity information. Regul. Toxicol. Pharmacol. 79, 12–24. [DOI] [PubMed] [Google Scholar]
- Smirnova L., Kleinstreuer N., Corvi R., Levchenko A., Fitzpatrick S. C., Hartung T. (2018). 3S – Systematic, systemic, and systems biology and toxicology. ALTEX 35, 139–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbisch D., Mehling A., Guth K., Ramirez T., Honarvar N., Kolle S., Landsiedel R., Jaworska J., Kern P. S., Gerberick F., et al. (2015). Assessing skin sensitization hazard in mice and men using non-animal test methods. Regul. Toxicol. Pharmacol. 71, 337–351. [DOI] [PubMed] [Google Scholar]
- van Ravenzwaay B., Jiang X., Luechtefeld T., Hartung T. (2017). The Threshold of Toxicological Concern for prenatal developmental toxicity in rats and rabbits. Regul. Toxicol. Pharmacol. 88, 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. S., Panko J., Paustenbach D. J. (2009). The European Union’s REACH regulation: a review of its history and requirements. Crit. Rev. Toxicol. 39, 553–575. [DOI] [PubMed] [Google Scholar]
- Zhu H., Bouhifd M., Kleinstreuer N., Kroese E. D., Liu Z., Luechtefeld T., Pamies D., Shen J., Strauss V., Wu S., et al. (2016). Supporting read-across using biological data. ALTEX 33, 167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.