Abstract
Despite advancements in symptom management for heart failure (HF), this devastating clinical syndrome remains the leading cause of death in the developed world. Studies using animal models have greatly advanced our understanding of the molecular mechanisms underlying HF; however, differences in cardiac physiology and the manifestation of HF between animals, particularly rodents, and humans necessitates the direct interrogation of human heart tissue samples. Nevertheless, an ever-present concern when examining human heart tissue samples is the potential for artefactual changes related to temperature changes during tissue shipment or sample processing. Herein, we examined the effects of temperature on the post-translational modifications (PTMs) of sarcomeric proteins, the proteins responsible for muscle contraction, under conditions mimicking those that might occur during tissue shipment or sample processing. Using a powerful top-down proteomics method, we found that sarcomeric protein PTMs were differentially affected by temperature. Specifically, cardiac troponin I and enigma homolog isoform 2 showed robust increases in phosphorylation when tissue was incubated at either 4 °C or 22 °C. The observed increase is likely due to increased cyclic AMP levels and activation of protein kinase A in the tissue. On the contrary, cardiac troponin T and myosin regulatory light chain phosphorylation decreased when tissue was incubated at 4 °C or 22 °C. Furthermore, significant protein degradation was also observed after incubation at 4 °C or 22 °C. Overall, these results indicate that temperature exerts various effects on sarcomeric protein PTMs and careful tissue handling is critical for studies involving human heart samples. Moreover, these findings highlight the power of top-down proteomics for examining the integrity of cardiac tissue samples.
Keywords: Top-down mass spectrometry, Sarcomeres, Post-translational modification, Phosphorylation
1. INTRODUCTION
Heart failure (HF) is a devastating condition that afflicts approximately 5 million Americans and accounted for 1 in 9 deaths in the year of 2013 [1–3]. Despite recent advancements in the treatment strategies available to manage the symptoms of HF, the number of deaths attributable to this devastating clinical syndrome remained nearly as high in 2013 as it was in 1995 [3]. Although animal models of HF have greatly advanced our understanding of the molecular mechanisms underlying this condition, differences in the manifestation of HF between animals, particularly rodents [4, 5], and humans necessitates the direct interrogation of human heart tissue samples to fully elucidate the mechanisms of HF pathogenesis in humans. However, an ever-present concern when examining human heart tissue samples is the potential for artefactual changes related to temperature changes due to prolonged storage of the tissue samples, sharing and shipping of the samples between laboratories, and handling of the tissue samples. Flash-freezing cardiac tissue in liquid nitrogen remains the best method for preserving the integrity of the tissue [6]; yet, tissue warm- up post-procurement may occur during shipping and/or when processing the cardiac tissue samples, with unknown consequences at the protein level.
Sarcomeres are the basic contractile units of striated muscle, and consist of myofilaments flanked on either side by complex protein structures known as Z-discs [7–9]. Myofilaments are composed of the myosin-based thick filaments and actin-based thin filaments [7, 10]. The thick filaments contain myosin heavy chain and cardiac myosin binding protein C (cMyBP-C), as well as the myosin regulatory and essential light chains (RLC and ELC, respectively). The thin filaments consist of actin, tropomyosin (Tpm), and the cardiac troponin complex, which is made up of troponin C (TnC), cardiac troponin I (cTnI), and cardiac troponin T (cTnT). Recent research has convincingly demonstrated that alterations in myofilament post-translational modifications (PTMs) can be causative in contractile dysfunction and HF [11–16]. In addition, emerging evidence indicates that Z-discs represent critical signaling nodes within cardiomyocytes [7, 17], and that the PTMs of Z-disc proteins can be altered in response to cardiac injury [10, 18].
Herein, we employed high-resolution top-down proteomics [9, 19–21] to evaluate the impact of temperature on tissue quality and sarcomeric protein PTMs. In contrast to the conventional bottom-up proteomics approach wherein proteins are digested into peptides prior to mass spectrometry (MS) analysis, intact proteins are analyzed in the top-down proteomics workflow, which provides a “bird’s eye” view of all proteoforms [22] (a term encompassing all protein variants of the same gene arising from genetic variation, alternative mRNA splicing and PTMs) and permits the comprehensive, and quantitative analysis of protein PTMs [9, 19, 21, 23]. Following MS analysis, a specific proteoform of interest can be “purified” in the gas phase and undergo fragmentation, known as tandem MS (MS/MS), for protein sequence characterization and PTM localization. Moreover, the top-down approach is the premier method for revealing small truncated species of proteins, which can be difficult to detect using bottom- up proteomics or conventional SDS-polyacrylamide gel electrophoresis. To gain insight into temperature-related changes in human cardiac tissue, we examined changes in sarcomeric protein PTMs at temperatures mimicking those that may occur during tissue storage, shipment, or sample processing, via a top-down proteomics.
2. METHODS
2.1. Chemicals and reagents
All reagents were purchased from Sigma-Aldrich, Inc. (St. Louis, MO, USA) unless otherwise noted. HPLC grade water, acetonitrile, and ethanol were purchased from Fischer Scientific (Fair Lawn, NJ, USA).
2.2. Human cardiac tissue collection
A donor heart characterized with normal cardiac function, but deemed unacceptable for transplantation, was obtained from the University of Wisconsin Hospital and Clinic. The procedure for the collection of human cardiac tissue has been approved by the Institutional Review Board of the University of Wisconsin-Madison. The donor heart was maintained in cold cardioplegic solution (4 °C) immediately after explanted and delivered to laboratory for dissection within 15 min as described previously [24]. Cardiac tissue dissection was carried out on a metal plate chilled with dry ice, and the dissection of the left ventricular tissue was carried out first and completed within 15 min. Individual pieces of tissue were wrapped with pre- labeled aluminum foil and snap-frozen in liquid nitrogen. The tissues were stored at −80 °C for subsequent analysis.
2.3. Time-course experiments and tissue storage condition test
One piece (~200 mg) of left ventricular tissue from the midwall region was used for all experiments. To assess time-related changes in sarcomeric protein PTMs, three portions of approximately 4–6 mg of tissue were excised on a metal plate pre-cooled with dry-ice and homogenized immediately to evaluate the baselines of sarcomeric protein PTMs (time 0). Additionally, 4–6 mg of tissue was excised and incubated at either 4 °C or 22 °C (room temperature) for 15 min, 30 min, 1 hr, 2 hrs, or 4 hrs, followed by tissue homogenization and protein extraction. Three small portions of the tissues were maintained in the HEPES buffer with protease and phosphatase inhibitors (25 mM HEPES pH 7.5, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 2.5 mM EDTA), and incubated at 4 °C for 30 min before protein extraction. The leftover tissue that remained frozen was rapidly cut into two parts (~80 mg each), and one portion was stored in liquid nitrogen while the other was stored at −80 °C. After approximately 18 months, about 4–6 mg of tissues from each portion were homogenized to extract sarcomeric proteins. The tissue that was stored in liquid nitrogen was then divided into 3 portions, and each were stored in liquid nitrogen, dry ice and wet ice, respectively, for 48 hrs to mimic shipment conditions. These tissues were then homogenized in the same way and the sarcomeric proteins were extracted.
2.4. Cardiac saromeric protein extraction
All sample processing steps were carried out in a cold room (4 °C), and the tissue portions were maintained completely frozen using dry ice prior to the temperature treatment. All homogenization steps were performed in the cold room, and homogenization of each samples took less than 20 sec. Extraction of sarcomeric proteins was carried out as reported previously [10, 25, 26]. Briefly, tissue was first homogenized rapidly in 10 vol. (μl/mg tissue) of HEPES buffer containing protease and phosphatase inhibitors (25 mM HEPES pH 7.5, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 2.5 mM EDTA) using a Teflon pestle (1.5 mL tube rounded tip; Scienceware, Pequannock, NJ, USA). The homogenate was centrifuged at 17,000 rcf for 15 min, and the pellet was washed with 10 vol. of the HEPES buffer, and further homogenized in 10 vol. of trifluoroacetic acid (TFA) solution (1% TFA, 1 mM Tris-(2-carboxyethyl)phosphine). The homogenate, which contains primarily sarcomeric proteins, was centrifuged at 17,000 rcf for 15 min and the supernatant was collected and centrifuged again at 17,000 rcf for 60 min to remove residual particulate matter prior to liquid chromatography (LC)-MS analysis.
2.5. Reverse phase chromatography (rpc) and top-down proteomics analysis
LC-MS analysis was carried out using a NanoAcquity Ultra-high Pressure LC system (Waters, Milford, MA, USA) coupled to a high-resolution Impact II quadrupole time-of- flight (Q-TOF) mass spectrometer (Bruker Daltonics, Bremen, Germany). To evaluate the reproducibility of protein extraction and the LC-MS method, three extraction replicates at the same time point (time 0) and three injection replicates of the same protein extract were tested. The sarcomeric proteins in each sample were eluted by a gradient of 5% to 95% mobile phase B (mobile phase A: 0.1% formic acid in water; mobile phase B: 0.1 % formic acid in 50:50 acetonitrile:ethanol) at a constant flow rate of 8 μL/min. Proteins eluted were delivered to the mass spectrometer via electrospray ionization. End plate offset and capillary voltage were 450 V and 4000 V, respectively. Nebulizer pressure was set to 0.5 Bar and dry gas flow rate was 4.0 L/min. In-source collisional energy was set to 10 V. Mass spectra were collected at a scan rate of 0.5 Hz over 500–2000 m/z range.
2.6. Offline fraction collection and top-down MS/MS analysis
The elution time of target proteins were identified and the eluents containing these proteins were collected and analyzed using a 12 Tesla solariX Fourier transform- ion cyclotron resonance (FT-ICR) mass spectrometer (Bruker Daltonics) via an automated chip-based nanoESI source (Triversa NanoMate; Advion Bioscience, Ithaca, NY, USA). The proteoforms of interest were first purified in the gas phase using the quadrupole with an 8–12 m/z isolation window, and in-cell isolation was performed to remove peaks other than the peak corresponding to the proteoform of interest. The proteoform of interest was fragmented by electron capture dissociation (ECD). The excitation power for in-cell isolation was set to 1% with a pulse time of 0.02 ms. The electron energy and reaction time for ECD were determined on a case-by-case basis to achieve optimal fragmentation. The transients from approximately 1000–4000 scans were summed to obtain high-quality spectra for the localization of protein PTMs.
2.7. Data analysis
All LC-MS data were processed and analyzed using the Data Analysis software ( version 3.2; Bruker Daltonics). Mass spectra were averaged over the retention time window wherein all proteoforms of the same protein eluted. The spectra were deconvoluted using the Maximum Entropy algorithm incorporated in the Data Analysis software. The resolving power for Maximum Entropy deconvolution was set to 60,000 for proteins below 50 kDa, which were isotopically resolved by the mass spectrometer. Most-abundant mass was reported for all MS data, and monoisotopic mass was reported for all MS/MS data. The abundance of a particular proteoform is reported as the ratio of the peak heights of the proteoform to the summed peak heights of all proteoforms of the same protein. The percentages of the mono- (%Pmono) and/or bis- (%Pbis) phosphorylated proteoforms were defined as the summed abundances of mono- and/or bis-phosphorylated species over the summed abundances of the entire protein population, respectively. Based on these percentages, the total amount of phosphorylation (Ptotal; mol of Pi/mol of protein) of a single protein was calculated using the following equation:
Tandem mass spectra were output from the Data Analysis software and analyzed using MASH Suite Pro [27, 28] software developed in- house. The spectra were deconvoluted with a signal- to-noise ratio of 3 and a cutoff fit score of 60%. All the program-processed data were manually validated to obtain accurate sequence and PTM information.
2.8. Western blotting analysis
Temperature treatment of the tissues and homogenization with HEPES extraction buffer were described above. The remaining pellets were homogenized using 10 vol. extraction buffer (50 mM Tris-HCl, 50 mM NaCl, 1% Triton X-100, 0.5% SDS, 1 mM PMSF, 1 mM Na3VO4, 2.5 mM EDTA). Homogenization was performed in the cold room and took less than 30 sec per sample. After centrifugation, the tissue extracts were analyzed by Western blotting to evaluate the phosphorylation of cTnI and cMyBP-C. The following primary antibodies were used for the analysis: rabbit polyclonal against phosphorylated Ser22/23 of cTnI (Cell Signaling Technology; cat. #4004), mouse monoclonal against cTnI (Abcam; cat. ab10231), rabbit polyclonal against PKA substrate (Cell Signaling Technology; cat. 9624) for probing cMyBP-C phosphorylation, and rabbit polyclonal against cMyBP-C (Thermo Fisher Scientific; cat. PA5-41993). Re-probing o f the PVDF membrane with different antibodies was performed as described previously [29]. The detection of cTnI and phosphorylated cTnI was performed using a bicolor fluorescent detection method with the following secondary antibodies: IRDye 800CW anti-mouse IgG (LI-COR Biosciences ; cat. 925–32210) and IRDye 680 RD anti-rabbit IgG (LI-COR Biosciences ; cat. 926–68070). The detection o f c MyBP-C and phosphorylated cMyBP-C (PKA substrate ) was by enhanced chemiluminesce nce. The images were taken using a LI-COR Odyssey imager (LI-COR Biosciences), and the quantification of signals were performed using the Image Studio Lite software (Version 5.2) (LI-COR Biosciences).
2.9. Enzyme-linked Immunosorbent assay (ELISA) for quantitative analysis of cAMP (cyclic adenosine monophosphate)
Approximately 4–6 mg of tissue was excised on a pre-cooled metal plate and incubated at 4 °C for 1 min, 5 min, 10 min, or 30 min. The tissue was immediately homogenized in 10 vol. of 0.1 M HCl and centrifuged at 19,000 rcf for 5 min. The supernatants were diluted 5-fold using 0.1 M HCl for cAMP analysis, or 50-fold using water for Bradford protein assay. cAMP analysis was performed using a direct cAMP ELISA kit in accordance with the manufacturer’s instructions (Enzo Life Sciences, Farmingdale, NY, USA). Briefly, cAMP standards of 0.78, 3.13, 12.5, 50, and 200 pmol/mLwere prepared in 0.1 M HCl. The cAMP standards and the tissue extracts were added to wells coated with a GxR IgG antibody. The solution of cAMP conjugated to alkaline phosphatase (AP) was added and followed by addition of rabbit polyclonal antibody to cAMP. The mixtures were incubated at room temperature for 2 hrs to allow for the binding of the cAMP antibody to cAMP in the samples or cAMP-AP conjugates (competitors). After washing, the AP substrate, para-Nitrophenylphosphate (pNpp), was added to assay the amount of cAMP-AP conjugates, followed by addition of the stopping solution and photometric measurement at 405 nm. The lower amount of cAMP-AP conjugates indicated a higher amount of free cAMP in the sample. The Bradford protein assay was carried out to measure the total protein concentration, and the cAMP concentration was normalized to total protein concentration.
2.10. Statistical analysis
Three extraction replicates at time 0 were used as controls (control group), and two-way ANOVA was performed to evaluate the statistical significance of variation between groups. Differences between means were considered to be statistically significant at p < 0.05.
3. RESULTS
3.1. Increased phosphorylation of cTnI and Enigma Homolog Isoform 2 (ENH2) within minutes following increased temperature
We first assessed the reproducibility of RPC separation of proteins and the stability of the instrument response. Injection and extraction replicates were nearly identical with little variability in the detected relative abundances of sarcomeric protein proteoforms (Supplemental Figure S1, S2), confirming the high reproducibility of the method. Next, to assess the impact of temperature on sarcomeric protein PTMs, we incubated pieces of tissue at temperatures mimicking those that might occur during tissue shipment (22 °C) or sample processing (4 °C). Following incubation at either 4 °C or 22 °C and extraction of the sarcomeric proteins, high-resolution top-down LC-MS analysis was carried out (Figure 1). These samples were compared to the samples extracted immediately before tissue defrosting (referred as the control group) to evaluate temperature-related changes in protein PTMs. Additionally, the effects of buffer and different storage conditions on the sarcomeric protein PTMs were investigated as outlined in Figure 1a.
Figure 1. Schematic for assessing the effect of temperature on sarcomeric protein PTM changes.
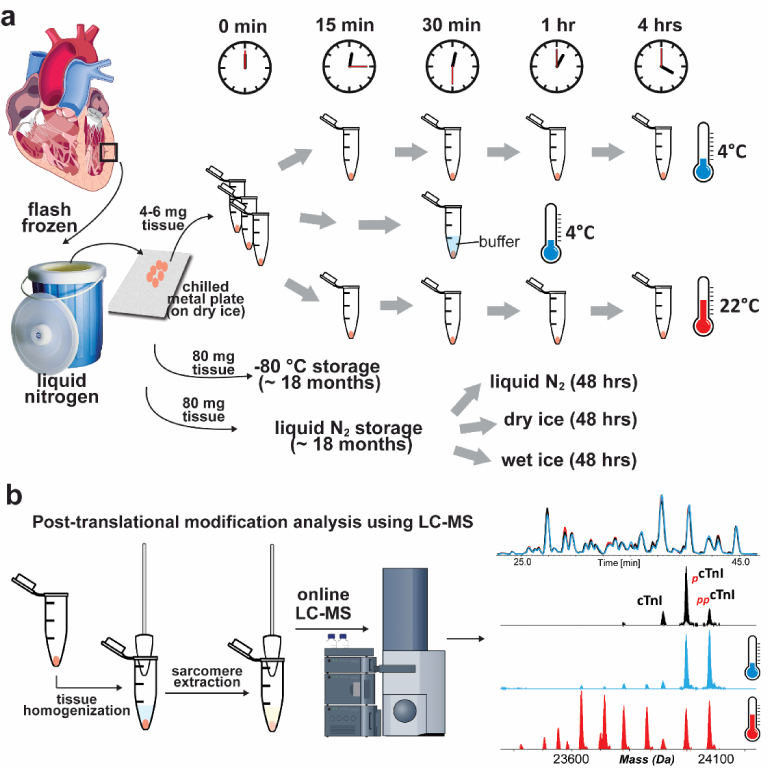
(a) Small pieces of cardiac tissue were kept at either 4 °C or 22 °C for various durations followed by tissue homogenization and protein extraction. Additionally, tissue was also stored in liquid nitrogen (N2) or at −80 °C for approximately 18 months to evaluate changes in protein PTMs under different storage conditions. Small pieces of tissues were also stored using liquid nitrogen (N2), dry ice and wet ice for 48 hrs to mimic the shipment and sample handling conditions. (b) Sarcomeric protein extracts were analyzed by high-resolution top-down LC-MS.
Following temperature treatment, prominent changes were observed in the phosphorylation of cTnI and ENH2, which is a Z-disc protein (Figure 2). Within 15 minutes of tissue incubation at 4 °C, the most abundant MS peak of cTnI changed from the mono-phosphorylated to bis-phosphorylated proteoform. The relative abundance of bis-phosphorylated cTnI increased by approximately 150% within 15 min, and by about 230% after 1 hr at 4 °C (Figure 2a). cTnI proteoforms resulting from removal of 2–4 amino acids from the C-terminus of the protein were detected within 30 min of incubation at 4 °C (Figure 2a). Similar to the tissue maintained at 4 °C, cardiac tissue thawed at 22 °C exhibited an increase in cTnI phosphorylation and degradation within 15 min ( Figure 2b). Degraded proteoforms of cTnI with removal of 2 or 3 C-terminal amino acids became the dominant proteoforms after incubating the tissue at 22 °C for 1 hr (Figure 2b). The high-resolution mass spectra of the cTnI proteoforms and their accurate molecular weights are shown in Supplemental Figure S3.
Figure 2. Rapid increase in the relative abundances of phosphorylated and degraded proteoforms of cTnI and ENH2 following increased temperature.
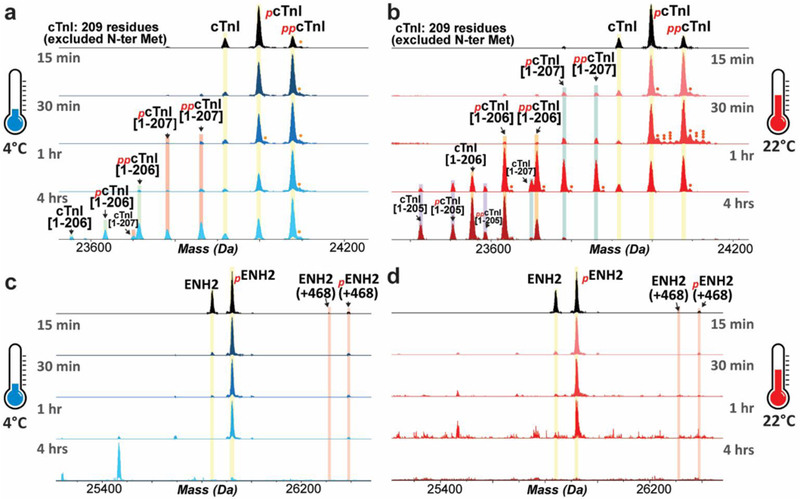
Top-down mass spectra show that hyper-phosphorylation and truncation of cTnI occurred after tissue incubation at 4 °C (a) or 22 °C (b). Increased phosphorylation and degradation of ENH2 upon tissue defrosting at 4 °C (c) or 22 °C (d). Single, double, and triple circles denote singly-, doubly- and triply-oxidated proteoforms, respectively.
In addition to increased cTnI phosphorylation, ENH2 phosphorylation also increased upon temperature increase to 4 °C or 22 °C (Figure 2c, d). The high-resolution mass spectra of the ENH2 proteoforms and their accurate molecular weights are shown in Supplemental Figure S4. Interestingly, even though degraded ENH2 proteoforms were not observed within the same elution time window (37.3–39.1 min), there was a progressive decline in the MS signal for ENH2 in the defrosted tissue after 1 hr at 4 °C or 30 min at 22 °C. The intact ENH2 became nearly undetectable after 4 hrs at 4 °C or 1 hr at 22 °C, indicating that ENH2 degradation occurred during the process of tissue defrosting. The degraded products of ENH2 may be small peptides that eluted at different retention time windows due to differences in hydrophobicity. We also observed two additional proteoforms of ENH2; one with an increase of 468 Da relative to the mass of the un-phosphorylated proteoform, and the second with a 468 Da mass increase relative to mono-phosphorylated ENH2 (Figure 2c, d). Due to the low abundance of these proteoforms, the sequences of these proteoforms were not further characterized.
3.2. Decreased phosphorylation of cTnT and RLC following increased temperature
In contrast to cTnI and ENH2 phosphorylation, which increased after incubation at either 4 °C or 22 °C, the phosphorylation of cTnT and RLC decreased in tissue kept at these temperatures (Figure 3). The decline in cTnT phosphorylation was mild when the tissue was maintained at 4 °C, but severe at 22 °C (Figure 3a, b). Particularly, cTnT phosphorylation in myocardial tissue decreased by approximately 70% when incubated at 22 °C for 4 hrs (Figure 3b). In addition to de-phosphorylation, degradation of cTnT was also observed in the cardiac tissue maintained at 22 °C for over 1 hr (Figure 3b, Supplemental Figure S5). The abundance of the C-terminally truncated cTnT proteoforms relative to the intact cTnT adult isoform was about 1:10 and 1:1 when the tissue was maintained at 22 °C for 1 hr and 4 hrs, respectively (Figure 3b).
Figure 3. Decrease in the relative abundances of phosphorylated proteoforms of cTnT and RLC following tissue incubation at either 4 °C or 22 °C.
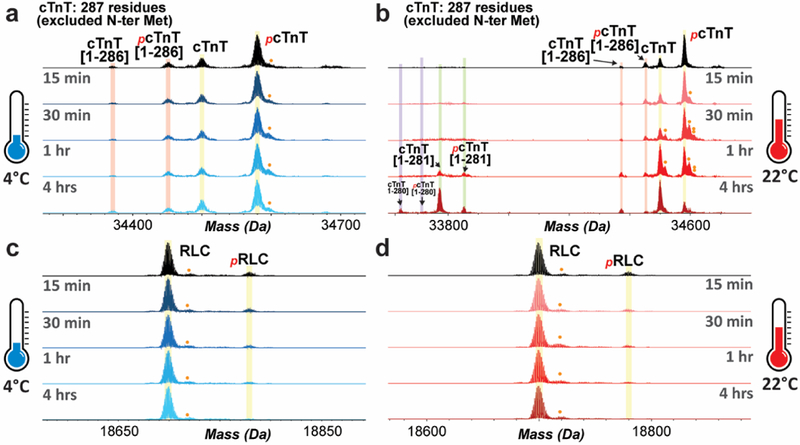
Top-down mass spectra of cTnT proteoforms that show slightly decreased phosphorylation upon tissue incubation at 4 °C (a), and severe de-phosphorylation and degradation of cTnT occurred at 22 °C (b). Decreased phosphorylation of RLC was observed with tissue warm- up at either 4 °C (c) or 22 °C (d). Single and double circle denote singly- and doubly-oxidized proteoforms, respectively.
Even though RLC phosphorylation was low (less than 10%) in the control group, the decline in RLC phosphorylation was apparent after 30 min of tissue incubation at 4 °C, or 15 min incubation at 22 °C (Figure 3c, d). The accurate molecular weights of RLC proteoforms are shown in Supplemental Figure S6. With prolonged temperature change (4 hrs), we also observed a slight increase in the phosphorylation of cardiac α- Tpm (formally known as Tpm1.1st [30]); however, the phosphorylation of cardiac β-Tpm (formally known as Tpm2.2st [30]) appeared to decrease to the extent that it was undetectable after tissue incubation for 4 hrs at either 4 °C or 22 °C (Supplemental Figure S7).
As summarized in Figure 4, the phosphorylation of major myofilament proteins were differentially affected by tissue incubation at either 4 °C or 22 °C. While incubation at either 4 °C or 22 °C had a significant effect on all the myofilament proteins analyzed, cTnI and ENH2 were affected to the greatest extent (Figure 4a, b). cTnT phosphorylation is relatively stable within 1 hr of temperature increase at 4 °C (Figure 4c), and RLC phosphorylation remains relatively unchanged within 15 min of tissue incubation at 4 °C (Figure 4d), but decreased significantly after 15 min at 22 °C.
Figure 4. Temperature-related changes in the relative abundances of un-phosphorylated and phosphorylated sarcomeric protein proteoforms.
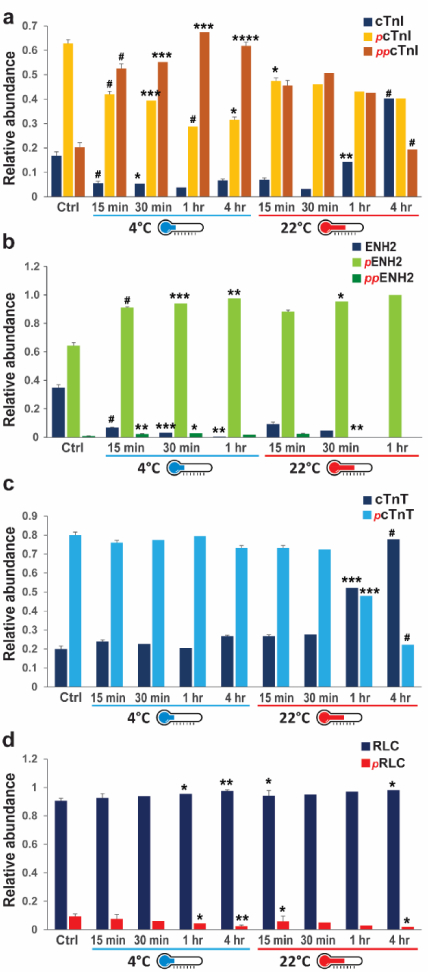
Quantification of the relative abundances of the cTnI (a), ENH2 (b), cTnT (c), and RLC (d) proteoforms during tissue incubation at either 4 °C or 22 °C. * p < 0.05, ** p < 0.01, *** p < 0.001, # p < 0.0001 by two-way ANOVA.
3.3. Localization of the sites of phosphorylation on cTnI by ECD
While the observed decreases in the phosphorylation of cTnT and RLC likely occurred at the canonical sites, Ser1 and Ser14, respectively [31, 32], increased phosphorylation of cTnI and ENH2 was intriguing. Particularly, cTnI has multiple sites of phosphorylation that can be targeted by various kinases, leading to dramatically different functional impacts on contractility [12]. In addition, ENH2 was only recently discovered as a phosphoprotein in a swine model of acute myocardial infarction [10], and the site of ENH2 in human cardiac tissue has not been identified. To localize the sites of phosphorylation in cTnI and ENH2, we performed MS/MS experiments using an ultra- high-resolution 12 Tesla FT-ICR mass spectrometer. Mono- and bis-phosphorylated proteoforms of cTnI, as well as the mono-phosphorylated proteoform of ENH2, were fragmented by ECD, which preserves labile PTMs such as phosphorylation [9, 19, 33].
In the case of pcTnI fragmentation, after taking into account the removal of N-terminal Met, the masses of the N-terminal fragment ions (c ions) prior to c20 exhibited an increase of 42.01 Da relative to the predicted masses for these fragment ions, indicating that the N-terminus is modified by acetylation (Supplemental Figure S8a), consistent with our previous reports [24]. The N-terminal fragments prior to c20 exclusively exhibit an increase of 42.01 Da without an additional 79.97 Da increase in mass (Supplemental Figure S8b), suggesting that the site of phosphorylation is localized to the C-terminus of amino acid 20. The N-terminal c22 ion has an increase of 79.97 Da in mass in addition to acetylation (Supplemental Figure S8c), suggesting that Ser22 is phosphorylated. In addition, the fact that both un-phosphorylated and mono-phosphorylated c22 were observed (Supplemental Figure S8d) suggests Ser22 is partially phosphorylated, and that pcTnI contains a mixed positional isomers with Ser22 being one of the phosphorylation sites. Given that c20 is exclusively un-phosphorylated, and c24 is exclusively mono-phosphorylated without detectable un-phosphorylated ion (Supplemental Figure S8b, d), all existing phosphorylation sites are between amino acid residues 20–24 (-Arg-Arg-Ser-Ser-Asn-), and therefore, the site of phosphorylation on pcTnI is localized to either Ser22 or Ser23 (Supplemental Figure S8e). A total of 59 c ions (N-terminal fragment ions) and 71 z· ions (C-terminal fragment ions) from a single tandem mass spectrum were matched to the sequence of pcTnI, which represents cleavage of 61% of all inter-residue bonds.
Similar to the localization of phosphorylation site on pcTnI, the sites of phosphorylation on ppcTnI was localized to Ser22 and Ser23 (Figure 5a–e) with a total of 45 c and 70 z· ions from a single tandem mass spectrum, representing cleavage of 56% intra-residue bonds. We further validated the increased cTnI phosphorylation by Western blotting using a specific antibody against phosphorylated (Ser22/23) cTnI (Figure 5f). We observed significant increase in the phosphorylation of cTnI at Ser22/23 following temperature treatment at 4 °C or 22 °C (Figure 5f, g), which was consistent with the MS results (Figure 4). However, the extent of cTnI phosphorylation change appeared different between the two quantification methods. This could result from different detection limits and linear range of the two methods. cTnI phosphorylation by Western blotting, when expressed in log scale, appeared more consistent with the quantification results by MS (Figure 5h).
Figure 5. Localization of the phosphorylation sites in ppcTnI by top-down MS/MS.

(a) and (b) Zoomed- in spectra for c9 and c20 ions, respectively, confirmed N-terminal acetylation of the ppcTnI. N-terminal fragments prior to c20 had no detectable mono-phosphorylated counterparts. (c) Zoomed-in spectrum for c22 ion showed only mono-phosphorylated ions, suggesting that amino acid sequence N-terminal to Ser22 is exclusively mono-phosphorylated. (d) Zoomed- in spectrum for c24 showed that c24 ions were exclusively bis-phosphorylated without detectable un-phosphorylated or mono- phosphorylated ion, suggesting that amino acid sequence prior to Ser24 contains both phosphorylation sites. Circles represent the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned mass. N.D. not detected. (e) Fragmentation map shows the fragment ions that were matched to the sequences of ppcTnI (45 c and 70 z· ions total). Ac denotes acetylation. p denotes phosphorylation. (f) Western blotting analysis to confirm increased of cTnI phosphorylation at sites of Ser22/23. (g) Histo gra m showing q ua ntifica tio n o f p hosp horylated cTnI b y Western b lotting. Two-wa y ANOVA test wa s performed to e va luate the sta tistica l significa nce o f d iffere nce. * p < 0.05, *** p < 0.001. (h) Co mpariso n o f re la tive ab unda nce o f b is-p hosp horylated c TnI q ua ntified by MS (red bars) and phosphoryla ted cTnI quantified by Western blotting in log scale (black dots).
3.4. Localization of the phosphorylation site in ENH2 by ECD
With the removal of the N-terminal Met, all c ions had a 42.01 Da increase in mass compared with the predicted fragment ions, indicating acetylation of the ENH2 N-terminus (Figure 6a), which is consistent with the finding in swine ENH2 [10]. All N-terminal fragments prior to c114 were exclusively un-phosphorylated (Figure 6a, b), suggesting that the site of phosphorylation is localized after residue 114. The fact that the c118 and c120 ions are exclusively mono-phosphorylated without detectable un-phosphorylated counterparts (Figure 6c, d) suggests that the phosphorylation site is exclusively localized to the N-terminus of residual 118. Therefore, the phosphorylation site in pENH2 is localized between amino acid residues 114 and 118 (-Gln-Arg-Arg-Gly-Ser-). Since Ser118 is the only possible site of phosphorylation, we assigned Ser118 as the sole site of phosphorylation in human ENH2, which is consistent with the ENH2 phosphorylation site observed in swine [10]. In addition, c118 is exclusively mono-phosphorylated without detectable un-phosphorylated ions. This further proved that Ser118 is the only site of phosphorylation in pENH2. A total of 78 c ions and 52 z· ions from two tandem mass spectra were matched to the sequence of pENH2, accounting for 52% of inter-residue bond cleavages.
Figure 6. Localization of the phosphorylation site in pENH2 by top-down MS/MS.
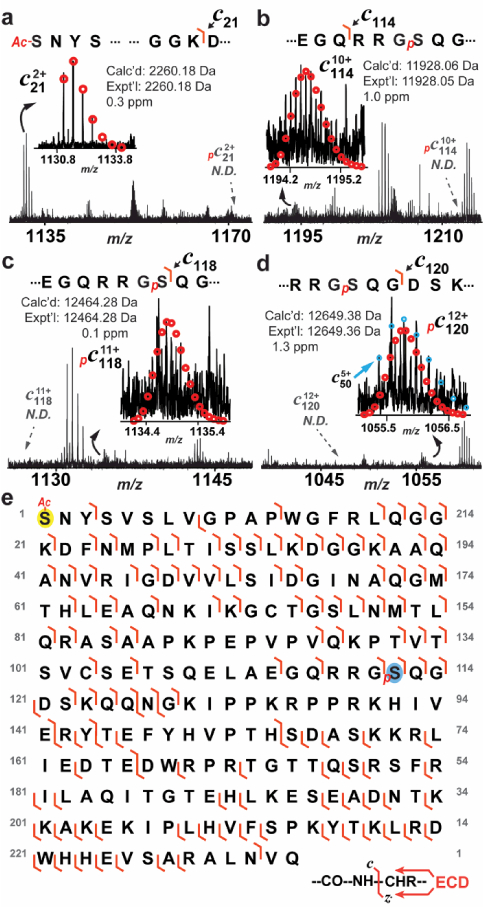
(a) Zoomed-in spectrum for c21 confirmed N-terminal acetylation of pENH2. (b) N-terminal fragments prior to c114 had no detectable mono-phosphorylated counterparts, suggesting that the phosphorylation site was located to the C-terminal of Gln114. (c) and (d) Zoomed- in spectra for c118 and c120 ion, respectively, showed only mono-phosphorylated ions, suggesting that amino acid sequence N-terminal to Ser118 is exclusively mono-phosphorylated. Circles represent the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned mass. N.D. not detected. (e) Fragmentation map shows the fragment ions that were matched to the sequences of pENH2 (78 c and 52 z· ions total). Ac denotes acetylation. p denotes phosphorylation.
3.5. cAMP increased within 5 min of tissue incubation at 4 °C
Given that Ser22/23 of cTnI are well-known target sites of protein kinase A (PKA), increased phosphorylation at these sites following tissue incubation at 4 °C or 22 °C suggests that PKA might be activated in the warm-up myocardium. To determine if PKA was activated, we examined the concentration of cAMP in tissue incubated at 4 °C. The tissue extracts were incubated with primary antibodies against cAMP, as well as cAMP-akaline phosphatase conjugates (cAMP-AP) (Figure 7a). Free cAMP in the sample competed with cAMP-AP for antibody binding in a concentration-dependent manner and, therefore, the free cAMP concentration was inversely proportional to the cAMP-AP concentration, which is measured via a colorimetric reaction with an AP substrate, para-Nitrophenylphosphate (pNpp), (Figure 7a). The cAMP concentration was normalized to total protein concentration prior to comparison between different groups.
Figure 7. The concentration of cAMP increased in tissue incubated at 4 °C.
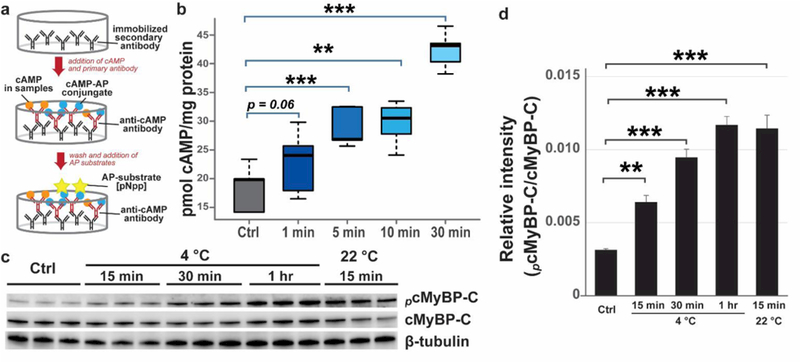
(a) Schematic of ELISA for quantitative analysis of cAMP concentration. AP, alkaline phosphatase. (b) Graph showing the concentration of cAMP during tissue defrosting at 4 °C. **p < 0.01, ***p < 0.001 by ANOVA. (c) Increased phosphorylation of cMyBP-C mediated by PKA after temperature treatment. Note tha t the the b lo t data are from the sa me b lot as F igure 5 and, thus, the load ing co ntro l is the sa me . (d) Histo gra m showing q ua ntificatio n o f p ho sp horylated c MyBP -C by Western b lo tting. Two-wa y ANOVA test was perfo rmed to e va lua te the sta tistica l signific a nce o f d iffere nce. ** p < 0.01, *** p < 0.001.
Based on the ELISA assays, cAMP concentration (nmol/ μg protein) increased dramatically upon tissue incubation at 4 °C (Figure 7b). The concentration of cAMP increased by nearly 50% within 5 min of tissue defrosting at 4 °C, and by about 100% after 30 min (Figure 7b). The increase in cAMP concentration in the myocardium supports activation of PKA in tissue incubated at 4 °C. This is highly consistent with the increased PKA-mediated phosphorylation of cTnI at Ser22/23 by both MS/MS and Western blotting analysis (Figure 5). To further validate increased PKA activity, we performed Western blotting analysis to evaluate the phosphorylation of cMyBP-C, a well-established substrate of PKA, using a PKA-substrate antibody. With 5 μg of total proteins being analyzed, this PKA-substrate antibody detected one major band between 130 and 250 kDa, and one weak band between 15 and 25 kDa (Supplemental Figure S9). Moreover, using a specific antibody against cMyBP-C, we found that the band detected by the PKA substrate antibody overlapped perfectly with the band detected by the cMyBP-C antibody (Supplemental Figure S9). Therefore, we inferred that the band between 130 and 250 kDa detected by the PKA-substrate antibody was phosphorylated cMyBP-C. Consistently, PKA-mediated phosphorylation of cMyBP-C also increased following temperature treatment (Figure 7c, d).
In addition, the phosphorylation of ENH2 and cTnI both increased with increased temperature, suggesting that the phosphorylation of ENH2 may be co-regulated with cTnI phosphorylation. Interestingly, the increase of cTnI and ENH2 phosphorylation and degradation can be partially suppressed by immersing the tissue in a buffer containing protease and phosphatase inhibitors, as well as 2.5 mM EDTA (Supplemental Figure S10). Nevertheless, the addition of a buffering solution containing protease and phosphatase inhibitors did not benefit the preservation of cTnT and RLC phosphorylation (Supplemental Figure S10). Further investigation of the buffer composition that can prevent artefactual changes of protein PTMs is critical for the accurate interrogation of cardiac protein PTMs changes associated with heart diseases.
We also found that frozen cardiac tissue can be stored in liquid nitrogen or −80 °C for up to 18 months with less than 10% changes in the phosphorylatio n of cTnI (Figure 8a–c), following the initial analysis (Figure 2). Even though the tissue stored at −80 °C appeared to have lower cTnI phosphorylation that was statistically significant, this difference ma y not result from the difference in the temperature of storage, because higher temperature was shown to increase cTnI phosphorylation (Figure 2). This may be because tissue from the initial analysis (control) wa spartially exposed to increased temperature and, therefore, the phosphorylation level was slightly higher. Additionally, we tried to mimic shipment conditions by storing the samples in liquid nitrogen, dry ice, and wet ice for 48 hrs. There was no significant difference in the cTnI phosphorylation observed after 48-hr “shipment” with liquid nitrogen or dry ice (Figure 8c–f). However, complete deterioration of the tissues occurred with wet ice storage after 48 hrs, and there was minimal amount of sarcomeric proteins recovered from the tissues, prohibiting proteomics analysis.
Figure 8. Investigation of the effects of different storage conditions on the phosphorylation of cTnI.
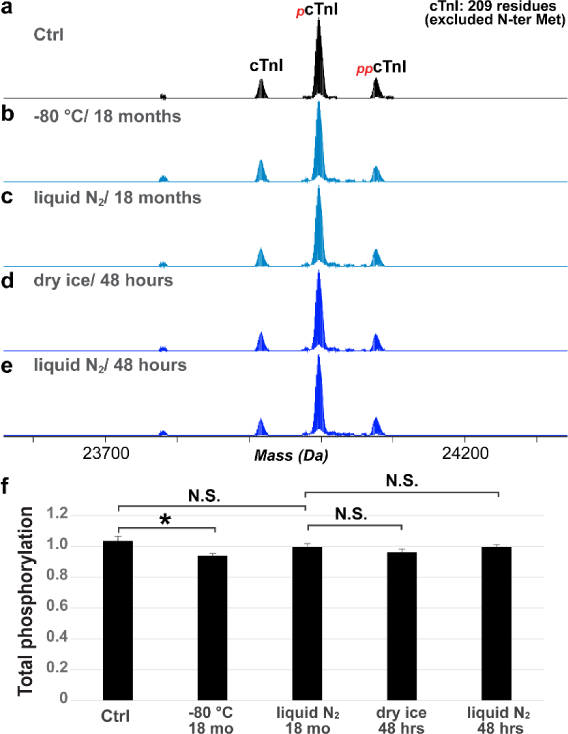
(a). Phosphorylation of cTnI in Ctrl (same as in Figure 2) compared to the cTnI phosphorylation level after storage at in −80 °C (b) and liquid nitrogen (c) for 18 months. (d, e) Phosphorylation of cTnI with storage in −80 °C and liquid nitrogen for 48 hrs after analysis shown in (c). (f) Quantification of the relative abundance of cTnI, mono- and bis-phosphorylated cTnI by MS. Two-way ANOVA was performed to evalute the statistical significance of difference. * p < 0.05, N.S. not statistically significant.
4. DISCUSSION
Protease and phosphatase inhibitors have been routinely included in buffer solutions for the preparation of biological samples to suppress protein degradation and de-phosphorylation and, therefore, it was expected to observe truncation and de-phosphorylation of a number of myofilament proteins upon incubation of myocardial tissue at either 4° or 22 °C. However, it was striking to find that cTnI and ENH2 phosphorylation actually increased upon tissue defrosting. cTnI is an important biomarker for myocardial infarction and a key regulator of muscle contraction and relaxation [10, 12, 34, 35]. cTnI forms a trimeric complex with cTnT and TnC for the positioning of Tpm on the actin filament in response to intracellular Ca2+ [36]. TnC binding of Ca2+ induces a conformational change in the troponin complex that leads to the repositioning of Tpm on the thin filament and relieves inhibition of actin- myosin interactions [36]. cTnI plays a central role in the regulation of contractility and the phosphorylation changes of cTnI at various sites have been implicated in animal models of heart disease and HF in humans [10, 12, 37].
cTnI is a well-established substrate of PKA in response to β-adrenergic activation in normal physiological response and disease states [34, 38]. PKA- mediated phosphorylation of cTnI at Ser22/23 decreases the Ca2+ sensitivity of the myofilament [34, 38]. Although Ser22/23 are the well-established sites phosphorylated by PKA, it has been found that these sites can also be phosphorylated by protein kinase G (PKG) or protein kinase C (PKC) [39–42]. Nevertheless, the finding that the concentration of cAMP, a direct PKA activator, increased rapidly in the cardiac tissue upon incubation of the tissue at 4 °C strongly supports the activation of PKA, which is likely responsible for the increased phosphorylation of cTnI at Ser22/23 upon increased temperature. PKA- mediated phosphorylation of cTnI represents an important mechanism for the regulation of contraction and relaxation in health and diseases. Studies have shown that phosphorylation of cTnI at Ser22/23 is decreased in human HF [14, 43–45]; however, others reported that cTnI phosphorylation increased rodent HF models [46]. While many factors can account for the differences in these findings, our study has demonstrated the temperature effect on cTnI phosphorylation, which may in part explain the discrepancy in the literature. In addition to cTnI, PKA also phosphorylates a number of proteins that are important regulators of contractility, including cMyBP-C and phospholamban (a regulator of the Ca2+ pump on the sarcoplasmic reticulum) [47–51]. Due to the large molecular weight of cMyBP-C, it remains challenging to directly study this protein using top-down proteomics. By employing Western blotting analysis, we also confirmed the increased PKA- mediated phosphorylated of cMyBP-C in the tissues following increased temperature. Recent advancements in multi-dimensional LC platform coupling serial size exclusion chromatography with reverse phase chromatography has demonstrated great promise in the top-down analysis of large proteins from the sarcomere [52].
In addition to PKA-mediated protein phosphorylation, PKC can phosphorylate cTnI at Ser22/23, Ser42/44, and Thr143 (excluded N-terminal Met) [39]. Depending on the PKC isoforms and the specific site, PKC-mediated phosphorylation of cTnI can exert various effects on the Ca2+ sensitivity, maximal force or cross-bridge cycling rate [53–56]. In addition, multiple studies have suggested that cTnI can be phosphorylated by PKG and p21-activated kinase 3 (PAK3) at Ser22/23 and Ser148 (excluded N-terminal Met), respectively [41, 42, 57]. However, Ser42/44, Ser148 and Thr143 were not identified in this study as the sites of phosphorylation, possibly due to low abundance of these PTMs. Previously, Zhang et al. reported an MS-based bottom- up method for identification of phosphorylation sites of cTnI, which were purified from the failing human hearts [58]. This study remains the current most sensitive cTnI phosphorylation assay, allowing for the identification of 14 phosphorylation sites [58].
ENH2 is a Z-disc protein that was just recently identified as a phosphoprotein [10]. In a swine model of acute myocardial infarction, Peng et al. identified concerted de-phosphorylation of cTnI, ENH2, and RLC following 90-min of ischemic injury [10]. The site of ENH2 phosphorylation was localized to Ser118 in the swine LV tissue [10], which is in agreement with the localization of ENH2 phosphorylation in human LV samples in the present study. In addition, Peng et al. identified a likely truncated proteoform of ENH2, the abundance of which increased following ischemic injury [10]. This suggests that ENH2 is unstable and prone to degradation, which is in high accordance with our finding that intact ENH2 became undetectable after 4 hr at 4 °C.
We also found that addition of a buffered solution containing protease/phosphatase inhibitors and 2.5 mM EDTA can partially suppress the increase of cTnI and ENH2 phosphorylation and degradation during the process of tissue defrosting. While protease inhibitors are likely to play a role in the inhibition of protein degradation, since EDTA is an Mg2+ chelating agent and Mg2+ is essential for the catalytic activity of protein kinases, including PKA [59, 60], the partial inhibition of PKA activity may be due to chelation of Mg2+ by EDTA.
In contrast to the phosphorylation of cTnI and ENH2, cTnT and RLC phosphorylation decreased in the myocardium with increased temperature. Previous studies have established Ser1 (excluding the N-terminal Met) as the site of phosphorylation on endogenous cTnT [31, 61, 62]. cTnT can also be phosphorylated at Thr203, Ser207, Thr212, Ser284, and Thr293 (excluded N-terminal Met) [31, 39, 63]. However, since multiply phosphorylated cTnT was not observed in our mass spectra, it is possible that cTnT proteoforms with these putative phosphorylation sites other than Ser1 were too low in abundance to be detected. The protein kinase responsible for cTnT phosphorylation at Ser1 was isolated and identified as casein kinase 2 (CK2) [61, 64]. It has been shown that cTnT phosphorylation at Ser1 has no effects on the Ca2+ sensitivity of actomyosin [65]. In skeletal muscle, troponin T (TnT) phosphorylation at Ser1 had no effects on the Ca2+ binding properties of the troponin complex [66], or on the interaction between TnT and Tpm [57]. The physiological role of constitutive phosphorylation of cTnT Ser1 therefore remains unclear; however, it has been suggested that phosphorylation of cTnT Ser1 may be involved in the stabilization of a contractile apparatus or its interaction with auxiliary proteins, rather than direct regulation of the contractile cycle [31].
Unlike cTnT phosphorylation at Ser1, phosphorylation of RLC at Ser14 has direct and significant impact on the contractile properties [67]. Phosphorylation of RLC at Ser14 has been shown to increase Ca2+ sensitivity, maximal force, and the rate of cross-bridge cycling [68]. The decreased phosphorylation of RLC has been reported in human end-stage failing hearts [69]. Recently, cardiac-specific myosin light chain kinase (cMLK) has been identified as the primary kinase responsible for cardiac RLC phosphorylation at Ser14 [70]. It remains unclear whether the decreased phosphorylation of RLC and cTnT is a result of decreased kinase expression/activity, or increased phosphatase expression/activity. While immersing the tissue in a buffer solution can partially suppress changes in cTnI and ENH2 PTMs, the decline of phosphorylation in cTnT and RLC persisted even though phosphatase inhibitors were included in the buffer. It is possible that 50 mM sodium fluoride, a Ser phosphatase inhibitor, is insufficient to suppress the artefactual decline in cTnT and RLC phosphorylation. Further studies on the composition of buffering solution for the preservation of protein PTMs are important for the accurate dissection of molecular changes in the failing and non- failing human hearts.
In addition to the evaluation of increased temperature, we also investigated the effects of tissue storage and shipment strategies on the phosphorylation of cTnI. We found that total phosphorylation of cTnI was not markedly affected with storage at −80 °C or liquid nitrogen for up to 18 months. There was also no significant changes in cTnI phosphorylation when stored in liquid nitrogen or dry ice for 48 hrs, mimicking most shipment conditions. As expected, storage of the tissue using wet ice led to complete tissue deterioration, prohibiting proteomics analysis. Even though −80 °C and liquid nitrogen storage did not have a profound effects on the PTM change, it is important to recognize that small pieces of tissue (< 10 mg) are likely more susceptible to change of temperature, and therefore, liquid nitrogen remains the best option for shipment of small tissues and prolonged tissue storage.
Herein, we have demonstrated the impact of temperature on the preservation of protein PTMs in human cardiac tissue. Importantly, this study underscores the power of top-down MS-based proteomics for the assessment of tissue quality. Based on the fact that defrosted tissue exhibited a various degree of protein degradation on cTnI, ENH2, and cTnT, the truncated proteoforms of multiple myofilament and associated proteins can collectively be indicative of poor tissue quality. It is important to point out that many truncated proteoforms of cTnI and cTnT have similar molecular weight as the intact proteoforms, therefore making the detection of such truncated proteoforms difficult and ambiguous via the gel electrophoresis approach. Moreover, although the conventional bottom- up approach is capable of identifying and quantifying thousands of proteins from a complex mixture [71, 72], due to limited sequence coverage as a result of proteolytic digestion, it remains challenging to distinguish highly homologous protein isoforms and proteolytic products with small truncation. On the contrary, top-down MS-based proteomics approach permits the analysis of intact proteins and unambiguously reveals the existing proteoforms including those with small truncation <1000 Da, as well as those with distinct sequence variations [73]. In addition, high-resolution top-down MS allows for the determination of accurate molecular weight with less than 10 ppm discrepancy. This permits the identification of the truncated proteoforms based on their accurate molecular weights and the mass differences from their intact counterparts.
In summary, our study highlights the importance of tissue handling in revealing accurate molecular alterations in the human failing versus non- failing hearts. To the best of our knowledge, this is the first study to investigate the impact of temperature on the PTM changes of the major myofilament and associated proteins. By employing a high-resolution top-down proteomics approach, we have observed artefactual increases of cTnI and ENH2 phosphorylation, and artefactual decreases of cTnT and RLC phosphorylation, which demonstrated that sarcomeric protein PTMs were differentially affected by temperature. The sites of protein increased phosphorylation on cTnI were localized to Ser22/23, which in accordance with increased cAMP concentration in the warmed-up myocardium, suggests the activation of PKA upon increased temperature. Moreover, this study revealed degraded cTnI, ENH2, and cTnT upon tissue increased temperature, which can be indicative of poor tissue quality for future studies employing human cardiac tissue.
Supplementary Material
HIGHLIGHTS.
Differential regulation of protein PTMs in human tissue with increased temperature.
Phosphorylation of cTnT and RLC decreased following increased temperature.
Phosphorylation of cTnI and ENH2 increased following increased temperature.
Increased cTnI phosphorylation resulted from increased cAMP and activation of PKA.
Top-down proteomics is a powerful method for assessing cardiac tissue quality.
ACKNOWLEDGEMENT
Financial support was kindly provided by NIH R01 HL109810 and R01 HL096971 (to Y.G.). Y. G. would also like to acknowledge NIH R01 GM117058, R01 GM125085, and S10 OD018475. W.C. would like to acknowledge AHA pre-doctoral fellowship (#17PRE33660224).
Abbreviations list
- AP
alkaline phosphatase
- cAMP
cyclic adenosine monophosphate
- CK2
casein kinase 2
- ECD
electron capture dissociation
- ELC
essential light chain
- ELISA
enzyme- linked Immunosorbent Assay
- ENH2
enigma homolog isoform 2
- pENH2
mono-phosphorylated ENH2
- ppENH2
bis-phosphorylated ENH2
- FT-ICR
Fourier transform- ion cyclotron resonance
- HF
heart failure
- HPLC
high pressure liquid chromatography
- LC
liquid chromatography
- LTQ
linear ion trap
- cMLK
cardiac-specific myosin light chain kinase
- cMyBP-C
cardiac myosin binding protein C
- MS
mass spectrometry
- MS/MS
tandem MS
- PKA
protein kinase A
- PKG
protein kinase G
- pNpp
para-Nitrophenylphosphate
- PP2A
protein phosphatase 2A
- PTMs
post-translational modifications
- Q-TOF
quadrupole time-of-flight
- RLC
regulatory light chain
- pRLC
mono-phosphorylated RLC
- RPC
reverse phase chromatography
- TFA
trifluoroacetic acid
- cTnI
cardiac troponin I
- pcTnI
mono-phosphorylated cTnI
- ppcTnI
bis-phosphorylated cTnI
- cTnT
cardiac troponin T
- pcTnT
mono-phosphorylated cTnT
- Tpm
tropomyosin
- pTpm
mono-phosphorylated Tpm
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
None.
REFERENCES
- [1].McMurray JJ, Pfeffer MA, Heart failure, Lancet 365(9474) (2005) 1877–89. [DOI] [PubMed] [Google Scholar]
- [2].Bui AL, Horwich TB, Fonarow GC, Epidemiology and risk profile of heart failure, Nat Rev Cardiol 8(1) (2011) 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jiménez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, N ichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB, A.H.A.S.C.a.S.S. Subcommittee, Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association, Circulation 133(4) (2016) e38–e360. [DOI] [PubMed] [Google Scholar]
- [4].Milani-Nejad N, Janssen PM, Small and large animal models in cardiac contraction research: advantages and disadvantages, Pharmacol Ther 141(3) (2014) 235–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Houser SR, Margulies KB, Murphy AM, Spinale FG, Francis GS, Prabhu SD, Rockman HA, Kass DA, Molkentin JD, Sussman MA, Koch WJ, Koch W, C.u.o.C.C. American Heart Association Council on Basic Cardiovascular Sciences, and Council on Functional Genomics and Translational Biology, Animal models of heart failure: a scientific statement from the American Heart Association, Circ Res 111(1) (2012) 131–50. [DOI] [PubMed] [Google Scholar]
- [6].Walker LA, Medway AM, Walker JS, Cleveland JC, Buttrick PM, Tissue procurement strategies affect the protein biochemistry of human heart samples, J Muscle Res Cell Motil 31(5–6) (2011) 309–14. [DOI] [PubMed] [Google Scholar]
- [7].Hwang PM, Sykes BD, Targeting the sarcomere to correct muscle function, Nat Rev Drug Discov 14(5) (2015) 313–28. [DOI] [PubMed] [Google Scholar]
- [8].Yuan C, Solaro RJ, Myofilament proteins: From cardia c disorders to proteomic changes, Proteomics Clin Appl 2(6) (2008) 788–99. [DOI] [PubMed] [Google Scholar]
- [9].Cai W, Tucholski TM, Gregorich ZR, Ge Y, Top-down Proteomics: Technology Advancements and Applications to Heart Diseases, Expert Rev Proteomics 13(8) (2016) 717–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Peng Y, Gregorich ZR, Valeja SG, Zhang H, Cai W, Chen Y-C, Guner H, Chen AJ, Schwahn DJ, Hacker TA, Liu X, Ge Y, Top-down Proteomics Reveals Concerted Reductions in Myofilament and Z-disc Protein Phosphorylation after Acute Myocardial Infarction, Molecular & Cellular Proteomics 13(10) (2014) 2752–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].de Tombe PP, Solaro RJ, Integration of cardiac myofilament activity and regulation with pathways signaling hypertrophy and failure, Ann Biomed Eng 28(8) (2000) 991–1001. [DOI] [PubMed] [Google Scholar]
- [12].Layland J, Solaro RJ, Shah AM, Regulation of cardiac contractile function by troponin I phosphorylation, Cardiovasc Res 66(1) (2005) 12–21. [DOI] [PubMed] [Google Scholar]
- [13].Yar S, Monasky MM, Solaro RJ, Maladaptive modifications in myofilament proteins and triggers in the progression to heart failure and sudden death, Pflugers Arch 466(6) (2014) 1189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Messer AE, Jacques AM, Marston SB, Troponin phosphorylation and regulatory function in human heart muscle: dephosphorylation of Ser23/24 on troponin I could account for the contractile defect in end-stage heart failure, J Mol Cell Cardiol 42(1) (2007) 247–59. [DOI] [PubMed] [Google Scholar]
- [15].Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, K levitsky R, Seidman CE, Seidman JG, Robbins J, Cardiac myosin-binding protein-C phosphorylation and cardiac function, Circ Res 97(11) (2005) 1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sheikh F, Ouyang K, Campbell SG, Lyon RC, Chuang J, Fitzsimons D, Tangney J, Hidalgo CG, Chung CS, Cheng H, Dalton ND, Gu Y, Kasahara H, Ghassemian M, O mens JH, Peterson KL, Granzier HL, Moss RL, McCulloch AD, Chen J, Mouse and computational models link Mlc2v dephosphorylation to altered myosin kinetics in early cardiac disease, J Clin Invest 122(4) (2012) 1209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pyle WG, Solaro RJ, At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function, Circ Res 94(3) (2004) 296–305. [DOI] [PubMed] [Google Scholar]
- [18].Kirk JA, Holewinski RJ, Kooij V, Agnetti G, Tunin RS, Witayavanitkul N, de Tombe PP, Gao WD, Van Eyk J, Kass DA, Cardiac resynchronization sensitizes the sarcomere to calcium by reactivating GSK-3β, J Clin Invest 124(1) (2014) 129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gregorich ZR, Ge Y, Top-down proteomics in health and disease: challenges and opportunities, Proteomics 14(10) (2014) 1195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McLafferty FW, Breuker K, Jin M, Han X, Infusini G, Jiang H, Kong X, Begley TP, Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics, FEBS J 274(24) (2007) 6256–68. [DOI] [PubMed] [Google Scholar]
- [21].Catherman AD, Skinner OS, Kelleher NL, Top Down proteomics: facts and perspectives, Biochem Biophys Res Commun 445(4) (2014) 683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Smith LM, Kelleher NL, C.f.T.D. Proteomics, Proteoform: a single term describing protein complexity, Nat Methods 10(3) (2013) 186–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Savaryn JP, Catherman AD, Thomas PM, Abecassis MM, Kelleher NL, The emergence of top-down proteomics in clinical research, Genome Med 5(6) (2013) 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang J, Guy MJ, Norman HS, Chen YC, Xu Q, Dong X, Guner H, Wang S, Kohmoto T, Young KH, Moss RL, Ge Y, Top-down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure, J Proteome Res 10(9) (2011) 4054–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gregorich ZR, Peng Y, Lane NM, Wolff JJ, Wang S, Guo W, Guner H, Doop J, Hacker TA, Ge Y, Comprehensive assessment of chamber-specific and transmural heterogeneity in myofilament protein phosphorylation by top-down mass spectrometry, J Mol Cell Cardiol 87 (2015) 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gregorich ZR, Peng Y, Cai W, Jin Y, Wei L, Chen AJ, McK iernan SH, Aiken JM, Moss RL, Diffee GM, Ge Y, Top-Down Targeted Proteomics Reveals Decrease in Myosin Regulatory Light-Chain Phosphorylation That Contributes to Sarcopenic Muscle Dysfunction, J Proteome Res (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cai W, Guner H, Gregorich ZR, Chen AJ, Ayaz-Guner S, Peng Y, Valeja SG, Liu X, Ge Y, MASH Suite Pro: A Comprehensive Software Tool for Top-down Proteomics, Mol Cell Proteomics (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Guner H, Close PL, Cai W, Zhang H, Peng Y, Gregorich ZR, Ge Y, MASH Suite: a user-friendly and versatile software interface for high-resolution mass spectrometry data interpretation and visualization, J Am Soc Mass Spectrom 25(3) (2014) 464–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yeung YG, Stanley ER, A solution for stripping antibodies from polyvinylidene fluoride immunoblots for multiple reprobing, Anal Biochem 389(1) (2009) 89–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Geeves MA, Hitchcock-DeGregori SE, Gunning PW, A systematic nomenclature for mammalian tropomyosin isoforms, J Muscle Res Cell Motil 36(2) (2015) 147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Katrukha IA, Gusev NB, Enigmas of cardiac troponin T phosphorylation, J Mol Cell Cardiol 65 (2013) 156–8. [DOI] [PubMed] [Google Scholar]
- [32].Scruggs SB, Reisdorph R, Armstrong ML, Warren CM, Reisdorph N, Solaro RJ, Buttrick PM, A novel, in-solution separation of endogenous cardiac sarcomeric proteins and identification of distinct charged variants of regulatory light chain, Mol Cell Proteomics 9(9) (2010) 1804–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zubarev RA, Electron-capture dissociation tandem mass spectrometry, Curr Opin Biotechnol 15(1) (2004) 12–6. [DOI] [PubMed] [Google Scholar]
- [34].Solaro RJ, Moir AJ, Perry SV, Phosphorylation of troponin I and the inotropic effect of adrenaline in the perfused rabbit heart, Nature 262(5569) (1976) 615–7. [DOI] [PubMed] [Google Scholar]
- [35].Jaffe AS, Ravkilde J, Roberts R, Naslund U, Apple FS, Galvani M, Katus H, It’s time for a change to a troponin standard, Circulation 102(11) (2000) 1216–20. [DOI] [PubMed] [Google Scholar]
- [36].Solaro RJ, Rarick HM, Troponin and tropomyosin: proteins that switch on and tune in the activity of cardiac myofilaments, Circ Res 83(5) (1998) 471–80. [DOI] [PubMed] [Google Scholar]
- [37].Sasse S, Brand NJ, Kyprianou P, Dhoot GK, Wade R, Arai M, Periasamy M, Yacoub MH, Barton PJ, Troponin I gene expression during human cardiac development and in end-stage heart failure, Circ Res 72(5) (1993) 932–8. [DOI] [PubMed] [Google Scholar]
- [38].Zhang R, Zhao J, Mandveno A, Potter JD, Cardiac troponin I phosphorylation increases the rate of cardiac muscle relaxation, Circ Res 76(6) (1995) 1028–35. [DOI] [PubMed] [Google Scholar]
- [39].Noland TA, Raynor RL, Kuo JF, Identification of sites phosphorylated in bovine cardiac troponin I and troponin T by protein kinase C and comparative substrate activity of synthetic peptides containing the phosphorylation sites, J Biol Chem 264(34) (1989) 20778–85. [PubMed] [Google Scholar]
- [40].Noland TA, Guo X, Raynor RL, Jideama NM, Averyhart-Fullard V, Solaro RJ, Kuo JF, Cardiac troponin I mutants. Phosphorylation by protein kinases C and A and regulation of Ca(2+)-stimulated MgATPase of reconstituted actomyosin S-1, J Biol Chem 270(43) (1995) 25445–54. [DOI] [PubMed] [Google Scholar]
- [41].Blumenthal DK, Stull JT, Gill GN, Phosphorylation of cardiac troponin by guanosine 3′:5′-monophosphate-dependent protein kinase, J Biol Chem 253(2) (1978) 324–6. [PubMed] [Google Scholar]
- [42].Yuasa K, Michibata H, O mori K, Yanaka N, A novel interaction of cGMP-dependent protein kinase I with troponin T, J Biol Chem 274(52) (1999) 37429–34. [DOI] [PubMed] [Google Scholar]
- [43].Purcell IF, Bing W, Marston SB, Functional analysis of human cardiac troponin by the in vitro motility assay: comparison of adult, foetal and failing hearts, Cardiovasc Res 43(4) (1999) 884–91. [DOI] [PubMed] [Google Scholar]
- [44].van der Velden J, Papp Z, Zaremba R, Boontje NM, de Jong JW, Owen VJ, Burton PB, Goldmann P, Jaquet K, Stienen GJ, Increased Ca2+-sensitivity of the contractile apparatus in end-stage human heart failure results from altered phosphorylation of contractile proteins, Cardiovasc Res 57(1) (2003) 37–47. [DOI] [PubMed] [Google Scholar]
- [45].Bodor GS, Oakeley AE, Allen PD, Crimmins DL, Ladenson JH, Anderson PA, Troponin I phosphorylation in the normal and failing adult human heart, Circulation 96(5) (1997) 1495–500. [DOI] [PubMed] [Google Scholar]
- [46].Belin RJ, Sumandea MP, Allen EJ, Schoenfelt K, Wang H, Solaro RJ, de Tombe PP, Augmented protein kinase C-alpha- induced myofilament protein phosphorylation contributes to myofilament dysfunction in experimental congestive heart failure, Circ Res 101(2) (2007) 195–204. [DOI] [PubMed] [Google Scholar]
- [47].Kranias EG, Garvey JL, Srivastava RD, Solaro RJ, Phosphorylation and functional modifications of sarcoplasmic reticulum and myofibrils in isolated rabbit hearts stimulated with isoprenaline, Biochem J 226(1) (1985) 113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].James P, Inui M, Tada M, Chiesi M, Carafoli E, Nature and site of phospholamban regulation of the Ca2+ pump of sarcoplasmic reticulum, Nature 342(6245) (1989) 90–2. [DOI] [PubMed] [Google Scholar]
- [49].Gautel M, Zuffardi O, Freiburg A, Labeit S, Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction?, EMBO J 14(9) (1995) 1952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].de Tombe PP, Myosin binding protein C in the heart, Circ Res 98(10) (2006) 1234–6. [DOI] [PubMed] [Google Scholar]
- [51].Colson BA, Bekyarova T, Locher MR, Fitzsimons DP, Irving TC, Moss RL, Protein kinase A-mediated phosphorylation of cMyBP-C increases proximity of myosin heads to actin in resting myocardium, Circ Res 103(3) (2008) 244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cai W, Tucholski T, Chen B, Alpert AJ, McIlwain S, Kohmoto T, Jin S, Ge Y, Top-Down Proteomics of Large Proteins up to 223 kDa Enabled by Serial Size Exclusion Chromatography Strategy, Anal Chem 89(10) (2017) 5467–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kentish JC, McCloskey DT, Layland J, Palmer S, Leiden JM, Martin AF, Solaro RJ, Phosphorylation of troponin I by protein kinase A accelerates relaxation and crossbridge cycle kinetics in mouse ventricular muscle, Circ Res 88(10) (2001) 1059–65. [DOI] [PubMed] [Google Scholar]
- [54].Takeishi Y, Chu G, Kirkpatrick DM, Li Z, Wakasaki H, Kranias EG, King GL, Walsh RA, In vivo phosphorylation of cardiac troponin I by protein kinase Cbeta2 decreases cardiomyocyte calcium responsiveness and contractility in transgenic mouse hearts, J Clin Invest 102(1) (1998) 72–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Westfall MV, Borton AR, Role of troponin I phosphorylation in protein kinase C-mediated enhanced contractile performance of rat myocytes, J Biol Chem 278(36) (2003) 33694–700. [DOI] [PubMed] [Google Scholar]
- [56].Bowling N, Walsh RA, Song G, Estridge T, Sandusky GE, Fouts RL, Mintze K, Pickard T, Roden R, Bristow MR, Sabbah HN, Mizrahi JL, Gromo G, King GL, Vlahos CJ, Increased protein kinase C activity and expression of Ca2+-sensitive isoforms in the failing human heart, Circulation 99(3) (1999) 384–91. [DOI] [PubMed] [Google Scholar]
- [57].Buscemi N, Foster DB, Neverova I, Van Eyk JE, p21-activated kinase increases the calcium sensitivity of rat triton-skinned cardiac muscle fiber bundles via a mechanism potentially involving novel phosphorylation of troponin I, Circ Res 91(6) (2002) 509–16. [DOI] [PubMed] [Google Scholar]
- [58].Zhang P, K irk JA, Ji W, dos Remedios CG, Kass DA, Van Eyk JE, Murphy AM, Multiple reaction monitoring to identify site-specific troponin I phosphorylated residues in the failing human heart, Circulation 126(15) (2012) 1828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Adams JA, Kinetic and catalytic mechanisms of protein kinases, Chem Rev 101(8) (2001) 2271–90. [DOI] [PubMed] [Google Scholar]
- [60].Knape MJ, Ahuja LG, Bertinetti D, Burghardt NC, Zimmermann B, Taylor SS, Herberg FW, Divalent Metal Ions Mg²⁺ and Ca²⁺ Have Distinct Effects on Protein Kinase A Activity and Regulation, ACS Chem Biol 10(10) (2015) 2303–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Villar-Palasi C, Kumon A, Purification and properties of dog cardiac troponin T kinase, J Biol Chem 256(14) (1981) 7409–15. [PubMed] [Google Scholar]
- [62].Zhang J, Zhang H, Ayaz-Guner S, Chen YC, Dong X, Xu Q, Ge Y, Phosphorylation, but not alternative splicing or proteolytic degradation, is conserved in human and mouse cardiac troponin T, Biochemistry 50(27) (2011) 6081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sumandea MP, Pyle WG, Kobayashi T, de Tombe PP, Solaro RJ, Identification of a functionally critical protein kinase C phosphorylation residue of cardiac troponin T, J Biol Chem 278(37) (2003) 35135–44. [DOI] [PubMed] [Google Scholar]
- [64].Dobrovol’skiĭ AB, Risnik VV, Gusev NB, [Troponin T kinase: possible relationship to casein kinases of the G type], Biokhimiia 46(6) (1981) 1006–14. [PubMed] [Google Scholar]
- [65].Tobacman LS, Lee R, Isolation and functional comparison of bovine cardiac troponin T isoforms, J Biol Chem 262(9) (1987) 4059–64. [PubMed] [Google Scholar]
- [66].Gusev NB, [Troponin from the myocardium and skeletal muscles: structure and properties], Biokhimiia 51(12) (1986) 1993–2009. [PubMed] [Google Scholar]
- [67].Scruggs SB, Solaro RJ, The significance of regulatory light chain phosphorylation in cardiac physiology, Arch Biochem Biophys 510(2) (2011) 129–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Olsson MC, Patel JR, Fitzsimons DP, Walker JW, Moss RL, Basal myosin light chain phosphorylation is a determinant of Ca2+ sensitivity of force and activation dependence of the kinetics of myocardial force development, Am J Physiol Heart Circ Physiol 287(6) (2004) H2712–8. [DOI] [PubMed] [Google Scholar]
- [69].van der Velden J, Papp Z, Boontje NM, Zaremba R, de Jong JW, Janssen PM, Hasenfuss G, Stienen GJ, The effect of myosin light chain 2 dephosphorylation on Ca2+ -sensitivity of force is enhanced in failing human hearts, Cardiovasc Res 57(2) (2003) 505–14. [DOI] [PubMed] [Google Scholar]
- [70].Chan JY, Takeda M, Briggs LE, Graham ML, Lu JT, Horikoshi N, Weinberg EO, Aoki H, Sato KR Chien, H. Kasahara, Identification of cardiac-specific myosin light chain kinase, Circ Res 102(5) (2008) 571–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chang YH, Ye L, Cai W, Lee Y, Guner H, Kamp TJ, Zhang J, Ge Y, Quantitative proteomics reveals differential regulation of protein expression in recipient myocardium after trilineage cardiovascular cell transplantation, Proteomics 15(15) (2015) 2560–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yang L, Gregorich ZR, Cai W, Zhang P, Young B, Gu Y, Zhang J, Ge Y, Quantitative Proteomics and Immunohistochemistry Reveal Insights into Cellular and Molecular Processes in the Infarct Border Zone One Month after Myocardial Infarction, J Proteome Res 16(5) (2017) 2101–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gregorich ZR, Cai W, Lin Z, Chen AJ, Peng Y, Kohmoto T, Ge Y, Distinct sequences and post-translational modifications in cardiac atrial and ventricular myosin light chains revealed by top-down mass spectrometry, J Mol Cell Cardiol 107 (2017) 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


