Abstract
Maize is one the most widely cultivated crops worldwide and an important model system for the study of genetics and cytogenetics. Although the availability of a genome sequence has enabled new quantitative genomic studies, developing methods to isolate specific types of cells will enable useful approaches for transcriptomic analysis in the crop plant. Fluorescent Activated Cell Sorting (FACS) is a powerful technique for cell isolation and the study of transcriptional profiles from specific cell populations. The use of FACS on plant cells requires the generation of protoplasts by tissue digestion and cell wall removal. Although some protocols are available, they are mainly focused on dicot species and obtaining sufficient protoplasts from inner tissue layers has been challenging in both monocots and dicots. Here, we report a new protocol that dramatically increases protoplast yield from maize for subsequent cell isolation by FACS. This protocol is efficient in generating protoplast from root and shoot inner layers and can also be applied successfully to Arabidopsis thaliana.
Keywords: Maize, Monocots, Transcriptome, FACS, Protoplasts
Introduction
Maize is a multipurpose crop used as food, fuel and fiber that constitutes a great share of the annual agricultural industry (USDA, 2018). There is a growing interest in applying new tools to characterize the genetic mechanisms behind productive traits to further improve crop yield. These traits often depend on the biological functions performed by a limited group of cells or tissues. Therefore, the isolation and characterization of specific cell and tissue types can greatly contribute to understanding the genetic mechanisms behind developmental processes influencing crop productivity. FACS is a powerful technique that is widely used for the isolation of cell populations based on their fluorescent properties. It has been used successfully in Arabidopsis thaliana to characterize the transcriptome of most root cell types (Birnbaum et al., 2003; Sena, Wang, Liu, Hofhuis, & Birnbaum, 2009).
Here, we describe a protocol that uses a cysteine pre-treatment to increase the efficiency of generating protoplasts from maize up to seven fold. Combined with fluorescent markers, isolation of specific cell types can be achieved using FACS. Cell isolation and sorting takes about three hours while all steps, including plant growth from seed, can be completed in one-week time. As an example, we show the efficient isolation of maize root endodermal cells.
Generation of maize root protoplasts (Basic protocol 1)
Protoplast isolation by enzymatic digestion of the cell wall is an important technique that enables the separation of individual cells for further treatment or analysis. For example, it constitutes the first step for many plant genetic transformation protocols, both transient and stable. Moreover, it is also a prerequisite for performing electrophysiological measurements on plant cells, cell sorting through FACS and other microfluidic devices, and many tissue culture techniques that involve plant regeneration from single cells.
Here, we provide a detailed description of the generation of maize protoplasts in two stages: a) seed germination and seedling growth, and; b) tissue dissection and protoplast preparation for subsequent cell sorting through FACS.
Materials
Maize seeds
3%(v/v) sodium hypochlorite
Heavy weight germination paper (Anchor Seed Germination Paper, 25 × 51 cm; Anchor Paper Co.)
Regular weight germination paper (Anchor Seed Germination Paper, 25 × 51 cm; Anchor Paper Co.)
Aluminum foil
Rubber bands assorted sizes
Forceps, tweezers and scalpel
Plastic 2–3 gallon plastic container (plastic bucket can be used)
Growth chamber. Settings: 27 °C, 16h light/24 °C, 8h dark; 50% humidity (Percival Scientific)
Small petri dishes (35 × 10 mm)
Shakers
50 ml Falcon conical tubes
40 μm cell strainer
1.5 ml microcentrifuge tubes
Microcentrifuge
Hemocytometer chamber
Stereomicroscope
Pretreatment solution (see reagents).
Enzyme mix solution (see reagents).
Washing solution (see reagents).
Seed germination and seedling growth
-
1
Incubate mature maize seeds in 3% (v/v) of sodium hypochlorite for 8 min in a glass beaker. Stir occasionally.
mix 250 ml of distilled water and 250 ml of commercial bleach (6% sodium hypochlorite) to sterilize up to 100 seeds.The purpose of the bleach treatment is to decrease the chance of infection by eliminating fungal or bacterial pathogens that could have been carried along in the seeds from growing facilities (field or greenhouse). However, the seeds are germinated in non-sterile conditions. -
2
Rinse the seeds 5 times with an equal volume of distilled water.
-
3
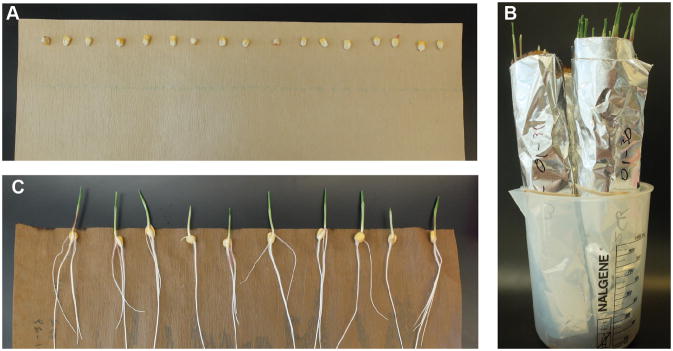
Place 20–25 seeds in a sheet of wetted heavy weight germination paper. Distribute the seeds as a row along the edge of the longest side of the paper. Put a second sheet of regular weight germination paper on top to cover the seeds (Fig. 1A).
-
4
Roll the germination paper sheets to form a cylinder. Try not to displace the seeds when rolling the paper. Place a rubber band just below the seeds to keep the paper roll tight, and one more at the bottom of the paper (the seeds should stay in place when the roll is placed in a vertical position).
-
5
Cover the paper cylinder with a layer of aluminum foil to prevent the roots receiving direct light. You can use another rubber band to keep the aluminum foil in place (Fig. 1B).
-
6
Place the germination paper with the seeds into a 2-gallon plastic container (a plastic bucket works fine). Fill bucket with tap water so it reaches up to 1/4 of the paper height. Incubate in a growth chamber at 27 °C, 16h light/24 °C, 8h dark and ≈ 50% humidity for 6 days.
-
7
On the morning of day 6, unroll the germination paper and assess seedling growth. By this time, you should see a well developed primary root and the presence of seminal and secondary roots. Use healthy looking plants to prepare protoplast as described in subsequent steps (Fig. 1C).
Figure 1.
Seedling germination and growth. After sterilization maize seeds are placed as a row in a sheet of wet germination paper (A). The germination paper is rolled over to form a cylinder and then covered with aluminum foil. A rubber band is used to hold the paper together (B). After 6 days, seedlings show well developed primary and seminal roots. Root tips can be collected for cell-wall digestion at this point (C).
Generation of root protoplasts
-
8
Prepare pretreatment, enzyme and washing solutions.
Prepare fresh on the same day, since enzymes can lose efficiency if stored. -
9
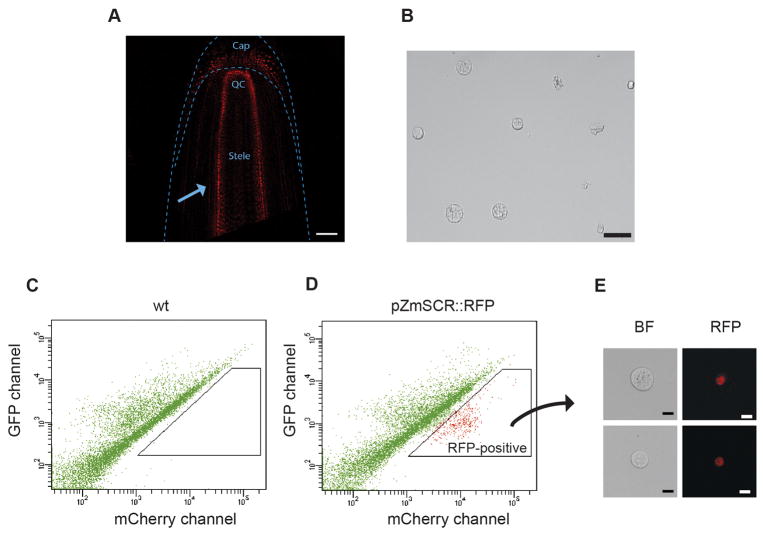
Identify the seedling’s primary and seminal roots and collect them cutting 4–5 mm above the tip of the root with a scalpel. If using a fluorescent reporter line for the first time or if the line is segregating the reporter gene, screen the roots for fluorescence under the microscope before collecting them (Fig. 2A).
-
10
Collect the cut root tips with tweezers and transfer them to a small petri dish containing 3 ml of pretreatment solution. Incubate at room temperature for 45 min with gentle shanking (90 rpm).
-
11
After the incubation, remove the pretreatment solution using a 1 ml micropipette. Press the pipette tip to the bottom of the dish to avoid taking any root tissue.
Tissue fragments are bigger than the pipette tip opening, so careful suction should prevent tissue to go into the pipette. -
12
Add 2 ml of distilled water to the dish to wash the root tips. Repeat step 3 to remove the water.
-
13
Add 3 ml of enzyme solution to the petri dish and further cut the tissue into smaller fragments (approximately 1 mm long) with the scalpel. Incubate for 90 min with gentle shaking (90 rpm).
-
14
After 90 min incubation, use a 1 ml pipette to mix the media several times until it becomes “cloudy”.
The cell wall is mostly digested at this time. However, the root exodermis prevents disaggregation unless is disturbed by a mild mechanical force. In this case, pipetting up and down several times is enough to release the protoplasts into the media, which will become “cloudy”. -
15
To remove undigested tissue and debris, use a 40 μm cell strainer to filter the protoplast solution. Place the cell strainer on top of a 50 ml Falcon tube and with the help of a 1 ml pipette pass the solution through the filter. Discard the strainer.
-
16
Transfer the filtered protoplasts solution to several 1.5 ml microcentrifuge tubes (easier to centrifuge) and centrifuge at 500 g for 3 minutes.
Microcentrifuge tubes are preferred because they are easier to manipulate, and the cell pellet attaches more firmly to the bottom of the tubes. However, centrifugation can be done in 50 ml Falcon tubes if dealing with greater volumes. -
17
Use a 1 ml pipette to carefully remove the supernatant. Remove as much as possible without disturbing the protoplast pellet and resuspend the pellet in 500–1000 μl of washing solution. You can adjust the volume of washing solution depending on the pellet size. For small pellets, resuspend in ≈ 300–400 μl. If performing FACS, use the resuspended protoplasts immediately (Fig. 2b).
Ideally, you want a concentration of 1–2 million protoplast per ml, however small variations won’t affect the cell sorting efficiency. It is important to start sorting as soon as the protoplasts are ready. In our experience, the protoplasting effect on gene expression remains minor up to 2 h after initiating the protocol.
Figure 2.
Sorting of RFP-positive maize endodermal cells. Transgenic reporter lines expressing RFP in the root endodermal nuclei are screened under the microscope to ensure only RFP-expressing plants are used for protoplasts preparation. The arrow points to the layer of fluorescent endodermal cells. Scale bar, 100 μm (A). Root tips are digested, and protoplasts are inspected and counted after filtration. Scale bar 50 μm (B). A control population of protoplasted root tips (No RFP expression) is loaded into the FACS machine. A scatter plot of red versus green emission channels is generated. The resulting pattern is due to the protoplasts autofluorescence (C). Protoplasts from RFP-expressing roots show a distinct pattern when viewed on the same scatter plot. An RFP-positive population clearly appears shifted higher in the red channel axis (D). A small sample of the collected RFP-positive protoplasts are examined under the confocal microscope to ensure they have the expected fluorescence. Scale bar 10 μm (E).
FACS collection of RFP-expressing cells (Basic protocol 2)
FACS enables the rapid isolation of plant protoplast subpopulations expressing a fluorescent protein. Thousands of cells can be quantified and collected in just a couple of minutes, and different populations can be harvested simultaneously. FACS has been used effectively for the isolation of Arabidopsis root protoplasts expressing Green Fluorescent Protein (GFP). Here we show how to isolate Maize root protoplasts prepared as described before using FACS. As an example, we show the collection of root endodermal cells expressing Red Fluorescent Protein (RFP), but the same method can be applied for different tissues expressing other fluorescent proteins.
Materials
BD FACSAria cell sorter or equivalent
RLT cell lysis buffer (RNA easy, QIAGEN) or similar
5 ml polystyrene round bottom tubes (Fisher Scientific)
1.5 ml microcentrifuges tubes
Transfer the protoplasts resuspended in washing solution to a 5 ml polystyrene tube and proceed immediately to load the tube into the FACS machine. Under the cytometer settings, set sample agitation speed at 200 to 300 rpm to avoid clogging the capillary tubes.
Set appropriate laser excitation and collection channels. For isolation of RFP expressing cells, excite with the yellow-green laser (561 nm) and set collection filters to 610/20 (mCherry channel). To quantify autofluorescence, use the GFP/FITC channel (excite at 488 nm, and set collection filters to 530/30). Generate a scatter plot with RFP channel vs GFP channel (auto fluorescence) In the FACSDiva software display.
Start the FACS run with an event flow rate of 4,000 to 10,000 events per second using a 100 μm nozzle and PBS (standard sheath fluid). As a negative control, run only non-fluorescent protoplast first. This will enable setting the collection gate for RFP positive protoplasts accurately (Fig. 2C).
Run the sample containing RFP fluorescent protoplasts. A new protoplast population should become apparent (when compared to the control) as a group with a higher RFP/FITC ratio in the scatter plot. Draw the collection gate around this population (Fig. 2D).
Start protoplast collection using the “purity only” mode for maximum stringency. Sort into 1.5 ml microcentrifuge tubes containing RLT buffer or similar (for RNA extraction) maintaining a ratio of up to 100 μl cell suspension (or less) to 350 μl RLT buffer (where pure water can be used to adjust to the proper cell suspension amount).
If sorting for the first time, examine a small sample of the collected protoplasts under the fluorescent microscope to confirm you recovered the desired protoplast population (Fig. 2E).
Freeze the collected samples immediately and store them at −80 °C until RNA is extracted. For RNA-seq, we found that using between 1000 to 10,000 cells gives good results. However, as few as 100 cells can be used when combined with, for example, the Smart-seq2 protocol (Illumina).
Reagents and solutions
Use mili-Q purified water in all recipes.
Pretreatment solution
Add the following to make 25 ml of solution.
0.75g Sorbitol
62μl of1M L-cysteine
Enzyme mix solution
Add the following to make 25 ml of solution. If processing less than four samples, we recommend scaling down to 12 ml due to the elevated cost of enzymes.
1.82g Mannitol
0.097g MES (2-morpholin-4-ylethanesulfonic acid)
0.5 ml of 1M KCl
0.3 g Cellulase “onozuka” RS (Yakult pharmaceutical industry CO.)
0.3 g Cellulase “onozuka” R10 (Yakult pharmaceutical industry CO.)
0.09 g Pectolyase Y-23 (MP Biomedicals)
0.1 g Macerozyme R-10 (Yakult pharmaceutical industry CO.)
Adjust pH solution to 5.7 with 1M TRIS (pH8) (Trizma)
0.25 ml of 1M CaCl2
0.025 g of BSA
Washing solution
Add the following to make 25 ml of solution.
1.82 g Mannitol
0.097 g MES
0.5 ml of 1M KCl
Adjust pH solution to 5.7 with 1M TRIS (pH8) (Trizma)
0.25 ml of 1M CaCl2
0.025 g of BSA
Commentary
Background information
A critical step for performing cell sorting in plants is the generation of protoplasts. Methods for the isolation of maize protoplasts from leaves and roots have existed for more than 40 years (Bala Bawa & G., 1971; Giles, 1974). However, one of the first comparative studies that tested the effect of different conditions like osmotic pressure, enzyme mixture and cysteine pretreatment was that of Senn and Pilet, (1980). An important conclusion drawn from this study was the fact that treating maize roots with L-cysteine significantly enhanced protoplast yields. Since L-cysteine (due to its thiol group) is known to reduce enzyme inactivation by phenol and quinones (Anderson, 1968), the authors suggest that yield enhancement is due to the inactivation of phenols produced by maize roots in large amounts.
A limitation of the mentioned study however, is the reduced number of enzymes tested. Since then, more enzymes for plant and fungi cell wall digestion have been made available. In particular, the enzyme cellulase RS is known to dissolve a wider range of plant cell walls and works well in monocots. A more recent study reported good protoplast yields from rice roots using an enzyme mix targeted for monocot cell walls, which included cellulase RS and Pectolyase, which are not normally used for Arabidopsis protoplast preparations (Evrard et al., 2012).
In the protocol reported here, we applied the L-cysteine pretreatment from Senn and Pilet (1980), followed by an enzymatic digestion step using the enzyme mix reported by Evrard et al. (2012) with minor modifications in the amount of pectolyase and macerozyme enzymes. This resulted in a 5- to 7-fold increase in protoplast yield when compared with either of the mentioned protocols above. In addition, a higher proportion of small cells were recovered (see Table 1). Since small cells are typically encountered in the meristematic zone which is enclosed by other root tissues, their presence is suggestive of a high efficiency in digesting inner cell layers. Our enzyme mix was also applied to the ear meristem resulting in high protoplast yields (see Table 2). However, in this case, yield did not increase with L-cysteine pretreatment. Therefore, it appears that at least for some shoot tissues incubation in L-cysteine is not required, which considerably shortens the protocol. Meristems are highly studied tissues for their central role in development. According to our quantification results, this protocol can facilitate their isolation which has been problematic in the past.
Table 1.
Protoplast yield (per root tip) and average size obtained using different enzyme cocktails and pretreatment combinations. The number of small protoplast was determined to assess the protocol efficiency in digesting inner meristematic tissue.
| No. of protplasts per root tip | Average protoplast diameter (μm) | % of small protoplast (<15 μm) | Absolute no. of small protoplasts (<15 μm) | |
|---|---|---|---|---|
| L-cysteine + Enzyme cocktail 1 (Seen et al.) | 4,524 | 24 | 10 | 456 |
| Enzyme cocktail 2 (Evrard et al.) | 3,151 | 18 | 30.3 | 956 |
| L-cysteine + Enzyme cocktail 2 (protocol reported in this unit) | 23,500 | 21 | 23.1 | 5,450 |
Table 2.
Ear meristem protoplast yield (per mg of tissue) and size obtained with enzyme cocktail 2.
| No. of protplasts per mg ear meristem tissue | Average protoplast diameter (μm) | % of small protoplast (<15 μm) | Absolute no. of small protoplasts (<15 μm) | |
|---|---|---|---|---|
| 15min | 1,967 | 14.8 | 55% | 1,082 |
| 0.5hr | 5,467 | 15.0 | 60% | 3,280 |
| 1hr | 7,133 | 16.6 | 58% | 4,137 |
| 2hr | 11,800 | 18.3 | 27% | 3,186 |
| 3hr | 10,800 | 16.9 | 36% | 3,888 |
| 4hr | 9,533 | 17.4 | 40% | 3,813 |
FACS was performed as described before for Arabidopsis protoplasts with very few modifications (Birnbaum et al., 2005). Although never tested in maize before, FACS has been used successfully in the isolation of Arabidopsis protoplasts from a variety of tissues, including rare cell sub-populations. In addition, it has been used for protoplast isolation in rice with good results (Evrard et al., 2012). Here we show that maize protoplasts are also amenable for downstream sorting using FACS. A difference between this and previously reported methods was the use of RFP as a signal for the detection of the cells of interest. Therefore, different excitation and collection channel properties were used. However, GFP and other fluorescent markers can be used with equally good results.
Critical parameters
Use fresh protoplasting solutions (prepare new every time). Enzymes can significantly lose activity if stored for long periods.
Ensure that the FACS instrument is calibrated and ready to run your samples immediately after the protoplast washing step. Collect protoplasts as soon as possible to avoid significant changes in gene expression as a result of being exposed to stress conditions.
Collect cells directly in RNA extraction buffer or trizol to avoid RNA degradation. This is especially important when collecting low cell numbers, since even a small amount of RNAses in the medium will cause complete degradation.
Troubleshooting
| Problem | Possible cause | Solution |
|---|---|---|
| Roots look intact after enzyme incubation. | Enzyme medium has not been sufficiently mixed | Mix gently with a 1ml pipette several times. It can sometimes take up to 5–8 minutes to disaggregate root tips |
| Protoplast solution has many cell debris. | Harsh pipetting technique, or insufficient filtering | Pipette the protoplast-containing solution gently Filter the protoplast containing solution twice using a 40 μm cell strainer |
| Low protoplast yield | Not enough root tips were used or solution osmolarity is incorrect | Cut more primary root tips and add them to the media (at least 10) Check solution osmolarity |
| Protoplast pellet is lost during washing step | Centrifugation time is too short, or supernatant aspiration was not done immediately | Centrifuge at 500 g for 5 extra minutes Aspirate supernatant and perform washing step immediately after centrifugation |
| FACS flow stream get clogged | Protoplast concentration is too high | Dilute your protoplast sample at least 5 five times and sort again |
| Too many sorting events are detected. | Sample has many cell debris that are being detected as a sorting event | Determine appropriate gates in the forward vs side scatter plot. A live cell staining dye can be used to differentiate living cells from cell debris. |
Anticipated results
Protoplasting 10 primary root tips (3–4 mm long) should yield approximately 200,000 healthy protoplasts. We have seen that collecting as few as 500 cells is enough to make good quality cDNA library if library amplification kits used, for example Smart-seq2.
Time considerations
If seed germination is started on Tuesday, protoplast preparation and FACS can be done on the following Monday.
Protoplast preparation, including pretreatment, digestion and washing steps, can be completed in 2 hours and 45 minutes.
Depending on the cell population being sorted, and the number of total protoplasts in the sample, FACS can take from 5 to 15 minutes.
Acknowledgments
We thank Pui-Leng Ip for handling samples during FACS. The work was funded by National Science Foundation grant 1445025 and NIH Grant R01-GM078279.
Literature cited
- Anderson JW. Extraction of enzymes and sucellular organelles from plant tissues. Phytochem. 1968;7:1973–1988. [Google Scholar]
- Bala Bawa S, GTJ «Budding» and nuclear division in cultured protoplasts of corn, convolvulus and onion. Bot Gaz. 1971;132:240–245. [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods. 2005;2(8):615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302(5652):1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Evrard A, Bargmann BO, Birnbaum KD, Tester M, Baumann U, Johnson AA. Fluorescence-activated cell sorting for analysis of cell type-specific responses to salinity stress in Arabidopsis and rice. Methods Mol Biol. 2012;913:265–276. doi: 10.1007/978-1-61779-986-0_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles KL. Complementation by protoplast fusion using mutant strains of maize. Plant & Cell Physiol. 1974;15:281–285. [Google Scholar]
- Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD. Organ regeneration does not require a functional stem cell niche in plants. Nature. 2009;457(7233):1150–1153. doi: 10.1038/nature07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn A, Pilet PE. Isolation and some Morphological Properties of Maize Root Protoplasts. Zeitschrift für Pflanzenphysiologie. 1980;100(4):299–310. doi: 10.1016/s0044-328x(80)80234-0. [DOI] [Google Scholar]
- USDA, F. a. s. World Agricultural Production (Report) 2018. (WAP 1-18). Retrieved January 2018, from USDA. [Google Scholar]
Key References
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods. 2005;2(8):615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- Evrard A, Bargmann BO, Birnbaum KD, Tester M, Baumann U, Johnson AA. Fluorescence-activated cell sorting for analysis of cell type-specific responses to salinity stress in Arabidopsis and rice. Methods Mol Biol. 2012;913:265–276. doi: 10.1007/978-1-61779-986-0_18. [DOI] [PMC free article] [PubMed] [Google Scholar]