Summary
The naive embryonic stem cells (nESCs) display unique characteristics compared with the primed counterparts, but the underlying molecular mechanisms remain elusive. Here we investigate the functional roles of Lncenc1, a highly abundant long noncoding RNA in nESCs. Knockdown or knockout of Lncenc1 in mouse nESCs leads to a significantly decreased expression of core pluripotency genes and a significant reduction of colony formation capability. Furthermore, upon the depletion of Lncenc1, the expression of glycolysis-associated genes is significantly reduced, and the glycolytic activity is substantially impaired, as indicated by a more than 50% reduction in levels of glucose consumption, lactate production, and extracellular acidification rate. Mechanistically, Lncenc1 interacts with PTBP1 and HNRNPK, which regulate the transcription of glycolytic genes, thereby maintaining the self-renewal of nESCs. Our results demonstrate the functions of Lncenc1 in linking energy metabolism and naive state of ESCs, which may enhance our understanding of the molecular basis underlying naive pluripotency.
Keywords: lncRNA, glycolysis, metabolism, pluripotent stem cells, naive pluripotency, Lncenc1, embryonic stem cell
Highlights
-
•
Lncenc1 is required for the self-renewal of naive mouse ESCs
-
•
Lncenc1 controls the glycolysis pathway in naive mouse ESCs
-
•
Lncenc1 activates the expression of glycolysis genes by recruiting HNRNPK and PTBP1
In this article, Bo Wen and colleagues show that Lncenc1, a long noncoding highly RNA abundant in naive ESCs (nESC), maintains self-renewal of nESCs by controlling glycolytic activity. Lncenc1 interacts with HNRNPK and PTBP1, which function as a complex activator of glycolysis gene expression. These results suggested roles of lncRNAs in linking energy metabolism and naive pluripotency.
Introduction
Pluripotent stem cells (PSCs) can self-renew indefinitely and harbor the capability to generate almost all cells of an organism, thus holding a great promise in medical applications (De Los Angeles et al., 2015, Martello and Smith, 2014). Mouse embryonic stem cells (ESCs), the first derived PSCs, were obtained from the inner cell mass of the blastocyst (Evans and Kaufman, 1981, Martin, 1981). Epiblast stem cells (EpiSCs), a newer type of PSCs, were derived from the post-implantation embryo of the mouse (Brons et al., 2007, Tesar et al., 2007). It was proposed that ESCs and EpiSCs represent two different states of pluripotency: the naive and primed state, corresponding to pre- and post-implantation epiblasts in vivo, respectively (Marks et al., 2012). ESCs cultured in conventional serum/feeder systems are heterogonous cells with multiple pluripotent states. By contrast, ESCs cultured in 2i/LIF, i.e., ground-state ESCs or naive ESCs (nESCs), are relatively homogeneous. The nESCs harbor unique characteristics including high capability of colony formation, expression of naive pluripotency genes, global DNA hypomethylation, reactivation of two X chromosomes in female cells, and uses of both glycolysis and oxidative metabolism, mostly reflecting naive pluripotency in vivo (Hackett and Surani, 2014). Furthermore, nESCs can be converted into epiblast-like cells (EpiLCs) in vitro (Hayashi et al., 2011), representing an intermediate state between nESCs and EpiSCs (Kalkan and Smith, 2014, Kurimoto et al., 2015).
The naive-state PSCs were initially derived in rodents including mouse (Li et al., 2008) and rat (Huang da et al., 2009). Recently, human naive-like PSCs were generated by overexpression of naive pluripotency factors (Takashima et al., 2014, Theunissen et al., 2014) or by isolating directly from early embryos (Guo et al., 2016). Human naive PSCs are similar to rodent PSCs on global transcriptional state, core transcription factor networks, and metabolism properties; however, there are still significant discrepancies regarding signaling profile and epigenetic identity (Bates and Silva, 2017), suggesting that our understanding of the molecular mechanisms of the naive state remains incomplete.
Long noncoding RNAs (lncRNAs), transcripts longer than 200 nucleotides without protein-coding capability, can regulate gene expression at transcriptional and post-transcriptional levels. They are emerging as important players in many biological processes including embryonic development, stem cell self-renewal, and differentiation (Flynn and Chang, 2014, Luo et al., 2016, Rosa and Ballarino, 2016). As one of the most commonly used cell models for lncRNA studies, PSCs have been extensively profiled for RNA expression, epigenetic modifications, and RNA-protein interactions (Guttman et al., 2009, Guttman et al., 2011, Kelley and Rinn, 2012). Functional studies have indicated that lncRNAs are involved in the self-renewal of PSCs through regulating key pluripotency factors (Kaneko et al., 2014, Perry and Ulitsky, 2016, Sheik Mohamed et al., 2010), mediating chromatin modifications (da Rocha et al., 2014, Jain et al., 2016, Lin et al., 2014), or sponging/counteracting microRNAs (Liu et al., 2017, Wang et al., 2013). However, the lists of potentially functional lncRNAs in ESCs that have been reported by different studies overlap poorly (Guttman et al., 2011, Kaneko et al., 2014), which suggests that the functions of lncRNAs in PSCs are still not yet fully understood. Here we profiled genome-wide lncRNA expressions in mouse nESCs and their derived EpiLCs, and investigated the functions of one of the highly expressed lncRNAs in nESCs.
Results
The Profiling of lncRNAs in Mouse nESCs and EpiLCs
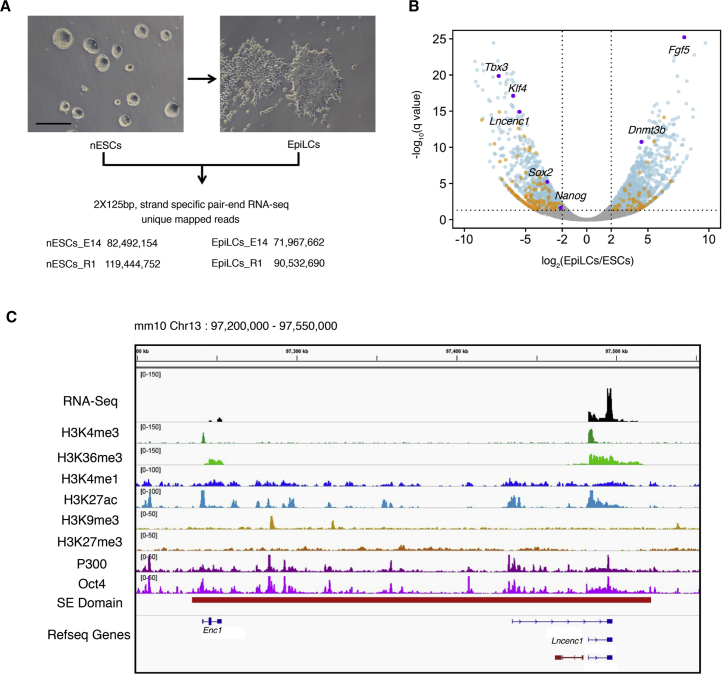
To investigate the transcripts of nESCs and EpiLCs, we performed strand-specific, ribosomal RNA-depleted RNA sequencing (RNA-seq) experiments in two independent mouse ESC lines (Figure 1A). Through differential expression (DE) analysis between nESCs and EpiLCs, 2,227 DE genes are detected (Figure 1B), including 2,025 mRNAs (1,196 upregulated and 829 downregulated) and 202 lncRNAs (67 upregulated and 135 downregulated). The full list of DE lncRNA genes is provided in Table S1.
Figure 1.
Profiling of Transcripts in nESC-to-EpiLC Transition
(A) Cell morphology changes during nESC-to-EpiLC transition and numbers of uniquely mapped reads from RNA-seq experiments conducted in two cell lines (E14 and R1).
(B) The volcano plot showing differentially expressed genes between nESCs and EpiLCs. Fold changes (log2) are plotted on the x axis, and q values (−log10 scale) are plotted on the y axis. Significantly changed genes (q < 0.05, log2 fold changes >2 or <−2) are indicated in blue (mRNAs) and orange (lncRNAs). Purple dots denote representative genes. See also Figure S1 and Table S1.
(C) Chromatin features in the Lncenc1 locus. The top track shows normalized reads densities of RNA-seq results of nESCs. Tracks below show signals of histone modifications, TF binding, and the super-enhancer domain. P300, Oct4, H3K27ac, and H3K4me1 ChIP-seq data were from GEO: GSE56138; H3K4me3, H3K27me3, H3K36me3, and H3K9me3 ChIP-seq data were from GEO: GSE23943. Coordinates of super-enhancer domains were obtained from Dowen et al. (2014). The lncRNA named lincenc1 (Sauvageau et al., 2013) is shown in red at the bottom track.
To examine the functions of lncRNAs preferentially expressed in nESCs, we tested seven lncRNAs in the lists through small hairpin RNA (shRNA)-based RNAi. We selected these lncRNAs because they are relatively enriched in nESCs, and their functions remain unknown. We achieved a significant depletion of greater than 40% with both shRNAs targeting four of seven lncRNAs tested (Lncenc1, Panct2, GM13110, and GM805). Depletion of each of these lncRNAs was associated with a significant reduction in Oct4 and/or Nanog mRNA levels, along with visible flatting of clone morphologies, suggesting that these lncRNAs may play roles in maintaining the state of nESCs (Figure S1).
Lncenc1 Is Specifically Expressed in nESCs and Is Highly Dynamic during Differentiation
Among these lncRNAs, we decided to focus on Lncenc1, as (1) it is the most abundant lncRNA that is differentially expressed in nESCs (Table S1); (2) its gene locus overlaps a super-enhancer domain (Figure 1C), suggesting that it is potentially functional; and (3) it is specifically expressed in nESCs relative to somatic tissues (Figure 2A). We noticed a published mouse work, which knocked out a lncRNA named as lincenc1 (Sauvageau et al., 2013), but the reported sequence transcribes from the opposite direction and does not overlap with the lncenc1 gene we studied here (Figure 1C).
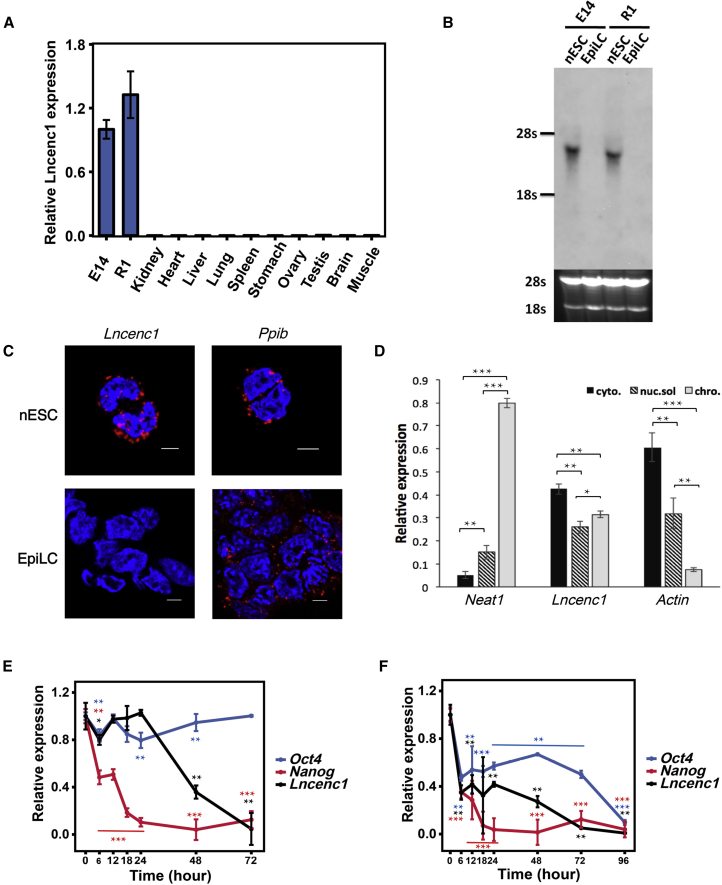
Figure 2.
Characterization of Lncenc1 and Its Expression Dynamics during ESC Differentiation
(A) Expression of Lncenc1 in mouse nESCs and somatic tissues as measured by qRT-PCR. Expression levels were normalized against Actb. Results are shown as means ± SD of three independent experiments.
(B) The Lncenc1 transcript detected by northern blot analysis. The agarose gel imaging (black background) underneath the blot (gray background) shows total RNA used for analysis.
(C) RNA in situ hybridization (RNAscope) targeting Lncenc1 in mouse nESCs and EpiLCs. Ppib was used as a positive control. Nuclei were stained with DAPI. Scale bars, 5 μm.
(D) Relative expression levels of Lncenc1 in cytoplasm, nucleoplasm, or chromatin fractions as determined by qRT-PCR analysis of RNA extracted from each fraction. Neat1 and Actb were used as controls. Data are the means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(E and F) Expression dynamics of two pluripotency genes (Oct4 and Nanog) and Lncenc1 during the nESC-to-EpiLC transition (E), and during spontaneous differentiation (F). Expression levels were normalized against Actb. Results are shown as means ± SD of three independent experiments. Data are compared with 0 hr. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figures S2B and S2C.
To test the completeness of the Lncenc1 transcript, we performed northern blot analysis. The data showed that the transcript is about 3,400 nucleotides in size in nESCs, but was almost undetectable in EpiLCs (Figure 2B). According to our RNA-seq data (Figure 1C) and RT-PCR analysis (Figure S2A), the shorter isoform (NR_110430) expresses in nESCs rather than the longer one (NR_110432). Furthermore, both RNA FISH and cellular fraction experiments indicated that transcripts of Lncenc1 located in both the nucleus and cytoplasm of mouse nESCs (Figures 2C and 2D).
To study dynamic changes of Lncenc1 expression during the nESC-to-EpiLC transition, we measured its expression over a 72-hr time window in E14 nESCs. The results showed that the expression level is relatively stable during the first 24 hr, but then decreases dramatically and becomes nearly undetectable after 72 hr (Figure 2E). Similar dynamics were observed in spontaneous differentiation induced by the withdrawal of 2i/LIF from the medium, and the decline of Lncenc1 expression happened earlier at around hour 6 (Figure 2F). Interestingly, the decline of Lncenc1 expression occurred after Nanog but before Oct4 in both cases; this suggests that Lncenc1 could be involved in pluripotency networks, consistent with a recent observation that the expression profile of Lncenc1 is associated with known pluripotency genes (Bergmann et al., 2015). Similar expression dynamics were also observed in the R1 cell line (Figures S2B and S2C), indicating a general role of Lncenc1 in nESCs. Functional experiments thereafter were performed only in E14 cells unless otherwise stated.
Lncenc1 Is Required for Self-Renewal in nESCs
To investigate the function of Lncenc1, we knocked down its expression in nESCs by two independent shRNAs, which can deplete 70%–80% of Lncenc1 transcripts as shown by qRT-PCR. As a result, the expression of six pluripotency genes decreases significantly, although the magnitude of deduction is usually less than 50% (Figure 3A). Consistently, protein levels of OCT4, NANOG, and SOX2 are decreased in Lncenc1 knockdown cells as shown by western blot and immunofluorescence (Figures S3A and S3B). Moreover, clone morphologies changed from “round and tight” to “flat and loose,” and alkaline phosphatase staining became weaker in Lncenc1-knockdown cells (Figures 3B and S3C). As Lncenc1 transcripts locate in both nucleus and cytosol, we also conducted antisense oligonucleotide (ASO)-mediated gene silencing, which showed higher knockdown efficiency compared with RNAi for nuclear RNAs (Lennox and Behlke, 2016). Consistently, when Lncenc1 was suppressed by two independent ASOs, expression levels of core pluripotency genes were significantly decreased (Figure S3D).
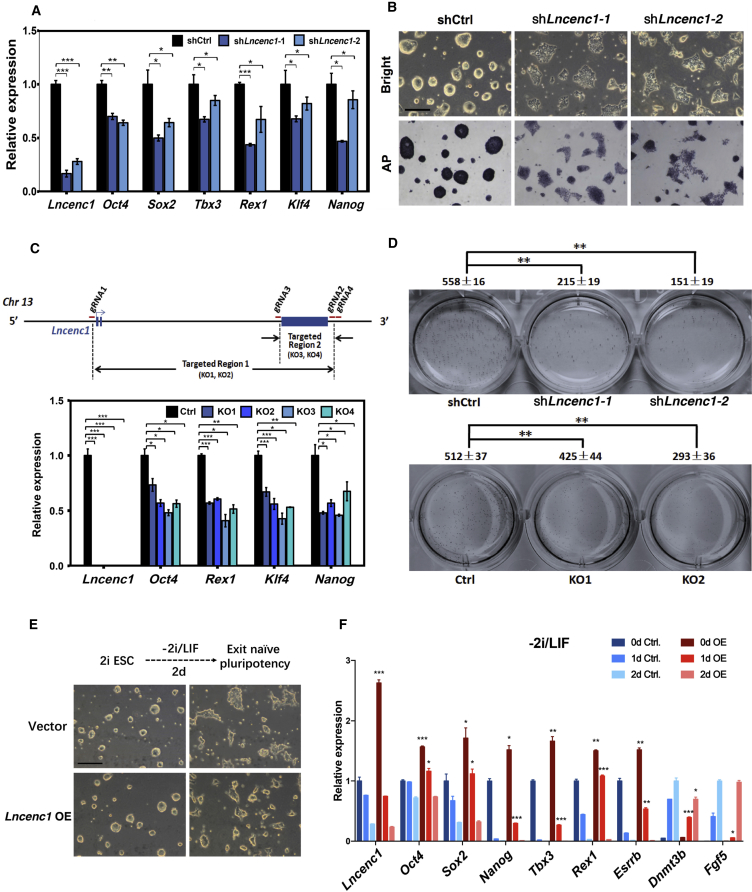
Figure 3.
Lncenc1 Is Required to Maintain the Self-Renewal of Mouse nESCs
(A) Expression levels of six pluripotency genes as measured by qRT-PCR in nESCs upon shRNA-mediated knockdown of Lncenc1. Expression levels were normalized against Actb. Results are shown as means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) Bright-field micrographs (upper panels) and alkaline phosphatase staining in control nESCs and Lncenc1 knockdown ESCs. Scale bars, 200 μm. See also Figure S3C.
(C) Expression of pluripotency genes in wild-type (Ctrl.) and Lncenc1 KO nESCs. Upper panel: the schematic representation of CRISPR/Cas9-mediated knockouts of the Lncenc1 locus. Four KO clones were obtained by depletion of the whole gene (KO1 and KO2) or depletion of the largest exon (KO3 and KO4). Expression levels were normalized against Actb. Results are shown as means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Results of colony formation experiments in control and Lncenc1 knockdown or KO nESCs. Clone numbers are shown as means ± SD of three replicates. Statistical significance of t test: ∗∗p < 0.01.
(E) Bright-field micrographs of control nESCs and Lncenc1-overexpressed ESCs in 2I/LIF (left) and then withdrawal of 2i/LIF for 2 days (right). Scale bars, 200 μm.
(F) Expression levels of pluripotency genes of control nESCs and Lncenc1-overexpressed ESCs in 2i/LIF (0d) and spontaneous differentiation (1d, 2d). Expression levels were normalized against Actb. Results are shown as means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To further rule out potential off-target effects of shRNA- or ASO-based gene silencing, we generated Lncenc1 knockout (KO) cell lines through CRISPR/Cas9-based genome editing technology. We successfully established four KO cell lines by deleting the entire gene locus or the largest exon, and Lncenc1 transcripts were almost completely depleted in these cells. Compared with wild-type cells, the expression of pluripotency genes is significantly reduced in all these KO cell lines (Figure 3C). More important, colony formation capability, a proxy for self-renewal efficiency (Martello and Smith, 2014), was significantly decreased in knockdown and KO cells (Figure 3D).
To test whether Lncenc1 is sufficient to maintain the naive state, we overexpressed Lncenc1 (OE) in nESCs and then withdrew 2i/LIF for 2 days. The morphology of OE cells is more “naive-like” compared with the control (Figure 3E). In the 2i/LIF culture (day 0), the expression levels of pluripotency genes tested are all significantly increased in the OE cells (Figure 3F). After the withdrawal of 2i/LIF for 1 day, the expression levels of pluripotency genes, especially naive-state genes (Nanog, Tbx3, Rex1, and Esrrb), remain significantly higher, whereas those of post-implantation genes (Fgf5 and Dnmt3b) are significantly lower in the OE cells, suggesting that the overexpression of Lncenc1 in nESCs delays their differentiation. On day 2, the expression levels of pluripotency genes decrease further and are comparable between OE and control cells. Therefore, Lncenc1 is required but not sufficient to keep the ESCs in a naive state.
Lncenc1 Controls the Glycolysis Pathway in nESCs
To reveal global gene expression changes upon Lncenc1 depletion, we conducted RNA-seq experiments with control and knockdown cells and identified 101 downregulated and 182 upregulated genes (Figure 4A; Table S2). Functional annotation of these affected genes indicated that they are highly enriched in the glycolysis/gluconeogenesis pathway (Figure 4B). We then examined mRNA expression with qRT-PCR in knockdown (by shRNAs and ASOs) and KO nESCs and confirmed that these glycolysis genes detected by RNA-seq are all significantly decreased upon depletion or deletion of Lncenc1 (Figures 4C, 4D, and S3E). Consistently, protein levels of these glycolytic enzymes are markedly decreased in Lncenc1 knockdown and KO cells as determined by western blotting (Figure 4E). Furthermore, upon the overexpression of Lncenc1 in nESCs, the mRNA levels of glycolysis genes are upregulated significantly; however, after spontaneous differentiation, the control and OE ESCs display similar dynamic changes on these glycolysis genes (Figure 4F).
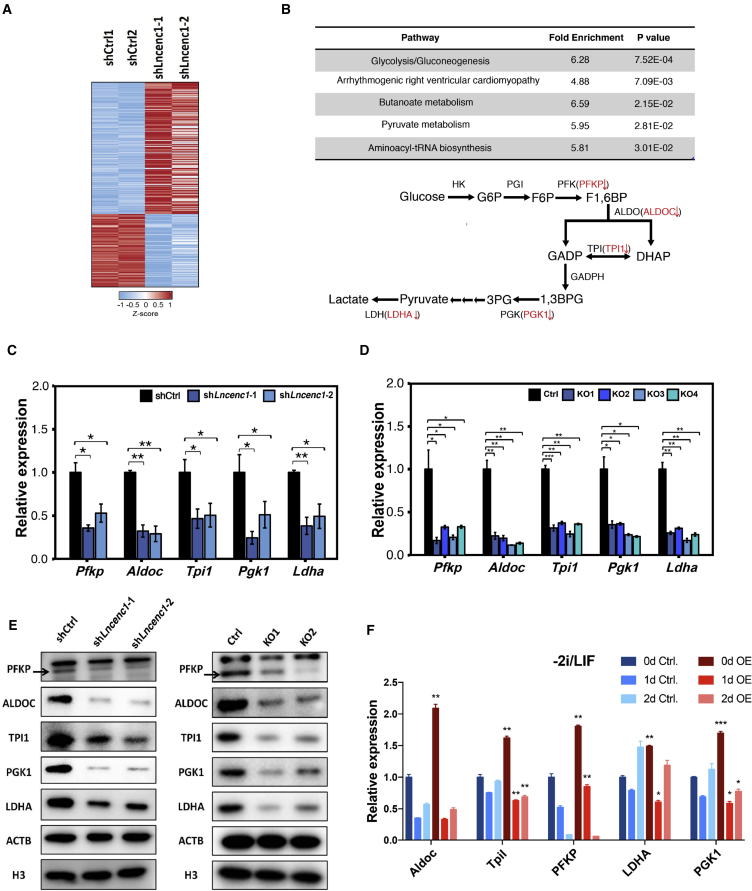
Figure 4.
Lncenc1 Regulates the Glycolysis Pathway in nESCs
(A) The heatmap of differentially expressed genes upon Lncenc1 knockdown as identified by RNA-seq experiments.
(B) Significantly enriched pathways (p < 0.05) of differentially expressed genes upon Lncenc1 knockdown as revealed by KEGG pathway analysis. Lower panel: the schematic showing of the glycolysis pathway and significantly downregulated genes (in red) upon Lncenc1 depletion.
(C and D) Expression of glycolysis genes in the Lncenc1 knockdown or KO nESCs. (C) knockdown by shRNAs; (D) KO by CRISPR/Cas9. Data were normalized against Actb and are shown as means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S3B.
(E) Protein levels of glycolytic enzymes in Lncenc1 knockdown (left) or KO (right) nESCs as detected by western blot analysis. ACTB and H3 were used as the loading controls.
(F) Expression levels of glycolysis genes in control and Lncenc1-overexpressed ESCs in 2i/LIF (0d) and spontaneous differentiation (1d, 2d). Data were normalized against Actb and are shown as means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To test whether the functions of Lncenc1 can be rescued in its depleted or deleted cells, we have re-expressed Lncenc1 in the knockout and knockdown ESCs. Unfortunately, although Lncenc1 can be re-expressed markedly, the expression of glycolysis and pluripotency genes still cannot be restored (Figure S4). These data suggest that increased expression alone might not be sufficient for Lncenc1 activity. Other factors might also contribute toward making Lncenc1 functional, such as the location of expression in the genome, and the localization of the ectopic Lncenc1 RNAs.
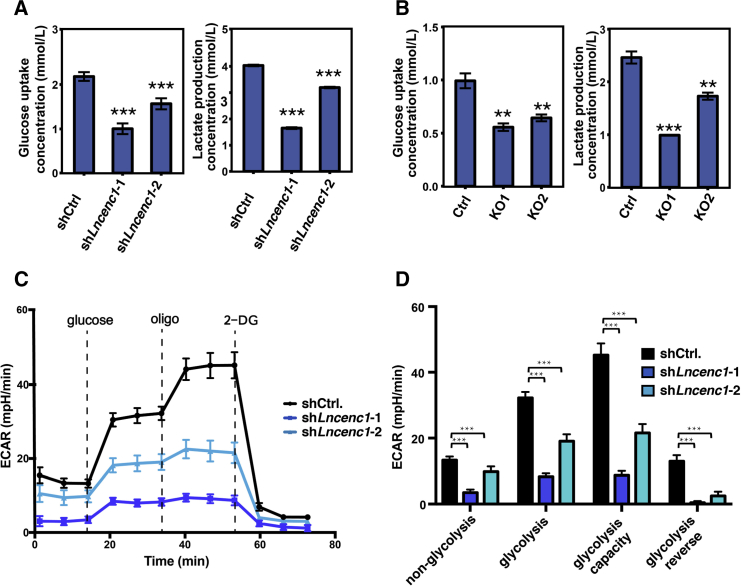
Next, we examined the glycolytic activity through functional assays. First we measured the glucose consumption and lactate production, the “in and out” of glycolysis, and found that they are both significantly decreased in Lncenc1 knockdown (Figure 5A) and KO (Figure 5B) cells compared with the controls. We further examined glycolytic activity through glycolysis stress tests. As expected, the level of glycolytic activity was significantly decreased in knockdown cells as indicated by the measurement of the extracellular acidification rate (ECAR), which is largely the result of glycolysis (Figure 5C). We measured the initial pH in glucose-free culture medium (non-glycolysis). Then, to activate glycolysis, we sequentially added oligomycin (a mitochondrial ATP synthase inhibitor) to the culture medium. Both before and after activation, the glycolytic rate of control cells was significantly higher than that of knockdown cells (Figure 5D). It is notable that, when knockdown experiments were conducted with shLncenc1-1, which displayed better efficiency, glucose consumption, lactate production, and ECAR were all reduced by 50% or more. These data indicate that the glycolysis activity was severely impaired after the downregulation of Lncenc1.
Figure 5.
Glycolic Activity Decreases upon Lncenc1 Depletion
(A and B) Measurements of glucose uptake and lactate production in Lncenc1 knockdown (A) or KO (B) nESCs. Shown are results of three replicates (means ± SD). Statistical significance of t test: ∗∗p < 0.01, ∗∗∗p < 0.001.
(C and D) Results of glycolysis stress tests as measured by the glucose-dependent extracellular acidification rate (ECAR). Shown are measurements of at least five replicates (means ± SD). Statistical significance of t test: ∗∗∗p < 0.001.
Lncenc1 Functionally Interacts with HNRNPK and PTBP1
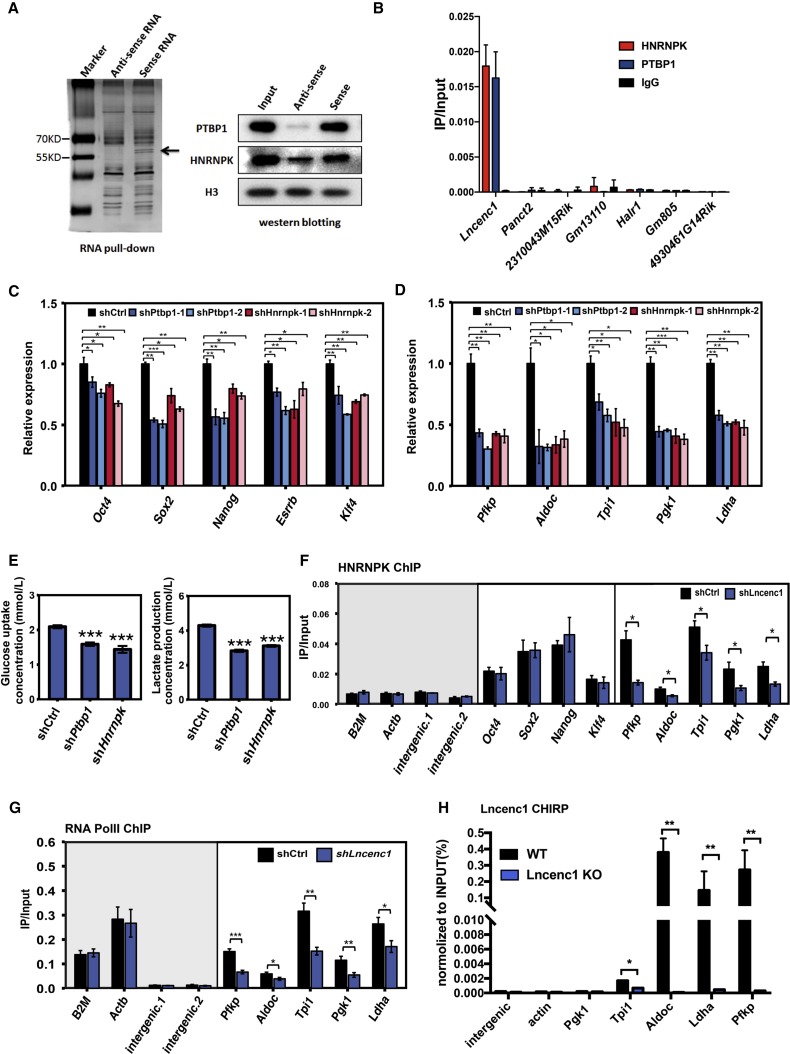
To identify potential partners of Lncenc1, we performed the RNA pull-down assay using biotin-labeled probes. Compared with the antisense control, the pull-down sample with sense probe showed two specific bands on the SDS-PAGE gel, which were identified as PTBP1 and HNRNPK by mass spectrometry and further validated by western blotting (Figure 6A). Moreover, results of RNA immunoprecipitation against PTBP1 and HNRNPK antibodies showed that Lncenc1 RNAs are highly enriched compared with the other six lncRNAs tested (Figure 6B), which further confirmed interactions between Lncenc1 and these two proteins. The yeast two-hybrid assay showed that PTBP1 and HNRNPK interact with each other (Kim et al., 2000), and they interact as part of a complex in murine ESCs (Lin et al., 2014). We then investigated whether PTBP1 and HNRNPK are functionally related to Lncenc1. As expected, knockdown of Ptbp1 or Hnrnpk resulted in a significant downregulation of pluripotency and glycolysis genes (Figures 6C and 6D) and a significant reduction of glucose consumption and lactate production (Figure 6E), which is consistent with phenotypes observed in the Lncenc1 knockdown or KO cells. These data indicated that Lncenc1, PTBP1, and HNRNPK are interacting physically and functionally.
Figure 6.
Lncenc1 Associates with HNRNPK and PTBP1 Functionally
(A) Results of RNA pull-down experiments. Biotinylated Lncenc1 sense or antisense RNAs were used for the experiments. Two specific bands (arrowheads) were excised and subjected to mass spectrometry analysis, and were further validated by western blotting.
(B) RNA immunoprecipitation (RIP) with anti-HNRNPK and anti-PTBP1 antibodies in E14 nESCs. Enrichments of lncRNAs, i.e., expression of RIP relative to input samples, were determined by qRT-PCR.
(C and D) Expression of pluripotency genes (C) or glycolysis genes (D) upon knockdown of Ptbp1 and Hnrnpk. Data were normalized against Actb, and shown as the means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(E) Glycolysis signatures of Hnrnpk and Ptbp1 knockdown cells. Data were presented as means ± SD.
(F–H) Chromatin immunoprecipitation (ChIP) experiments with HNRNPK (F) or RNA polymerase II (G) antibodies; CHIRP experiment with specific anti-Lncenc1 probes (H). Relative enrichment of ChIP or CHIRP and input DNA were determined by qPCR. Primers were designed at promoters of glycolysis genes, pluripotency genes, and control genes (B2m and Actb), and two intergenic regions were used as negative controls. Data are shown as means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To test whether the Lncenc1-PTBP1-HNRNPK complex regulates the expression of pluripotency and/or glycolysis genes, we performed chromatin immunoprecipitation (ChIP) experiments in Lncenc1 knockdown and control nESCs. Indeed, HNRNPK binds to the promoters of pluripotency and glycolysis genes. However, only the occupancy of HNRNPK on glycolytic genes, but not pluripotency genes, were significantly decreased in knockdown cells compared with the control cells (Figure 6F). Furthermore, the occupancy of RNA polymerase II on glycolysis gene promoters decreased significantly upon Lncenc1 or PTBP1 knockdown (Figures 6G and S5). In addition, we performed chromatin isolation by RNA purification (ChIRP) assay in Lncenc1 knockout and control nESCs. Consistent with our ChIP data, Lncenc1 occupied the promoters at four of the five glycolytic genes (Figure 6H). These data suggested that Lncenc1 guides HNRNPK and PTBP1 binding to glycolysis genes; therefore promoting their levels of transcription.
Interplays among Lncenc1, Core Pluripotency Networks, and Glycolytic Genes
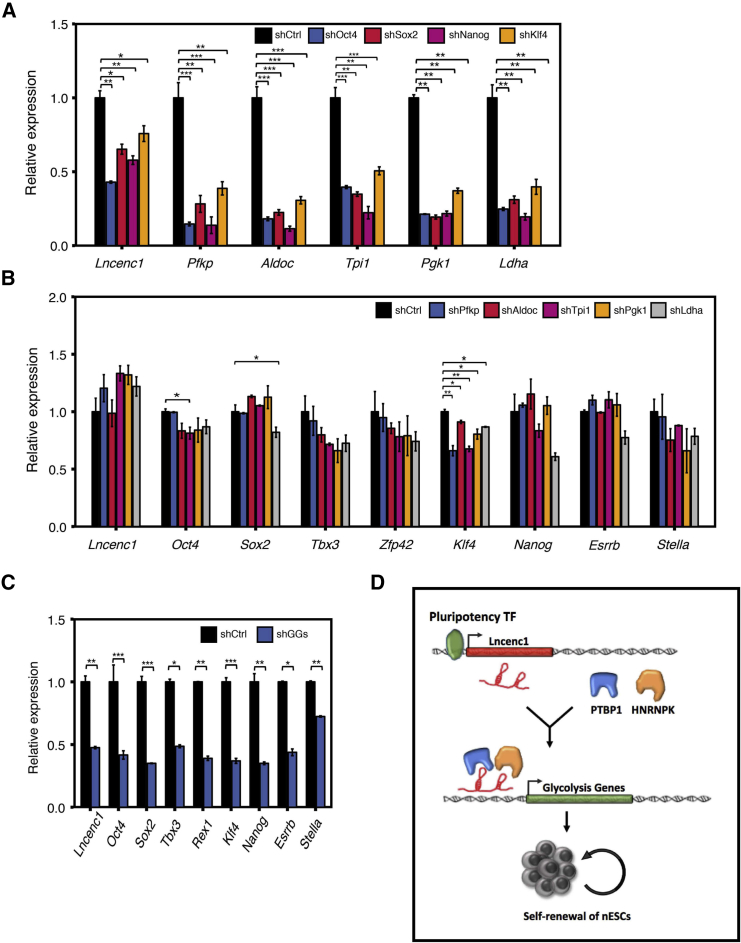
Given that Lncenc1 can affect self-renewal and glycolysis, we were curious about the regulatory relationships among them. First, upon the knockdown of the individual pluripotency gene (Oct4, Sox2, Nanog, or Klf4) (Figure S6A), Lncenc1 and all examined glycolysis genes were downregulated consistently (Figure 7A). Second, a recent study showed that core pluripotency factors directly regulate expression of glycolysis genes (Kim et al., 2015), and that the Lncenc1 locus is enriched for the binding of OCT4 (Figure 1C), indicating that Lncenc1 and glycolysis genes are regulated directly by core pluripotency factors.
Figure 7.
Regulatory Relationships among Lncenc1, Pluripotency Genes, and Glycolysis Genes
(A–C) Expression of Lncenc1 and glycolysis genes when individual pluripotency genes were knocked down (A); expression of Lncenc1 and pluripotency genes when individual glycolysis genes were knocked down (B); and expression of Lncenc1 and pluripotency genes upon knockdown of all five glycolysis genes (GGs) (C). For qRT-PCR experiments, expression levels were normalized against Actb, and data are shown as means ± SD of three independent experiments. Statistical significance of t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Working model of Lncenc1 in nESCs.
In contrast, knockdown of individual glycolysis genes failed to downregulate Lncenc1 and pluripotency genes, except for Klf4 (Figures 7B and S6B). However, when all these five glycolysis genes were knocked down together (with two sets of shRNA pools), Lncenc1 and pluripotency genes were all markedly declined (Figures 7C, S6C, and S6D). Furthermore, the adding of 2-deoxy-D-glucose (2-DG) (a glycolysis inhibitor) to the 2i/LIF medium leads to “flatter” clone morphology and significant decreases of pluripotency gene expression (Figure S6E). These data suggested that the glycolysis pathway, rather than individual genes, regulates the core pluripotency genes.
In summary, our data suggested that Lncenc1, PTBP1, and HNRNPK form functional complexes, which promote expression of glycolysis genes, thereby maintaining the glycolytic activity, which is required for the self-renewal of naive ESCs (Figure 7D).
Discussion
Functional studies indicated that some lncRNAs regulate self-renewal or pluripotency through integrating or disturbing core pluripotency networks. For example, Gomafu regulates Oct4 and Nanog expression through the formation of a positive feedback loop with Oct4 (Sheik Mohamed et al., 2010), and TUNA maintains self-renewal of mESCs through promoting transcriptional activities of pluripotent genes including Nanog, Sox2, and Fgf4 (Kaneko et al., 2014). However, an Oct4 pseudogene lncRNA guides SUV39H1, a repressive chromatin modifier, to the promoter and epigenetically silences the Oct4 gene in differentiated cells (Scarola et al., 2015). Similarly, in somatic cells, lincRNA-p21 interacts with SETDB1, which silences pluripotency genes by depositing histone H3K9me3 on their promoters (Bao et al., 2015). Furthermore, linc-ROR promotes reprogramming by repressing pathways including P53 responses (Loewer et al., 2010), and this lncRNA maintains self-renewal of human ESCs through sponging miR-145, which targets mRNAs of core pluripotency genes (Wang et al., 2013). Recently, Austin Smith and colleagues reported that the lncRNA Ephemeron modulates the exit from naive pluripotency by connecting microRNA and de novo DNA methylation (Bates and Silva, 2017). In this study, we demonstrate that Lncenc1 maintains self-renewal by regulating the glycolysis pathway. To our knowledge, Lncenc1 is the first lncRNA linking self-renewal and energy metabolism in PSCs.
We showed that Lncenc1 interacts with two RNA binding proteins (RBPs), PTBP1 and HNRNPK. Interestingly, the lncRNA TUNA also interacts with PTBP1 and HNRNPK, activating pluripotency genes directly (Kaneko et al., 2014). However, linc-p21 interacts with HNRNPK and SETBD1, silencing rather than activating its target genes in somatic cells (Bao et al., 2015). PTBP1 is required for embryonic development (Suckale et al., 2011), and HNRNPK could interact with multiple proteins through its K interactive region to play various roles in diverse cellular processes (Geuens et al., 2016). Based on these data, we propose that a class of RBPs such as HNRNPK could form complexes with different lncRNAs and/or proteins that regulate specific classes of genes in different cell types. Further investigations are needed to reveal the functions of lncRNA-RBP complexes in various biological systems.
Recently, energy metabolism has been closely linked to stem cell fates. For example, during the ESC-to-EpiSC transition, energy metabolism switched from bivalent (utilization of both glycolysis and oxidative phosphorylation) to exclusively glycolytic, and hypoxia inducible factor 1a was sufficient to drive ESC to an EpiSC-like stage (Zhou et al., 2012). A recent metabolome analysis further indicated that, although early metabolites are increased, downstream glycolysis metabolites are decreased in primed hESCs (Sperber et al., 2015). Interestingly, when mESCs adapted from serum to 2i conditions, the glycolysis pathway was significantly upregulated as detected by RNA-seq (Marks et al., 2012). Similarly, protein levels of glycolysis enzymes are significantly increased in naive ESCs compared with ESCs cultured in serum (Taleahmad et al., 2015). Our data further demonstrate that the downregulation of glycolysis by lncenc1 depletion impairs self-renewal, which suggests that a high level of glycolysis is vital for naive ESCs. However, it remains unclear how glycolysis regulates naive pluripotency. It is possible that a faster generation of ATPs by glycolysis benefits highly proliferative cells including stem cells and cancer cells, or that glycolysis-generated metabolites are essential for establishing the epigenetic modifications required for the maintenance of PSCs (Lin et al., 2016, Moussaieff et al., 2015, Sperber et al., 2015). Further investigations are required to decipher the detailed mechanisms underlying glycolysis and naive pluripotency states. Moreover, it would be possible to optimize culture systems for PSCs by manipulating metabolic status by adding or removing specific glycolysis metabolites.
Nevertheless, it remains elusive how Lncenc1 functions in vivo, and developing Lncenc1-deficient mouse models will be critical in addressing this question. Lncenc1 RNAs locate both in nucleus and cytoplasm. In this study, we investigated the functions of the nuclear Lncenc1, yet the activity of cytoplasmic Lncenc1 remains to be explored in the future. Finally, as exemplified by Lncenc1 and other lncRNAs regulating glycolysis in cancer (Bao et al., 2015, Dowen et al., 2014, Geuens et al., 2016, Rupaimoole et al., 2015), it would be interesting to explore further functional roles of different lncRNAs in regulating energy metabolism in stem cells, development, and diseases.
Experimental Procedures
Cell Culture
To maintain ESCs in ground state or naive state, cells from two mouse ESC lines (E14TG2a and R1) were cultured in 2i /LIF conditions as described (Li et al., 2008). The nESC-to-EpiLC transition was conducted as described (Hayashi et al., 2011) with minor modifications. For spontaneous differentiation, nESC cells were plated at a density of 1 × 104 per cm2 in gelatin-coated plates in N2B27 medium without 2i/LIF for 4 days. For the 2-DG treatment, nESCs were cultured in 2i/LIF medium for 1 day and then treated with 2-DG (100 mM) for 2 days.
RNA-Seq and Data Analysis
Total RNA was prepared with TRIzol reagents (Life Technologies). For RNA-seq experiments, rRNA-depleted and strand-specific libraries were constructed with the Ribo-zero gold (Epicentre) and TruSeq Stranded Total RNA Sample Prep Kit (Illumina), and the libraries were sequenced using the HighSeq 2000 system (Illumina). The analyses of RNA-seq data, including raw data processing, mapping, and gene expression, were conducted as described previously (Fan et al., 2018). For differential gene expression analysis, we analyzed raw read counts for GENCODE M7 gene models using the HTSeq software (Anders et al., 2015), and then calculated statistics of DE via DESeq2 (Love et al., 2014) with default parameters. To define differentially expressed genes, we used a false discovery rate of 0.05 (ESC-EpiLC comparison) or 0.1 (Lncenc1 knockdown studies) as thresholds. We performed KEGG pathway analysis using DAVID bioinformatics tools as described (Huang da et al., 2009).
Northern Blot Analysis
For northern blotting, 10 μg of total RNAs was mixed with 2× loading buffer (Fermentas), incubated at 55°C for 5 min, and resolved by denaturing agarose gel electrophoresis; the RNAs were then transferred to a nylon membrane and crosslinked by UV irradiation. Digoxigenin-labeled RNA probe preparation, hybridization, and detection were conducted via a DIG Northern Starter Kit (Roche) according to the manufacturer's protocol.
Cellular Fractionation
Cellular fractionation experiments were performed with 1 × 107 nESCs as described previously (Bhatt et al., 2012).
Gene Knockdown
The shRNA-based knockdown experiments were conducted as described previously (Bao et al., 2015). The validated shRNA sequences were provided in Table S3. For ASO-mediated knockdown, we used phosphorothioate-modified oligodeoxynucleotides (Ideue et al., 2009). Two independent ASOs targeting Lncenc1 (mUmCmCmUmUAAGGTGCTTCmAmGmTmTmC, and mGmUmCmAmUGCTGATGCTGmGmUmGmCmA), and one control ASO (mCmCmUmUmCCCTGAAGGTTmCmCmUmCmC) were synthesized (BioSune). Then 2 × 105 nESCs were transfected with 100 μM of each ASO with 6 μL RNAiMAX (Life Technologies) and harvested 48 hr after the transfection.
Gene Knockout
To deplete the Lncenc1 locus in E14 ESCs, we applied CRISPR/Cas9-based assays as described (Cong et al., 2013). Two pairs of guide RNAs (pair1: GAGCCAATCTCTAGGCAAGT and GTATGACAAATGCTTATTGA; pair2: GTGCTTTTGTGTTATCCCGG and GGTCCATTATGTAACCACCT) were used to target the Lncenc1 genomic locus, deleting regions about 15 kb (pair1) and 3 kb (pair2), respectively. Lncenc1 knockouts were verified by genomic DNA PCR and sequencing.
Overexpression
The Lncenc1 cDNA was cloned into the pcDNA3.1 vector, and the overexpression experiments were performed with electroporation by the Neon system (Invitrogen), according to the manufacturer's instructions.
qRT-PCR
The qRT-PCR experiments were conducted as described (Bao et al., 2015). Primer sequences for qRT-PCR are listed in Table S4.
Western Blotting
Total cell lysates were prepared in 1× SDS buffer. An equal amount of proteins were separated by SDS-PAGE gel electrophoresis and transferred onto polyvinylidene difluoride membranes. The following antibodies were used for western blotting: anti-HNRNPK (Abcam, ab39975), anti-PTBP1 (Invitrogen, 324800), anti-PFKP (Proteintech, 13,389), anti-ALDOC (Proteintech, 14,884), anti-TPI1 (Proteintech, 10,713), anti-LDHA (Proteintech, 14,824), anti-PGK1 (Proteintech, 17,811), anti-SOX2 (Proteintech, 11,064), anti-H3 (Abcam, ab1791), anti-ACTB (Proteintech, 60,008-1-Ig), OCT4 (Santa Cruz, sc-8628). The blots were developed with ECL Plus Western Blotting Substrate (Thermo Scientific), and imaged by the FluorChem M System (ProteinSimple).
RNA Fluorescence In Situ Hybridization
RNA fluorescence in situ hybridization for the detection of Lncenc1 and Ppib (positive control) was performed using an RNAscope (Advanced Cell Diagnostics, Hayward, CA, 323100), according to the protocol provided by the manufacturer.
Alkaline Phosphatase Detection
ESCs were fixed with 4% paraformaldehyde and stained with the Alkaline Phosphatase Staining Kit (Millipore) according to the manufacturer's protocol.
Colony Formation
ESCs were plated at a density of 100 per cm2 in laminin-coated 6-well plates in 2i/LIF. The medium was changed every 3 days. After 6 days, colonies were fixed and stained for alkaline phosphatase. Colonies were counted using the ImageJ software.
The Measurement of Glucose Uptake and Lactate Production
The same amount (1 × 106) of control and knockdown or knockout ESCs were seeded on a well of 12-well plates in 0.5 mL of N2B27 medium containing 2i/LIF. After 6 hr, the culture medium was collected for glucose and lactate measurement, using the Cobas C311 Chemistry Analyzer (Roche), according to the manufacturer's instructions. The intracellular glucose consumption and lactate production were calculated by the concentration differences between cultured medium and the blank medium.
Seahorse XF Bioenergetic Analysis
SeaHorse plates were pre-treated by coating with 0.1% gelatin. ECAR in mESCs were measured using the Seahorse XF96 bioanalyzer according to the manufacturer's protocols. Cells were seeded in an XF96 microplate at a density of 20,000 cells per well. Culture medium was changed to unbuffered XF assay medium at pH 7.4 (Seahorse Biosciences), supplemented with 5.5 mM glucose (Sigma) and 1 mM pyruvate for assay of glycolysis rates. Before assay, the microplate was transferred into a non-CO2 incubator at 37°C for 60 min. For the glycolysis stress test, after collecting baseline acidification rate data, cells were sequentially treated with 10 mM glucose, 1 mM oligomycin, and 50 mM 2-DG, and the subsequent changes were quantified. All metabolic profiles were normalized to cell numbers.
RNA Pull-Down Assay
RNA pull-down experiments were performed as described previously (Tsai et al., 2010). Specific gel bands were excised and subject to mass spectrometry analysis.
RNA Immunoprecipitation
Mouse nESCs (1 × 107) were subjected to nuclei isolation as described (Doi et al., 2009). Nuclei were then resuspended in 1 mL of cold RNA immunoprecipitation (RIP) buffer (150 mM KCl, 25 mM Tris [pH 7.4], 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, 100 U/mL RNAase inhibitor, 1× Complete) and sheared by sonication. For immunoprecipitation (IP), 2 μg of specific antibody (HNRNPK [Abcam, ab39975]; PTBP1 [Invitrogen, 324800]) or the control IgG (Cell Signaling Technologies, 2,729) was incubated with 6–10 mg supernatant at 4°C for 2 hr, then 20 μL of protein G magnetic beads (Invitrogen) were added to each IP sample and incubated at 4°C for 1 hr. The beads were collected and washed three times with RIP buffer. RNAs were extracted with TRIzol, and enrichment for each gene was determined by qRT-PCR.
ChIP
ChIP experiments were performed with 1 × 107 mESCs as described previously (Irizarry et al., 2008). The following antibodies were used for ChIP: anti-HNRNPK (Abcam, ab39975) and anti-RNA PolII (Abcam, ab817). Primers for ChIP-PCR were provided in Table S5.
ChIRP
ChIRP experiments were performed as described previously (Chu et al., 2011). Probe sequences are provided in Table S6.
Author Contributions
Z.S., M.Z., L.C., and B.W. conceived and designed the study. Z.S., M.Z., L.C., Q.W., P.T., and Z.Y. performed the experiments. P.L. performed bioinformatics analysis. All authors reviewed and approved the manuscript.
Acknowledgments
We thank Hao Wu and Yun Liu for critical reading of the manuscript, and Dan Ye for suggestions on metabolism assays. We thank the Biomedical Core Facility, Fudan University, for mass spectrometry analysis, and Lizhi Jiang for assistance with metabolism assays. This work was supported by the National Basic Research Program of China (2015CB943000 to B.W.), and the National Natural Science Foundation of China (31771435 to B.W).
Published: August 30, 2018
Footnotes
Supplemental Information includes six figures and six tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.08.001.
Accession Numbers
The RNA-seq datasets from this study have been submitted to the NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GEO: GSE116602.
Supplemental Information
References
- Anders S., Pyl P.T., Huber W. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Wu H., Zhu X., Guo X., Hutchins A.P., Luo Z., Song H., Chen Y., Lai K., Yin M. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25:80–92. doi: 10.1038/cr.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates L.E., Silva J.C. Reprogramming human cells to naive pluripotency: how close are we? Curr. Opin. Genet. Dev. 2017;46:58–65. doi: 10.1016/j.gde.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann J.H., Li J., Eckersley-Maslin M.A., Rigo F., Freier S.M., Spector D.L. Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 2015;25:1336–1346. doi: 10.1101/gr.189027.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt D.M., Pandya-Jones A., Tong A.J., Barozzi I., Lissner M.M., Natoli G., Black D.L., Smale S.T. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha S.T., Boeva V., Escamilla-Del-Arenal M., Ancelin K., Granier C., Matias N.R., Sanulli S., Chow J., Schulz E., Picard C. Jarid2 is implicated in the initial xist-induced targeting of PRC2 to the inactive x chromosome. Mol. Cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- De Los Angeles A., Ferrari F., Xi R., Fujiwara Y., Benvenisty N., Deng H., Hochedlinger K., Jaenisch R., Lee S., Leitch H.G. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- Doi A., Park I.H., Wen B., Murakami P., Aryee M.J., Irizarry R., Herb B., Ladd-Acosta C., Rho J., Loewer S. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schuijers J., Lee T.I., Zhao K. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fan H., Lv P., Huo X., Wu J., Wang Q., Cheng L., Liu Y., Tang Q.Q., Zhang L., Zhang F. The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res. 2018;28:192–202. doi: 10.1101/gr.224576.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., von Meyenn F., Santos F., Chen Y., Reik W., Bertone P., Smith A., Nichols J. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports. 2016;6:437–446. doi: 10.1016/j.stemcr.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J.A., Surani M.A. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ideue T., Hino K., Kitao S., Yokoi T., Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Ladd-Acosta C., Carvalho B., Wu H., Brandenburg S.A., Jeddeloh J.A., Wen B., Feinberg A.P. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A.K., Xi Y., McCarthy R., Allton K., Akdemir K.C., Patel L.R., Aronow B., Lin C., Li W., Yang L. LncPRESS1 is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell. 2016;64:967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Smith A. Mapping the route from naive pluripotency to lineage specification. Philos. Trans. R. Soc. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Bonasio R., Saldana-Meyer R., Yoshida T., Son J., Nishino K., Umezawa A., Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D., Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Jang H., Kim T.W., Kang B.H., Lee S.E., Jeon Y.K., Chung D.H., Choi J., Shin J., Cho E.J. Core pluripotency factors directly regulate metabolism in embryonic stem cell to maintain pluripotency. Stem Cells. 2015;33:2699–2711. doi: 10.1002/stem.2073. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Hahm B., Kim Y.K., Choi M., Jang S.K. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 2000;298:395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Hayashi K., Ohta H., Kiyonari H., Mitani T., Moritoki Y., Kohri K., Kimura H., Yamamoto T. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell. 2015;16:517–532. doi: 10.1016/j.stem.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Lennox K.A., Behlke M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y., Maxson R.E., Schulze E.N., Song H., Hsieh C.L. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Li C., Xing Z., Hu Q., Liang K., Han L., Wang C., Hawke D.H., Wang S., Zhang Y. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat. Cell. Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N., Chang K.Y., Li Z., Gates K., Rana Z.A., Dang J., Zhang D., Han T., Yang C.S., Cunningham T.J. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li Y., Lin B., Sheng Y., Yang L. HBL1 is a human long noncoding RNA that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev. Cell. 2017;42:333–348 e35. doi: 10.1016/j.devcel.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H., Garber M., Curran M., Onder T., Agarwal S. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Lu J.Y., Liu L., Yin Y., Chen C., Han X., Wu B., Xu R., Liu W., Yan P. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016;18:637–652. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Smith A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Perry R.B., Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development. 2016;143:3882–3894. doi: 10.1242/dev.140962. [DOI] [PubMed] [Google Scholar]
- Rosa A., Ballarino M. Long noncoding RNA regulation of pluripotency. Stem Cells Int. 2016;2016:1797692. doi: 10.1155/2016/1797692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R., Lee J., Haemmerle M., Ling H., Previs R.A., Pradeep S., Wu S.Y., Ivan C., Ferracin M., Dennison J.B. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Reports. 2015;13:2395–2402. doi: 10.1016/j.celrep.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarola M., Comisso E., Pascolo R., Chiaradia R., Marion R.M., Schneider C., Blasco M.A., Schoeftner S., Benetti R. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat. Commun. 2015;6:7631. doi: 10.1038/ncomms8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik Mohamed J., Gaughwin P.M., Lim B., Robson P., Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z., Fischer K.A., Devi A., Detraux D., Gu H. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckale J., Wendling O., Masjkur J., Jager M., Munster C., Anastassiadis K., Stewart A.F., Solimena M. PTBP1 is required for embryonic development before gastrulation. PLoS One. 2011;6:e16992. doi: 10.1371/journal.pone.0016992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleahmad S., Mirzaei M., Parker L.M., Hassani S.N., Mollamohammadi S., Sharifi-Zarchi A., Haynes P.A., Baharvand H., Salekdeh G.H. Proteome analysis of ground state pluripotency. Sci. Rep. 2015;5:17985. doi: 10.1038/srep17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., Wu M., Xiong J., Guo X., Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C.A., Horwitz M.S., Hockenbery D., Ware C. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.