Abstract
Objectives
We evaluated the automatic prediction of obstructive disease from myocardial perfusion imaging (MPI) by deep learning as compared to total perfusion deficit (TPD).
Background
Deep convolutional neural networks trained with large multi-center population may provide improved prediction of per-patient and per-vessel coronary artery disease (CAD) from SPECT MPI.
Methods
1638 patients (67% males) without known CAD, undergoing stress 99mTc-sestamibi or tetrofosmin MPI with new generation solid-state scanners in 9 different sites, with invasive coronary angiography performed within 6 months of MPI, were studied. Obstructive disease was defined as ≥ 70% narrowing of coronary arteries (≥ 50% for left main). Left ventricular (LV) myocardium was segmented using clinical nuclear cardiology software and verified by an expert reader. Stress TPD was computed using gender- and camera-specific normal limits. Deep learning was trained using raw and quantitative polar maps and evaluated for prediction of obstructive stenosis in a stratified 10-fold cross validation procedure.
Results
1018 (62%) patients and 1797 out of 4914 (37%) arteries had obstructive disease. Area under the receiver operating characteristics curve (AUC) for disease prediction by deep learning was higher than for TPD (per-patient: 0.80 vs. 0.78, per-vessel 0.76 vs. 0.73, P < 0.01). With deep learning threshold set to the same specificity as TPD, per-patient sensitivity improved from 79.8% (TPD) to 82.3% (deep learning) (P < 0.05), and per-vessel sensitivity improved from 64.4% (TPD) to 69.8% (deep learning) (P < 0.01).
Conclusions
Deep learning has the potential to improve automatic interpretation of MPI as compared to current clinical methods.
Keywords: Obstructive coronary artery disease, SPECT myocardial perfusion imaging, deep learning, convolutional neural network
INTRODUCTION
SPECT myocardial perfusion imaging (MPI) is a widely used technique for the diagnosis of coronary artery disease (CAD), with more than 7 million scans performed annually in the United States alone (1). Quantification of relative perfusion using normal databases allows to define the total perfusion deficit (TPD) at stress and rest (2). TPD quantification has been established to be equivalent to visual expert reading for the detection of obstructive CAD (3,4), while providing near automation (5–7).
Machine learning helps computers learn and develop rules, without having to be instructed every step of the way by human programmers (8). Deep learning is a powerful new machine learning tool, with breakthrough applications in disease detection and classification (9–14). It may play an important role in emerging artificial intelligence tools for cardiovascular medicine (15–17). A class of artificial neural network algorithms, deep learning uses more layers than traditional approaches (9,12), and thus is especially well suited for large, diverse, and complex datasets (15,17). In contrast to conventional machine learning that typically require pre-defined image measurements to characterize the information contained in the raw input images, deep learning can absorb the image measurement engineering directly into a learning step, while processing the data in its natural form (9,13). In this study, we aimed to explore the potential of deep learning for prediction of obstructive coronary artery disease from SPECT MPI, as compared to the standard quantitative analysis (2). SPECT MPI was acquired in a large multicenter registry on latest generation scanners.
METHODS
Study Population
The study population comprised of 1638 patients (67% males) referred for SPECT MPI from 2008 to 2015, at 9 national and international (Canada, Switzerland, Israel) sites. All patients were without prior myocardial infarction, percutaneous coronary intervention or coronary artery bypass graft surgery and all underwent a clinically indicated invasive coronary angiography (ICA) within 180 days of MPI. De-identified images and ICA correlations from each site were transferred to Cedars-Sinai Medical Center. Clinical patient characteristics are listed in Table 1. The study was approved by the institutional review boards at each participating institution. The data has been collected under NIH sponsored REgistry of Fast Myocardial Perfusion Imaging with NExt generation SPECT (REFINE SPECT).
Table 1.
Baseline characteristics of the studied population.
| No obstructive coronary artery disease | Obstructive coronary artery disease | P-value | |
|---|---|---|---|
|
| |||
| Number, n | 620 | 1018 | |
| One-vessel disease, n (%) | - | 466 (46) | |
| Two-vessel disease, n (%) | - | 325 (32) | |
| Triple-vessel disease, n (%) | - | 227 (22) | |
|
| |||
| Age (years) | 62.6 ± 11.7 | 65.2 ± 11.0 | < 0.001 |
|
| |||
| Male (%) | 55.7 | 73.7 | < 0.01 |
|
| |||
| Female (%) | 44.3 | 26.7 | < 0.01 |
|
| |||
| Diabetes Mellitus (%) | 25.8 | 31.8 | < 0.01 |
|
| |||
| Hypertension (%) | 66.3 | 71.6 | < 0.05 |
|
| |||
| Dyslipidemia (%) | 58 | 65.3 | < 0.01 |
|
| |||
| Smoking (%) | 21.1 | 23.5 | = 0.27 |
|
| |||
| Exercise MPI (%) | 39.2 | 38.9 | = 0.91 |
|
| |||
| Pharmacological MPI (%) | 60.8 | 61.1 | = 0.91 |
Image Acquisition
We analyzed only stress MPI images—99mTc-sestamibi (1469 patients [90%]) or 99mTc-tetrofosmin (169 [10%])—performed with high-efficiency solid-state SPECT scanners. Four sites (1104 [67%]) used the D-SPECT scanner (Spectrum-Dynamics, Haifa, Israel) (18), and five sites used Discovery NM530c or NM/CT570c scanners (534 [33%]) (GE Healthcare, Haifa, Israel) (19). Among the study patients, 1526 (93%) underwent same day stress and rest image acquisition, 65 (4%) had stress and rest images acquired in different days, and 47 (3%) had only stress images. The mean ± standard deviation weight-adjusted stress dose was 673 ± 458 MBq (18.2 ± 12.4 mCi).
Patients underwent either symptom-limited Bruce protocol treadmill exercise testing (639 [39%]) or pharmacologic stress (999 [61%]), with radiotracer injection at peak exercise or during maximal hyperemia, respectively. Upright (DSPECT) and supine (GE 530c/570c) stress imaging began 15–60 min after stress, and lasted 4–6 min. Images in other positions were not used in this analysis as the goal of this study was to utilize the default simplest imaging protocol used in clinical routine. Reconstructed images were generated from the list mode data by vendor-recommended iterative reconstruction optimized for each scanner (18,20). No attenuation or scatter correction was applied.
Invasive Coronary Angiography
ICA was performed according to standard clinical protocols within 6 months of the MPI examination. All coronary angiograms were visually interpreted by an on-site cardiologist. Luminal diameter narrowing of 50% or greater of the left main artery, or of 70% or greater in the left anterior descending (LAD), left circumflex (LCx) or right coronary arteries (RCA), were considered significant and were used as the gold standard for obstructive CAD. Left main stenosis was attributed to both LAD and LCx vessels.
Image processing
Left ventricular (LV) myocardial contours were computed using standard Quantitative Perfusion SPECT (QPS) software at Cedars-Sinai Medical Center (2). LV contours were verified by one nuclear medicine technologist with more than 15 years of dedicated experience in nuclear cardiology, who was blinded to any clinical findings. When needed, the reader corrected the gross initial LV localization, the LV mask (region containing the left ventricle), and the valve plane position (7,21).
Automated myocardial perfusion SPECT quantification
An ellipsoidal model and contour of the myocardium derived from quality-controlled LV contours were used by the standard QPS algorithm to extract polar map samples from the raw images with 10° angular resolution, and were used to generate tracer count distributions normalized to the maximal counts (raw polar map). Images were quantified separately using their respective tracer, gender and camera-specific normal limits. Subsequently, two quantitative perfusion polar maps were computed: blackout map and total perfusion deficit (TPD) map.
Blackout polar map
To generate the blackout polar map, the perfusion defect extent was outlined by blacking-out raw polar map samples that were below the abnormality threshold of 2.5 standard deviations (5).
TPD polar map
To generate the quantitative polar TPD map, comparison to normal limits was used to estimate the perfusion deficit per polar map sample. The perfusion deficit map displays individual hypoperfusion severity for each polar sample normalized to 0–4 range (2). The total perfusion deficit is subsequently defined as the ratio (expressed as a percentage) of the summed normalized severities for the whole myocardium (per-patient) or for the vascular region (per-vessel) divided by the total theoretic maximum total perfusion deficit (no visible radiotracer uptake in the myocardium), for the whole polar map (2).
Images for the raw polar map and for the two quantitative perfusion polar maps were automatically generated and exported in 64 × 64 resolution to evaluate the deep learning approach. Standard clinical TPD measures (per-patient and per-vessel) were obtained for comparison to deep learning.
Deep learning model
The overall process is illustrated in Figure 1. Prediction of obstructive CAD was accomplished by a deep convolutional neural network –a type of neural network inspired by the organization of visual cortex and proposed for image analysis (9). The network was arranged in two stages. The first stage extracts features from images. The network connects directly with polar map pixels via one convolutional layer (bank of image filters), followed by maximum pooling filters -filters that retain only the maximum value from a 2×2 input, in order to compensate for small image shifts and distortions (9). These operations are repeated 3 times in sequence to extract image features; the first pass detects low-level features, e.g. the local count changes for each image pixel, while the two subsequent passes allow for identification of higher-level features, e.g. describing regional perfusion defects. The feature maps created by these filtering sequences, reflected in the network by three feature extraction units, are then passed along to the second stage. This stage comprises three fully connected layers (arrays of neurons connected to each neuron in the previous layer), followed by three parallel outputs—one per coronary territory. The fully connected layers transform the image features into the final LAD, LCx and RCA scores, by adjusting weights for neuron activations during training.
FIGURE 1. Deep learning prediction of obstructive coronary artery disease from MPI.
A deep convolutional neural network trained from obstructive stenosis correlations by invasive coronary angiography was used to estimate the probability of obstructive coronary artery disease in LAD, LCx and RCA territories. The maximum probability was used as the probability of patient disease. FC: fully connected layer, MPI: SPECT myocardial perfusion imaging, ReLU: rectified linear unit (linear function mapping input to output values with a threshold).
Prediction of per-vessel/per-patient disease
Deep learning computes a probability of obstructive CAD in each vessel without pre-defined subdivision of the polar map. During training, the feature extraction units learn to recognize key polar map features, and the fully connected layers learn how to combine these features to predict per-vessel disease, by minimizing the average error between the predicted per-vessel probabilities and the disease location (Online Appendix 1) as defined by invasive angiography. Therefore, multivessel disease prediction is based on the patterns of predicted probabilities for each vessel. During testing, the maximum probability per artery territory is used as the per-patient score (Figure 1).
Implementation
The deep learning model was implemented using the Caffe (caffe.berkeleyvision.org) deep learning toolkit in the Python programming language, version 2.7.12 (22). The architecture is further detailed in the Online Appendix. The choice of Caffe over other available tools was dictated mostly by technical implementation considerations. These include fast hardware graphics support, open source licensing, and performance benchmarks (23). Model training was performed on graphical processor units (GTX 1080 Ti, Nvidia, Santa Clara, CA).
Cross validation
The performance and general error estimation of the deep learning prediction was assessed using a stratified 10-fold cross-validation procedure to reduce the variance in the prediction error, maximize the use of data while preventing model overfitting, and guard against testing hypotheses suggested by arbitrarily split data, a known limitation of the single split validation approach (24–26). The procedure randomly divided the study population into 10 non-overlapping groups of patients of approximately the same size. These 10 groups were stratified to have the same proportion of gender, camera type, and LAD, LCx and RCA obstructive disease as the studied population. Ten train/validation folds were built, with each group being used in turn as the test set and the remaining 9 groups being used as training sets. Ten deep learning models were then trained, validated separately, and then concatenated to provide an overall multi-center performance estimate of the deep learning scores for per-patient and per-vessel CAD prediction. Therefore, none of the data points used in the receiver operating characteristics (ROC) creation was used for model training and model evaluation at the same time. This approach allows an unbiased estimate of how deep learning performs in studies not used in model training, and removes the uncertainty due to random division into one test and one validation groups.
Added value of quantitative MPI maps
The added value of quantitative perfusion maps was assessed by evaluating the deep learning model performance trained including two quantitative perfusion polar maps in addition to raw polar maps. The two methods (with and without quantitative maps) were evaluated into the same cross validation procedure, using strictly the same folds.
Statistical Analysis
The diagnostic performance of deep learning and TPD was evaluated using ROC analysis and pairwise comparisons of area under the ROC curves (AUC) according to DeLong et al (27). Chi-squared McNemar’s test was used to assess the improvement in sensitivity. The per-patient and per-vessel improvement in sensitivity was computed for the deep learning thresholds resulting in the same specificity as previously established diagnostic cutoff values of per-patient TPD = 5% (4), and per-vessel TPD = 2% (3). A two-tailed P value < 0.05 was considered statistically significant. Statistical calculations were performed in R software version 3.4 using the base package for McNemar’s test and the pROC package for DeLong analysis (28).
RESULTS
LV contours were manually corrected in 204 (12.5%) of the images. Overall, the 10-fold cross validation (10 deep learning models) training/validation loop took 30 min using graphical processor units processing. In the testing phase, prediction of obstructive disease was completed in < 0.5 seconds for each patient, using CPU computation only.
Per-patient analysis
In total, 1018 (62%) patients had obstructive disease. The inclusion of quantitative perfusion maps significantly improved per-patient AUC of the deep learning model, compared to using raw polar maps only (P < 0.01) (Figure 2A). The per-patient AUC by deep learning using raw and quantitative polar maps was also significantly higher than the AUC by TPD (P < 0.001). In contrast, the AUC by deep learning from raw polar maps only, was similar to the AUC by TPD (P = 0.98). When operating with the same specificity and at a previously established threshold of per-patient TPD = 5%, deep learning significantly improved the sensitivity of per-patient prediction from 79.8% to 82.3% (+2.5% absolute improvement, P < 0.05).
FIGURE 2. Deep learning prediction of obstructive disease.
Deep learning prediction of obstructive stenosis on a per-patient or per-vessel basis from combined raw and quantitative polar maps (DL: Raw+TPD, light red) had higher AUC than deep learning from raw polar maps only (DL: Raw, dark red), or TPD (TPD, blue). (A) DL score had significantly higher sensitivity than TPD when its abnormality threshold was set to match the specificity of TPD with standard threshold of 5%. (B) Per-vessel DL score had significantly higher sensitivity than per-vessel TPD when its abnormality threshold was set to match per-vessel TPD specificity with standard threshold of 2%. AUC: area under ROC curve, DL: deep learning, ROC: receiver operating characteristic, TPD: total perfusion deficit.
Per-vessel analysis
Per vessel, 1797 out of 4914 (37%) territories had obstructive disease, distributed into 716 (43.7%) LAD, 540 (33%) LCx, and 541 (33%) RCA territories (on average, 599 [36.6%] diseased territories and 1039 [63.4%] non-diseased territories). The inclusion of quantitative perfusion maps significantly improved the per-vessel AUC of the deep learning model, compared to using raw polar maps only (P < 0.001) (Figure 2B). The per-vessel AUC by deep learning using raw and quantitative polar maps was also significantly higher than the AUC by per-vessel TPD (P < 0.001). In contrast, the AUC by deep learning from raw polar maps only was similar to the AUC by per-vessel TPD (P = 0.64). When operating with the same specificity and at a previously established threshold of per-vessel TPD = 2%, deep learning significantly improved the sensitivity of per-vessel prediction from 64.4% to 69.8% (+5.4% absolute improvement, P < 0.01).
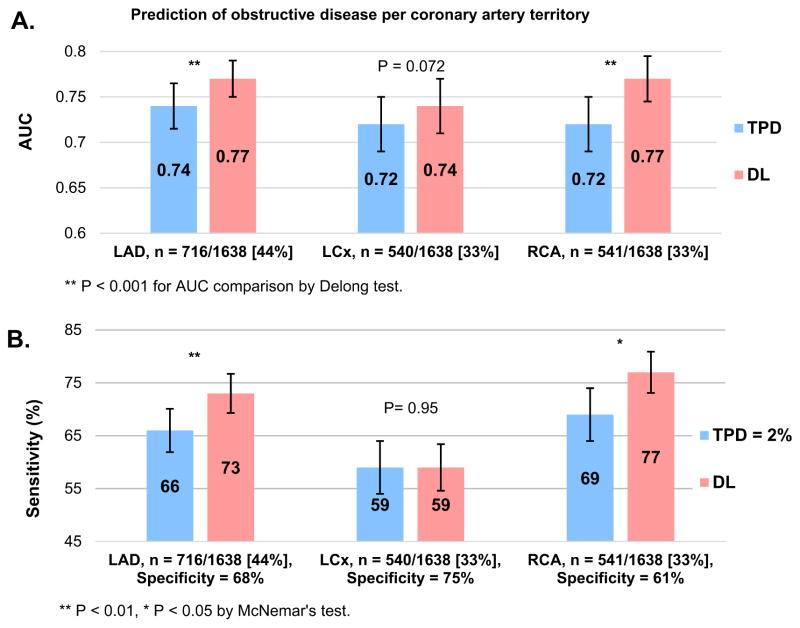
The prediction of LAD and RCA stenosis was better for deep learning than for per-vessel TPD both in terms or AUC and sensitivity improvement (Figure 3). In contrast, AUC and sensitivity for the prediction of LCx disease were similar.
FIGURE 3. Prediction of obstructive coronary artery disease for LAD, LCx and RCA territories.
(A) AUC (bars) and 95% CI (whiskers) for LAD, LCx and RCA artery disease prediction by per-vessel TPD and by deep learning using raw and quantitative perfusion polar maps. (B) Sensitivity (bars) and 95% CI (whiskers) for prediction of artery disease by per-vessel TPD = 2% and by the deep learning approach. The operating point for deep learning was matched to obtain the same specificity as the per-vessel TPD threshold of 2%. AUC: area under the receiver operating characteristic curve, DL: deep learning, TPD: total perfusion deficit.
Case example
Figure 4 illustrates two cases with discrepancy in prediction by TPD and deep learning: a case where deep learning prediction of disease outperformed the per-patient prediction by TPD (Figure 4A), and a case with triple vessel disease where the prediction of per-vessel obstructive disease by deep learning outperformed the prediction by per-vessel TPD (Figure 4B).
FIGURE 4. Discrepancies in prediction of obstructive disease from stress MPI by deep learning and by TPD.
The short/long axis views (left), raw polar map (top) and blackout polar map (bottom) are shown for two patiens with obstructive disease. Also shown are deep learning prediction of obstructive disease (DL+, red) or non-disease (DL−, green), as well as corresponding TPD prediction of obstructive disease per artery territory (per-vessel TPD ≥ 2%, red; per-vessel TPD < 2%, green), or per patient (per-patient TPD ≥ 5%, red; per-patient TPD < 5%, green). (A) Per-patient deep learning outperformed TPD prediction in a 77-year-old male with BMI = 28 kg/m2, hypertension, diabetes and dyslipidemia, who underwent pharmacological stress MPI. The patient had 99% narrowing of both the mid LCx and first obtuse marginal arteries. (B) Per-vessel deep learning prediction outperformed TPD in an 82-year-old male with BMI = 35 kg/m2, hypertension, diabetes and dyslipidemia who underwent pharmacological stress MPI. Patient had triple-vessel disease with 95% narrowing of the left main artery, 80% of the proximal LCx artery and 85% of the distal RCA artery. BMI: body mass index, DL: deep learning, MPI: SPECT myocardial perfusion imaging, TPD: total perfusion deficit.
DISCUSSION
We applied deep learning to the automatic prediction of obstructive coronary artery disease from MPI polar maps and compared it to clinically established standard perfusion quantification by TPD. The MPI data was collected in a large multicenter registry (to our knowledge, the largest to date with invasive angiography), with the number of examples for each coronary vessel obstruction similar to the number of images per category in well-known computer vision datasets (500–1000 images) as reported by Deng et al. (29).
We demonstrated that deep learning utilizing a combination of raw and quantitative perfusion polar maps outperforms standard TPD in prediction of obstructive disease. We also observed that the addition of quantitative polar maps to raw maps improves CAD prediction, on both a per-patient and a per-vessel basis. In addition, we observed that CAD prediction using raw polar maps and gender information was similar to CAD prediction by standard TPD. These findings suggest that the quantitative maps provide important supplementary information. This could be because TPD and blackout polar maps use additional normal low-likelihood population information (used for TPD computation in the normal database files) for the prediction of obstructive disease.
Classical machine learning approaches, requiring the extraction and integration of pre-identified imaging measurements, have shown automated and near-to-expert performance in cardiac applications (26,30–33). In a previous single-center study, machine learning using quantitative measures and clinical measures could predict obstructive disease better than TPD alone (31); however, the information used included complementary clinical features. In this study, we demonstrate the improvement by deep learning over standard clinical approach, utilizing quantitative perfusion polar map data only. It is possible that further significant improvements with deep learning could be achieved by the integration of additional imaging and clinical data. It has been hypothesized that the ability of deep learning to quantify relevant measurements from the raw data will play an important role in cardiovascular medicine e.g. in precision cardiology (15) or as an unsupervised tool for echocardiography imaging (8,16,17). In our study, we show that deep learning can enhance already highly-refined automatic quantification of MPI by direct analysis of polar map image data. We could leverage the availability of large heterogenous population from multiple sites in our registry. We also could leverage the existing capabilities of the conventional software to transform raw 3D MPI images to simpler low-dimension polar maps, which nevertheless retain key perfusion information. The direct diagnosis from raw 3D images by deep learning could be also possible but may require much larger sample sizes and could be more demanding for the deep learning architecture.
We observed improved overall prediction of obstructive disease by deep learning as compared to TPD. The comparison to expert visual reading was not performed in current study due to the differing standards for clinical visual reading across the participating sites. Standard TPD processing has previously shown to be equivalent to expert visual reading in a large study of 997 patients (3). The overall baseline TPD prediction of obstructive disease in the current study as compared to previous studies was lower; the likely reason is that the population studied is different in this multi-center study as compared to previous published studies (3,31).
The improvement in the per-vessel disease prediction by deep learning as compared to TPD was more pronounced than the per-patient improvement. The per-vessel TPD is obtained from a predefined subdivision of the polar perfusion map into fixed vascular territories, based on the standard definition by the 17-segment American Heart Association (AHA) model. In contrast, per-vessel disease prediction by deep learning was performed by a computational model trained on large number of abnormal cases. Thus, deep learning could leverage the large database of CAD hypoperfusion patterns, which likely explains the improved performance as compared to standard normal database and fixed vascular territory analysis.
Limitations
This initial study of deep learning applied to MPI has several limitations. First, the degree of stenosis from invasive angiography was interpreted visually, since quantitative angiography is not routinely performed in many sites; however, we did use a 70% diameter stenosis cutoff which has shown to be a greater discriminator of functionally significant lesions than a 50% cutoff (34). Second, we used polar maps from stress static images only, acquired in only one position to ensure applicability to simplest imaging protocol (lowest common denominator) used in clinical routine. Improvements could be potentially achieved by using additional positions (prone, supine) as is common practice (4), gated acquisitions, imaging measurements and clinical variables. Further improvements could be accomplished by optimizing the deep learning model, similar to work in the classification of natural images (29). Rest scans and ischemia could be also considered; however, in this study we included consecutive patients without previously known CAD. Possibly, larger set of training data could allow further improvement; nevertheless, we demonstrate already that deep learning trained with the sample available to us, provides improved performance compared to standard analysis by TPD.
CONCLUSION
Deep learning improved automatic prediction of obstructive coronary artery disease compared to the current standard method and has the potential to enhance automatic interpretation of MPI. To evaluate the feasibility of clinical application of deep learning for MPI analysis, further studies should focus on evaluating the generalizability of this new approach to other camera types, and on the extension to incorporate additional clinical and imaging data.
Supplementary Material
Clinical Perspectives.
Competency in Medical Knowledge
We have evaluated a deep learning algorithm using convolutional neural networks and demonstrated higher prediction and improved sensitivity than standard total perfusion deficit for obstructive disease prediction from MPI.
Translational Outlook
MPI is a highly automated imaging procedure for the assessment of coronary artery disease. Deep learning improves the automatic prediction and localization of obstructive CAD compared to current standard methods, without additional effort or increased time burden for the reader physician.
Acknowledgments
Funding:
This research was supported in part by grant R01HL089765 from the National Heart, Lung, and Blood Institute/National Institutes of Health (NHLBI/NIH) (PI: Piotr Slomka). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We want to thank all the people whose efforts allowed us to collect, process and analyze the data in the NIH sponsored REFINE SPECT registry. We thank Joanna Liang for proofreading the text of the manuscript.
ABBREVIATIONS
- SPECT
single-photon emission computed tomography
- MPI
SPECT myocardial perfusion imaging
- CAD
coronary artery disease
- QPS
quantitative perfusion SPECT
- TPD
total perfusion deficit
- MI
myocardial infarction
- ROC
receiver operating characteristic
- ICA
invasive coronary angiography
- AUC
area under ROC curve
Footnotes
Financial disclosure:
Daniel Berman, Guido Germano and Piotr Slomka participate in software royalties for QPS software at Cedars-Sinai Medical Center. Other authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Einstein AJ. Effects of Radiation Exposure From Cardiac Imaging: How Good Are the Data? J Am Coll Cardiol. 2012;59:553–565. doi: 10.1016/j.jacc.2011.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slomka PJ, Nishina H, Berman DS, et al. Automated quantification of myocardial perfusion SPECT using simplified normal limits. J Nucl Cardiol. 2005;12:66–77. doi: 10.1016/j.nuclcard.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Arsanjani R, Xu Y, Hayes SW, et al. Comparison of Fully Automated Computer Analysis and Visual Scoring for Detection of Coronary Artery Disease from Myocardial Perfusion SPECT in a Large Population. J Nucl Med. 2013;54:221–228. doi: 10.2967/jnumed.112.108969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakazato R, Tamarappoo BK, Kang X, et al. Quantitative Upright–Supine High-Speed SPECT Myocardial Perfusion Imaging for Detection of Coronary Artery Disease: Correlation with Invasive Coronary Angiography. J Nucl Med. 2010;51:1724–1731. doi: 10.2967/jnumed.110.078782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germano G, Kavanagh PB, Fish MB, et al. “Same-Patient Processing” for multiple cardiac SPECT studies. 1”. Improving LV segmentation accuracy. J Nucl Cardiol. 2016;23:1435–1441. doi: 10.1007/s12350-016-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motwani M, Leslie WD, Goertzen AL, et al. Fully automated analysis of attenuation-corrected SPECT for the long-term prediction of acute myocardial infarction. J Nucl Cardiol. 2017 doi: 10.1007/s12350-017-0840-0. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betancur J, Rubeaux M, Fuchs T, et al. Automatic Valve Plane Localization in Myocardial Perfusion SPECT/CT by Machine Learning: Anatomical and Clinical Validation. J Nucl Med. 2017;58:961–967. doi: 10.2967/jnumed.116.179911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tajik AJ. Machine Learning for Echocardiographic Imaging. J Am Coll Cardiol. 2016;68:2296–2298. doi: 10.1016/j.jacc.2016.09.915. [DOI] [PubMed] [Google Scholar]
- 9.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 10.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 12.Greenspan H, Ginneken Bv, Summers RM. Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans Med Imaging. 2016;35:1153–1159. [Google Scholar]
- 13.Shen D, Wu G, Suk H-I. Deep Learning in Medical Image Analysis. Annual review of biomedical engineering. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial Intelligence in Precision Cardiovascular Medicine. J Am Coll Cardiol. 2017;69:2657–2664. doi: 10.1016/j.jacc.2017.03.571. [DOI] [PubMed] [Google Scholar]
- 16.Krittanawong C, Tunhasiriwet A, Zhang H, Wang Z, Aydar M, Kitai T. Deep Learning With Unsupervised Feature in Echocardiographic Imaging. J Am Coll Cardiol. 2017;69:2100–2101. doi: 10.1016/j.jacc.2016.12.047. [DOI] [PubMed] [Google Scholar]
- 17.Narula S, Shameer K, Salem Omar AM, Dudley JT, Sengupta PP. Reply. J Am Coll Cardiol. 2017;69:2101–2102. doi: 10.1016/j.jacc.2017.01.062. [DOI] [PubMed] [Google Scholar]
- 18.Gambhir SS, Berman DS, Ziffer J, et al. A Novel High-Sensitivity Rapid-Acquisition Single-Photon Cardiac Imaging Camera. J Nucl Med. 2009;50:635–643. doi: 10.2967/jnumed.108.060020. [DOI] [PubMed] [Google Scholar]
- 19.Herzog BA, Buechel RR, Katz R, et al. Nuclear Myocardial Perfusion Imaging with a Cadmium-Zinc-Telluride Detector Technique: Optimized Protocol for Scan Time Reduction. J Nucl Med. 2010;51:46–51. doi: 10.2967/jnumed.109.065532. [DOI] [PubMed] [Google Scholar]
- 20.Buechel RR, Herzog BA, Husmann L, et al. Ultrafast nuclear myocardial perfusion imaging on a new gamma camera with semiconductor detector technique: first clinical validation. Eur J Nucl Med Mol Imaging. 2010;37:773–778. doi: 10.1007/s00259-009-1375-7. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Kavanagh P, Fish M, et al. Automated Quality Control for Segmentation of Myocardial Perfusion SPECT. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2009;50:1418–1426. doi: 10.2967/jnumed.108.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia Y, Shelhamer E, Donahue J, et al. Caffe: Convolutional architecture for fast feature embedding. MM’ 14, Proceedings of the 22nd ACM international conference on Multimedia; Orlando, FL: ACM; 2014. pp. 675–678. [Google Scholar]
- 23.Bahrampour S, Ramakrishnan N, Schott L, Shah M. Comparative Study of Caffe, Neon, Theano, and Torch for Deep Learning. CoRR. 2016 abs/1511.06435. [Google Scholar]
- 24.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21:3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 25.Motwani M, Dey D, Berman DS, et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. 2017;38:500–507. doi: 10.1093/eurheartj/ehw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betancur J, Otaki Y, Motwani M, et al. Prognostic Value of Combined Clinical and Myocardial Perfusion Imaging Data Using Machine Learning. JACC Cardiovasc Imaging. 2017 doi: 10.1016/j.jcmg.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 28.Team RC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2017. [Google Scholar]
- 29.Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition; 2009; pp. 248–255. [Google Scholar]
- 30.Arsanjani R, Dey D, Khachatryan T, et al. Prediction of revascularization after myocardial perfusion SPECT by machine learning in a large population. J Nucl Cardiol. 2015;22:877–884. doi: 10.1007/s12350-014-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arsanjani R, Xu Y, Dey D, et al. Improved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population. J Nucl Cardiol. 2013;20:553–562. doi: 10.1007/s12350-013-9706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narula S, Shameer K, Salem Omar AM, Dudley JT, Sengupta PP. Machine-Learning Algorithms to Automate Morphological and Functional Assessments in 2D Echocardiography. J Am Coll Cardiol. 2016;68:2287–2295. doi: 10.1016/j.jacc.2016.08.062. [DOI] [PubMed] [Google Scholar]
- 33.Sengupta PP, Huang Y-M, Bansal M, et al. Cognitive Machine-Learning Algorithm for Cardiac Imaging: A Pilot Study for Differentiating Constrictive Pericarditis From Restrictive Cardiomyopathy. Circ Cardiovasc Imaging. 2016;9:e004330. doi: 10.1161/CIRCIMAGING.115.004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonino PAL, Fearon WF, De Bruyne B, et al. Angiographic Versus Functional Severity of Coronary Artery Stenoses in the FAME Study: Fractional Flow Reserve Versus Angiography in Multivessel Evaluation. J Am Coll Cardiol. 2010;55:2816–2821. doi: 10.1016/j.jacc.2009.11.096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.