Abstract
Microorganisms have a remarkable capacity to evolve resistance to antimicrobial agents, threatening the efficacy of the limited arsenal of antimicrobials and becoming a dire public health crisis. This is of particular concern for fungal pathogens, which cause devastating invasive infections with treatment options limited to only three major classes of antifungal drugs. The paucity of antifungals with clinical utility is in part due to close evolutionary relationships between these eukaryotic pathogens and their human hosts, which limits the unique targets to be exploited therapeutically. This review highlights the mechanisms by which fungal pathogens of humans evolve resistance to antifungal drugs, which provide crucial insights to enable development of novel therapeutic strategies to thwart drug resistance and combat fungal infectious disease.
Keywords: antifungal, resistance, fungal pathogen, stress responses, intrinsic resistance, evolution
Graphical Abstract
Introduction
Fungal pathogens have a profound impact on global human health, food security, and biodiversity. Advances in modern medicine have improved the treatment of diverse human diseases and restricted infectious disease outbreaks, thereby extending human lifespan. As a consequence, opportunistic fungal pathogens have emerged as a leading cause of human mortality, particularly in individuals with underlying health conditions or undergoing immunosuppressive treatments, with attributable mortalities estimated at ~1.5 million per year [1]. The predominant causative agents include species of Candida, Aspergillus and Cryptococcus [1]. Candida albicans is a leading cause of nosocomial infections, with mortality rates often exceeding ~40% despite treatment [2]. Non-albicans Candida (NAC) species are also problematic with drug-resistant isolates becoming increasingly more common [3]. Aspergillus fumigatus is a ubiquitous saprophytic fungus estimated to cause over 200,000 cases of invasive aspergillosis occur each year, with mortality rates often exceeding 50% [1]. Finally, cryptococcosis, caused by species such as Cryptococcus neoformans and Cryptococcus deuterogattii, affects over one million individuals annually, often resulting in severe central nervous system infections in vulnerable individuals [1]. While cryptococcosis caused by C. neoformans is an AIDS-defining illness, C. deuterogattii is distinguished by its propensity to cause infections in otherwise healthy hosts [4]. Fungal pathogens not only impact humans directly, they jeopardize food security through mass devastation of crops that feed billions, as well as producing toxins that contaminate food supplies and lead to the development of cancers [5]. In recent years, there has been an unprecedented number of fungal diseases causing extinctions in wild species, with mass mortalities of bats and amphibians threatening biodiversity. Climate change is poised to exacerbate the problem as increasing global temperatures are being accompanied by pests and pathogens moving northward, with fungi leading the way [6].
The impact of fungi on human health is amplified by the fact that only three classes of antifungal drugs are available to treat systemic fungal infections [3,7]. These include the azoles that target ergosterol biosynthesis, the echinocandins that inhibit fungal cell wall biosynthesis, and the polyenes that bind to ergosterol in the fungal cell membrane leading to cell lysis (Figure 1) [3,7]. Our limited arsenal of antifungals is further threatened by the development of multidrug-resistant strains of fungi and the emergence of intrinsically resistant pathogens. In this review, we highlight mechanisms of antifungal resistance with a focus on fungal pathogens that infect human hosts. We also touch on promising new therapeutic strategies that may be employed in the future to address this global health crisis.
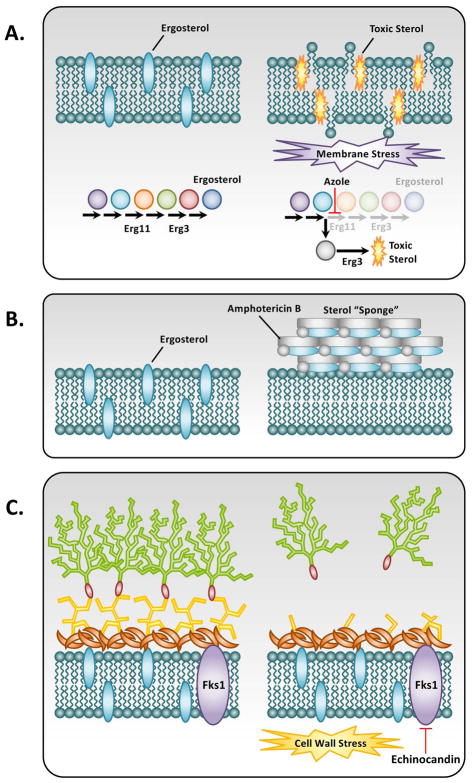
Figure 1. Antifungal drug mode of action.
A) The azoles function by targeting the ergosterol biosynthetic enzyme lanosterol demethylase, encoded by ERG11 (C. albicans and C. neoformans) or cyp51A and cyp51B (A. fumigatus), causing a block in the production of ergosterol and the accumulation of toxic sterol intermediates produced by ERG3. This toxic sterol induces a severe membrane stress on the cell and ultimately inhibits growth of the fungi. B) The polyenes such as amphotericin B act primarily by forming large, extramembranous aggregates that extract ergosterol from lipid bilayers. C) Fungal cell walls are composed of (1,3)-β-D-glucans covalently linked to (1,6)-β-D-glucans as well as chitin, mannans, and cell wall proteins. The echinocandins act as non-competitive inhibitors of (1,3)-β-D-glucan synthase, encoded by FKS1 (C. albicans, C. neoformans, and A. fumigatus), thereby causing a loss of cell wall integrity and severe cell wall stress. (Adapted from Cowen LE 2008. Nat. Rev. Micro. 6(3):187–98).
Acquired Resistance Mechanisms
Our frequent and prophylactic use of antifungal agents has led to the development of robust resistance in medically important fungi. Numerous adaptive mechanisms of antifungal drug resistance have been identified, including drug target alteration or overexpression, upregulation of multidrug transporters, and activation of stress responses (Figure 2) [8]. Although resistance to the polyenes remains extremely rare [9], resistance to the azoles and echinocandins is readily documented and their modes of acquired resistance will be elaborated on below.
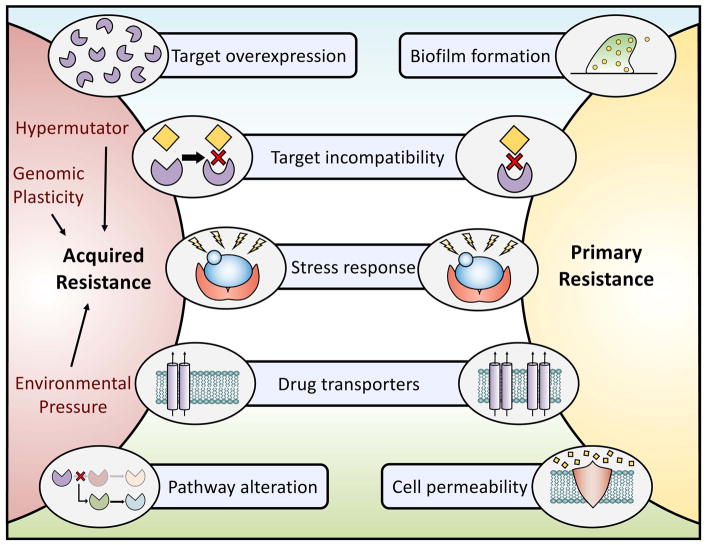
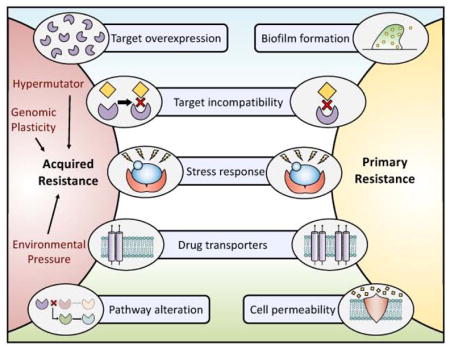
Figure 2. Exploring the relationships between and mechanisms governing intrinsic and acquired resistance.
The development of acquired resistance can occur through several mechanisms. Examples include overexpression of the drug target, amino acid substitutions in the drug target that impede drug binding, signaling through stress response pathways, upregulation of efflux pumps, or alterations in cellular pathways. Acquired resistance in fungal pathogens can be accelerated via multiple factors including but not limited to an organisms’ genetic plasticity, the existence of hypermutator strains, or environmental pressures that result in strains becoming resistant to agricultural fungicides leading to cross resistance in clinical isolates. Primary resistance is achieved through several mechanisms overlapping with those implicated in acquired resistance including target incompatibility, stress response signaling, and efflux pump overexpression. In addition, the formation of fungal biofilms decreases overall fungal drug susceptibility, and differences in cellular permeability may prevent a drug from reaching its target. The combined effect of these contributing mechanisms leads the selection of increasingly resistant organisms.
Resistance to Azoles
Azole antifungals inhibit the ergosterol biosynthetic pathway by targeting the cytochrome P450-dependent enzyme lanosterol 14-α-demethylase, encoded by Erg11 in yeasts, and Cyp51A/Cyp51B in molds. Inhibiting this pathway disrupts the production of ergosterol and results in an accumulation of toxic sterol intermediates that perturb membrane stability and impede fungal growth [7]. One of the most prevalent mechanisms of azole resistance involves alteration or overexpression of the drug target gene, ERG11/cyp51A/cyp51B, with Candida and Aspergillus resistant isolates often having amino acid substitutions in regions close to the heme-binding site of the enzyme [10,11]. Furthermore, constitutive overexpression of ERG11 via gain-of-function mutations in the transcriptional activator Upc2 is commonly found in resistant isolates of C. albicans [12]. In other human fungal pathogens, distinct sterol regulatory elements such as the transcription factor Sre1 in C. neoformans and SrbA in A. fumigatus, have also been implicated in responses to antifungal drugs and virulence [13,14]. Alterations in other components of the ergosterol biosynthetic pathway, such as loss of function of the Δ-5,6-desaturase enzyme Erg3, can also enable azole resistance. ERG3 mutations lead to the depletion of ergosterol and the accumulation of alternative sterols, often resulting in cross-resistance to azoles and polyenes [15]. ERG3-mediated azole resistance intimately depends on key stress response regulators such as the protein phosphatase calcineurin [16], the protein kinase Pkc1 [17], the molecular chaperone Hsp90 [16], and likely other regulators that remain to be identified. In fact, functional genomic screens have identified several genes encompassing diverse cellular processes important for mediating azole tolerance in both C. albicans and C. neoformans [18–20].
Another common mechanism of acquired azole resistance involves the upregulation of multidrug transporters. The ATP-binding cassette (ABC) transporters Cdr1 and Cdr2, as well as the major facilitator Mdr1 have all been implicated in clinical azole resistance of many Candida species [21,22]. Upregulation of Mdr1 has been shown to simultaneously enable C. albicans azole resistance as well as escape from intrinsic host defenses through the efflux of antimicrobial peptides, such as histatin 5 [23]. The expression of CDR1 and CDR2 is regulated by the transcription factor Tac1 in C. albicans, with TAC1 alleles harboring gain-of-function mutations readily identified in resistant isolates [24]. Similarly, mutations in the transcription factor gene MRR1 lead to upregulation of Mdr1 in azole-resistant isolates of C. albicans [25]. In C. neoformans and A. fumigatus, the ABC transporters responsible for azole efflux are Afr1 and AtrF, respectively [26,27].
Genomic alterations that lead to an increased dosage of drug transporters provide an alternative route to enhance drug efflux. Fungal species are capable of remarkable genomic plasticity in response to diverse environmental stresses. Studies examining the genome composition of C. albicans azole-resistant isolates identified a duplication of the left arm of chromosome 5 (termed i(5L)) as a common aneuploidy [28]. The formation of i(5L) results in increased dosage of both ERG11 and TAC1, enabling a dual mechanism of azole resistance [28,29]. Recently, other aneuploid lineages of C. albicans harboring increased copy numbers of chromosome 3 and chromosome 6 were determined to have reduced susceptibility to azoles [30]. Although the molecular basis remains elusive, these findings further support the notion that genomic diversity promotes stress adaptation and survival in C. albicans. Finally, in C. neoformans, duplication of chromosome 1, the genomic location of the azole target gene ERG11, was discovered as an adaptive mechanism to confer azole resistance [31]. Thus, genomic plasticity appears to be a conserved and central adaptive mechanism (Figure 2).
Given the large-scale deployment of azoles in agriculture, the potential for pathogens that acquired resistance in the field to be transmitted to human hosts is cause for grave concern. A. fumigatus with environmentally acquired resistance has been identified in azole-naïve patients without previous azole exposure [32]. Specifically, in a study of 144 soil samples collected from greenhouses in China, 5.8% of the analyzed samples displayed cross-resistance with agro-chemicals and medical azoles [33]. Intriguingly, exposure to environmental azoles has also been shown to induce cross-resistance to azole antifungals in both C. neoformans and C. gattii, mostly due to mutations in ERG11 [34].
Resistance to the Echinocandins
The echinocandins are the newest class of antifungal drug released into the clinic. They target the fungal cell wall by inhibiting (1,3)-β-D-glucan synthase, encoded by FKS1 (and FKS2 in C. glabrata), inducing a severe cell wall stress and leading to a loss of cell wall integrity [7]. These first-line drugs have the advantage of being fungicidal against the majority of Candida species, although they are predominantly fungistatic against A. fumigatus [7]. Echinocandin resistance is primarily conferred via amino acid substitutions within highly conserved regions of the Fks subunits of glucan synthase [35]. These hot spot regions are conserved across different Candida species, with levels of resistance varying depending on the mutations and expression level of these genes [15]. In C. glabrata, FKS2 expression is calcineurin dependent, therefore resistance conferred by FKS2 can be reversed upon administration of calcineurin inhibitors such as FK506 [15,36].
Complex cellular circuitry orchestrating responses to cell wall stressors can also confer echinocandin tolerance and resistance. Cell wall integrity signaling mediated via protein kinase C (PKC), the protein phosphatase calcineurin, and the molecular chaperone Hsp90, is vital in enabling echinocandin drug tolerance and compensatory mechanisms such as upregulation of chitin synthesis [16,37,38]. Additionally, a recent study implicated the C. albicans transcription factor Cas5 in governing echinocandin tolerance and resistance [39]. Similar to the azoles where diverse cellular processes modulate susceptibility, diverse genes have been implicated in echinocandin tolerance in fungal pathogens [20,40,41]. The genes required to respond to different drug classes appear largely distinct and species restrictive, suggesting specificity in the circuitry governing cellular responses to stress.
More recently, strains with acquired resistance to multiple classes of antifungal drugs have become of grave concern. One mechanism by which this occurs is via the emergence of hypermutator lineages. One poignant example of this has been reported in C. glabrata where one third of the 1,300 isolates analyzed in one study were identified as non-susceptible to both an echinocandin and an azole [42]. This multidrug resistance phenotype was likely attributable to a hypermutator phenotype [42,43], as approximately 55% of resistant clinical isolates had loss-of-function mutations in the DNA repair gene MSH2, accelerating the emergence of multidrug resistance [43]. Recently, a hypermutator lineage was also identified in C. deuterogattii where a mutation in MSH2 enabled elevated mutation rates [44]. Interestingly, while hypermutators in bacterial populations often produce offspring that are generally less fit, only modest fitness defects have been observed with hypermutators in fungi [43,44]. Further, although in C. deuterogattii loss of function of the mismatch repair component MSH2 does not appear to directly impact virulence, it may provide a means to promote loss of virulence over time as mutations accumulate in critical pathways [44]. Although relatively little is known about hypermutators in eukaryotes, these examples suggest that this may be a prevalent adaptive strategy in eukaryotic pathogens (Figure 2).
The Increasing Prevalence of Primary Resistance
In addition to the ability to acquire resistance through mutations or genomic alterations, fungal species that are inherently resistant to antifungals have become a growing problem in medicine and agriculture. Inherent resistance, or primary resistance, is used to describe species in which all known isolates possess an innate resistance to an antifungal (Figure 2). A classic example of this is the resistance of Cryptococcus species to echinocandins. This has remained enigmatic as C. neoformans not only possesses a β-glucan synthase encoded by FKS1, but echinocandins inhibit the enzyme effectively in vitro [45]. Recently, a screen of over 7,000 C. neoformans mutants identified mutations in CDC50 that rendered the strain sensitive to echinocandins [46]. CDC50 encodes the β-subunit of a lipid flippase involved in phospholipid translocation and trafficking, and is required for membrane integrity, stress resistance and virulence [46].
Variation in resistance phenotypes are observed not only between species and among isolates of the same species, but also between distinct growth states for the same strain. Microbial biofilms are surface-associated communities that exhibit remarkable levels of drug resistance relative to their planktonic counterparts [47]. The glucan matrix that surrounds fungal biofilms can act as a physical barrier to impede the bioavailability of antimicrobial compounds. Other factors such as alterations in efflux pump expression, changes in cell membrane and wall composition, and alterations in stress response profiles also contribute to the altered drug resistance profiles of these communities [47].
Finally, the emergence of pathogenic species with elevated resistance to current antifungal drugs is concerning. This is apparent with the increased prevalence of resistant NAC species, such as C. tropicalis, C. glabrata, and C. parapsilosis, and with the recent global emergence of C. auris [48]. Since its first isolation in 2009, there have been several clinical outbreaks of C. auris suggesting that this pathogen has exceptional adaptive mechanisms to persist in hospital environments that remain to be identified [46]. The most comprehensive assessment of C. auris resistance to date found that 93% of isolates have extremely high levels of resistance to the azole fluconazole (≥32μg/mL) [49]. Here, the distinction between intrinsic and acquired resistance becomes less clear, as independently arising mutations suggest an acquired mechanism, however, no sensitive pools of C. auris, in which azole treatment would be effective have been identified. This intrinsic resistance could be explained by the combined effect of an expansion of multidrug transporters in the genome and nine amino acid changes in Erg11, which are known to confer significant resistance in C. albicans [49,50]. Even more concerning, 4% of C. auris isolates were resistant to all three classes of antifungals, leaving no therapies for these formidable infections [49].
Conclusion: Outlook for Future Antifungal Development
A major obstacle in antifungal discovery is identifying essential targets in fungi that can be specifically engaged by molecules that lack host toxicity. A promising strategy to expand the drug target space is to explore combination therapy. This approach is fundamental to the treatment of AIDS, malaria, and tuberculosis and provides multiple advantages, including reducing the rate of resistance, increasing potency, lowering host toxicity and broadening the therapeutic range [7]. Drug combinations can also be employed to target resistance mechanisms themselves, particularly those involved in drug tolerance. For example, C. glabrata azole resistance is frequently achieved through substitutions in the transcription factor Pdr1, resulting in upregulation of efflux. By inhibiting the interaction of the Pdr1 activation domain and the mediator Gal11A in resistant C. glabrata isolates, the compound iKIX1 can restore azole sensitivity and reduce fungal proliferation in murine systemic infection models [51]. Targeting stress response regulators such as the molecular chaperone Hsp90 has also been shown to have vast therapeutic potential to increase the efficacy of both azoles and echinocandins while impairing the emergence of resistance [16,38,52]. Advances in high throughput screening and chemical genomic approaches in the model yeast Saccharomyces cerevisiae, as well as the fungal pathogens C. albicans and C. neoformans have enabled powerful strategies to identify new chemical matter and targets for antifungal drug development [53–55]. Although antifungal resistance continues to emerge, recent advances suggest that an expansion of the current arsenal of antifungal treatments is on the horizon.
Highlights.
Our limited arsenal of antifungals is threatened by drug-resistant fungal pathogens.
Inherent resistance is when all strains of a species show antifungal resistance.
Target alteration and drug efflux are canonical adaptive resistance mechanisms.
Stress responses are important mediators of resistance to diverse antifungals.
Agricultural use of antifungals may drive the development of resistance.
Acknowledgments
We thank members of the Cowen lab for helpful discussions. L.E.C. is supported by the Canadian Institutes of Health Research Operating Grants (MOP-86452 and MOP-119520) and Foundation Grant (FDN-154288), the Natural Sciences and Engineering Council (NSERC) of Canada Discovery Grants (06261 and 462167), an NSERC E.W.R. Steacie Memorial Fellowship (477598), and a National Institutes of Health NIAID R01 (1R01AI127375-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–63. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Perfect JR. The antifungal pipeline: a reality check. Nat Rev Drug Discov. 2017;16:603–616. doi: 10.1038/nrd.2017.46. A comprehensive review summarizing the latest research being conducted to expand our antifungal armamentarium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci U S A. 2004;101:17258–63. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–94. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fones HN, Fisher MC, Gurr SJ. Emerging fungal threats to plants and animals challenge agriculture and ecosystem resilience. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.funk-0027-2016. FUNK-0027–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Robbins N, Caplan T, Cowen LE. Molecular evolution of antifungal drug resistance. Annu Rev Microbiol. 2017;71:753–75. doi: 10.1146/annurev-micro-030117-020345. A comprehensive review summarizing mechanisms of antifungal resistance with a focus on fungal pathogens of humans. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev. 2011;75:213–67. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 2013;11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FCS, Odds FC, Vanden Bossche H. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology. 1999;145:2701–13. doi: 10.1099/00221287-145-10-2701. [DOI] [PubMed] [Google Scholar]
- 11.Balashov SV, Gardiner R, Park S, Perlin DS. Rapid, high-throughput, multiplex, real-time PCR for identification of mutations in the cyp51A gene of Aspergillus fumigatus that confer resistance to itraconazole. J Clin Microbiol. 2005;43:214–22. doi: 10.1128/JCM.43.1.214-222.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver PM, Oliver BG, White TC. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot Cell. 2004;3:1391–7. doi: 10.1128/EC.3.6.1391-1397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64:614–29. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 14.Willger SD, Puttikamonkul S, Kim K-H, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med. 2015;5:a019752. doi: 10.1101/cshperspect.a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–89. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 17.Lafayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, Leslie Gunatilaka AA, Perfect JR, Cowen LE. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 2010;6:e1001069. doi: 10.1371/journal.ppat.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Mallick J, Maqnas A, Sun Y, Choudhury BI, Cote P, Yan L, Ni T-J-H, Li Y, Zhang D, et al. Chemogenomic profiling of the fungal pathogen Candida albicans. Antimicrob Agents Chemother. 2017;62 doi: 10.1128/AAC.02365-17. pii e02365–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Lee KT, So YS, Yang DH, Jung KW, Choi J, Lee DG, Kwon H, Jang J, Wang LL, Cha S, et al. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat Commun. 2016;7:1–7. doi: 10.1038/ncomms12766. This study described the generation of 264 signature-tagged gene-deletion strains for 129 putative kinases in C. neoformans, and used this genetic library to identify potential anticryptococcal or antifungal drug targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung KW, Yang DH, Maeng S, Lee KT, So YS, Hong J, Choi J, Byun HJ, Kim H, Bang S, et al. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun. 2015;6:1–14. doi: 10.1038/ncomms7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanglard D, Kuchler K, Ischer F, Pagani JL, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–86. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–16. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 23.Hampe IAI, Friedman J, Edgerton M, Morschhäuser J. An acquired mechanism of antifungal drug resistance simultaneously enables Candida albicans to escape from intrinsic host defenses. PLoS Pathog. 2017;13:e1006655. doi: 10.1371/journal.ppat.1006655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coste A, Turner V, Ischer F, Morschhäuser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics. 2006;172:2139–56. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morschhäuser J, Barker KS, Liu TT, Blaß-Warmuth J, Homayouni R, Rogers PD. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 2007;3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Posteraro B, Sanguinetti M, Sanglard D, La Sorda M, Boccia S, Romano L, Morace G, Fadda G. Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Mol Microbiol. 2003;47:357–71. doi: 10.1046/j.1365-2958.2003.03281.x. [DOI] [PubMed] [Google Scholar]
- 27.Slaven JW, Anderson MJ, Sanglard D, Dixon GK, Bille J, Roberts IS, Denning DW. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet Biol. 2002;36:199–206. doi: 10.1016/s1087-1845(02)00016-6. [DOI] [PubMed] [Google Scholar]
- 28.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–70. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68:624–41. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 30.Hirakawa MP, Chyou DE, Huang D, Slan AR, Bennett RJ. Parasex generates phenotypic diversity de novo and impacts drug resistance and virulence in Candida albicans. Genetics. 2017;207:1195–1211. doi: 10.1534/genetics.117.300295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sionov E, Lee H, Chang YC, Kwon-Chung KJ. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snelders E, Huis In ’t Veld RAG, Rijs AJMM, Kema GHJ, Melchers WJG, Verweij PE. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl Environ Microbiol. 2009;75:4053–7. doi: 10.1128/AEM.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger S, Chazli Y, El Babu AF, Coste AT. Azole resistance in Aspergillus fumigatus: a consequence of antifungal use in agriculture? Front Microbiol. 2017;8:1–6. doi: 10.3389/fmicb.2017.01024. This review summarizes several studies that suggest agricultural azoles are responsible for medical treatment failure in azole-naïve patients infected with A. fumiagtus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bastos RW, Carneiro HCS, de Oliveira LVN, Rocha KM, Freitas GJC, Costa MC, Magalhães TFF, de Carvalho VSD, Rocha CE, Ferreira GF, et al. Environmental triazole induces cross-resistance to clinical drugs and affects morphophysiology and virulence of Cryptococcus gattii and C. neoformans. Antimicrob Agents Chemother. 2017;62 doi: 10.1128/AAC.01179-17. pii: e01179–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park S, Kelly R, Kahn JN, Robles J, Hsu M-J, Register E, Li W, Vyas V, Fan H, Abruzzo G, et al. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob Agents Chemother. 2005;49:3264–73. doi: 10.1128/AAC.49.8.3264-3273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katiyar SK, Alastruey-Izquierdo A, Healey KR, Johnson ME, Perlin DS, Edlind TD. Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob Agents Chemother. 2012;56:6304–9. doi: 10.1128/AAC.00813-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh-Babak SD, Babak T, Diezmann S, Hill JA, Xie JL, Chen Y-L, Poutanen SM, Rennie RP, Heitman J, Cowen LE. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 2012;8:e1002718. doi: 10.1371/journal.ppat.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5:e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Xie JL, Qin L, Miao Z, Grys BT, Diaz JDLC, Ting K, Krieger JR, Tong J, Tan K, Leach MD, et al. The Candida albicans transcription factor Cas5 couples stress responses, drug resistance and cell cycle regulation. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00547-y. This study revealed that Cas5 governs distinct transcriptional programs that switch targets in response to different environmental conditions and described circuitry governing antifungal drug resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiederhold N. Antifungal resistance: current trends and future strategies to combat. Infect Drug Resist. 2017;10:249–59. doi: 10.2147/IDR.S124918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Healey KR, Zhao Y, Perez WB, Lockhart SR, Sobel JD, Farmakiotis D, Kontoyiannis DP, Sanglard D, Taj-Aldeen SJ, Alexander BD, et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms11128. This manuscript demonstrated that a mutator phenotype is prevalent in C. glabrata clinical isolates, which at least partially explains the elevated rates of triazole and multi-drug resistance associated with C. glabrata. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Billmyre RB, Clancey SA, Heitman J. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. eLife. 2017;6:e28802. doi: 10.7554/eLife.28802. This study identified a hypermutator lineage in C. deuterogattii and suggested that pathogenic eukaryotic microbes may experience selection pressures on mutation rate, particularly during long periods of clonal growth or while expanding into new environments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maligie MA, Selitrennikoff CP. Cryptococcus neoformans resistance to echinocandins: (1,3)beta-glucan synthase activity is sensitive to echinocandins. Antimicrob Agents Chemother. 2005;49:2851–6. doi: 10.1128/AAC.49.7.2851-2856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang W, Liao G, Baker GM, Wang Y, Lau R, Paderu P, Perlin DS, Xue C. Lipid flippase subunit Cdc50 mediates drug resistance and virulence in Cryptococcus neoformans. mBio. 2016;7:e00478–16. doi: 10.1128/mBio.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.d’Enfert C. Biofilms and their role in the resistance of pathogenic Candida to antifungal agents. Curr Drug Targets. 2006;7:465–70. doi: 10.2174/138945006776359458. [DOI] [PubMed] [Google Scholar]
- 46.Chowdhary A, Sharma C, Meis JF. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–40. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee S, Alampalli SV, Nageshan RK, Chettiar ST, Joshi S, Tatu US, Klevens R, Edwards J, Richards C, Horan T, et al. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics. 2015;16:686. doi: 10.1186/s12864-015-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Nishikawa JL, Boeszoermenyi A, Vale-Silva LA, Torelli R, Posteraro B, Sohn Y-J, Ji F, Gelev V, Sanglard D, Sanguinetti M, et al. Inhibiting fungal multidrug resistance by disrupting an activator–mediator interaction. Nature. 2016;530:485–89. doi: 10.1038/nature16963. This paper demonstrated the feasibility of using small-molecules to target a transcription factor-binding site in Mediator as a novel therapeutic strategy in fungal infectious disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cowen LE, Singh SD, Köhler JR, Collins C, Zaas AK, Schell WA, Aziz H, Mylonakis E, Perfect JR, Whitesell L, et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci U S A. 2009;106:2818–23. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piotrowski JS, Li SC, Deshpande R, Simpkins SW, Nelson J, Yashiroda Y, Barber JM, Safizadeh H, Wilson E, Okada H, et al. Functional annotation of chemical libraries across diverse biological processes. Nat Chem Biol. 2017;13:982–93. doi: 10.1038/nchembio.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol. 2003;50:167–81. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 53.Brown JCS, Nelson J, VanderSluis B, Deshpande R, Butts A, Kagan S, Polacheck I, Krysan DJ, Myers CL, Madhani HD. Unraveling the biology of a fungal meningitis pathogen using chemical genetics. Cell. 2014;159:1168–87. doi: 10.1016/j.cell.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]