Summary
MicroRNAs (miRNAs) are the effectors of a conserved gene-silencing system with broad roles in post-transcriptional regulation. Due to functional overlaps, assigning specific functions to individual miRNAs has been challenging. DICER1 cleaves pre-miRNA hairpins into mature miRNAs, and previously Dicer1 knockout mouse embryonic stem cells have been generated to study miRNA function in early mouse development. Here we report an essential requirement of DICER1 for the self-renewal of human embryonic stem cells (hESCs). Utilizing a conditional knockout approach, we found that DICER1 deletion led to increased death receptor-mediated apoptosis and failure of hESC self-renewal. We further devised a targeted miRNA screening strategy and uncovered essential pro-survival roles of members of the mir-302-367 and mir-371-373 clusters that bear the seed sequence AAGUGC. This platform is uniquely suitable for dissecting the roles of individual miRNAs in hESC self-renewal and differentiation, which may help us better understand the early development of human embryos.
Keywords: human embryonic stem cells (hESCs), human pluripotent stem cells (hPSCs), DICER1, microRNAs (miRNAs), self-renewal, apoptosis, ESCC miRNAs
Graphical Abstract
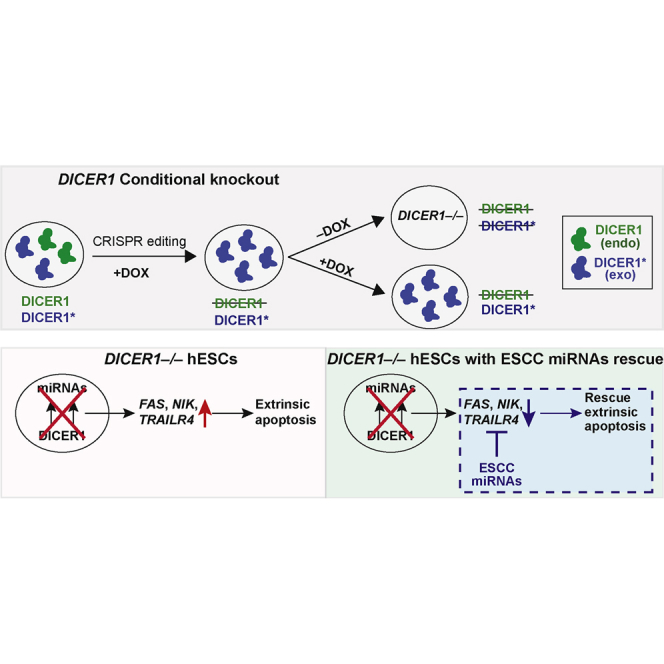
Highlights
-
•
Conditional DICER1 knockouts enable the study of its essential requirement in hESCs
-
•
DICER1 deletion leads to increased extrinsic (receptor-mediated) apoptosis
-
•
miRNAs rescue the DICER1 knockout apoptotic phenotype
-
•
This work provides a platform for interrogating miRNA function in hESCs
Huangfu and colleagues report an unexpected requirement for DICER1 in preventing death-receptor-mediated apoptosis in hESCs. This essential pro-survival role is largely mediated by mir-302-367 and mir-371-373 cluster members that bear the seed sequence “AAGUGC” shared by the ESC-specific cell-cycle-regulating family of miRNAs. This work provides a robust platform for interrogating miRNA function in hESC pluripotency and differentiation.
Introduction
MicroRNAs (miRNAs) are ∼20- to 24-nucleotide long noncoding RNAs that bind target mRNAs through partial complementarity with their 3′ UTRs to promote target mRNA decay and/or inhibit translation (Ebert and Sharp, 2012). The RNase III enzyme Dicer1 is required for the processing of precursor miRNA hairpins into their mature functional products (Yang and Lai, 2011). Dicer1 knockout mouse embryos exhibit post-implantation morphological abnormalities at around embryonic day 7.5 (E7.5) (Bernstein et al., 2003, Spruce et al., 2010), emphasizing the importance of miRNAs for early mammalian development. Due to overlapping miRNA requirements, no individual miRNA mouse knockouts have recapitulated the Dicer1 knockout phenotype (Kuhnert et al., 2008, Liu et al., 2008, Park et al., 2010, Ventura et al., 2008, Zhao et al., 2007). Mouse embryonic stem cells (mESCs) have been used to gain further understanding of miRNA functions in the pre-implantation period. mESCs lacking either Dicer1 or Dgcr8, factors essential for miRNA biogenesis, can self-renew but show reduced proliferation and a relative accumulation of cells in the G1 phase of the cell cycle (Kanellopoulou et al., 2005, Murchison et al., 2005, Wang et al., 2007, Wang et al., 2008). miRNA rescue experiments in Dgcr8 knockout mESCs have shown that the rapid cell cycle of mESCs is regulated by members of the mir-290-295 (homologous to human mir-371-373), mir-302-367, and mir-17-92b clusters, known collectively as the ESC-specific cell-cycle-regulating (ESCC) family of miRNAs (Wang et al., 2008).
Thus far the exact requirements for DICER1 in human embryonic stem cells (hESCs) have not been established. One study used a knockdown approach in hESCs but DICER1 mRNA was only reduced about 5-fold (Qi et al., 2009). Compared with mESCs, hESCs reflect a later developmental stage (Brons et al., 2007, Tesar et al., 2007), and there could be human-specific DICER1 and miRNA requirements. Here we report the generation and characterization of DICER1-deficient hESCs. We show that in hESCs, unlike in mESCs, DICER1 is essential for self-renewal and inhibition of death receptor-mediated apoptosis. Furthermore, ESCC seed sequence-containing members of the mir-302-367 and mir-371-373 clusters could reverse the apoptotic phenotype. Our study provides a valuable platform for the discovery of individual miRNA functions during early development of human embryos and hESC differentiation.
Results
Generation of DICER1 Conditional Knockout hESCs
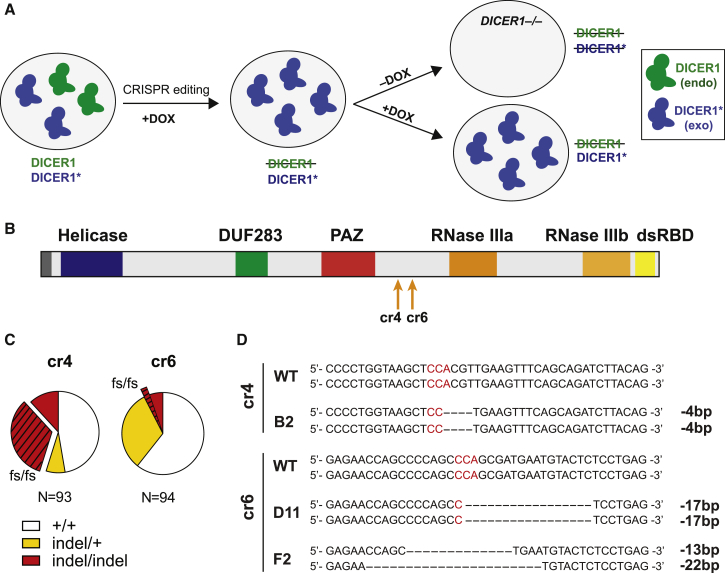
To investigate the role of DICER1 in human pluripotency, we attempted to generate DICER1 knockout hESC lines. Despite using the highly efficient iCRISPR platform (Gonzalez et al., 2014), we were unable to generate homozygous null mutant hESC lines after screening 464 lines generated with nine guide RNAs (gRNAs) (Figures S1A–S1C). We retargeted a line already carrying a heterozygous DICER1 frameshift mutation, but still failed to generate a frameshift mutation in the second allele (Figure S1D). These results suggest that DICER1-deficient hESCs have self-renewing defects. To overcome the apparent cell lethality issues, we designed a conditional knockout strategy. We first selected two CRISPRs (cr4 and cr6) based on their ability to efficiently generate indel (insertion/deletion) mutations (Figure S1C). We then generated two Tet-On transgenic lines expressing DICER1 transgenes that have a silent mutation in the PAM sequence and thus are immune to cr4 or cr6 gRNA targeting (referred to as TRE-DICER1∗ lines) (Figures S1E and S1F). We next used CRISPR/Cas9 to knock out endogenous DICER1 while maintaining expression of exogenous DICER1∗ by doxycycline treatment (Figure 1A). This approach enabled us to create 32 conditional biallelic frameshift knockout lines out of 187 clonal lines analyzed (Figure 1C). We focused further analysis on three conditional knockout lines: B2 (generated using cr4), and D11 and F2 (generated using cr6) (Figures 1B and 1D). When maintained with doxycycline treatment, these conditional DICER1 knockout hESC lines were indistinguishable from wild-type hESCs and exhibited normal karyotypes (Figure S1G).
Figure 1.
Generation of DICER1 Conditional Knockout hESC Lines
(A) Schematic of the procedure to generate conditional DICER1 knockout hESCs. CRISPR/Cas9 was used to generate DICER1 knockout mutations in cells infected with a gRNA-immune doxycycline (DOX)-inducible TRE-DICER1∗ transgene. Cas9 and DICER1∗ protein expression was induced by doxycycline treatment. DICER1 knockout mutants were maintained in doxycycline until the start of experiments. An asterisk is used to distinguish the exogenous modified DICER1∗ from endogenous DICER1.
(B) Schematic representation of DICER1 protein with domains. Orange arrows represent gRNA-targeting loci upstream of the RNase IIIa functional domain. cr, CRISPR; DUF, domain of unknown function; dsRBD, double-stranded RNA-binding domain.
(C) CRISPR/Cas9 targeting results of conditional DICER1 hESC lines. White, wild-type lines; yellow, monoallelic mutant lines; red, biallelic mutant lines. Stripes indicate DICER1 knockout lines with frameshift mutations in both alleles. fs, frameshift allele.
(D) Representative endogenous DICER1 sequences of three selected conditional DICER1 knockout clones with PAM sequences labeled in red. WT, wild-type.
Unless otherwise indicated, scale bars represent 100 μm in all figures. See also Figure S1.
DICER1 Is Essential for hESC Self-Renewal
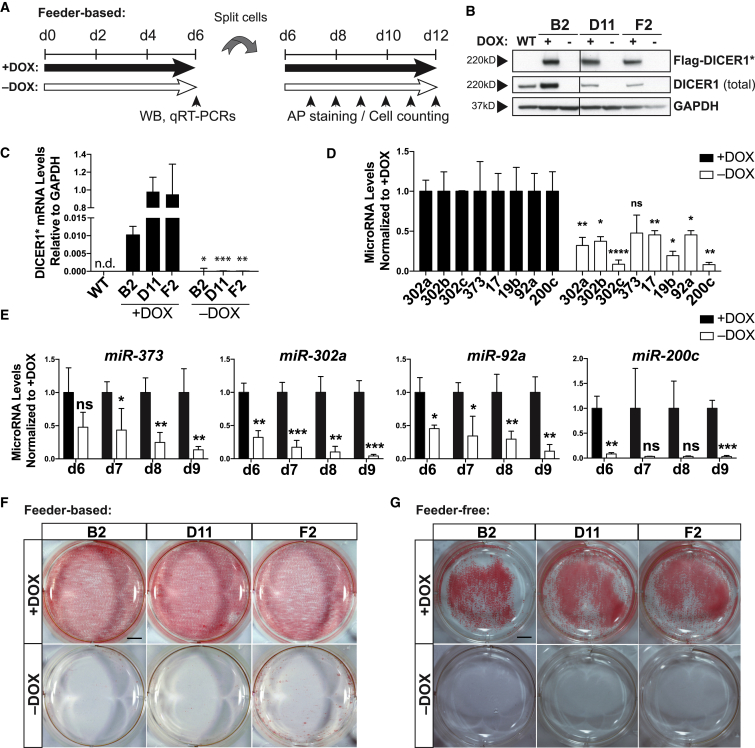
We removed doxycycline to investigate the effects of DICER1 deletion in conditional DICER1 knockout hESCs (Figure 2A). Six days after doxycycline withdrawal, no DICER1∗ mRNA or protein expression was detected, nor did we observe significant expression of the endogenous DICER1 protein (Figures 2B and 2C). The DICER1-depleted hESCs survived and maintained comparable protein levels of pluripotency markers OCT4 (POU5F1), NANOG, and SOX2 based on immunofluorescence staining, although some reduction of mRNA levels was observed by qRT-PCR (Figures S2A and S2B). We also tested the levels of several mature miRNAs that are abundant in wild-type hESCs including miR-373 and miR-302a. All miRNAs examined were significantly reduced in DICER1-depleted hESCs by qRT-PCR and northern blot, and there was a concomitant increase in precursor miRNAs (Figures 2D and S2C). These results confirmed that miRNA processing was compromised in DICER1 null hESCs. However, miRNAs were not fully depleted after 6 days, which is consistent with the notion that miRNAs are highly stable molecules (Guo et al., 2015).
Figure 2.
DICER1 Is Essential for hESC Self-Renewal
(A) Schematic representation of experimental procedure in feeder-based condition. Cells were split at 28,000 cells/cm2. DOX, doxycycline; WB, western blot; AP, alkaline phosphatase.
(B) Western blot of FLAG-DICER1∗ and total DICER1 levels of three conditional DICER1 knockout lines (B2, D11, F2) in feeder-based condition with and without DOX treatment for 6 days. WT, wild-type.
(C) qRT-PCR analysis of DICER1∗ transgene expression in three conditional DICER1 knockout lines in feeder-based condition with and without doxycycline treatment for 6 days. Levels relative to GAPDH. Statistical analysis was performed in comparison with +DOX samples. n = 3 independent experiments. n.d., not detected.
(D) Mature miRNA qRT-PCR analysis of eight hESC-expressed miRNAs in DICER1 conditional knockout line B2 after 6 days of DOX withdrawal in feeder-based condition. Levels are normalized to +DOX. Statistical analysis was performed in comparison with +DOX samples. n = 3 independent experiments.
(E) qRT-PCR analysis of four hESC-abundant miRNAs (miR-373, miR-302a, miR-92a, and miR-200c) at days 6 through 9 of DOX withdrawal. miRNA levels in the +DOX samples are used for comparison. Analysis was done on the B2 DICER1 conditional knockout line in feeder-based condition. For each time point, statistical analysis was performed in comparison with +DOX samples. Day-6 data are reused from (D) for comparison purposes. n = 3 independent experiments.
(F) AP staining of three conditional DICER1 knockout lines in feeder-based conditions with and without DOX treatment for 12 days. n = 6 independent experiments. Scale bar, 5 mm.
(G) AP staining of three conditional DICER1 knockout lines in feeder-free condition with and without doxycycline treatment for 8 days. n = 3 independent experiments. Scale bar, 5 mm.
Error bars indicate SD, and significance is indicated as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001; ns, not significant (p ≥ 0.05). See also Figure S2.
After passaging these DICER1-depleted cells and maintaining them in the absence of doxycycline for another 6 days (Figure 2A), we observed further reduction in the accumulation of mature miRNAs (Figure 2E). Coinciding with this reduction in miRNA levels, although the DICER1-depleted cells were properly attached to the dish after replating, they began to show compromised growth 10 days after doxycycline withdrawal as determined by staining for alkaline phosphatase activity and live cell counting (Figures S2D and S2E). By day 12 of doxycycline withdrawal there were barely any cells detected in all three conditional DICER1 knockout lines (Figures 2F and S2F). The few remaining DICER1-depleted cells could not be propagated without doxycycline treatment.
In addition to the original feeder-based condition, we also tested the requirements for DICER1 in the Essential 8 (E8) chemically defined feeder-free culture condition (Chen et al., 2011). Upon doxycycline withdrawal, DICER1-depleted cells could not be propagated in the E8 condition either, and almost no cells were detected by alkaline phosphatase staining after 8 days of doxycycline withdrawal (Figure 2G). Therefore, DICER1 is required for self-renewal of hESCs cultured in feeder-based and feeder-free conditions. DICER1-depleted cells survived longer in the feeder-based condition presumably due to pro-survival effects of the feeder cells. In comparison, Dicer1-depleted mESCs are capable of self-renewal although they proliferate at a slower rate than wild-type mESCs (Bernstein et al., 2003, Kanellopoulou et al., 2005, Murchison et al., 2005, Wang et al., 2007, Wang et al., 2008).
Loss of DICER1 in hESCs Causes Apoptosis
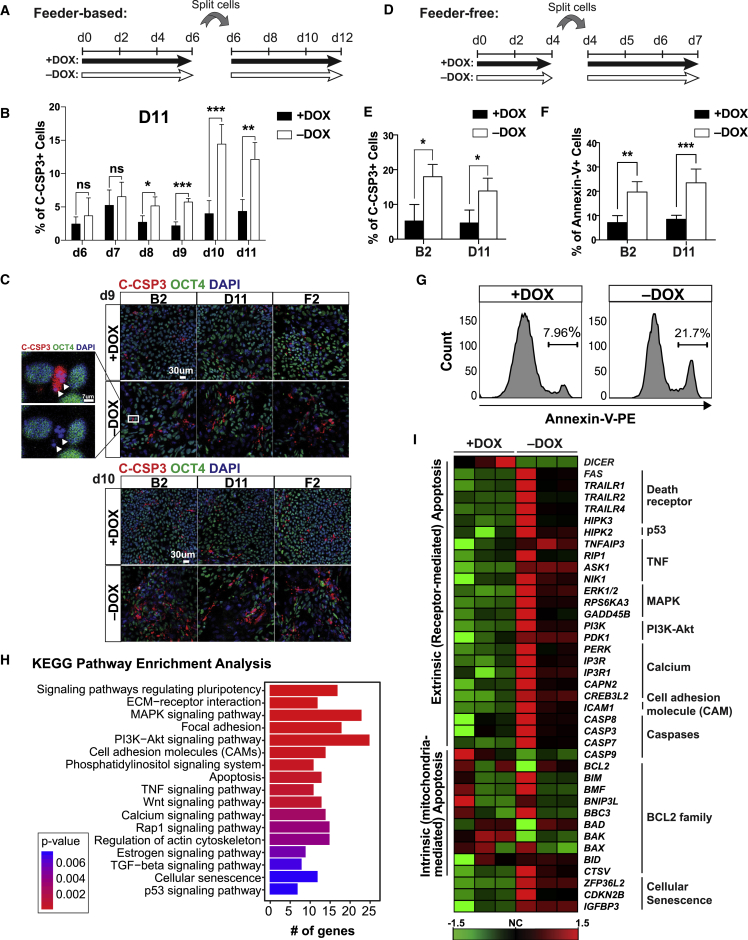
Unlike Dicer1-deficient mESCs, we did not observe an apparent mitotic defect based on flow-cytometry analysis of the mitotic marker phosphohistone H3 (Figures S3A and S3B), nor defects in cell-cycle progression based on cell-cycle analysis by propidium iodide staining (Figure S3C). Instead, we detected significantly increased numbers of cleaved caspase-3 (C-CSP3)-positive cells by flow cytometry starting from day 8 after doxycycline removal and peaking at day 10 in both DICER1 conditional knockout lines (B2 and D11) cultured in the feeder-based condition (Figures 3A, 3B, S3D, and S3E). The increased apoptotic rate coincided with the compromised cell growth that first became apparent around days 9–10 (Figures S2D and S2E). C-CSP3-positive cells lost OCT4 expression and showed nuclear condensation and fragmentation typical of apoptotic cells when examined by immunofluorescence staining at days 9 and 10 (Figure 3C). Similarly, DICER1 conditional knockout cells in the E8 condition also showed significantly increased numbers of apoptotic cells by flow cytometry for C-CSP3 expression and annexin-V staining (another apoptosis assay) after doxycycline removal (Figures 3E–3G and S3F). Therefore, unlike its pro-proliferation role in mESCs, DICER1's main function in hESCs is pro-survival.
Figure 3.
DICER1 Depletion in hESCs Causes Caspase-3-Mediated Apoptosis
(A) Schematic representation of the experimental design in feeder-based condition. Cells were split at 28,000 cells/cm2. DOX, doxycycline.
(B) Flow cytometry of cleaved caspase-3 (C-CSP3) from days 6 through 11 in the D11 conditional DICER1 knockout line with and without DOX in feeder-based condition. n = 3 independent experiments.
(C) Representative immunofluorescence pictures of C-CSP3 and OCT4 in the D11 conditional DICER1 knockout line maintained with and without DOX at days 9 and 10 in feeder-based condition. Inset represents a close-up view showing fragmentation (arrowheads) of nuclei and loss of OCT4 in a C-CSP3-positive cell. Scale bars, 30 μm (main images) and 7 μm (close-up). n = 3 independent experiments.
(D) Schematic representation of the experimental design in feeder-free condition. Cells were split at a density of 14,000 cells/cm2.
(E) Flow-cytometry detection of C-CSP3 expression in B2 and D11 conditional DICER1 knockout lines in feeder-free condition maintained with and without DOX for 7 days. n = 4 independent experiments.
(F) Flow cytometry of annexin-V in B2 and D11 conditional DICER1 knockouts cultured in feeder-free condition maintained with or without DOX for 7 days. n = 4 independent experiments for B2, and n = 5 independent experiments for D11.
(G) Representative histograms of annexin-V flow cytometry on D11 conditional DICER1 knockout line maintained with or without DOX in feeder-free condition. n = 5 independent experiments.
(H) KEGG pathway enrichment analysis was performed in R (v3.4.2) with clusterProfiler (v3.6.0) on RNA-seq data of B2 conditional DICER1 knockout line cultured in feeder-based condition with or without DOX for 9 days. n = 3 independent experiments.
(I) Heatmap of apoptosis-related genes. Data are derived from RNA-seq of B2 conditional DICER1 knockout hESCs with and without DOX for 9 days in feeder-based condition. The heatmap was generated by cluster analysis using the online analysis tool CARMAweb (https://carmaweb.genome.tugraz.at/genesis) with the methods normalized gene. The variation between the biological repeats could be due to the rapid nature of apoptosis. n = 3 independent experiments.
Error bars indicate SD, and significance is indicated as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant (p ≥ 0.05). See also Figure S3.
RNA sequencing (RNA-seq) analyses identified 692 significantly upregulated and 277 significantly downregulated genes in DICER1-depleted hESCs on day 9 of doxycycline withdrawal in the feeder-based condition compared with the doxycycline-treated control (Figure S3G). As expected, DICER1 was the most significantly downregulated gene (Figure S3G). KEGG pathway enrichment analysis of these differentially expressed genes identified pathways involved in apoptosis, pluripotency, and cell adhesion (Figure 3H), while gene ontology analysis revealed mostly development-related terms (Figure S3I). In particular, DICER1 deletion led to increased expression of pro-apoptotic genes of the extrinsic (receptor-mediated) pathway, notably FAS, NIK (MAP3K14), and TRAILR4 (TNFRSF10D) as verified by qRT-PCR analyses (Figures 3I and S3H). Key genes involved in the intrinsic (mitochondria-mediated) apoptotic pathway were generally not upregulated in our RNA-seq analysis or qRT-PCR validation, except for BIM (Figures 3I and S3H). DICER1-depleted hESCs did not show increased expression of BNIP3L, an intrinsic pathway pro-apoptotic factor upregulated upon miR-302/367 knockdown in hESCs (Figures 3I and S3H) (Zhang et al., 2015). This difference could be due to perturbing global versus specific miRNAs, or the knockdown versus the knockout approach. Together, these results demonstrate a previously unappreciated role for DICER1 in preventing receptor-mediated apoptosis in hESCs.
miRNA Rescue of DICER1 Deficiency
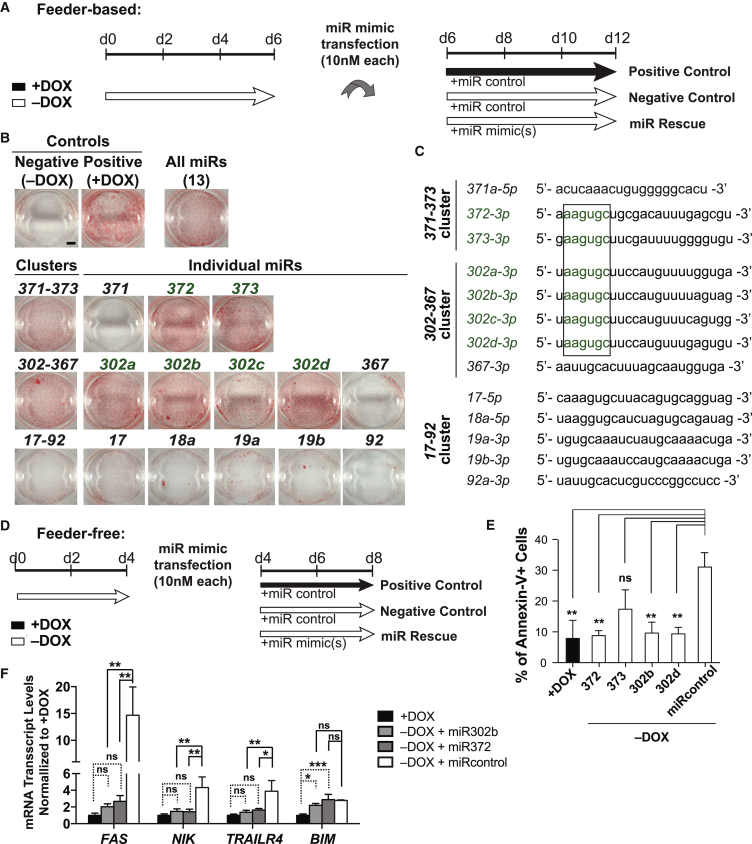
To further understand the DICER1-dependent hESC survival, we attempted to rescue the DICER1 knockout phenotype by transfecting DICER1-depleted hESCs with selected miRNA families (Figure 4A). We tested the ESCC-family of miRNAs (mir-302-367, mir-371-373, and mir-17-92) since they have been previously implicated in mESC proliferation and they are among the most abundant miRNAs in both mouse and human ESCs (Greve et al., 2013, Wang et al., 2008). The most striking rescues were observed for members of the mir-302-367 and mir-371-373 clusters based on alkaline phosphatase staining and cell number counts (Figures 4B and S4B). The transfected cells expressed pluripotency markers OCT4 and NANOG (Figure S4A). Although members of the mir-17-92 cluster were sufficient to rescue the proliferation defect in miRNA-depleted mESCs (Wang et al., 2008), their effects were relatively mild in hESCs (Figures 4B and S4B), suggesting differences between mouse and human ESCs in miRNA requirements. Interestingly, not all members of the mir-302-367 and mir-371-373 clusters rescued the DICER1 knockout hESC phenotype. In the mir-302-367 cluster, miR-302a, miR-302b, miR-302c, and miR-302d showed significant rescue but miR-367 did not (Figures 4B and S4B). Likewise, in the mir-371-373 cluster, miR-372 and miR-373 but not miR-371 showed significant rescue (Figures 4B and S4B). Notably, the miRNAs with significant rescue effects all shared the ESCC seed sequence “AAGUGC” (Figure 4C).
Figure 4.
ESCC Seed Sequence-Containing miRNAs Rescue DICER1 Knockout Defects
(A) Schematic representation of miRNA mimic transfection procedure in feeder-based condition. DOX, doxycycline.
(B) Representative AP-stained D11 conditional DICER1 knockout hESCs with or without transfection of miRNAs in feeder-based condition. In green are the six miRNAs that partially rescue the DICER1 knockout phenotype. n = 5 independent experiments. Scale bar, 5 mm.
(C) List of mature miRNAs (3p or 5p arms) transfected in D11 conditional DICER1 knockout line. In green are the mature miRNAs that significantly rescued the DICER1 knockout phenotype. The box encloses the seed sequence shared among those mature miRNAs that rescue the DICER1 knockout phenotype.
(D) Schematic representation of miRNA mimic transfection procedure in feeder-free condition.
(E) Flow cytometry of annexin-V in D11 conditional DICER1 knockout line cultured in feeder-free conditions for 8 days and transfected with mature miRNAs or control miRNA as shown in (D). For comparison between multiple experimental groups in (E) and (F), one-way ANOVA was used followed by multiple comparisons with Tukey correction. n = 3 independent experiments.
(F) qRT-PCR analysis of representative pro-apoptotic genes belonging to the extrinsic apoptotic pathway (FAS, NIK, TRAILR4) as well as intrinsic marker BIM in feeder-free condition with and without doxycycline treatment for 8 days and transfected with miR-302b, miR-372, or miR control as shown in (D). n = 3 independent experiments.
Error bars indicate SD, and significance is indicated as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; ns, not significant (p ≥ 0.05). See also Figure S4.
We also observed similar rescue of cell survival in the E8 feeder-free condition, and annexin-V staining showed a significant reduction of apoptotic rates in DICER1-deficient hESCs transfected with miR-372, miR-373, miR-302b, and miR-302d (Figures 4D, 4E, and S4C). In silico analysis using TargetScan and MirTarget (Agarwal et al., 2015, Wong and Wang, 2015) showed that FAS, NIK, TRAILR4, and BIM genes, significantly upregulated in DICER1-depleted hESCs, were predicted targets of the ESCC seed sequence-bearing miRNAs (Table S1). Therefore, we examined whether their upregulation could be reversed in hESCs transfected with miR-372 or miR-302b. By qRT-PCR analysis, the expression levels of FAS, NIK, and TRAILR4, but not BIM, were significantly rescued to levels close to those of doxycycline-treated control hESCs expressing DICER1 (Figure 4F). These results suggest that the ESCC seed sequence-bearing members of mir-302-367 and mir-371-373 clusters are responsible for preventing apoptosis in DICER1 knockout hESCs primarily through the regulation of death receptor-mediated apoptosis pathway genes such as FAS.
Discussion
Overlapping functions of miRNAs pose a technical challenge to fully understanding their exact roles during early embryonic development (Greve et al., 2013). Our conditional DICER1 knockout hESCs provide an excellent platform to investigate the roles of individual miRNAs (Park et al., 2010). For instance, we find that reintroduction of ESCC seed sequence-containing miRNAs of the mir-302-367 and mir-371-373 clusters is sufficient to promote cell survival in DICER1 knockout hESCs, whereas mir-17-92 does not play a significant role in hESCs in this context despite its prominent role in mESC cell-cycle progression (Wang et al., 2008). Our platform is a valuable tool for the discovery of novel miRNA requirements for both hESC pluripotency maintenance and stem cell differentiation.
In contrast to the essential requirement for DICER in hESCs, Dicer1 is not required for mESC survival. This could be due to the different developmental stages thought to be represented by mouse and human ESCs. Supporting this idea, recent studies have uncovered essential requirements for EZH2 and DNMT1 in hESCs, which are not observed in mESCs (Collinson et al., 2016, Liao et al., 2015, Shen et al., 2008, Tsumura et al., 2006). In this regard, it may be more appropriate to compare DICER1 knockout phenotypes in hESCs with those observed in mouse epiblast stem cells (mEpiSCs). In mEpiSCs, Dicer1 deletion causes increased Bim expression, leading to activation of the intrinsic apoptotic pathway (the mitochondrial pathway); these effects are mitigated by reintroduction of miR-20, miR-92, and miR-302 family members into Dicer1-depleted cells (Pernaute et al., 2014). Although we also observed some upregulation of BIM expression in DICER1-depleted hESCs, there was no clear rescue of BIM expression upon reintroduction of miR-302b or miR-372. Instead, DICER1-depleted hESCs upregulate a cohort of extrinsic apoptotic pathway genes. Reintroduction of miR-302b or miR-372 in DICER1 knockout cells rescued the cell-survival defect and reversed the aberrant upregulation of extrinsic pathway genes FAS, NIK, and TRAILR4. It is of interest to note that the human, but not the mouse, FAS and TRAILR4 3′ UTR sequences were predicted by in silico analyses to contain the ESCC targets (Table S1). These differences may explain the differing roles for DICER1 in hESCs compared with mESCs or mEpiSCs.
In addition to the human-mouse species difference, the pro-survival requirement for DICER1 also depends on the cellular context. Mouse studies have shown that Dicer1 is essential for the survival of a number of developmental cell types such as limb mesoderm, αβ T cells, and melanocyte stem cells and differentiated melanocytes (Cobb et al., 2005, Harfe et al., 2005, Levy et al., 2010). On the other hand, Dicer1 is not required for survival of the γδ T cells (Cobb et al., 2005), and it is dispensable in the specification and survival of the fetal adrenal cortex (Krill et al., 2013). In HCT116 human colorectal cancer cells, DICER1 is also not essential for survival (Kim et al., 2016). Combined with directed differentiation, our DICER1 conditional knockout hESC lines can be used in the future to determine cell-context-dependent DICER1 and miRNA functions, and inform us about human-specific aspects of post-implantation development.
Experimental Procedures
hESC Culture
All experiments were performed on HUES8 hESCs (NIHhESC-09-0021). HUES8 iCas9 cells were generated as previously described (Gonzalez et al., 2014). For experiments conducted in the feeder-based condition, irradiated mouse embryonic fibroblasts were used as feeder cells, and hESCs were cultured in DMEM/F12 (without HEPES) supplemented with 20% KnockOut Serum Replacement, 1× nonessential amino acids, 1× GlutaMAX, 100 U/mL penicillin/100 μg/mL streptomycin (Gemini), 0.055 mM 2-mercaptoethanol, and 10 ng/mL recombinant human basic fibroblast growth factor. Cultures were passaged at a 1:12 split ratio every 4–6 days using TrypLE to dissociate the cells. Rho-associated protein kinase inhibitor Y-27632 (Selleck Chemicals, S1049) (5 μM) was added to the culture medium when passaging cells.
For experiments conducted in the feeder-free condition, hESCs were cultured in the complete E8 medium (Thermo Fisher Scientific, A1517001) on vitronectin (VTN-N; Thermo Fisher Scientific, A14700) pre-coated plates. Cells were replated at 125,000 cells/mL in 6-well plates and passaged every 4 days after cell dissociation using 0.5 mM EDTA (KD Medical, RGE-3130). The hESC culture work was conducted as per NIH guidelines and approved by the Tri-SCI Embryonic Stem Cell Research Oversight Committee.
miRNA Mimics Rescue Assay
For the miRNA mimics rescue experiments, DICER1 knockout hESCs (B2 and D11 lines) were cultured with or without doxycycline for 6 days (feeder-based condition) or 4 days (feeder-free condition). They were replated at 125,000 cells/mL in 24-well plates in feeder-based conditions and at 125,000 cells/mL in 12-well plates in feeder-free conditions, and transfected while in suspension with miRNA mimics (mirVana miRNA mimics; Thermo Fisher, 4464066) using Lipofectamine RNAiMAX (Thermo Fisher Scientific, 13778150) as per manufacturer's instructions. For the positive control, doxycycline was added back. For the negative control, a control miRNA mimic that contains a nonspecific sequence that does not bind to any mRNAs was transfected. The final concentration of each miRNA mimic was 10 nM, as previously described (Wang et al., 2008). The transfected cells were allowed to grow without doxycycline for 6 days (feeder-based condition) or 4 days (feeder-free condition), at which point the cells were harvested for analyses.
Statistical Analysis
All values are shown as mean ± SD. Data were analyzed using unpaired two-tailed Student's t test in GraphPad Prism unless otherwise indicated. p values of ≥0.05 were considered not significant (ns) and p values of <0.05 (∗), <0.01 (∗∗), <0.001 (∗∗∗), and <0.0001 (∗∗∗∗) were considered significant.
Author Contributions
V.T. led, designed, and performed most experiments; D.Y. performed most of the revision experiments and assisted with flow cytometry; S.M. performed northern blots; F.G. cloned the DICER1∗ vectors; R.K. and C.X. analyzed the RNA-seq data; R.W.R. and N.V. assisted with additional experiments; D.H. and E.C.L. supervised the study; V.T. and D.H. wrote the manuscript; all other authors provided editorial advice.
Acknowledgments
We thank Evan Darling at the MSKCC Molecular Cytology Core for help with the imaging; Dr. Gouri Nanjangud at the MSKCC Molecular Cytogenetics Core for performing karyotype analysis; and Agnes Viale at the MSKCC Integrated Genomics Core for next-generation sequencing. We thank members of the Huangfu lab for insightful discussion and critical reading of the manuscript. This study was funded in part by an MSKCC Functional Genomics Initiative grant to E.C.L. and D.H., NIH/NIDDK (R01DK096239) to D.H., and an MSKCC Cancer Center support grant (P30CA008748). V.T. was supported by an NIH-supported T32 Training Grant in Developmental and Stem Cell Biology (T32HD060600) and an R01 Supplement grant (3R01DK096239-03S1). E.C.L.’s group was supported by the NIH/NIGMS (R01-GM083300) and NIH/NHLBI (R01-HL135564) grants.
Published: August 23, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.07.013.
Accession Numbers
All sequencing datasets are available from the Gene Expression Omnibus under accession number GEO: GSE113375.
Supplemental Information
References
- Agarwal V., Bell G.W., Nam J., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb B.S., Nesterova T.B., Thompson E., Hertweck A., O'Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson A., Collier A.J., Morgan N.P., Sienerth A.R., Chandra T., Andrews S., Rugg-Gunn P.J. Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in human embryonic stem cells. Cell Rep. 2016;17:2700–2714. doi: 10.1016/j.celrep.2016.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Sharp P.A. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve T.S., Judson R.L., Blelloch R. microRNA control of mouse and human pluripotent stem cell behavior. Annu. Rev. Cell Dev. Biol. 2013;29:213–239. doi: 10.1146/annurev-cellbio-101512-122343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Liu J., Elfenbein S.J., Ma Y., Zhong M., Qiu C., Ding Y., Lu J. Characterization of the mammalian miRNA turnover landscape. Nucleic Acids Res. 2015;43:2326–2341. doi: 10.1093/nar/gkv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Kim B., Kim V.N. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA. 2016;113:E1881–E1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krill K.T., Gurdziel K., Heaton J.H., Simon D.P., Hammer G.D. Dicer deficiency reveals microRNAs predicted to control gene expression in the developing adrenal cortex. Mol. Endocrinol. 2013;27:754–768. doi: 10.1210/me.2012-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert F., Mancuso M.R., Hampton J., Stankunas K., Asano T., Chen C.Z., Kuo C.J. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- Levy C., Khaled M., Robinson K.C., Veguilla R.A., Chen P.H., Yokoyama S., Makino E., Lu J., Larue L., Beermann F. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Karnik R., Gu H., Ziller M.J., Clement K., Tsankov A.M., Akopian V., Gifford C.A., Donaghey J., Galonska C. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Bezprozvannaya S., Williams A.H., Qi X., Richardson J.A., Bassel-Duby R., Olson E.N. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Choi Y.S., McManus M.T. Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 2010;19:R169–R175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernaute B., Spruce T., Smith K.M., Sanchez-Nieto J.M., Manzanares M., Cobb B., Rodriguez T.A. MicroRNAs control the apoptotic threshold in primed pluripotent stem cells through regulation of BIM. Genes Dev. 2014;28:1873–1878. doi: 10.1101/gad.245621.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Yu J.Y., Shcherbata H.R., Mathieu J., Wang A.J., Seal S., Zhou W., Stadler B.M., Bourgin D., Wang L. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Liu Y., Hsu Y.J., Fujiwara Y., Kim J., Mao X., Yuan G.C., Orkin S.H. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruce T., Pernaute B., Di-Gregorio A., Cobb B.S., Merkenschlager M., Manzanares M., Rodriguez T.A. An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Dev. Cell. 2010;19:207–219. doi: 10.1016/j.devcel.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H.R. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Baskerville S., Shenoy A., Babiarz J.E., Baehner L., Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N., Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.S., Lai E.C. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hong Y., Xiang D., Zhu P., Wu E., Li W., Mosenson J., Wu W.S. MicroRNA-302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Reports. 2015;4:645–657. doi: 10.1016/j.stemcr.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.