Summary
The development of midbrain dopaminergic (mDA) neurons is controlled by multiple morphogens and transcription factors. However, little is known about the role of extracellular matrix proteins in this process. Here we examined the function of roof plate-specific spondins (RSPO1-4) and the floor plate-specific, spondin 1 (SPON1). Only RSPO2 and SPON1 were expressed at high levels during mDA neurogenesis, and the receptor LGR5 was expressed by midbrain floor plate progenitors. Surprisingly, RSPO2, but not SPON1, specifically promoted the differentiation of mDA neuroblasts into mDA neurons in mouse primary cultures and embryonic stem cells (ESCs). In addition, RSPO2 was found to promote not only mDA differentiation, but also mDA neurogenesis in human ESCs. Our results thus uncover an unexpected function of the matricellular protein RSPO2 and suggest an application to improve mDA neurogenesis and differentiation in human stem cell preparations destined to cell replacement therapy or drug discovery for Parkinson disease.
Keywords: extracellular matrix, RSPO2, progenitors, dopaminergic neurons, differentiation, neurogenesis, human embryonic stem cells, Wnt, cell replacement, Parkinson disease
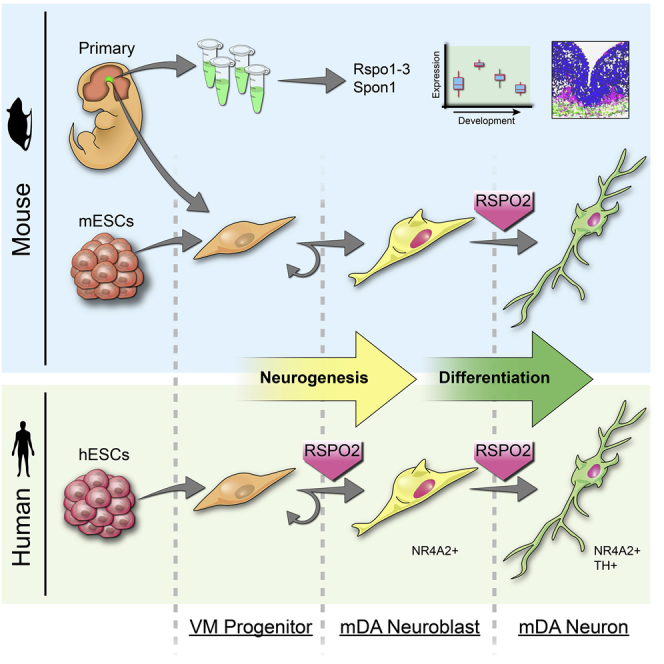
Graphical Abstract
Highlights
-
•
Rspo2 is dynamically expressed during midbrain dopaminergic neuron development
-
•
RSPO2 promotes the dopaminergic differentiation of mouse neurons in culture
-
•
RSPO2 increases dopaminergic neurogenesis and differentiation of human ESCs
Gyllborg and colleagues report on the function of the matricellular protein R-Spondin 2 (RSPO2) in dopaminergic neuron development. RSPO2 is dynamically expressed during midbrain development and promotes the dopaminergic differentiation of mouse and human neuroblasts. Furthermore, they show that RSPO2 induced dopaminergic neurogenesis in human stem cell cultures, suggesting a possible application of RSPO2 in cell replacement strategies for Parkinson disease.
Introduction
The development of ventral midbrain (VM) dopaminergic (DA) neurons involves multiple parallel, controlled processes encompassing many signaling molecules and transcription factors tightly coordinated in order to produce functional midbrain DA (mDA) neurons (reviewed in Arenas et al., 2015). Wnt signaling has been shown to play many essential functions in VM DA neuron development both in vivo and in vitro, by activating both Wnt/β-catenin-dependent and/or -independent pathways (Arenas, 2014, Inestrosa and Arenas, 2010). In brief, Wnt1/β-catenin-dependent signaling in the midbrain is responsible for the specification of the mDA progenitor domain, progenitor proliferation, and survival at late stages (Castelo-Branco et al., 2003, Prakash et al., 2006, Andersson et al., 2013). Wnt5a/β-catenin-independent signaling is responsible for regulating midbrain morphogenesis, neurogenesis, and mDA precursor differentiation (Andersson et al., 2008, Andersson et al., 2013, Castelo-Branco et al., 2003). Other VM Wnt/β-catenin activators include Wnt2 responsible for increasing progenitor proliferation (Sousa et al., 2010) and Wnt7a for progenitor proliferation, neurogenesis, and axon morphogenesis (Fernando et al., 2014). Wnt modulators and inhibitors including Sfrp1/2 have a role in potentiating the Wnt/PCP signal (Kele et al., 2012), and Dickkopf members such as Dkk1 regulate mDA differentiation and morphogenesis (Ribeiro et al., 2011), while Dkk3 promotes mDA neuron differentiation (Fukusumi et al., 2015).
The roof plate-specific spondin (RSPO) family of secreted matricellular Wnt modulator proteins (RSPO1-RSPO4) show ∼60% amino acid homology and share similar domain structure (de Lau et al., 2012). RSPOs contain a thrombospondin type I repeat domain (Chen et al., 2002, Kazanskaya et al., 2004) that is shared with spondin 1 (SPON1; floor plate spondin), which promotes neural cell adhesion, neurite outgrowth, and nerve precursor differentiation (Feinstein et al., 1999, Klar et al., 1992, Schubert et al., 2006). RSPOs also contain two furin-like cysteine-rich domains that are necessary to activate the Wnt/β-catenin pathway (Kim et al., 2008) and to maintain the stem cell compartments in organs such as the intestine, liver, pancreas, kidney, and hair follicle among others (Barker et al., 2007, Barker et al., 2012, Huch et al., 2013a, Huch et al., 2013b, Jaks et al., 2008).
RSPOs act synergistically to activate Wnt/β-catenin (Kazanskaya et al., 2004, Kim et al., 2005) by binding to the leucine-rich repeat-containing G protein-coupled receptor 4, 5, or 6 (LGR4-6) instead of the typical WNT receptors, frizzleds (FZDs), or the co-receptor, low-density lipoprotein receptor-related protein 5 or 6 (LRP5/6) (Carmon et al., 2011, Glinka et al., 2011, de Lau et al., 2011). Upon RSPO binding to LGR, the transmembrane E3 ubiquitin ligases, ring finger protein 43 (RNF43), or the zinc and ring finger 3 (ZNRF3), bind to RSPO (Chen et al., 2013) and become part of the RSPO-LGR complex (Hao et al., 2012, Koo et al., 2012). This prevents RNF43 and ZNRF3 from interacting with the WNT-FZD complex and leads to the ubiquitination and degradation of RSPO-LGR instead of the WNT-FZD-LRP signaling complex, thus increasing Wnt/β-catenin signaling (de Lau et al., 2014, Zebisch and Jones, 2015). However, RSPOs have also been reported to inhibit Wnt/β-catenin signaling (Rong et al., 2014) and to activate Wnt/PCP signaling by binding to the heparan-sulfateproteoglycan, syndecan 4 (Ohkawara et al., 2011), suggesting that their activity may be cell context dependent.
Rspo2 has been shown to play roles in morphogenesis of the respiratory tract as well as limb development (Bell et al., 2008). Within the nervous system, Rspo2 expression is regulated by the LIM homeodomain transcription factor, Lmx1a (Hoekstra et al., 2013), a transcription factor required for the specification of the midbrain floor plate and mDA neuron development (Andersson et al., 2006, Deng et al., 2011). In addition, miR135a2 has been predicted to target Rspo2 and Lmx1b. While deletion of the Lmx1b results in reduced Wnt/β-catenin signaling and the mDA progenitor pool (Anderegg et al., 2013), deletion of Rspo2 resulted only in a modest decrease in the immunoreactivity of mDA neuron markers, such as TH (tyrosine hydroxylase; the rate-limiting enzyme for dopamine synthesis) and PITX3 (paired-like homeodomain transcription factor 3; a transcriptional regulator of mDA neuron differentiation [Maxwell et al., 2005, Nunes et al., 2003]). It is thus at present unknown whether the development of mDA neurons can be controlled by proteins of the RSPO family or SPON1 and whether they can control stem cell behavior as described in other tissues.
Here we report that Rspo2, but not Rspo1, 3, 4, or Spon1, is dynamically expressed in the VM during the critical time window of mDA neurogenesis. Moreover, analysis of the function of recombinant RSPO2 protein revealed robust positive effects on mDA differentiation that resulted in a 2-fold increase in the number of mDA neurons, not only in primary VM cultures but also in mouse and human embryonic stem cell (ESC) preparations. Thus, our results identify RSPO2 as an efficient factor capable of promoting mDA neuron differentiation in both mouse and human stem cell differentiation protocols. This knowledge can be applied to improve current human ESC-based models and cell replacement strategies for Parkinson disease focusing on the loss of mDA neurons.
Results
Expression of Spondins in the Developing Mouse Midbrain Floor Plate
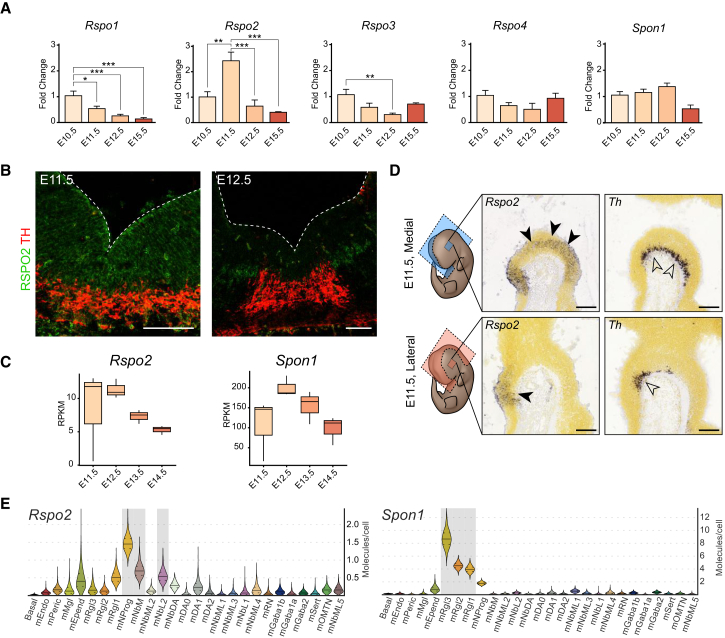
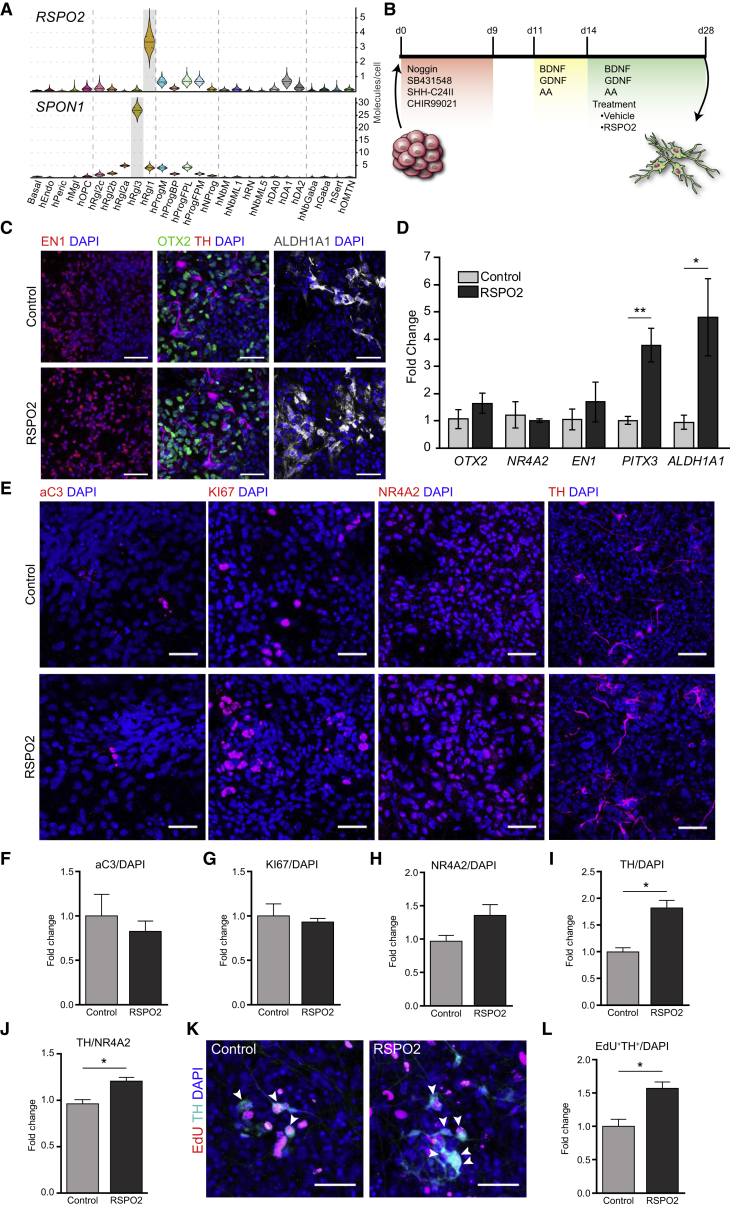
We first determined the expression of all the roof plate family of Spondins, Rspo1-4, and the related floor plate Spon1 (hereafter collectively referred to as spondins) in the developing VM. A clear differential upregulation of the expression of only Rspo2 was detected by qPCR at embryonic day 11.5 (E11.5), during mDA neurogenesis and differentiation. All the other spondins either decreased over time or remained relatively constant from E10.5 to E15.5 (Figure 1A). Immunohistochemistry (IHC) showed diffuse expression of RSPO2 in the midbrain floor plate at E11.5, which later concentrated in patches at E12.5 (Figure 1B). TruSeq RNA sequencing (RNA-seq) of the VM between E11.5 and E14.5 (Figure S1A) (Toledo et al., 2017a) confirmed that Rspo2 is expressed early in mDA development and falls at later stages. This technique also revealed very low expression levels of Rspo1, Rspo3, and Rspo4 (less than 1.5 reads per million, Figure S1B). In contrast, Rspo2 and Spon1 were expressed at one or two orders of magnitude higher, respectively (Figure 1C). The expression pattern of Rspos in the VM were also examined in the Allen Developing Mouse Brain Atlas (Allen Institute for Brain Science, 2017), where we found that Rspo2 is clearly expressed in mDA progenitors adjacent to mDA neurons, identified by Th expression in the marginal zone (Figures 1D and S1C, arrowheads). Lower expression of Rspo2 was detected at E13.5, while other RSPOs were undetectable in the VM, but present in other structures (Figures S1D and S1E). Notably, Rspo2 was also detected in a subset of mDA neurons of the substantia nigra pars compacta region, at postnatal day 56 (Figure S1F) (Lein et al., 2007). Moreover, analysis of our single-cell RNA-seq dataset of the developing mouse VM from E11.5 to E18.5 (La Manno et al., 2016) revealed that Rspo2 is significantly expressed in early neural progenitor cells and medial neuroblasts, two cells in the mDA neuron lineage, while Spon1 was expressed in radial glia-like cells (Figure 1E). Other spondins were either undetected, expressed uniformly or in varying cell types at low copy number (Figure S1G). We thus can conclude that Spon1 and Rspo2 are expressed at significant levels in the midbrain floor plate, Rspo2 being specifically expressed in cells of the mDA lineage and dynamically regulated during mDA neurogenesis.
Figure 1.
Expression of Spondins in the Developing Ventral Midbrain
(A) Expression of Rspo(1–4) and Spon1 measured by qPCR in VM tissue from E10.5 to E15.5. Statistical analysis compared with E11.5 for Rspo2, the rest compared with E10.5. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by ANOVA. Data presented as means ± SEM measured in a.u., normalized to starting time point, E10.5.
(B) Detection of RSPO2 and TH in the VM by IHC, at E11.5 and E12.5. Dashed lines delineate the ventricular cavity. Scale bars, 100 μm.
(C) Expression levels of Rspo2 and Spon1 by TruSeq RNA-seq analysis of VM tissue obtained from TH-GFP mice over time points E11.5 to E14.5 (Toledo et al., 2017a). See also Figure S1A.
(D) E11.5 mouse VM in situ hybridization (image data from Allen Institute for Brain Science: Allen Developing Mouse Brain Atlas) for Rspo2 (solid arrowheads) and Th (open arrowheads) as a reference. Sagittal sections through the midline (upper panels) and lateral (lower panels). Scale bars, 200 μm.
(E) Violin plots generated from single-cell RNA-seq data of the developing mouse VM. Rspo2 and Spon1 expression levels are shown across all known cell types. Right axis shows absolute molecule counts. Gray, enriched over baseline with posterior probability >99.8%. Cell types with enriched expression: mNProg, neuronal progenitor; mNbM, neuroblast medial; mNbML2, neuroblast mediolateral 2; mRgl1-3, radial glia-like cells 1-3. For the rest of the nomenclature, see La Manno et al. (2016).
See also Figure S1.
Expression of Spondin Receptors in the Developing Mouse Midbrain Floor Plate
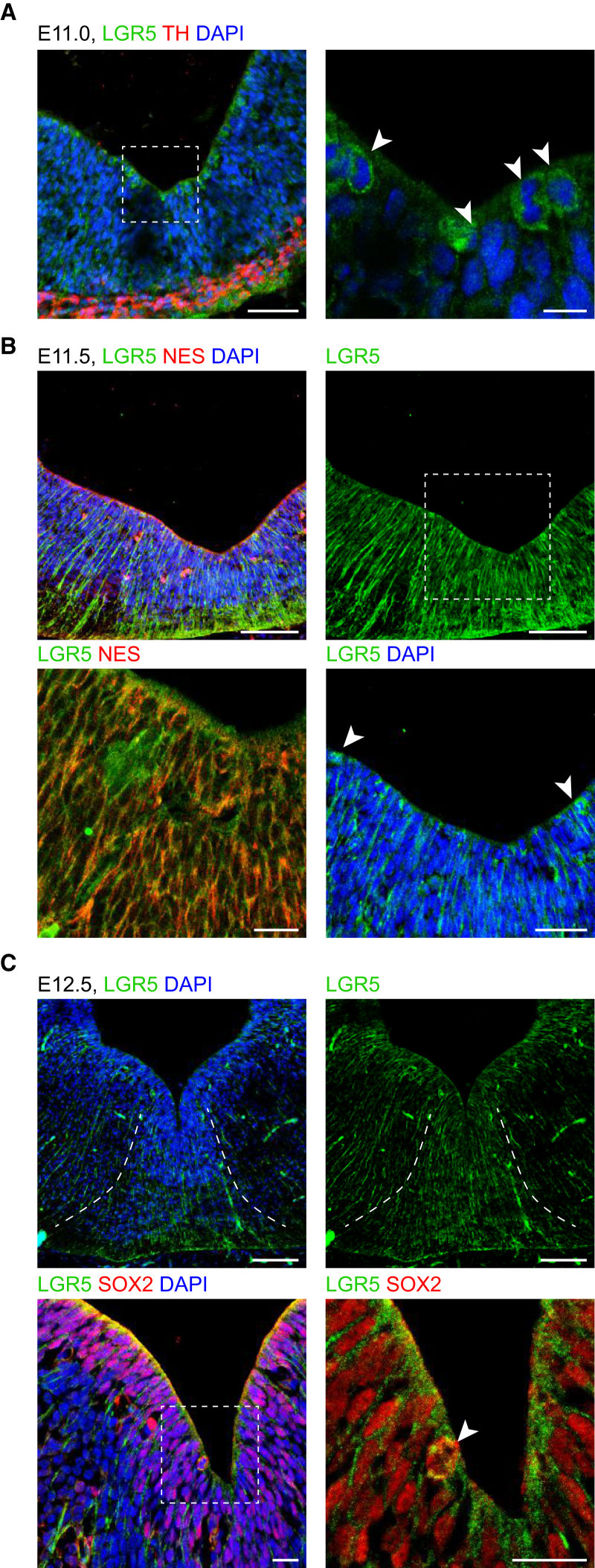
The expression of Lgr4-6 and associated receptors were first examined in bulk RNA-seq data obtained from the developing VM. We found that Lgr6 is expressed at very low levels and that Lgr4 and Lgr5 were not developmentally regulated between E11.5 and E14.5 (Figure S2A). Single-cell RNA-seq analysis revealed significant expression of both receptors in basal plate cells, such as radial glia 2 (Lgr4 and Lgr5) as well as lateral neuroblast 2 and GABA2 neurons (Lgr4) (Figure S2B). This result was confirmed by IHC, which revealed the presence of immunoreactive cells in the basal plate (Figure S2C). However, IHC for LGR5 revealed a more dynamic distribution of this protein than expected (Figure 2). At E11, individual LGR5+ cells sparsely lining the ventricle were detected (Figure 2A, arrowheads). At E11.5, LGR5 IHC was clearly detected in their processes, which co-localized with the radial glia marker Nestin (NES) (Figure 2B), but its expression decreased in cell bodies lining the ventricle (Figure 2B, arrowheads). At E12.5, the levels of LGR5 decreased, resulting in less LRG5+ radial glia processes in the basal plate than in the floor plate (Figure 2C), and very few LRG5+ cell bodies (Figure 2C, arrowhead). Analysis of the expression of the two additional RSPO receptors, revealed low levels of Rnf43, peaking at E11.5–E12.5, and very low or undetectable levels of Znrf3 by either bulk or single-cell RNA-seq (Figures S2D and S2E). Receptors of SPON1, APP, and APOER2/LRP8 (Ho and Südhof, 2004, Hoe et al., 2005) were also expressed, as observed through the RNA-seq data and in situ hybridization images (Figures S2F and S2G). Thus, our results show that the developing midbrain expresses Spondins receptors, suggesting that the ligands may play a role in development.
Figure 2.
Identification of LGR5-Positive Cells in Early VM Development
(A) IHC showing LGR5+ cells lining the ventricle in the VM at E11. Right panel shows a higher magnification of LRG5+ cells in the left panel, identified with arrowheads. Scale bars, 50 μm (left panel) and 10 μm (right panel).
(B) LGR5 IHC at E11.5 showed less positive cells bodies (arrowheads) and a strong staining radial processes co-stained with Nestin (NES), visible at intermediate (upper right) and higher magnification (lower right). Scale bars, 100 and 20 μm (lower left panel).
(C) LGR5 and SOX2 IHC in the VM at E12.5. Arrowhead shows sporadic double-positive cells in the ventricular zone (VZ), while LRG5 fibers are found to be more restricted to the VM domain. Dashed lines delineate the floor plate. Scale bars, 100 μm (upper panels) and 20 μm (lower panels).
See also Figure S2.
RSPO2, but Not RSPO1 or SPON1, Promotes the Differentiation of Neuroblasts into mDA Neurons
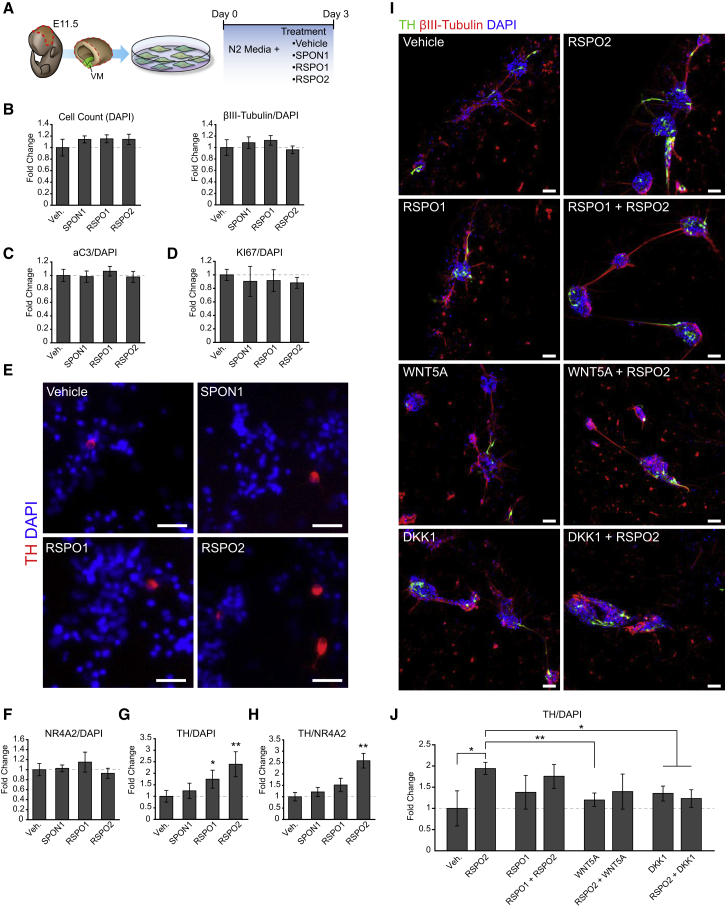
We next examined the function of the two spondins expressed at higher levels during VM development, RSPO2 and SPON1. E11.5 VM primary cultures were treated for 3 days with recombinant RSPO1, RSPO2, or SPON1 (Figure 3A). We first found that the total number of cells, identified by nuclear DAPI staining, or the total number of neurons in the culture, visualized by immunocytochemistry (ICC) with the pan-neuronal marker βIII-tubulin, did not change in the culture (Figures 3B and S3A). Accordingly, we did not detect any change in cell death, as assessed by active caspase-3 staining, or in the number of proliferating cells as determined by Ki67 (Figures 3C and 3D, respectively). We next examined whether the proportion of cells in the mDA lineage changed with the treatments. Postmitotic cells in the mDA lineage (neuroblast and neurons), marked by the presence of NR4A2 (nuclear receptor subfamily 4, group A, member 2; also known as Nurr1), showed no significant difference between the treatments (Figure 3F). However, in cultures treated with recombinant RSPO1 or RSPO2, but not with SPON1, a significant increase in the number of TH+ mDA neurons was detected (∼1.75- and ∼2.5-fold increase, respectively) treatment (Figures 3G and 3E). We then examined whether this effect was due to an increase in the proportion of postmitotic NR4A2+ neuroblasts that differentiate into TH+ mDA neurons and found that this was the case for RSPO2 (Figure 3H). We also performed expression analysis of DA genes by qPCR in RSPO2-treated cultures (Figure S3B) and found a significant increase in Aldh1a1 expression, a marker for the substantia nigra pars compacta and VTA type 2 cell types (La Manno et al., 2016), but no changes in Otx2, Lmx1a, Nr4a2, En1, or Pitx3, suggesting a very specific role of RSPO2 in late differentiation. Combined, our results indicate that RSPO2 is the only spondin capable of promoting the development of mDA neurons, and that it does so by promoting the differentiation of NR4A2+ neuroblasts into a subset of TH+ mDA neurons.
Figure 3.
RSPO2 Promotes Midbrain Dopaminergic Neurons Differentiation in Primary Cultures
(A) Schematic representation of the mouse primary cell culture protocol.
(B) Quantification of the total cell numbers (DAPI) and proportion of neurons (βIII-tubulin/DAPI) normalized to control (Vehicle).
(C) Quantification of ongoing cell death, measured by active caspase-3 ICC (aC3+/DAPI), showed no difference compared with Vehicle.
(D) Quantification of KI67 ICC (KI67+/DAPI) showed no difference compared with Vehicle.
(E) ICC staining showing and increase in TH+ neurons compared with DAPI after RSPO2 treatment, but not RSPO1 or SPON1. Scale bars, 50 μm.
(F) Quantification of postmitotic cells in the DA lineage, measured by NR4A2 ICC, showed no difference compared with Vehicle.
(G) Bar plots showing that RSPO2 treatment increases the total number of TH+ cells (TH/DAPI), compared with control (Vehicle).
(H) Normalized ratio of TH+ cells/NR4A2+ cells revealed an increase in the proportion of postmitotic dopaminergic cells (NR4A2+) becoming mDA neurons (TH+).
(I) ICC staining of cells treated with various Wnt signaling pathway modifiers, stained with TH and βIII-tubulin. Scale bars, 50 μm.
(J) Quantification of TH+ cells in various treatments from (I) as compared with Vehicle.
All data normalized to control (Vehicle) presented as means ± SEM. n = 3, t test; ∗p < 0.05, ∗∗p < 0.01. See also Figure S3.
We also examined whether the activity of RSPO2 in primary cultures could be potentiated or inhibited by co-treatment with RSPO1 or modulators or Wnt signaling. Surprisingly we found that RSPO1 reduced the increase in the number of TH+ neurons by RSPO2 (Figures 3I and 3J), indicating that RSPO1 and RSPO2 have clearly distinct activities. Moreover, we found that co-treatment of primary cultures with either WNT5A or Dkk1, both of which inhibit Wnt/β-catenin signaling in the VM (for review see Arenas, 2014), reduced the effects of RSPO2 on the number of TH+ cells (Figures 3I and 3J). This result suggests that the effect of RSPO2 on mDA neurons involves, at least partially, the activation of Wnt/β-catenin signaling.
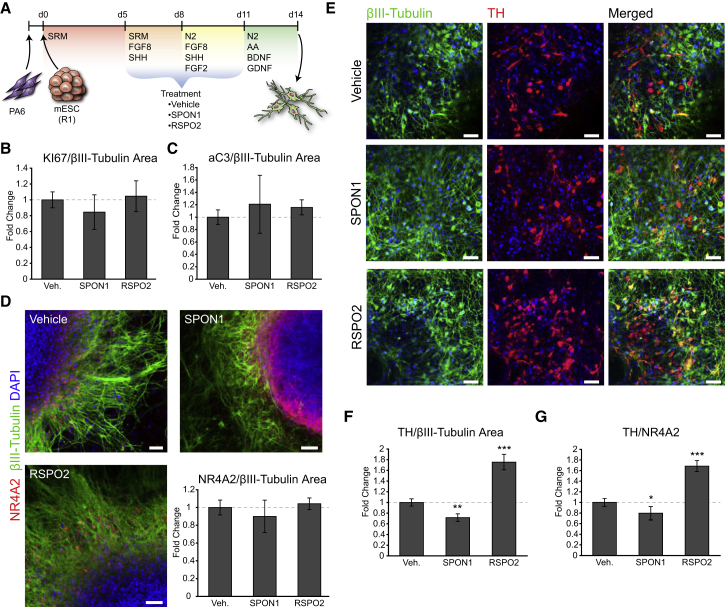
To rule out an early effect of spondins on mDA progenitors, we examined earlier stages of mDA neuron development in mouse ESCs. We used a 14-day differentiation protocol and treated the cultures with either SPON1 or RSPO2 between day 5 and 11 (Figure 4A). However, neither SPON1 nor RSPO2 showed any significant effect on cell proliferation or cell death as shown by active caspase-3 and Ki67 immunostainings (Figures 4B and 4C). Similar to the primary cultures, the number of postmitotic NR4A2+ neuroblasts did not change by SPON1 or RSPO2 treatment (Figure 4D), and a significant 1.8-fold increase in TH+ neurons was detected (Figures 4E and 4F). Although, SPON1 induced a significant 30% decrease in the number of TH+ cells (Figures 4E and 4F). Analysis of the proportion of TH+ neurons being generated from NR4A2+ neuroblasts in differentiated mouse ESCs also revealed a modest decrease in DA differentiation by SPON1 and a clear increase by RSPO2 (Figure 4G). These results thus confirm the specific role of RSPO2 in promoting the late differentiation of mDA neuroblasts into neurons in the developing mouse VM.
Figure 4.
RSPO2 Promotes the Dopaminergic Differentiation of Mouse ESCs
(A) Cartoon showing the protocol for the differentiation of mouse ESCs into mDA neurons and the time line (d, days) of spondin treatment, between days 5 and 11.
(B) Quantification of proliferation (KI67) in areas undergoing neurogenesis (βIII-tubulin+) as assessed by ICC, normalized to control.
(C) Quantification of ongoing cell death by active caspase-3 (aC3) ICC in βIII-tubulin+ areas.
(D) ICC for NR4A2 and βIII-tubulin after 14 days in vitro and bar plots showing no difference in the number of NR4A2+ cells per βIII-tubulin area. Scale bars, 50 μm.
(E) ICC staining for TH and βIII-tubulin after 14 days in vitro in vehicle-, SPON1-, and RSPO2-treated cultures. Scale bars, 50 μm.
(F) The number of TH+ cells per βIII-tubulin area by ICC, normalized to control, increased by RSPO2 treatment and decreased by SPON1.
(G) The proportion of NR4A2+ cells that become TH+ increased by RSPO2 treatment and decreased by SPON1.
All data normalized to control (Vehicle) presented as means ± SEM. n = 3, t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
RSPO2 Promotes the Differentiation of Human ESCs into DA Neurons and mDA neurogenesis
We next examined whether the function of RSPO2 is conserved in human DA neuron development and whether RSPO2 could be used to improve the differentiation of human ESCs into mDA neurons. We first investigated the expression of RSPO2 and SPON1 in our single-cell RNA-seq dataset from human VM development (La Manno et al., 2016). Interestingly, we found higher levels of expression of RSPO2 and SPON1 in the human VM compared with mouse and significant differences in the cell types expressing them. While RSPO2 is exclusively expressed in radial glia type 1 (hRgl1), SPON1 is exclusively expressed in radial glia type 3 (hRgl3, Figure 5A). Analysis of the expression of the Rspo receptors by single-cell RNA-seq revealed very low levels of LGR4-6, RNF43, and ZNRF3 (mean molecules/cell) and higher levels of LGR5 in different cells, including a floor plate progenitor (hProgFPM, Figure S4A), a cell type thought to give rise to mDA neurons (La Manno et al., 2016). We thus decided to examine the possible role of RSPO2 on mDA neuron development in human ESC cultures.
Figure 5.
Role of RSPO2 in Differentiation of Human ESCs
(A) Violin plots generated from single-cell RNA-seq data of the developing human VM. RSPO2 and SPON1 expression levels are shown across all cell types in the human VM. Right axis shows absolute molecule counts. Gray, enriched over baseline with posterior probability >99.8%. Cell types with enriched expression: hRgl1,3, radial glia-like cells 1,3. For the rest of the cell-type nomenclature, see La Manno et al. (2016).
(B) Protocol for the dopaminergic differentiation of human ESCs.
(C) Human ESC cultures after 28 days stained for various DA lineage markers: EN1, OTX2, TH, and ALDH1A1. Scale bars, 50 μm.
(D) Expression of DA markers (OTX2, NR4A2, EN1, PITX3, and ALDH1A1) by qPCR between Vehicle- and RSPO2-treated cultures after 28 days (unpaired t test, n = 3, ∗p < 0.05, ∗∗p < 0.01).
(E) Human ESC cultures after 28 days stained for markers of apoptosis (aC3), proliferation (KI67), markers of DA lineage (NR4A2 and TH). Scale bars, 50 μm.
(F) The number of apoptotic cells (aC3+/DAPI) was no modified by RSPO2 treatment.
(G) The number of proliferating cells (KI67+/DAPI) was the same in both conditions.
(H) The number of NR4A2+ cells were not significantly different in RSPO2-treated and control cultures (t test, n = 3, p = 0.09).
(I) A significant increase in the total number of DA neurons (TH+/DAPI) was detected in RSPO2-treated human ESC cultures (t test, n = 3, ∗p = 0.01).
(J) RSPO2 treatment increases the proportion of NR4A2+ cells that become TH+ in human ESC cultures (t test, n = 3, ∗p = 0.02).
(K) RSPO2 increases DA neurogenesis in human ESC cultures, as assessed by the incorporation of EdU in TH+ cells at day 28, arrowheads. Scale bars, 50 μm.
(L) Quantification of the number of EdU/TH double-positive cells showed that RSPO2 significantly increases DA neurogenesis in human ESC cultures (t test, n = 3, ∗p = 0.03).
See also Figure S4.
Human ESCs were differentiated for 28 days as described recently (Nolbrant et al., 2017), using a protocol that includes the use of GSK3β inhibitors to enhance the induction of human midbrain floor plate (for review see Arenas et al., 2015). Human ESCs were treated with recombinant RSPO2 from day 14 to 28 (Figure 5B) and characterized by ICC and qPCR with various mDA markers (Figures 5C and 5D). While no change was detected for OTX2, NR4A2, or EN1, we observed a significant increase in PITX3 and ALDH1A1 expression. We also found that the levels of ALDH1A1 protein increased by RSPO2 treatment, complementing thus the previous finding of decreased levels in Rspo2−/− mice (Hoekstra et al., 2013). Also, in line with both our primary and mouse ESC cultures, we found that RSPO2 induced no significant differences in the total number of cells (DAPI), cell death (active caspase-3), or proliferation (Ki67) in the human ESC cultures (Figures 5E–5G). Notably, quantification of the number of postmitotic cells in the DA lineage revealed no significant difference in the number of NR4A2+ cells (Figures 5E and 5H), but a significant ≈1.8-fold increase in the number of TH+ neurons treated with RSPO2 (Figures 5E and 5I). A significant ≈1.2-fold increase in the proportion of NR4A2+ cells that became TH+ was detected (Figure 5J), suggesting that RSPO2 also promotes the differentiation of neuroblasts into mDA neurons. However, this increase was not sufficient to explain the ≈1.8-fold increase in TH+ cells, and suggested the involvement of an additional upstream mechanism. To mark proliferating progenitors, we performed a 5-ethynyl-2'-deoxyuridine (EdU) pulse-chase experiment and examined whether they undergo neurogenesis as identified by the presence of cells double-positive for EdU and TH. Our results show that RSPO2 induced a significant increase in the EdU+ cells that undergo neurogenesis and become TH+ (Figures 5K and 5L). Thus, our data indicate that RSPO2, in addition of promoting the differentiation of neuroblasts into mDA neurons in mouse and human, it promotes mDA neurogenesis in human ESC cultures.
Discussion
In this study we examine the expression and function of the extracellular matrix (ECM) proteins of the RSPO family and of SPON1, in the mouse and human VM and ESCs. Our analysis revealed that RSPO2, SPON1, and LGR5 are the most significantly expressed spondins and receptors in the VM in both the developing mouse and human VM. However, we identified substantial differences in the cell types expressing spondins. In the mouse, Rspo2 was found in neuronal progenitors and neuroblasts and Spon1 in radial glia 1–3; whereas, in the human VM, RSPO2 and SPON1 were exclusively expressed in radial glia 1 and 3, respectively. An additional difference between the two species was that human RSPO2 was expressed in earlier cell types (hRgl1), compared with mouse Rspo2, which was expressed in a rapid amplifying type of progenitor (mNProg) and the medial postmitotic neuroblast (mNbM) (La Manno et al., 2016). In agreement with this, we found an earlier function of RSPO2 to promote mDA neurogenesis in human ESCs and a shared late function of RSPO2 in mouse and human, to promote the differentiation of postmitotic neuroblasts into mDA neurons. Unlike other systems in which RSPOs control stem cell or progenitor maintenance (de Lau et al., 2014) and SPON1 regulates differentiation (Schubert et al., 2006), we did not observe any effect of RSPOs on the number of Ki67+ cells or the total number of EdU+ cells in human ESC cultures, or of SPON1 to promote differentiation. This is not totally surprising as the midbrain floor plate expresses genes otherwise present in completely different compartments (i.e., roof plate genes, such as Lmx1a, Msx1, Wnt1, and Rspo2, together with floor pate genes such as Shh, Foxa2, Wnt5a, and Spon1), resulting in different regulatory networks and functions compared with other brain regions or structures. One such example is the presence of neurogenesis in the midbrain floor plate, which represents an exception to the entire neural tube. Notably our results indicate that roof plate RSPO2, as other “roof plate factors” in the VM (i.e., Lmx1a, Msx1, and Wnt1; see Arenas et al., 2015 for review) also regulates mDA neurogenesis.
Understanding human mDA neuron development in vivo and implementing such knowledge to protocols for the differentiation of human ESCs into mDA neurons is currently considered as the basis for refining and improving existing protocols to prepare cells for cell replacement therapy. In the past, the implementation of correct levels of Wnt signaling for the specification of midbrain floor plate progenitors has been critical for the development of protocols allowing the transplantation of mDA progenitors capable of efficiently differentiating into functional mDA neurons in vivo (Kriks et al., 2011, Nolbrant et al., 2017). These protocols are still susceptible to improvement by implementing new factors and knowledge, such as additional components of the Wnt signaling pathway recently reported in the developing mouse and human VM (Toledo et al., 2017b) or the Wnt modulator that we hereby report, RSPO2. As an ECM protein, RSPO2 may function to define, together with other ECM proteins, the niche where human mDA neurogenesis takes place. Indeed, our results indicate that by manipulating the extracellular compartment it is possible to control essential intracellular functions such as mDA neurogenesis and differentiation in human ESC cultures. Moreover, RSPO2 may also play a role in mDA neuron subtype specification, as suggested by the increase in ALDH1A1 in mouse and human cell expression by RSPO2 treatment (our results) or the decrease in its protein levels in RSPO2−/− mice (Hoekstra et al., 2013). While much attention has been devoted to the role of transcription factors and morphogens in development and stem cell biology, only recently have we started to implement the knowledge of what can be considered as the third pillar of development: the ECM. Indeed, ECM proteins are gaining increasing interest as the accrued knowledge of their diversity (reviewed in Murphy-Ullrich and Sage, 2014) and functions begin to accumulate (Ganapathy et al., 2016, Long et al., 2016, Wiese and Faissner, 2015). In this context, our study identifies RSPO2 as a matricellular component capable of promoting mDA neurogenesis and differentiation. Our results thus pave the way for RSPO2 to be used to improve current human ESC preparations for regenerative medicine or drug discovery for Parkinson disease.
Experimental Procedures
Animals
Wild-type (WT) CD-1 (Charles River, Germany) or WT Swiss (Janvier Labs) mice were mated overnight, and the noon of the day of a plug was considered E0.5. Mice were shipped as pregnant females, and then housed and treated in accordance with the guidelines of the local ethics committee (Stockholm's Norra Djurförsöksetisks Nämd. Ethical numbers N145/09, N273/11, N326/12, and N158/15).
Tissue Collection
After CD-1 mice were killed and embryos collected at different stages, embryonic heads were fixed in 4% paraformaldehyde (PFA) for at least 6 hr at +4°C. Then tissue was washed briefly in PBS and processed to 30% sucrose/PBS solution overnight at +4°C. Tissue was then embedded in O.C.T. (TissueTek) and stored at −80°C until sectioning. Sectioning was performed on a Leica cryostat at 16 μm sections used for IHC analysis.
qPCR
Mice
RNA was isolated from dissected WT CD-1 mice VM from various time points. RNA was extracted using RNeasy Mini Kit (QIAGEN). SYBR green real-time qPCR assay was performed as described previously (Rawal et al., 2006, Sacchetti et al., 2009).
Mouse Primary Culture
Total RNA was isolated using RNeasy Kit (QIAGEN), and cDNA was made with cDNA SuperScript II Reverse Transcriptase Kit (Thermo Fisher Scientific). Specific genes were amplified using Fast SYBR Green Master Mix Kit (Thermo Fisher Scientific). Real-time PCR was performed using fast protocols on a 7500 Fast Real-Time PCR system (Thermo Fisher Scientific). GAPDH was used to normalize the expression of mRNA.
Human ESC Culture
Total RNA was extracted from cells at day 28 of culture using the NucleoSpin RNA extraction kit (Macherey-Nagel, cat. no. 740955.10) as per the manufacturer's instructions. Quantity and quality of RNA was determined using a NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized from 500 ng of RNA using SuperScript II Reverse Transcriptase (Invitrogen). SYBR green (QIAGEN) qPCR assays were performed using a LightCycler 480II (Roche) and primers purchased from Sigma (sequences in table). A 5-fold serial dilution standard curve was generated for each primer to generate efficiency curves and relative expression levels were obtained using the Pfaffl method (Pfaffl, 2001). HPRT was used as the housekeeping gene.
Primer sequences in Table S1.
RNA-Seq Datasets and Gene Expression Profiles
Gene expression data were obtained through previous datasets. For tissue TruSeq datasets, see Toledo et al. (2017a), and for single-cell RNA-seq datasets see La Manno et al. (2016).
Primary Cell Culture
Plates were coated with poly-D-lysine and VMs were dissected out from E11.5 WT embryos into ice-cold PBS. VMs were dissociated by trituration using flame-polished glass Pasteur pipettes. Cells were plated at a density of 150,000 cells/cm2. They were grown in N2 medium (1:1 F12:MEM, HEPES, N2 supplement, and glutamine; Thermo Fisher Scientific) with additional 6 mg/mL glucose and 3 mg/mL AlbuMAXI BSA. Cells were treated with for 3 days with different recombinant proteins (recombinant human F-spondin protein 80 ng/mL [cat. no. 3135-SP], recombinant human R-spondin 2 protein 160 ng/mL [cat. no. 3266-RS], recombinant mouse R-spondin 1 protein 160 ng/mL [cat. no. 3474-RS], recombinant mouse Dkk-1 protein 100 ng/mL [cat. no. 5897-DK], and recombinant human/mouse Wnt-5a protein 100 ng/mL [cat. no. 645-WN], all resuspended in 0.1% BSA/PBS; R&D Systems) or vehicle (0.1% BSA/PBS). Cells were then fixed with 4% PFA and processed for staining.
Mouse ESC Culture
Mouse ESC (R1) cultures were co-cultured with immortalized PA6 feeder cells. Differentiation of mouse ESCs was done following a 14-day in vitro protocol (Barberi et al., 2003). Mouse ESCs were maintained in proliferation medium (Knockout-DMEM, 15% Knockout Serum Replacement, 2 mM L-glutamine, 10,000 U/mL penicillin/streptomycin [all from Life Technologies], 1% non-essential amino acids [VWR], 0.1 mM β-mercaptoethanol [Sigma], and 1,000 U/mL leukemia inhibitory factor [LIF] [ESGRO; Chemicon/Millipore]). Mouse ESCs were plated (150 cells/cm2) on a confluent mitomycin C-treated PA6 feeder cell layer in 24-well plates in proliferation medium without LIF. At day 5, sonic hedgehog (SHH) (200 ng/mL) and fibroblast growth factor 8 (FGF8) (25–100 ng/mL) were added to the medium. After 8 days, N2 medium was used in the presence of SHH, FGF8, and basic FGF (10 ng/mL). From day 11 to 14, N2 medium containing ascorbic acid (AA) (200 μM) (Sigma), brain-derived neurotrophic factor (BDNF) (20 ng/mL) (R&D Systems), and glial cell line-derived neurotrophic factor (GDNF) (10 ng/mL) (R&D Systems) was used. Treatments with recombinant spondins or vehicle started at day 5 and lasted up until day 11, concentrations the same as primary cultures. Cells were then fixed with 4% PFA and processed for staining.
Human ESC Culture
Undifferentiated RC17 ESCs (passages 33–54, Roslin Cells, hPSC reg. no. RCe021-A) were maintained in E8 medium (A1517001) on Geltrex (1%, 12760021)-coated plates and passaged weekly with EDTA (0.5 mM). To start differentiation (day 0), human ESC colonies were detached using EDTA (0.5 mM) and placed in non-treated 60-mm culture dishes in differentiation medium consisting of DMEM:F12/Neurobasal (1:1), N2 supplement (1:100), B27 supplement (1:50), SB431542 (10 μM, Tocris Biosciences), rhnoggin (100 ng/mL, R&D Systems), SHH-C24II (200 ng/mL, R&D Systems), and CHIR99021 (0.9 μM, Tocris Biosciences). Medium was changed once on day 2. The resultant embryoid bodies were collected on day 4 and placed on polyornithine (PO)-, fibronectin (Fn)-, and laminin (Lm)-coated plates in reduced N2 (1:200) and B27 (1:100) condition. Growth and patterning factors were removed on day 9 with the cultures kept in DMEM:F12/Neurobasal (1:1), N2 supplement (1:200), and B27 supplement (1:100). On day 11 of differentiation, the cell clusters were dissociated to single cells with accutase and replated onto dry PO/Fn/Lm-coated plates in Neurobasal, B27 (1:50), BDNF (20 ng/mL), GDNF (10 ng/mL), AA (200 μM), and db-cAMP (0.5 mM, Sigma). From day 14, cultures were treated with RSPO2 (1 μg/mL, R&D Systems) up until day 28, when they were fixed in 3.7% PFA for 30 min. EdU pulse was administered on day 23 for 24 hr to capture all the dividing cells. Cells were then fixed 4 days later on day 28. All culture reagents are from Invitrogen unless otherwise stated here or previously.
IHC/ICC
Cells and tissue were fixed in 4% PFA, washed in PBS and blocked in PBTA (PBS, 5% normal goat/donkey serum [Jackson ImmunoResearch], 0.1% Triton X-100, and 1% BSA) for 1 hr at room temperature (RT). Primary antibodies were diluted in 0.1% Triton X-100 and 1% BSA/PBS, and incubations were carried out overnight at 4°C.
Antibodies used were: βIII-tubulin (1:1,000, Promega, G7121), cleaved caspase-3 (1:200, Cell Signaling Technology, 9661S), NR4A2 (1:100, Santa Cruz, sc-991), TH (1:1,000, Millipore, AB152), TH (1:1,000, Pel-Freez, P40101), NES (1:500, BD Biosciences, 556309), LGR5 (1:200, Atlas Antibodies, HPA012530), LGR4 (1:50, Santa Cruz Biotechnology, sc-292344), RSPO2 (1:500, Atlas Antibodies, HPA025764), SOX2 (1:200, R&D Systems, MAB2018), Ki67 (1:200, Thermo Fisher Scientific, MA5-14520), EN1 (Abcam, ab70993), OTX2 (R&D Systems, AF1979), ALDH1A1 (Abcam, ab23375), and click-iT EdU (Invitrogen, C10337). Corresponding secondary antibodies were Alexa Fluor Dyes (Invitrogen) (1:1,000) and incubated at RT for 1 hr. Cells were counterstained with DAPI (Thermo Fisher Scientific, D1306) or Hoechst 33258 (Sigma). Cells were then washed and kept in PBS and sections were washed and mounted with Mounting Medium (Dako).
IHC/ICC were captured on an Olympus FV1000 confocal microscope, a Zeiss LSM510 confocal microscope, or a Zeiss Axioplan microscope. Human ESC images were captured on either an operetta high-content imaging system (PerkinElmer) or an SP8 confocal microscope (Leica). Human ESCs were counted using an automated software package (Columbus). Images and figure layouts were processed in FIJI (Schindelin et al., 2012), Adobe Photoshop, and Adobe Illustrator.
Cell Counts and Statistical Analyses
All experiments were performed as technical duplicates (two wells per treatment) and three independent biological replicates. Analysis of images was performed from at least five images of each well. For mouse ESC cultures, cells were counted within a βIII-tubulin area (defined as an area just outside of cell clusters where nuclei could be distinguished) with the aid of FIJI image processing software (Schindelin et al., 2012). Statistical test performed include two-tailed t test and/or ANOVA, deviations are SDs unless otherwise stated.
Author Contributions
D.G. performed most of the mouse experiments, analysis, and wrote the manuscript. M.A. performed the human ESC cultures and analysis. E.M.T. provided the bioinformatical analysis. S.T. and S.Y. contributed to some of the mouse experiments. C. ffrench-Constant and E.A. supervised and aided in the experimental design, conclusions, and writing of the manuscript. All authors reviewed the manuscript.
Acknowledgments
We thank Linda Adlerz for her support with the qPCR experiments, members of the Arenas lab for their help and suggestions, and Alessandra Nanni for technical and secretarial assistance. Financial support was obtained from Swedish Research Council (VR projects: DBRM, 2011-3116, 2011-3318, and 2016-01526), Swedish Foundation for Strategic Research (SRL and SB16-0065), European Commission (NeuroStemCellRepair and DDPD-Genes), Karolinska Institutet, Hjärnfonden (FO2015:0202, FO2017:0059), Cancerfonden (CAN 2016/572), and SFO Strat Regen (SG-2018) to E.A. E.M.T. received a fellowship from VR. M.A. and C. ffrench-Constant were supported by NeuroStemCellRepair and Wellcome Trust Senior Investigator Award.
Published: August 23, 2018
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.07.014.
Accession Numbers
The NCBI GEO accession numbers for the TruSeq RNA-seq raw data reported in this paper is GEO: GSE82099 for E12.5, and GSE117394 for E11.5/13.5/14.5 time points.
Supplemental Information
References
- Allen Institute for Brain Science. (2017). Allen developing mouse brain atlas. Website: http://developingmouse.brain-map.org.
- Anderegg A., Lin H.-P., Chen J.-A., Caronia-Brown G., Cherepanova N., Yun B., Joksimovic M., Rock J., Harfe B.D., Johnson R. An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet. 2013;9:e1003973. doi: 10.1371/journal.pgen.1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E., Tryggvason U., Deng Q., Friling S., Alekseenko Z., Robert B., Perlmann T., Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Andersson E.R., Prakash N., Cajanek L., Minina E., Bryja V., Bryjova L., Yamaguchi T.P., Hall A.C., Wurst W., Arenas E. Wnt5a regulates ventral midbrain morphogenesis and the development of A9-A10 dopaminergic cells in vivo. PLoS One. 2008;3:e3517. doi: 10.1371/journal.pone.0003517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E.R., Saltó C., Villaescusa J.C., Cajanek L., Yang S., Bryjova L., Nagy I.I., Vainio S.J., Ramirez C., Bryja V. Wnt5a cooperates with canonical Wnts to generate midbrain dopaminergic neurons in vivo and in stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:E602–E610. doi: 10.1073/pnas.1208524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas E. Wnt signaling in midbrain dopaminergic neuron development and regenerative medicine for Parkinson’s disease. J. Mol. Cell Biol. 2014;6:42–53. doi: 10.1093/jmcb/mju001. [DOI] [PubMed] [Google Scholar]
- Arenas E., Denham M., Villaescusa J.C. How to make a midbrain dopaminergic neuron. Development. 2015;142:1918–1936. doi: 10.1242/dev.097394. [DOI] [PubMed] [Google Scholar]
- Barberi T., Klivenyi P., Calingasan N.Y., Lee H., Kawamata H., Loonam K., Perrier A.L., Bruses J., Rubio M.E., Topf N. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat. Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N., Rookmaaker M.B., Kujala P., Ng A., Leushacke M., Snippert H., van de Wetering M., Tan S., Van Es J.H., Huch M. Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep. 2012;2:540–552. doi: 10.1016/j.celrep.2012.08.018. [DOI] [PubMed] [Google Scholar]
- Bell S.M., Schreiner C.M., Wert S.E., Mucenski M.L., Scott W.J., Whitsett J.A. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development. 2008;135:1049–1058. doi: 10.1242/dev.013359. [DOI] [PubMed] [Google Scholar]
- Carmon K.S., Gong X., Lin Q., Thomas A., Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco G., Wagner J., Rodriguez F.J., Kele J., Sousa K., Rawal N., Pasolli H.A., Fuchs E., Kitajewski J., Arenas E. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc. Natl. Acad. Sci. USA. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-Z., Wang S., Tang R., Yang Q.-S., Zhao E., Chao Y., Ying K., Xie Y., Mao Y.-M. Cloning and identification of a cDNA that encodes a novel human protein with thrombospondin type I repeat domain, hPWTSR. Mol. Biol. Rep. 2002;29:287–292. doi: 10.1023/a:1020479301379. [DOI] [PubMed] [Google Scholar]
- Chen P.-H., Chen X., Lin Z., Fang D., He X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013;27:1345–1350. doi: 10.1101/gad.219915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q., Andersson E., Hedlund E., Alekseenko Z., Coppola E., Panman L., Millonig J.H., Brunet J.-F., Ericson J., Perlmann T. Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development. 2011;138:3399–3408. doi: 10.1242/dev.065482. [DOI] [PubMed] [Google Scholar]
- Feinstein Y., Borrell V., Garcia C., Burstyn-Cohen T., Tzarfaty V., Frumkin A., Nose A., Okamoto H., Higashijima S., Soriano E. F-spondin and mindin: two structurally and functionally related genes expressed in the hippocampus that promote outgrowth of embryonic hippocampal neurons. Development. 1999;126:3637–3648. doi: 10.1242/dev.126.16.3637. [DOI] [PubMed] [Google Scholar]
- Fernando C.V., Kele J., Bye C.R., Niclis J.C., Alsanie W., Blakely B.D., Stenman J., Turner B.J., Parish C.L. Diverse roles for Wnt7a in ventral midbrain neurogenesis and dopaminergic axon morphogenesis. Stem Cells Dev. 2014;23:1991–2003. doi: 10.1089/scd.2014.0166. [DOI] [PubMed] [Google Scholar]
- Fukusumi Y., Meier F., Götz S., Matheus F., Irmler M., Beckervordersandforth R., Faus-Kessler T., Minina E., Rauser B., Zhang J. Dickkopf 3 promotes the differentiation of a rostrolateral midbrain dopaminergic neuronal subset in vivo and from pluripotent stem cells in vitro in the mouse. J. Neurosci. 2015;35:13385–13401. doi: 10.1523/JNEUROSCI.1722-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy K., Sowmithra S., Bhonde R., Datta I. By changing dimensionality, sequential culturing of midbrain cells, rather than two-dimensional culture, generates a neuron-glia ratio closer to in vivo adult midbrain. Cells Tissues Organs. 2016;201:445–463. doi: 10.1159/000446424. [DOI] [PubMed] [Google Scholar]
- Glinka A., Dolde C., Kirsch N., Huang Y.-L., Kazanskaya O., Ingelfinger D., Boutros M., Cruciat C.-M., Niehrs C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H.-X., Xie Y., Zhang Y., Charlat O., Oster E., Avello M., Lei H., Mickanin C., Liu D., Ruffner H. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- Ho A., Südhof T.C. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc. Natl. Acad. Sci. USA. 2004;101:2548–2553. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe H., Wessner D., Beffert U., Becker A.G., Matsuoka Y., Rebeck G.W. F-spondin interaction with the apolipoprotein E receptor ApoEr2 affects processing of amyloid precursor protein. Mol. Cell. Biol. 2005;25:9259–9268. doi: 10.1128/MCB.25.21.9259-9268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra E.J., von Oerthel L., van der Heide L.P., Kouwenhoven W.M., Veenvliet J.V., Wever I., Jin Y.-R., Yoon J.K., van der Linden A.J.A., Holstege F.C.P. Lmx1a encodes a rostral set of mesodiencephalic dopaminergic neurons marked by the Wnt/B-catenin signaling activator R-spondin 2. PLoS One. 2013;8:e74049. doi: 10.1371/journal.pone.0074049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S.W., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J. In vitro expansion of single Lgr5(+) liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:5–10. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Bonfanti P., Boj S.F., Sato T., Loomans C.J.M., van de Wetering M., Sojoodi M., Li V.S.W., Schuijers J., Gracanin A. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inestrosa N.C., Arenas E. Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- Jaks V., Barker N., Kasper M., van Es J.H., Snippert H.J., Clevers H., Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Kazanskaya O., Glinka A., del Barco Barrantes I., Stannek P., Niehrs C., Wu W. R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev. Cell. 2004;7:525–534. doi: 10.1016/j.devcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Kele J., Andersson E.R., Villaescusa J.C., Cajanek L., Parish C.L., Bonilla S., Toledo E.M., Bryja V., Rubin J.S., Shimono A. SFRP1 and SFRP2 dose-dependently regulate midbrain dopamine neuron development in vivo and in embryonic stem cells. Stem Cells. 2012;30:865–875. doi: 10.1002/stem.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-A., Kakitani M., Zhao J., Oshima T., Tang T., Binnerts M., Liu Y., Boyle B., Park E., Emtage P. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- Kim K., Wagle M., Tran K., Zhan X., Dixon M.A., Liu S., Gros D., Korver W., Yonkovich S., Tomasevic N. R-spondin family members regulate the Wnt pathway by a common mechanism. Mol. Biol. Cell. 2008;19:2588–2596. doi: 10.1091/mbc.E08-02-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A., Baldassare M., Jessell T.M. F-spondin: a gene expressed at high levels in the floor plate encodes a secreted protein that promotes neural cell adhesion and neurite extension. Cell. 1992;69:95–110. doi: 10.1016/0092-8674(92)90121-r. [DOI] [PubMed] [Google Scholar]
- Koo B.-K., Spit M., Jordens I., Low T.Y., Stange D.E., van de Wetering M., van Es J.H., Mohammed S., Heck A.J.R., Maurice M.M. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- Kriks S., Shim J.-W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W., Barker N., Low T.Y., Koo B.-K., Li V.S.W., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- de Lau W.B.M., Snel B., Clevers H.C. The R-spondin protein family. Genome Biol. 2012;13:242. doi: 10.1186/gb-2012-13-3-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W., Peng W.C., Gros P., Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28:305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Long K., Moss L., Laursen L., Boulter L., Ffrench-Constant C. Integrin signalling regulates the expansion of neuroepithelial progenitors and neurogenesis via Wnt7a and Decorin. Nat. Commun. 2016;7:10354. doi: 10.1038/ncomms10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., Borm L.E., Stott S.R.W., Toledo E.M., Villaescusa J.C. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016;167:566–580.e19. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell S.L., Ho H.-Y., Kuehner E., Zhao S., Li M. Pitx3 regulates tyrosine hydroxylase expression in the substantia nigra and identifies a subgroup of mesencephalic dopaminergic progenitor neurons during mouse development. Dev. Biol. 2005;282:467–479. doi: 10.1016/j.ydbio.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Murphy-Ullrich J.E., Sage E.H. Revisiting the matricellular concept. Matrix Biol. 2014;37:1–14. doi: 10.1016/j.matbio.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolbrant S., Heuer A., Parmar M., Kirkeby A. Generation of high-purity human ventral midbrain dopaminergic progenitors for in vitro maturation and intracerebral transplantation. Nat. Protoc. 2017;12:1962–1979. doi: 10.1038/nprot.2017.078. [DOI] [PubMed] [Google Scholar]
- Nunes I., Tovmasian L.T., Silva R.M., Burke R.E., Goff S.P. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc. Natl. Acad. Sci. USA. 2003;100:4245–4250. doi: 10.1073/pnas.0230529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawara B., Glinka A., Niehrs C. Rspo3 binds syndecan 4 and induces Wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev. Cell. 2011;20:303–314. doi: 10.1016/j.devcel.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N., Brodski C., Naserke T., Puelles E., Gogoi R., Hall A., Panhuysen M., Echevarria D., Sussel L., Weisenhorn D.M.V. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development. 2006;133:89–98. doi: 10.1242/dev.02181. [DOI] [PubMed] [Google Scholar]
- Rawal N., Castelo-Branco G., Sousa K.M., Kele J., Kobayashi K., Okano H., Arenas E. Dynamic temporal and cell type-specific expression of Wnt signaling components in the developing midbrain. Exp. Cell Res. 2006;312:1626–1636. doi: 10.1016/j.yexcr.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Ribeiro D., Ellwanger K., Glagow D., Theofilopoulos S., Corsini N.S., Martin-Villalba A., Niehrs C., Arenas E. Dkk1 regulates ventral midbrain dopaminergic differentiation and morphogenesis. PLoS One. 2011;6:e15786. doi: 10.1371/journal.pone.0015786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong X., Chen C., Zhou P., Zhou Y., Li Y., Lu L., Liu Y., Zhou J., Duan C. R-spondin 3 regulates dorsoventral and anteroposterior patterning by antagonizing Wnt/β-catenin signaling in zebrafish embryos. PLoS One. 2014;9:e99514. doi: 10.1371/journal.pone.0099514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti P., Sousa K.M., Hall A.C., Liste I., Steffensen K.R., Theofilopoulos S., Parish C.L., Hazenberg C., Richter L.A., Hovatta O. Liver X receptors and oxysterols promote ventral midbrain neurogenesis in vivo and in human embryonic stem cells. Cell Stem Cell. 2009;5:409–419. doi: 10.1016/j.stem.2009.08.019. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D., Klar A., Park M., Dargusch R., Fischer W.H. F-spondin promotes nerve precursor differentiation. J. Neurochem. 2006;96:444–453. doi: 10.1111/j.1471-4159.2005.03563.x. [DOI] [PubMed] [Google Scholar]
- Sousa K.M., Villaescusa J.C., Cajanek L., Ondr J.K., Castelo-Branco G., Hofstra W., Bryja V., Palmberg C., Bergman T., Wainwright B. Wnt2 regulates progenitor proliferation in the developing ventral midbrain. J. Biol. Chem. 2010;285:7246–7253. doi: 10.1074/jbc.M109.079822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo E.M., Manno G.L., Rivetti P., Gyllborg D., Villaescusa C., Linnarsson S., Arenas E. Molecular analysis of the midbrain dopaminergic niche during neurogenesis. bioRxiv. 2017 [Google Scholar]
- Toledo E.M., Gyllborg D., Arenas E. Translation of WNT developmental programs into stem cell replacement strategies for the treatment of Parkinson’s disease. Br. J. Pharmacol. 2017;174:4716–4724. doi: 10.1111/bph.13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese S., Faissner A. The role of extracellular matrix in spinal cord development. Exp. Neurol. 2015;274:90–99. doi: 10.1016/j.expneurol.2015.05.018. [DOI] [PubMed] [Google Scholar]
- Zebisch M., Jones E.Y. Crystal structure of R-spondin 2 in complex with the ectodomains of its receptors LGR5 and ZNRF3. J. Struct. Biol. 2015;191:149–155. doi: 10.1016/j.jsb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.