Summary
Pelizaeus-Merzbacher disease (PMD) is a fatal X-linked disorder caused by loss of myelinating oligodendrocytes and consequent hypomyelination. The underlying cellular and molecular dysfunctions are not fully defined, but therapeutic enhancement of oligodendrocyte survival could restore functional myelination in patients. Here we generated pure, scalable quantities of induced pluripotent stem cell-derived oligodendrocyte progenitor cells (OPCs) from a severe mouse model of PMD, Plp1jimpy. Temporal phenotypic and transcriptomic studies defined an early pathological window characterized by endoplasmic reticulum (ER) stress and cell death as OPCs exit their progenitor state. High-throughput phenotypic screening identified a compound, Ro 25–6981, which modulates the ER stress response and rescues mutant oligodendrocyte survival in jimpy, in vitro and in vivo, and in human PMD oligocortical spheroids. Surprisingly, increasing oligodendrocyte survival did not restore subsequent myelination, revealing a second pathological phase. Collectively, our work shows that PMD oligodendrocyte loss can be rescued pharmacologically and defines a need for multifactorial intervention to restore myelination.
Keywords: iPSC disease modeling, high-throughput screening, oligodendrocyte progenitor cells, oligodendrocytes, myelin, Pelizaeus-Merzbacher disease, proteolipid protein 1, endoplasmic reticulum stress, PLP1, rare disease
Highlights
-
•
Oligodendrocyte differentiation triggers ER stress and early cell loss in jimpy
-
•
HTS reveals Ro 25–6981, an enhancer of jimpy oligodendrocytes in vitro and in vivo
-
•
Ro 25–6981 increases human mutant oligodendrocytes in 3D cortical spheroids
-
•
Rescued oligodendrocytes fail to myelinate, revealing a second phase of pathology
Tesar and colleagues interrogate the severe genetic myelin disorder Pelizaeus-Merzbacher disease (PMD) using iPSC-derived oligodendrocyte progenitor cells (OPCs). Using high-throughput phenotypic screening, they rescue cell death of mutant oligodendrocytes immediately after initiation of differentiation with chemical modulators, including Ro 25–6981. Interestingly oligodendrocyte restoration did not lead to widespread myelination, unmasking a new phase of the disease.
Introduction
Leukodystophies are a class of rare, heritable disorders characterized by a loss of central nervous system (CNS) myelin. Collectively these diseases impact approximately 1 in 7,500 live births, and are typified by extensive neurological impairment with reduced life expectancy (Bonkowsky et al., 2010, Parikh et al., 2015). While the causative mutations for these disorders are well established, a comprehensive understanding of the molecular and cellular phenotypes of each mutation remains elusive. Genotype-specific, molecular, and cellular phenotyping of oligodendrocytes—the myelinating glia of the CNS—during key pathological events would be invaluable for filling this deficiency and underpin future precision medicine initiatives.
We sought to establish the technology to systemically interrogate the archetypal, cell-autonomous leukodystrophy called Pelizaeus-Merzbacher disease (PMD) (MIM no. 312080) (Garbern, 2007, van der Knaap and Bugiani, 2017). PMD is a fatal X-linked disorder of the CNS caused by mutations in proteolipid protein 1 (PLP1). PLP and its splice isoform DM20 are almost exclusively expressed in oligodendrocytes and oligodendrocyte progenitor cells (OPCs), respectively (Mallon et al., 2002). While the disease is thought to be mediated through mutant PLP-induced endoplasmic reticulum (ER) stress (Garbern, 2007), complete appreciation of the cellular and molecular pathology in PMD remains elusive, in part due to complexities in accessing or appropriately modeling the disease-affected oligodendrocyte lineage during critical developmental windows (Bradl et al., 1999, Cerghet et al., 2001, Duncan et al., 2011, Gow et al., 1998, Knapp et al., 1986, Knapp et al., 1987, Southwood et al., 2002, Williams and Gard, 1997). Our group and others has recently begun to address these tractability issues using human PMD patient, induced pluripotent stem cell (iPSC)-based approaches (Nevin et al., 2017, Numasawa-Kuroiwa et al., 2014), but these cellular models are still constrained by multi-month differentiations and are confounded by contaminating cell types.
Our mouse pluripotent stem cell-based technology enables the rapid generation of high-purity, expandable OPCs that are capable of coordinated differentiation to oligodendrocytes in vitro over a defined 3-day period (Najm et al., 2011). This system provides precise developmental control to examine the oligodendrocyte lineage without artifact from contaminating cell types (Najm et al., 2011, Najm et al., 2015). Notably, these tools and similiar techniques have recently been coupled to high-throughput compound screening investigations and have identified novel therapeutics for autoimmune-mediated myelin disorders (Green et al., 2017, Helman et al., 2015, Hubler et al., 2018, Lee et al., 2013, Najm et al., 2015).
Here we extend this cellular technology to a well-established animal model for the severe, connatal form of PMD called the “jimpy” (Plp1jimpy) mouse (Duncan et al., 2011, Phillips, 1954, Yool et al., 2000), providing scalable access to the previously restricted PMD oligodendrocyte lineage. Using iPSC-derived jimpy OPCs, we first establish critical cellular and molecular deficits at key developmental stages during oligodendrocyte differentiation. We overcome this pathology using high-throughput chemical screening, identifying compounds capable of rescuing mutant oligodendrocyte survival in jimpy and human PMD cultures in vitro, and in jimpy mice in vivo. Surprisingly, these studies revealed that enhancing mutant oligodendrocyte survival does not, on its own, confer an expected restoration of myelin, unmasking a second stage of pathology and informing future phenotyping and therapeutic strategies for PMD.
Results
Jimpy iPSC-Derived OPCs Reveal Early Differentiation Pathology
We established three independent iPSC lines from jimpy and isogenic wild-type littermate mice (see Experimental Procedures). iPSC lines exhibited typical iPSC morphology and expressed the canonical pluripotency markers Oct3/4 and Nanog (Figures S1A–S1D). Sanger sequencing confirmed jimpy mutation status (Figure S1E) and all iPSC lines displayed grossly normal karyotypes (Figure S1F). We directed all six iPSC lines to form high-purity OPCs (see Experimental Procedures) expressing the canonical OPC transcription factors Sox10 and Olig2 (Figures S2A–S2E), which had no overt morphological differences (Figure S2F).
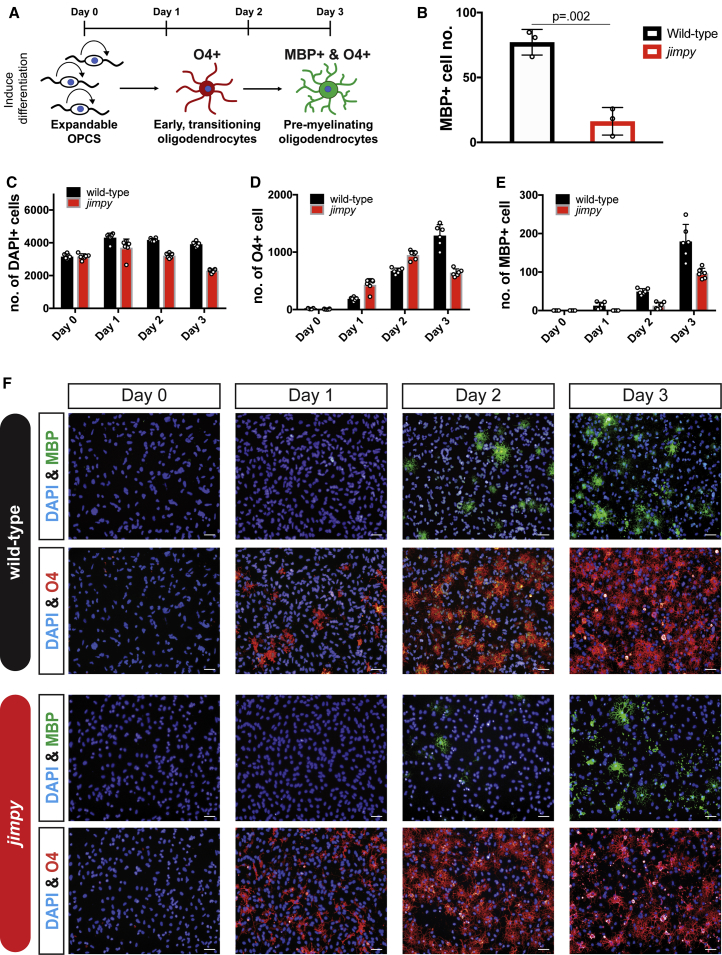
Previous reports have documented cellular phenotypes at discrete stages during oligodendrogenesis in PMD cellular and animal models (Gow et al., 1998, Nevin et al., 2017, Numasawa-Kuroiwa et al., 2014, Readhead et al., 1994). We directed all iPSC-derived jimpy and wild-type control OPC lines toward an oligodendrocyte fate (Figure 1A) and noted a striking absence of MBP+ oligodendrocytes in all jimpy lines (Figure 1B). We further parsed this cellular defect with time-course immunocytochemistry. As expected, jimpy MBP+ oligodendrocytes failed to accumulate across all time points (Figures 1E and 1F). Surprisingly we noted substantial cell death well before MBP+ oligodendrocyte formation, within 1 day after inducing differentiation (Figures 1C and 1F), which we further confirmed by time-lapse imaging (Video S1). Intriguingly, jimpy OPCs showed increased propensity to acquire the late OPC/immature oligodendrocyte marker O4 early in the differentiation process (Figures 1D and 1F), which may reflect a feedback loop to promote stem cell exit in the context of severe cell loss during oligodendrocyte differentiation. Together these results precisely define an early developmental susceptibility period during which transitioning jimpy OPCs die as they exit their progenitor cell state.
Figure 1.
Temporal Dissection of Cellular Phenotypes Reveals Cell Death Just as Jimpy OPCs Commit to an Oligodendrocyte Fate
(A) Schematic summary of the experimental assay.
(B) Quantification of wild-type and jimpy MBP+ oligodendrocytes after oligodendrocyte differentiation. n = 3 distinct cell lines per genotype, with each replicate value (comprising a mean of n = 4 replicate wells per cell line) represented by a white circle. p values calculated using a two-way, unpaired t test between genotypes.
(C–E) Wild-type and jimpy oligodendrocyte differentiation time-course data showing quantification of total (C) DAPI+ cells, (D) O4+ cells, and (E) MBP+ oligodendrocytes. n = 6 replicate wells per genotype, with each replicate value represented by a white circle. (F) Representative immunocytochemistry images from time course showing MBP+ oligodendrocytes (green), O4+ cells (red), and total cells (DAPI, blue).
Scale bars, 50 μm. Error bars represent mean ± SD. See also Figures S1 and S2, and Video S1.
Transcriptome Profiling Reveals ER Stress in Jimpy Oligodendrocytes Immediately after the Onset of OPC Differentiation
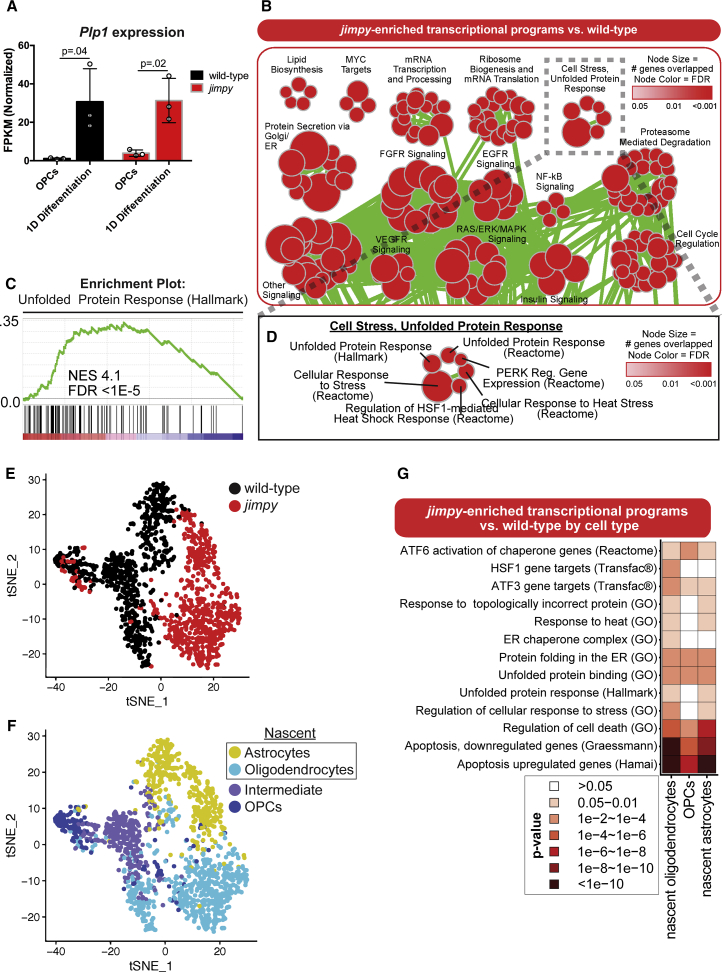
We analyzed transcriptomic changes between jimpy and wild-type cultures after 1 day of oligodendrocyte differentiation by gene set enrichment analysis (GSEA) (Table S1), at the onset of jimpy cell death and upregulation of the disease-causative gene, Plp1 (Figure 2A). Jimpy cultures showed multiple dysregulated pathways comprising several gene sets linked to the ER stress response (Figure 2B; Table S1), including the unfolded protein response (UPR) pathway (Figures 2C and 2D).
Figure 2.
RNA-Seq and scRNA-Seq Reveal Transcriptome Signatures of ER Stress and Cell Death in Early, Differentiating Jimpy Oligodendrocytes
(A) Plp1 mRNA expression in wild-type and jimpy OPCs and in cultures during oligodendrocyte (at day 1). Error bars represent mean ± SD. n = 3 sequenced cell lines per genotype, with each replicate represented by a white circle. p values calculated using a two-way, unpaired t test for each genotype.
(B) Gene ontology enrichment map for RNA-seq data showing enriched pathways in jimpy after 1 day of oligodendrocyte differentiation. Node size indicates the number genes represented in the pathway. Color intensity indicates false discovery rate (FDR). Node overlap indicates shared pathway genes. n = 3 cell lines, per genotype.
(C) Jimpy enrichment plots for the hallmark unfolded protein response pathway genes relative to wild-type. The upper plot (green line) details the running sum statistic of the ranked list of genes with the normalized enrichment score (NES) and FDR. The lower plot displays a gene's position in the ranked list of genes for the pathway.
(D) Enlargement of the “cell stress, unfolded protein response” node group.
(E and F) tSNE plots showing (E) genotype and (F) cell-type distributions. n = 1 cell line per genotype.
(G) Heatmap showing pathway enrichment in jimpy cell types relative to their wild-type counterparts. Scale corresponds to p value.
We next sought to precisely define the cell types exhibiting these pathological stress signatures using single-cell RNA sequencing (scRNA-seq). Unsupervised clustering separated jimpy and wild-type cells in t-distributed stochastic neighbor embedding (tSNE) space (Figure 2E), while also arranging OPCs, nascent oligodendrocytes, nascent astrocytes, and transitioning cells (between OPC and nascent oligodendrocyte) into distinct populations defined by marker expression (Figures 2F and S3B–S3G). We then generated a list of differentially expressed genes between genotypes for each cell type present in both cultures (Table S2). GSEA on the nascent oligodendrocyte populations revealed a significant enrichment of ER stress and cell death pathways in this early jimpy oligodendrocyte population (Figures 2G; Table S3), confirming the presence of canonical PMD molecular pathology in the disease-affected cell type.
Interestingly we also noted enrichment for similar pathways in nascent jimpy astrocytes, indicating possible astrocyte contribution to disease in PMD (Skoff, 1976). However these transcriptional changes could simply represent a response to the substantial cell death in the cultures. To test this, we first directed iPSC-derived jimpy and wild-type OPCs toward an astrocyte fate. We observed no difference in the percentage of GFAP+ astrocytes in wild-type and jimpy cultures (Figures S3H–S3J). Next we performed astrocyte co-culture experiments to examine the effect of jimpy and wild-type astrocytes on oligodendrocyte survival. This analysis revealed no astrocyte-mediated effect on mutant or wild-type oligodendrocytes (Figures S3K–S3P). Together these data demonstrate that astrocytes are not the primary mediators of oligodendrocyte death in jimpy cultures and that pathological changes in jimpy astrocytes are likely secondary to primary oligodendrocyte death.
High-Throughput Screening Reveals Chemical Enhancers of Jimpy Oligodendrocyte Survival
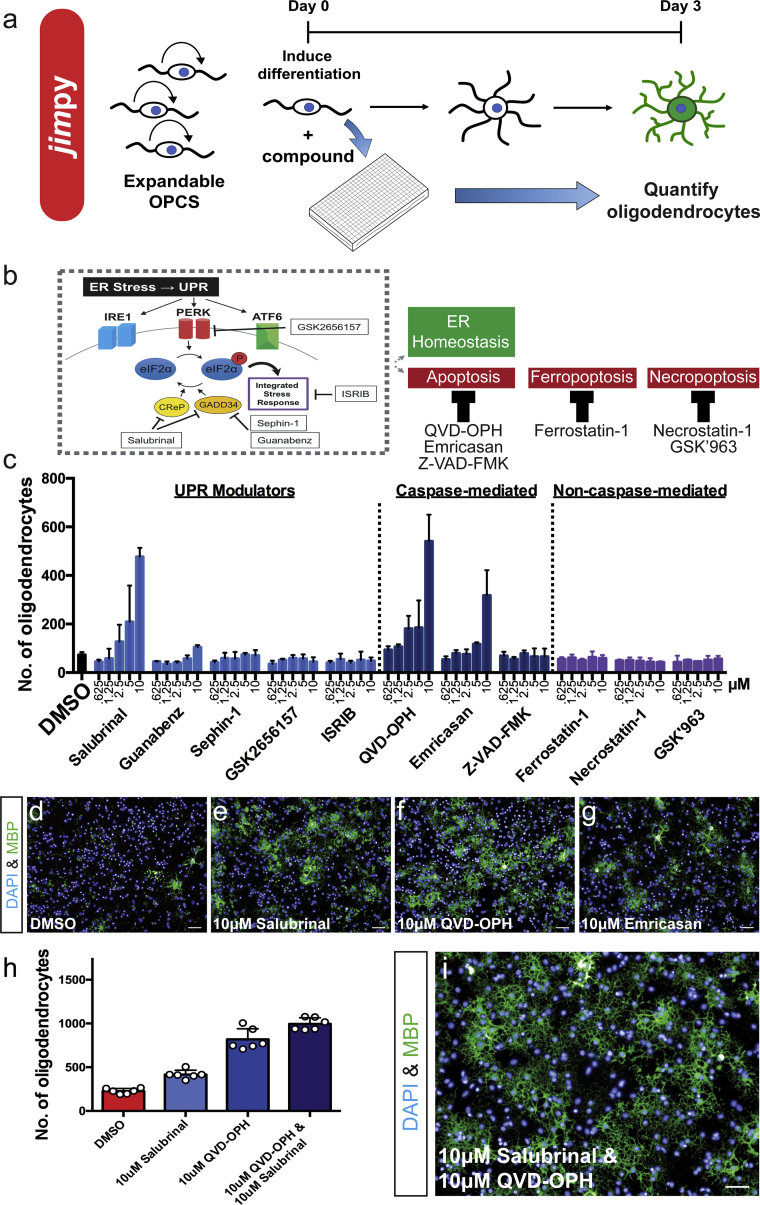
Our stem cell platform provides scalable access to a defined developmental window for potential therapeutic intervention in jimpy. To explore whether small molecules could be used to resolve the molecular and cellular pathology in early differentiating jimpy OPCs, we developed a chemical screening pipeline to identify compounds that improved the survival of MBP+ jimpy oligodendrocytes in conditions promoting oligodendrocyte differentiation (Figure 3A). We first performed a hypothesis-driven compound screen to interrogate the protein kinase R-like ER kinase (PERK) arm of the UPR, as well as a variety of cell death pathways that could drive oligodendrocyte loss during unresolved ER stress (Figure 3B) (Nevin et al., 2017). Using jimpy iPSC-derived OPCs we applied each compound in triplicate over a 5-point dose curve (10 μM to 625 nM) with simultaneous initiation of oligodendrocyte differentiation with thyroid hormone and then quantified MBP+ oligodendrocyte number after 3 days. Out of five compounds that modulate distinct targets in the PERK pathway, only salubrinal (Figures 3C–3E), a dual GADD34 and CReP inhibitor (Boyce et al., 2005), showed a dose-dependent ability to enhance jimpy oligodendrocyte survival. Examination of cell death-modulating compounds identified the high-efficacy pan-caspase inhibitors, emricasan and Q-VD-OPh, as effective mediators of jimpy oligodendrocyte death (Figures 3C, 3D, 3F, and 3G). Strikingly, the combination of Q-VD-OPh and salubrinal demonstrated an additive ability to restore jimpy cell number back to wild-type levels and enhance jimpy oligodendrocyte survival (Figures 3H, 3I, and S4). Collectively, these data show that loss of jimpy oligodendrocytes can be prevented by modulating ER stress and caspase-mediated cell death during the immediate transition from OPC to oligodendrocyte.
Figure 3.
High-Throughput Chemical Screening Identifies Cellular and Molecular Modulators of Jimpy Pathology
(A) Chemical screening schematic.
(B) Chemical-targeting of ER stress-related sequelae by UPR modulation or inhibition of caspase-mediated apoptosis.
(C) Quantification of MBP+ oligodendrocytes per compound dose (five doses, 10 μM to 625 nM) after 3-day oligodendrocyte differentiation of iPSC-derived jimpy OPCs. n = 3 replicate wells per dose, per compound.
(D–G) Representative immunocytochemistry images after oligodendrocyte differentiation of jimpy OPCs treated with (D) DMSO vehicle, (E) 10 μM salubrinal, (F) 10 μM Q-VD-OPh, and (G) 10 μM emricasan, showing MBP+ oligodendrocytes (green) and total DAPI+ cells (blue).
(H) Quantification of MBP+ jimpy oligodendrocytes for DMSO vehicle (red), 10 μM salubrinal (light blue), 10 μM Q-VD-OPh (blue), and 10 μM salubrinal and 10 μM Q-VD-OPh (dark blue). n = 6 replicate wells per treatment, with each replicate value represented by a white circle.
(I) Representative immunocytochemistry images after oligodendrocyte differentiation of jimpy OPCs treated with 10 μM salubrinal and 10 μM Q-VD-OPh, showing MBP+ oligodendrocytes (green) and total DAPI+ cells (blue).
Error bars represent mean ± SD. Scale bars, 50 μm. See also Figure S4.
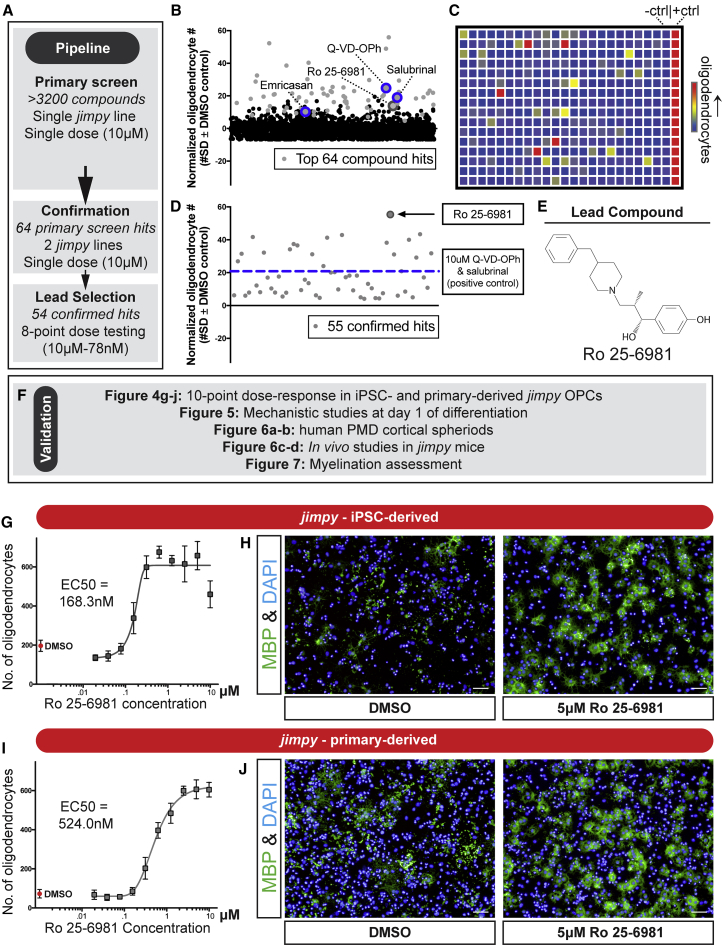
We scaled this screening pipeline to an unbiased collection of >3,000 bioactive compounds to identify additional nodes of intervention for jimpy pathology. To gauge screening performance we included the combination of salubrinal plus Q-VD-OPh as a positive control and a DMSO vehicle negative control. We performed a high-throughput primary screen at a single dose of 10 μM followed by rigorous secondary screening to filter to a lead compound (Figure 4A). We evaluated the performance of our primary screen based on the dynamic range of quantified oligodendrocytes between our positive and negative controls using the Z factor (Z’ = 0.566 ± 0.23 [mean ± SD]) (Figures S5A–S5C), indicating a high quality and reproducible screening assay based on established performance analytics (Zhang et al., 1999). Automated image analysis scored each well based on enhancement of MBP+ oligodendrocyte number on a per-plate basis (Figure 4C). As an orthogonal filter we also used total cell number as an overall marker of increased cell survival. All compounds were ranked (Table S4) and our top 64 hits (Figure 4B) were defined as being at least four SDs above the negative control for oligodendrocyte number and two SDs above negative control for total cell number. Of note, three of the top compound hits included Q-VD-OPh, salubrinal, and emricasan (Figure 4B), which were identified and validated in our previous hypothesis-driven chemical screen.
Figure 4.
High-Throughput Chemical Screening Identifies Cellular and Molecular Modulators of Jimpy Pathology
(A) Overview of screening pipeline with filtering steps for lead compound selection.
(B) Plot of primary screen results showing top hit (gray circles) and non-hit (black circles) compounds listed by alphabetical order with their corresponding SD divergence from DMSO vehicle on the MBP+ jimpy oligodendrocyte metric after 3-day oligodendrocyte differentiation. n = 1 well per tested compound.
(C) Heatmap of a representative primary screen plate depicting high (red) or low (blue) quantified MBP+ jimpy oligodendrocytes for each compound, as well as negative and positive controls.
(D) Plot of confirmed hits (gray circles) listed in alphabetical order showing their maximal SD divergence from DMSO vehicle for MBP+ jimpy oligodendrocytes in an 8-point (10 μM to 78 nM) dose response. n = 1 well per compound dose. n = 16 replicate wells for the positive and negative control samples.
(E) Chemical structure of lead compound Ro 25–6981.
(F) Overview of validation pipeline for lead compound Ro 25–6981.
(G and I) Quantification of MBP+ jimpy oligodendrocytes after differentiation of (G) iPSC-derived and (I) cerebral cortical-derived OPCs, treated with Ro 25–6981 (10-point dose response) or DMSO vehicle. Individual points represent mean ± SD. n = 3 replicate wells for each dose, n = 16 replicate wells for the DMSO vehicle control.
(H and J) Representative immunocytochemistry images after oligodendrocyte differentiation of (H) iPSC-derived or (J) cerebral cortical-derived OPCs treated with DMSO vehicle or 5 μM Ro 25–6981 showing MBP+ oligodendrocytes (green) and total DAPI+ cells (blue).
Next we used a stringent secondary screening pipeline to prioritize these hits to a single lead compound (Figure 4A). First, all hits were tested in OPCs generated from two additional independent jimpy iPSC lines using the same primary screen dose of 10 μM and primary screen hit selection criteria. The efficacy of 55 out of 64 hits was confirmed in at least one additional jimpy cell line (Table S5). Next, each confirmed compound was examined over an 8-point dose response (from 10 μM to 78 nM), and compounds were ranked by their potency to enhance oligodendrocyte survival (Figure 4D; Table S5). This analysis highlighted the lead compound Ro 25–6981 (Figures 4D and 4E; Table S5), which showed efficacy far superior even to the combination of salubrinal and Q-VD-OPh (Figure 4D), our screening positive control.
Next we initiated a rigorous validation procedure to comprehensively define Ro 25–6981's molecular and cellular effects in multiple, orthogonal contexts (Figure 4F). We first sought to assess the potency of Ro 25–5981 using a 10-point dose response (from 10 μM to 20 nM). This analysis revealed strong efficacy extending to nanomolar doses (half maximal effective concentration [EC50] = 168.3 nM) (Figures 4G and 4H). To rule out possible iPSC-derived OPC artifacts we demonstrated that Ro 25–6981 had remarkably similar dose-dependent efficacy and potency to enhance oligodendrocyte survival in primary cultures of OPCs isolated from postnatal jimpy mice (EC50 = 524.0 nM) (Figures 4I and 4J).
To examine if the cellular or molecular effects of Ro 25–6981 were specific to mutant cells we first assessed oligodendrocyte enhancement using Ro 25–5981 in a 10-point dose response (from 10 μM to 20 nM) or DMSO vehicle on cultures of differentiating wild-type and jimpy OPCs. In addition we also examined the effects of our jimpy cellular pathology positive control compounds, salubrinal and Q-VD-OPh. As expected all compounds robustly (∼3- to 5-fold) enhanced jimpy oligodendrocytes (Figure S6B). In contrast these same compounds showed only a modest (∼1.5- to 2-fold) enhancement of wild-type oligodendrocytes (Figure S6A).
Mechanistic Studies Reveal a Non-canonical, UPR-Mediator Role for Ro 25–6981 in Differentiating Jimpy OPCs
Ro 25–6981 is an NR2B-selective N-methyl-D-aspartate (NMDA) receptor antagonist (Fischer et al., 1997). However, we noted absent expression in our RNA-seq data profiling OPC and early oligodendrocyte differentiation in all iPSC-derived wild-type and jimpy cell cultures (fragments per kilobase of transcript per million mapped reads < 0.15; Table S1), consistent with publicly available datasets (Zhang et al., 2014). Still, a handful of studies have documented expression of the NR2B subunit in oligodendrocytes and astrocytes in normal and pathological contexts (Karadottir et al., 2005, Krebs et al., 2003, Salter and Fern, 2005).
To address the possibility of an NR2B-mediated effect we first sought to investigate Grin2b (NR2B subunit gene) expression throughout the OPC to oligodendrocyte transition using time course qRT-PCR in iPSC-derived jimpy and wild-type cultures. This comprehensive profiling failed to detect Grin2b expression at any time point (Figure S6C). We also performed functional testing of 29 unique NMDA receptor modulators (including NR2B-selective and pan-NMDA receptor antagonist drugs with high potency to their annotated targets) in 10-point dose response (from 10 μM to 20 nM) on iPSC-derived jimpy OPCs. These data failed to establish a correlation between NR2B-NMDA receptor antagonism and the enhancement of jimpy oligodendrocyte survival (Table S6). Furthermore, structure-activity relationship analysis revealed that only the Ro 25–6981 scaffold could potently increase jimpy oligodendrocyte survival. Together, these data eliminated the NR2B-NMDA receptor as the functional target in jimpy cultures.
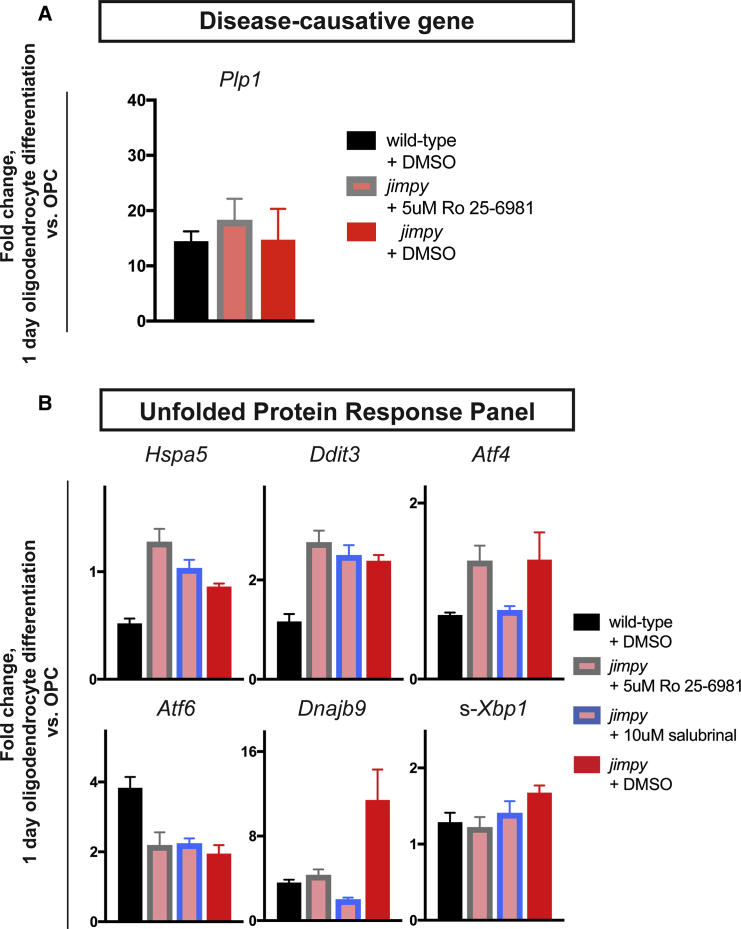
To investigate alternative mechanisms for Ro 25–6981's action on jimpy oligodendrocyte survival in vitro we employed targeted qRT-PCR assessment during jimpy and wild-type oligodendrocyte differentiation. First we assessed Ro 25–6981's ability to alter Plp1 expression levels as reduction of mutant PLP is beneficial to PMD cellular and animal models (Karim et al., 2010, Prukop et al., 2014). We found no effect of Ro 25–6981 on Plp1 induction in jimpy (Figure 5A). We next examined Ro 25–6981's ability to modulate jimpy-dysregulated UPR-related factors curated from our transcriptome data including Ddit3, Hspa5, Dnajb9, spliced-Xbp1 (s-XBP1), Atf4, and Atf6. As a comparator we also assessed the effect of the known UPR modulator salubrinal. Both compounds showed reductions of Dnajb9 and s-Xbp1 to wild-type levels, no change in Atf6, and increased expression of Hspa5 (Figure 5B). Of note, salubrinal restored Atf4 to wild-type levels while Ro 25–6981 showed no change. In contrast Ro 25–6981 modestly increased Ddit3, above the salubrinal-mediated effect (Figure 5B). Interestingly, while these compounds produced remarkably similar profiles, their points of divergence may provide a clue to their vastly different efficacies in jimpy oligodendrocyte restoration. The Ddit3 and Hspa5 results are especially intriguing as depletion of DDIT3 has been shown to exacerbate disease in PMD animal models (Southwood et al., 2002), while reduction of HSPA5 is deleterious to oligodendrocyte survival in both normal and autoimmune disease contexts (Hussien et al., 2015) and is depleted in PMD models (Numata et al., 2013).
Figure 5.
Ro 25–6981 Mechanistic Studies Reveal UPR Modulation in Jimpy
(A) qRT-PCR of Plp1 for wild-type and jimpy at day 1 of oligodendrocyte differentiation with indicated treatment. Vehicle-treated controls same as Figure S6D. For Atf6 n = 3 technical replicates per sample. n = 4 technical replicates per sample.
(B) qRT-PCR of UPR-related panel for wild-type and jimpy at day 1 of oligodendrocyte differentiation with indicated treatment. Vehicle-treated controls same as Figure S6D. For Atf6 n = 3 technical replicates per sample. For all other probes n = 4 technical replicates per sample.
Error bars represent mean ± SD. See also Figure S6 and Table S6.
We further examined these UPR expression effects testing Ro 25–6981 as well as salubrinal on differentiating wild-type cultures. Surprisingly the vast majority of the transcriptional changes were jimpy specific, with Atf6 and Ddit3 even showing completely opposite trends in the Ro 25–6981 condition (Figure S6D). Together these data suggested that Ro 25–6981 functioned by modulating the UPR in a genotype-specific manner that was highly specific to jimpy cultures.
Ro 25–6981 Increases Oligodendrocytes in Human PMD Patient iPSC-Derived Cultures and Jimpy Mice
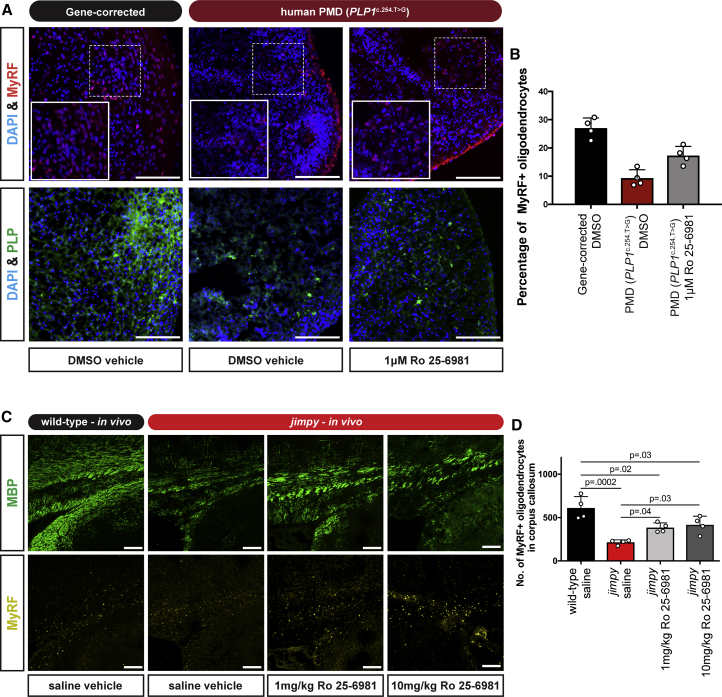
To test if Ro 25–6981's effect was mutation or organism specific, we applied this compound to human PMD oligocortical spheroids (Madhavan et al., 2018) generated using iPSCs from a severe connatal PMD patient with a PLP1c.254T>G point mutation (Nevin et al., 2017). We applied 1 μM Ro 25–6981 or DMSO vehicle to mutant spheroids during a 20-day period of oligodendrocyte generation and quantified oligodendrocytes as a percentage of total cell number using an antibody to the oligodendrocyte-specific transcription factor, myelin regulatory factor (MyRF). In addition, we administered DMSO vehicle to control, isogenic gene-corrected spheroids. Ro 25–6981 treatment profoundly increased the percentage of mutant oligodendrocytes in the human PMD spheroids (Figures 6A and 6B), confirming the ability to target common cellular pathology and enhance mutant oligodendrocyte survival across distinct PMD genotypes and species. Of note these levels of oligodendrocytes were still below those of the isogenic control, suggestive of remaining pathology, potentially also reflected by the unrestored PLP expression pattern (Figure 6A).
Figure 6.
Ro 25–6981 Enhances Mutant Oligodendrocyte Survival in Human Patient PMD Spheroids and in Jimpy Mice
(A) Representative immunocytochemistry images of human PMD patient (PLP1c.254.T>G) and wild-type iPSC-derived oligocortical spheroids showing MyRF+ (red) and PLP+ (green) oligodendrocytes, as well as total DAPI+ cells (blue) after treatment.
(B) Percentage of MyRF+ oligodendrocytes relative to total DAPI+ cells in human PMD patient (PLP1c.254.T>G) and wild-type iPSC-derived oligocortical spheroids after treatment. n = 4 independent spheroids per treatment.
(C) Representative immunocytochemistry images of the medial corpus callosum of wild-type and jimpy mice after treatment showing MBP+ oligodendrocytes (green) and MyRF+ oligodendrocytes (yellow).
(D) Quantification of MyRF+ oligodendrocytes within the medial corpus callosum of wild-type and jimpy mice after treatment. n = 4 injected animals per treatment.
Scale bars, 100 μm. Error bars represent mean ± SD, with each replicate value indicated by a white circle. p values calculated using a one-way ANOVA with Holm-Sidak correction for multiple comparisons.
Next we examined if Ro 25–6981 treatment could increase oligodendrocytes in jimpy mice, in vivo. We administered 1 or 10 mg/kg Ro 25–6981 or saline vehicle to jimpy mice and saline vehicle to wild-type mice from postnatal days 5–14, and quantified MyRF+ oligodendrocytes along the medial rostral caudal axis of the corpus callosum. Ro 25–6981 treatment significantly increased jimpy oligodendrocytes in a dose-dependent manner, but below wild-type levels (Figures 6C and 6D). These results confirmed that Ro 25–6981's effect was not limited to in vitro contexts and could enhance oligodendrocyte survival in jimpy mice in vivo.
Increasing Jimpy Oligodendrocyte Survival Uncovers a Second Pathological Phase during Myelination
Oligodendrogenesis is thought to be closely coupled to myelination (Mayoral and Chan, 2016)—a process which involves the generation and extension of specialized oligodendrocyte membrane around neuronal axons. Given the strong enhancement of early oligodendrocyte survival across multiple contexts we expected to see a frank restoration of myelinating oligodendrocytes. Surprisingly we found only a slight increase in myelinating cells in Ro 25–6981-treated jimpy mice as marked by MBP (Figure 6C).
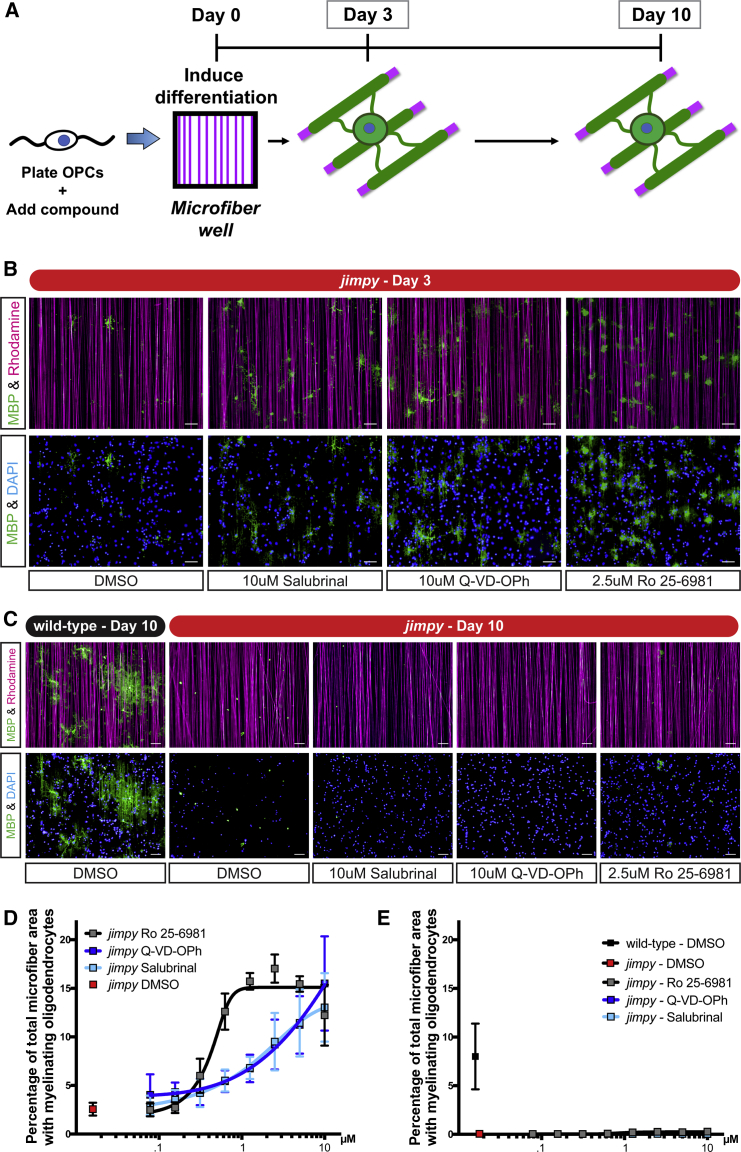
To appreciate why increased oligodendrocyte survival did not translate to a broad restoration of myelin we employed a defined system using non-biological poly-lactide microfibers. These microfibers serve as a robust, reproducible, and scalable myelination substrate (Bechler et al., 2015). We plated jimpy OPCs in the presence of Ro 25–6981 in an 8-point (10 μM to 78 nM) dose-response format or DMSO vehicle. To account for Ro 25-6981-specific effects on myelination we also tested our jimpy cellular pathology positive control compounds, salubrinal and Q-VD-OPh. We induced oligodendrocyte differentiation, allowed for microfiber ensheathment (in vitro “myelination”), and quantified MBP+ myelinating oligodendrocytes (Figure 7A) using an automated image analysis pipeline (Figures S7A–S7E). Despite showing a robust, dose-dependent increase in jimpy oligodendrocyte survival and early myelination (Figures 7B and 7D), Ro 25–6981, salubrinal, and Q-VD-OPh were unable maintain myelin by day 10 across all tested doses. This result was specific to jimpy, as wild-type DMSO vehicle-treated cultures showed extensive myelination at day 10 (Figures 7C and 7E). The ability to restore jimpy oligodendrocytes without a subsequent restoration of mutant myelin was unexpected and revealed that there are two distinct stages of disease pathology: (1) ER stress-mediated cell death as cells transition from OPCs to oligodendrocytes and (2) dysfunctional myelination due to the presence of mutant PLP protein in surviving oligodendrocytes. Collectively, our results show that the first phase can be resolved with small molecules and that effective clinical intervention for PMD will need to address both phases of pathology.
Figure 7.
Increased Jimpy Oligodendrocyte Survival during Differentiation Unmasks a Second Pathological Phase during Myelination
(A) Schematic of the in vitro myelination assay using 4-μm diameter synthetic microfiber fibers detailing the examination of myelinating oligodendrocytes at day 3 and 10.
(B and C) Representative immunocytochemistry images of MBP+ myelinating oligodendrocytes (green), rhodamine+ microfibers (purple), and total DAPI+ cells (blue) at (B) day 3 and (C) day 10. Compound-treated cultures shown at their maximum effective concentration.
(D and E) Quantification of percent total microfiber area overlap with MBP+ myelinating oligodendrocytes at (D) day 3 and (E) day 10 with indicated treatment and genotype. Scale bars, 50 μm. n = 3 replicate wells per compound treatment. n = 16 replicate wells per vehicle control.
Error bars represent mean ± SD. See also Figure S7.
Discussion
In this study we established a genotype-specific, scalable, and tractable system to investigate a severe PMD point mutation modeled in the jimpy mouse. Coupled with stringent control over oligodendrocyte differentiation, this system enabled precise, temporal dissection of jimpy pathology. Using a simple developmental trigger (induction of oligodendrocyte differentiation) we recapitulated known cellular and molecular PMD phenotypes including ER stress and oligodendrocyte death (Ikeda et al., 2018, Southwood et al., 2002), and defined a surprisingly early susceptibility period. Chemical screening demonstrated a reversal of jimpy oligodendrocyte loss by inhibiting specific targets in the UPR and apoptosis pathways. Furthermore, this system allowed us to identify Ro 25–6981, a highly effective and potent modulator of jimpy pathology with a previously undefined effect on the UPR.
While our studies were designed to identify cellular and molecular phenotypes as well as compound modulators on a genotype-specific basis, we reasoned that UPR effectors could counteract the sequelae seen in alternate, human PMD mutations by targeting common pathology in this disease (Nevin et al., 2017). We tested Ro 25–6981 on PMD patient-derived human oligocortical spheroids and demonstrated a robust increase in mutant oligodendrocytes, confirming efficacy in human disease-relevant tissue and suggesting modulation of shared disease components.
Given the striking enhancement of mutant oligodendrocytes across multiple contexts (cellular, genotypic, and species), we expected to observe a substantial increase in myelin after Ro 25–6981 treatment, in vivo. While Ro 25–6981 increased the quantity of jimpy oligodendrocytes, it did not restore myelination to wild-type levels. As oligodendrogenesis and myelination are thought to be distinct processes (Mayoral and Chan, 2016), we examined myelination in jimpy with Ro 25–6981 treatment using non-biologic, synthetic microfibers. These data revealed that Ro 25–6981 was unable to restore myelination in jimpy, revealing a second stage of disease, refractory to treatment that enhances early oligodendrocyte survival. As such, our results establish a dual therapeutic paradigm: (1) restore oligodendrocyte loss during differentiation and then (2) enhance myelination capacity of surviving, mutant oligodendrocytes. Importantly, approaches that simply target these rare, surviving oligodendrocytes are unlikely to be successful without first enhancing the survival during the early OPC to oligodendrocyte transition. This understanding has substantial impact on therapeutic development for PMD and potentially other disorders where ER stress is implicated.
The dichotomy of compound-mediated increased mutant oligodendrocyte survival without increased myelination could be explained by the unique demands of the latter process, which involves immense synthesis of myelin lipids and proteins (Baron and Hoekstra, 2010), including PLP which has been estimated to constitute approximately 50% of the total myelin protein (Baumann and Pham-Dinh, 2001). As such the myelination stage may necessitate engagement of completely separate targets for pharmacological interventions. Prior studies by us and others have shown, paradoxically, that modulating opposing responses in the UPR pathway, including inhibition of PERK or GADD34, leads to phenotypic benefit in PMD and other myelinating disorders (D'Antonio et al., 2013, Musner et al., 2016, Nevin et al., 2017). Future studies will need to parse the precise target of Ro 25–6981's UPR-modulating effect and assess the potential use of combinatorial therapies.
In addition, the presence mutant PLP protein could lead to specific sequelae (Koizume et al., 2006, Kramer-Albers et al., 2006, Roussel et al., 1987), distinct from the pathology that develops immediately after jimpy OPCs begin to differentiate to oligodendrocytes. Interestingly, PLP1 duplications, which constitute ∼70% of human patients (Inoue, 2005), may be especially amendable to UPR-modulating compounds such as Ro 25–6981, as these cases are typified by overly abundant but normal PLP protein. As such, moderating ER overload may be a viable strategy to restore oligodendrocytes and myelin in these patients.
More broadly, given that ER stress is a fundamental component of many diseases of myelin including other leukodystrophies and multiple sclerosis (Clayton and Popko, 2016, Way et al., 2015), jimpy modulators such as Ro 25-6981 may provide effects beyond PMD, to other genetic disorders of myelin and multiple sclerosis.
Experimental Procedures
Animal Studies
All animal procedures and experimentation was approved by Case Western Reserve University’s Institutional Animal Care and Use Committee.
Generation of Jimpy iPSCs
Tail-tip fibroblasts were derived from jimpy mutant and wild-type littermates reprogrammed to iPSCs using a lentivirus encoding a doxycycline-inducible polycistronic Oct4, Sox2, Klf4, and c-Myc construct as described previously (Somers et al., 2010, Sommer et al., 2009).
Generation of jimpy OPCs
iPSCs were differentiated to high purity, expandable OPCs as described previously (Najm et al., 2011) (Lagar et al., 2018). OPCs were propagated in DMEM/F12 (11320082, Thermo Fisher Scientific), 1× N2 supplement (AR009, R&D Systems), 1× B-27 without vitamin A supplement (12,587-010, Thermo Fisher Scientific), and 1× Glutamax, supplemented with 20 ng/mL fibroblast growth factor 2 (233-FB, R&D Systems) and 20 ng/mL platelet-derived growth factor-AA (221-AA, R&D Systems).
Differentiation of Jimpy OPCs to Oligodendrocytes
OPCs were differentiated to oligodendrocytes with differentiation medium that consisted of DMEM/F12, 1× N2 supplement, 1× B-27 without vitamin A supplement, supplemented with 100 ng/mL noggin (3344-NG, R&D Systems), 10 ng/mL neurotrophin-3 (NT-3) (267-N3, R&D Systems), 50 μM cAMP (D0260, Sigma), 100 ng/mL insulin-like growth factor-1 (291-G1, R&D Systems) NT-3, and 40 ng/mL triiodothyronine (thyroid hormone; T-6397, Sigma).
Compound Screening
OPCs were seeded on 384-well poly-D-lysine CellCarrier Ultra plates (6057500, PerkinElmer), compound was added, and oligodendrocyte differentiation was induced. Three days later plates were fixed with 4% paraformaldehyde (PFA), and immunostained using rat anti-MBP and goat anti-Sox10, followed by counterstaining with DAPI. Images were captured and quantified using the Operetta High Content Imaging and Analysis system (PerkinElmer), Harmony software (PerkinElmer), and Acapella software (PerkinElmer).
qRT-PCR
RNA was prepared with the RNeasy Mini Kit (74104, QIAGEN). qRT-PCR was performed using pre-designed TaqMan gene expression assays (Thermo Fisher Scientific) and probes values were normalized to an Actb endogenous control.
Human PMD Cultures
Human PMD patient (PLP1c.254T>G) and gene-corrected (isogenic control) iPSCs were previously generated as described (Nevin et al., 2017) and used to generate oligocortical spheroids (Madhavan et al., 2018). Spheroids were treated with DMSO vehicle or Ro 25–6981 from days 60 to 90. Spheroids were fixed with 4% PFA, and immunostained using rabbit anti-MyRF and rat anti-PLP, followed by counterstaining with DAPI. Images were captured and quantified using Adobe Photoshop (Abobe Systems).
In Vivo Jimpy Studies
Jimpy and wild-type control mice were administered Ro 25–6981 (1594, R&D Systems) or saline vehicle from postnatal day 5 to 14. Mice were sacrificed and tissue sections were immunostained using rabbit anti-MyRF and rat anti-MBP antibody. Complete corpus callosum (medial, sagittal sections) images were captured and quantified using Adobe Photoshop.
In Vitro Myelination Assay
OPCs were seeded on Mimetix 384-well plates containing synthetic microfibers (AMS-TECL-015, AMSBio), compound was added, and oligodendrocyte differentiation was induced. At day 3 and 10 after seeding, plates were fixed with 4% PFA, and immunostained using rat anti-MBP, followed by counterstaining with DAPI. Images were captured and quantified using the Operetta High Content Imaging and Analysis system, Harmony software, and Acapella software.
Quantitation and Statistical Analyses
GraphPad Prism was used to perform all statistical analyses unless otherwise indicated. Statistical tests as well as replicate descriptions are detailed in each figure legend. A p value < 0.05 was considered significant for all analyses, unless otherwise denoted.
Author Contributions
M.S.E. and P.J.T. conceived and initiated the project. H.E.S. and M.S.E. harvested animal tissues. D.C.F. and T.E.M. generated and analyzed the RNA-seq data, and generated the associated figures. C.W., Y.L., and F.J. performed Drop-Seq, data analysis, and generated the associated figures. B.L.L.C. and M.S.E. executed the astrocyte studies. K.C.A., L.B., and M.S.E. performed qRT-PCR and analyzed the data. H.E.S., M.M., and M.S.E. isolated primary cell cultures. D.J.A. and M.S.E. designed the high-throughput primary compound screen and analyzed the data. M.M., L.B., and B.S.N. carried out immunohistochemistry and quantifications. M.M., Z.S.N, M.S.E., and H.E.S. cultured human spheroids and generated associated figures. M.S.E. and P.J.T. wrote the manuscript with feedback from all authors.
Acknowledgments
This research was supported by grants from: the European Leukodystrophy Association (to P.J.T.); the NIH R01NS093357 (to P.J.T.), R01HG009658 (to F.J.), and T32GM007250 (to M.S.E.); the New York Stem Cell Foundation (to P.J.T); and philanthropic support from the Peterson, Fakhouri, Long, Goodman, Geller, Galbut/Heil, and Weidenthal families and the Research Institute for Children's Health. Additional support was provided by the Cytometry & Microscopy, Genomics, Imaging, and Small Molecule Drug Development core facilities of the Case Western Reserve University (CWRU) Comprehensive Cancer Center (P30CA043703), the High Performance Computing Resource in the Core Facility for Advanced Research Computing at CWRU, the Cleveland Clinic Lerner Research Institute Imaging Core, and the CWRU Light Microscopy Core Facility (NIH grant S10-OD016164). We are grateful to Zita Hubler, Yuriy Fedorov, and Brian Popko for technical assistance and/or discussion. P.J.T. and D.J.A. are co-founders and consultants for Convelo Therapeutics, which has licensed patents from CWRU inventors (P.J.T., D.J.A., and M.S.E.). P.J.T., D.J.A., and CWRU hold equity in Convelo Therapeutics. D.C.F. became an employee of Convelo Therapeutics subsequent to completion of these studies. P.J.T. is a consultant and on the Scientific Advisory Board of Cell Line Genetics. P.J.T. is Chair of the Scientific Advisory Board (volunteer position) for the Pelizaeus-Merzbacher Disease Foundation.
Published: August 23, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, six tables, and one video and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.07.015.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE111605.
Supplemental Information
GSEA of bulk culture RNA-seq showing enriched pathways in jimpy OPCs relative to wild-type OPCs, after 1 day of oligodendrocyte differentiation.
scRNA-seq data showing differentially expressed genes (p < 01) between wild-type and jimpy OPCs, oligodendrocytes, and astrocytes, respectively, after 1 day of oligodendrocyte differentiation.
GSEA of scRNA-seq showing enriched pathways and corresponding p values in jimpy OPCs, oligodendrocytes, and astrocytes relative to their respective wild-type counterparts.
Primary (>3,000) bioactive compound screening data showing results of each tested compound.
References
- Baron W., Hoekstra D. On the biogenesis of myelin membranes: sorting, trafficking and cell polarity. FEBS Lett. 2010;584:1760–1770. doi: 10.1016/j.febslet.2009.10.085. [DOI] [PubMed] [Google Scholar]
- Baumann N., Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol. Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Bechler M.E., Byrne L., Ffrench-Constant C. CNS myelin sheath lengths are an intrinsic property of oligodendrocytes. Curr. Biol. 2015;25:2411–2416. doi: 10.1016/j.cub.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowsky J.L., Nelson C., Kingston J.L., Filloux F.M., Mundorff M.B., Srivastava R. The burden of inherited leukodystrophies in children. Neurology. 2010;75:718–725. doi: 10.1212/WNL.0b013e3181eee46b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M., Bryant K.F., Jousse C., Long K., Harding H.P., Scheuner D., Kaufman R.J., Ma D., Coen D.M., Ron D. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Bradl M., Bauer J., Inomata T., Zielasek J., Nave K.A., Toyka K., Lassmann H., Wekerle H. Transgenic Lewis rats overexpressing the proteolipid protein gene: myelin degeneration and its effect on T cell-mediated experimental autoimmune encephalomyelitis. Acta Neuropathol. 1999;97:595–606. doi: 10.1007/s004010051035. [DOI] [PubMed] [Google Scholar]
- Cerghet M., Bessert D.A., Nave K.A., Skoff R.P. Differential expression of apoptotic markers in jimpy and in Plp overexpressors: evidence for different apoptotic pathways. J. Neurocytol. 2001;30:841–855. doi: 10.1023/a:1019697506757. [DOI] [PubMed] [Google Scholar]
- Clayton B.L., Popko B. Endoplasmic reticulum stress and the unfolded protein response in disorders of myelinating glia. Brain Res. 2016;1648:594–602. doi: 10.1016/j.brainres.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antonio M., Musner N., Scapin C., Ungaro D., Del Carro U., Ron D., Feltri M.L., Wrabetz L. Resetting translational homeostasis restores myelination in Charcot-Marie-Tooth disease type 1B mice. J. Exp. Med. 2013;210:821–838. doi: 10.1084/jem.20122005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan I.D., Kondo Y., Zhang S.C. The myelin mutants as models to study myelin repair in the leukodystrophies. Neurotherapeutics. 2011;8:607–624. doi: 10.1007/s13311-011-0080-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G., Mutel V., Trube G., Malherbe P., Kew J.N., Mohacsi E., Heitz M.P., Kemp J.A. Ro 25-6981, a highly potent and selective blocker of N-methyl-D-aspartate receptors containing the NR2B subunit. Characterization in vitro. J. Pharmacol. Exp. Ther. 1997;283:1285–1292. [PubMed] [Google Scholar]
- Garbern J.Y. Pelizaeus-Merzbacher disease: genetic and cellular pathogenesis. Cell. Mol. Life Sci. 2007;64:50–65. doi: 10.1007/s00018-006-6182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A., Southwood C.M., Lazzarini R.A. Disrupted proteolipid protein trafficking results in oligodendrocyte apoptosis in an animal model of Pelizaeus-Merzbacher disease. J. Cell Biol. 1998;140:925–934. doi: 10.1083/jcb.140.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A.J., Gelfand J.M., Cree B.A., Bevan C., Boscardin W.J., Mei F., Inman J., Arnow S., Devereux M., Abounasr A. Clemastine fumarate as a remyelinating therapy for multiple sclerosis (ReBUILD): a randomised, controlled, double-blind, crossover trial. Lancet. 2017;390:2481–2489. doi: 10.1016/S0140-6736(17)32346-2. [DOI] [PubMed] [Google Scholar]
- Helman G., Van Haren K., Escolar M.L., Vanderver A. Emerging treatments for pediatric leukodystrophies. Pediatr. Clin. North Am. 2015;62:649–666. doi: 10.1016/j.pcl.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubler Z., Allimuthu D., Bederman I., Elitt M.S., Madhavan M., Allan K.C., Shick H.E., Garrison E., T Karl M. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature. 2018 doi: 10.1038/s41586-018-0360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussien Y., Podojil J.R., Robinson A.P., Lee A.S., Miller S.D., Popko B. ER chaperone BiP/GRP78 is required for myelinating cell survival and provides protection during experimental autoimmune encephalomyelitis. J. Neurosci. 2015;35:15921–15933. doi: 10.1523/JNEUROSCI.0693-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Hossain M.I., Zhou L., Horie M., Ikenaka K., Horii A., Takebayashi H. Histological detection of dynamic glial responses in the dysmyelinating Tabby-jimpy mutant brain. Anat. Sci. Int. 2018;93:119–127. doi: 10.1007/s12565-016-0383-5. [DOI] [PubMed] [Google Scholar]
- Inoue K. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 2005;6:1–16. doi: 10.1007/s10048-004-0207-y. [DOI] [PubMed] [Google Scholar]
- Karadottir R., Cavelier P., Bergersen L.H., Attwell D. NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature. 2005;438:1162–1166. doi: 10.1038/nature04302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S.A., Barrie J.A., McCulloch M.C., Montague P., Edgar J.M., Iden D.L., Anderson T.J., Nave K.A., Griffiths I.R., McLaughlin M. PLP/DM20 expression and turnover in a transgenic mouse model of Pelizaeus-Merzbacher disease. Glia. 2010;58:1727–1738. doi: 10.1002/glia.21043. [DOI] [PubMed] [Google Scholar]
- Knapp P.E., Bartlett W.P., Skoff R.P. Cultured oligodendrocytes mimic in vivo phenotypic characteristics: cell shape, expression of myelin-specific antigens, and membrane production. Dev. Biol. 1987;120:356–365. doi: 10.1016/0012-1606(87)90238-7. [DOI] [PubMed] [Google Scholar]
- Knapp P.E., Skoff R.P., Redstone D.W. Oligodendroglial cell death in jimpy mice: an explanation for the myelin deficit. J. Neurosci. 1986;6:2813–2822. doi: 10.1523/JNEUROSCI.06-10-02813.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizume S., Takizawa S., Fujita K., Aida N., Yamashita S., Miyagi Y., Osaka H. Aberrant trafficking of a proteolipid protein in a mild Pelizaeus-Merzbacher disease. Neuroscience. 2006;141:1861–1869. doi: 10.1016/j.neuroscience.2006.05.067. [DOI] [PubMed] [Google Scholar]
- Kramer-Albers E.M., Gehrig-Burger K., Thiele C., Trotter J., Nave K.A. Perturbed interactions of mutant proteolipid protein/DM20 with cholesterol and lipid rafts in oligodendroglia: implications for dysmyelination in spastic paraplegia. J. Neurosci. 2006;26:11743–11752. doi: 10.1523/JNEUROSCI.3581-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C., Fernandes H.B., Sheldon C., Raymond L.A., Baimbridge K.G. Functional NMDA receptor subtype 2B is expressed in astrocytes after ischemia in vivo and anoxia in vitro. J. Neurosci. 2003;23:3364–3372. doi: 10.1523/JNEUROSCI.23-08-03364.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager A.M., Corradin O., Cregg J.M., Elitt M.S., Shick H.E., Clayton B.L., Allan K.C., Olsen H.E., Madhavan M., Tesar P.J. Rapid functional genetics of the oligodendrocyte lineage using pluripotent stem cells. Nat. Commun. 2018 doi: 10.1038/s41467-018-06102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Chong S.Y., Tuck S.J., Corey J.M., Chan J.R. A rapid and reproducible assay for modeling myelination by oligodendrocytes using engineered nanofibers. Nat. Protoc. 2013;8:771–782. doi: 10.1038/nprot.2013.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan M., Nevin Z.S., Shick H.E., Garrison E., Clarkson-Paredes C., Karl M., Clayton B.L.L., Factor D.C., Allan K.C., Barbar L. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat. Methods. 2018 doi: 10.1038/s41592-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon B.S., Shick H.E., Kidd G.J., Macklin W.B. Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J. Neurosci. 2002;22:876–885. doi: 10.1523/JNEUROSCI.22-03-00876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayoral S.R., Chan J.R. The environment rules: spatiotemporal regulation of oligodendrocyte differentiation. Curr. Opin. Neurobiol. 2016;39:47–52. doi: 10.1016/j.conb.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musner N., Sidoli M., Zambroni D., Del Carro U., Ungaro D., D'Antonio M., Feltri M.L., Wrabetz L. Perk ablation ameliorates myelination in S63del-Charcot-Marie-Tooth 1B neuropathy. ASN Neuro. 2016;8 doi: 10.1177/1759091416642351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm F.J., Madhavan M., Zaremba A., Shick E., Karl R.T., Factor D.C., Miller T.E., Nevin Z.S., Kantor C., Sargent A. Drug-based modulation of endogenous stem cells promotes functional remyelination in vivo. Nature. 2015;522:216–220. doi: 10.1038/nature14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm F.J., Zaremba A., Caprariello A.V., Nayak S., Freundt E.C., Scacheri P.C., Miller R.H., Tesar P.J. Rapid and robust generation of functional oligodendrocyte progenitor cells from epiblast stem cells. Nat. Methods. 2011;8:957–962. doi: 10.1038/nmeth.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevin Z.S., Factor D.C., Karl R.T., Douvaras P., Laukka J., Windrem M.S., Goldman S.A., Fossati V., Hobson G.M., Tesar P.J. Modeling the mutational and phenotypic landscapes of Pelizaeus-Merzbacher disease with human iPSC-derived oligodendrocytes. Am. J. Hum. Genet. 2017;100:617–634. doi: 10.1016/j.ajhg.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numasawa-Kuroiwa Y., Okada Y., Shibata S., Kishi N., Akamatsu W., Shoji M., Nakanishi A., Oyama M., Osaka H., Inoue K. Involvement of ER stress in dysmyelination of Pelizaeus-Merzbacher disease with PLP1 missense mutations shown by iPSC-derived oligodendrocytes. Stem Cell Reports. 2014;2:648–661. doi: 10.1016/j.stemcr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata Y., Morimura T., Nakamura S., Hirano E., Kure S., Goto Y.I., Inoue K. Depletion of molecular chaperones from the endoplasmic reticulum and fragmentation of the Golgi apparatus associated with pathogenesis in Pelizaeus-Merzbacher disease. J. Biol. Chem. 2013;288:7451–7466. doi: 10.1074/jbc.M112.435388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S., Bernard G., Leventer R.J., van der Knaap M.S., van Hove J., Pizzino A., McNeill N.H., Helman G., Simons C., Schmidt J.L. A clinical approach to the diagnosis of patients with leukodystrophies and genetic leukoencephelopathies. Mol. Genet. Metab. 2015;114:501–515. doi: 10.1016/j.ymgme.2014.12.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R.J. Jimpy, a new totally sexlinked gene in the house mouse. Z. Indukt. Abstamm. Vererbungsl. 1954;86:322–326. doi: 10.1007/BF00312228. [DOI] [PubMed] [Google Scholar]
- Prukop T., Epplen D.B., Nientiedt T., Wichert S.P., Fledrich R., Stassart R.M., Rossner M.J., Edgar J.M., Werner H.B., Nave K.A. Progesterone antagonist therapy in a Pelizaeus-Merzbacher mouse model. Am. J. Hum. Genet. 2014;94:533–546. doi: 10.1016/j.ajhg.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Readhead C., Schneider A., Griffiths I., Nave K.A. Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12:583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- Roussel G., Neskovic N.M., Trifilieff E., Artault J.C., Nussbaum J.L. Arrest of proteolipid transport through the Golgi apparatus in Jimpy brain. J. Neurocytol. 1987;16:195–204. doi: 10.1007/BF01795303. [DOI] [PubMed] [Google Scholar]
- Salter M.G., Fern R. NMDA receptors are expressed in developing oligodendrocyte processes and mediate injury. Nature. 2005;438:1167–1171. doi: 10.1038/nature04301. [DOI] [PubMed] [Google Scholar]
- Skoff R.P. Myelin deficit in the Jimpy mouse may be due to cellular abnormalities in astroglia. Nature. 1976;264:560–562. doi: 10.1038/264560a0. [DOI] [PubMed] [Google Scholar]
- Somers A., Jean J.C., Sommer C.A., Omari A., Ford C.C., Mills J.A., Ying L., Sommer A.G., Jean J.M., Smith B.W. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood C.M., Garbern J., Jiang W., Gow A. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron. 2002;36:585–596. doi: 10.1016/s0896-6273(02)01045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap M.S., Bugiani M. Leukodystrophies: a proposed classification system based on pathological changes and pathogenetic mechanisms. Acta Neuropathol. 2017;134:351–382. doi: 10.1007/s00401-017-1739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way S.W., Podojil J.R., Clayton B.L., Zaremba A., Collins T.L., Kunjamma R.B., Robinson A.P., Brugarolas P., Miller R.H., Miller S.D. Pharmaceutical integrated stress response enhancement protects oligodendrocytes and provides a potential multiple sclerosis therapeutic. Nat. Commun. 2015;6:6532. doi: 10.1038/ncomms7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W.C., 2nd, Gard A.L. In vitro death of jimpy oligodendrocytes: correlation with onset of DM-20/PLP expression and resistance to oligodendrogliotrophic factors. J. Neurosci. Res. 1997;50:177–189. doi: 10.1002/(SICI)1097-4547(19971015)50:2<177::AID-JNR7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Yool D.A., Edgar J.M., Montague P., Malcolm S. The proteolipid protein gene and myelin disorders in man and animal models. Hum. Mol. Genet. 2000;9:987–992. doi: 10.1093/hmg/9.6.987. [DOI] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O'Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GSEA of bulk culture RNA-seq showing enriched pathways in jimpy OPCs relative to wild-type OPCs, after 1 day of oligodendrocyte differentiation.
scRNA-seq data showing differentially expressed genes (p < 01) between wild-type and jimpy OPCs, oligodendrocytes, and astrocytes, respectively, after 1 day of oligodendrocyte differentiation.
GSEA of scRNA-seq showing enriched pathways and corresponding p values in jimpy OPCs, oligodendrocytes, and astrocytes relative to their respective wild-type counterparts.
Primary (>3,000) bioactive compound screening data showing results of each tested compound.