Summary
Mutations in the NPHS1 gene, which encodes NEPHRIN, cause congenital nephrotic syndrome, resulting from impaired slit diaphragm (SD) formation in glomerular podocytes. However, methods for SD reconstitution have been unavailable, thereby limiting studies in the field. In the present study, we established human induced pluripotent stem cells (iPSCs) from a patient with an NPHS1 missense mutation, and reproduced the SD formation process using iPSC-derived kidney organoids. The mutant NEPHRIN failed to become localized on the cell surface for pre-SD domain formation in the induced podocytes. Upon transplantation, the mutant podocytes developed foot processes, but exhibited impaired SD formation. Genetic correction of the single amino acid mutation restored NEPHRIN localization and phosphorylation, colocalization of other SD-associated proteins, and SD formation. Thus, these kidney organoids from patient-derived iPSCs identified SD abnormalities in the podocytes at the initial phase of congenital nephrotic disease.
Keywords: iPSCs, kidney, nephrotic syndrome, podocyte, slit diaphragm, NEPHRIN, NPHS1
Graphical Abstract

Highlights
-
•
iPSCs are established from a patient with a missense NPHS1 mutation
-
•
The induced podocytes exhibit impaired NEPHRIN localization
-
•
The slit diaphragm is lacking in the mutant podocytes
-
•
Genetic correction of the point mutation restores the phenotypes
Nishinakamura and colleagues reveal the pathogenesis of congenital nephrotic disease using iPSCs derived from a patient with an NPHS1 missense mutation. The mutant kidney podocytes show impaired NEPHRIN localization and slit diaphragm formation, which are restored by genetic correction of the point mutation.
Introduction
The glomerulus is the filtering apparatus of the kidney. In the glomerulus, podocytes (glomerular epithelial cells) cover the endothelial cells and play a major role in the filtration process (Quaggin and Kreidberg, 2008, Schell et al., 2014). Podocytes possess multiple cytoplasmic protrusions called foot processes that interdigitate with those from neighboring podocytes. The gaps between these foot processes (filtration slits) are bridged by the slit diaphragm (SD), which prevents serum proteins of high molecular weight from leaking into the urine (Patrakka and Tryggvason, 2007, Ruotsalainen et al., 1999). The major components of the SD include NEPHRIN (encoded by NPHS1), PODOCIN (encoded by NHPS2), and NEPH1 (encoded by KIRREL1). NEPHRIN and NEPH1 are transmembrane proteins that intercalate with those from neighboring cells to form a molecular mesh: the SD. PODOCIN binds to the cytoplasmic region of NEPHRIN and is considered to stabilize the SD. Thus, mutations in these genes cause proteinuria in humans and/or mice.
Finnish-type congenital nephrotic syndrome is an autosomal recessive disorder, and affected patients manifest severe proteinuria immediately after birth in most cases (Kestilä et al., 1998). Investigation of the mutated gene in this disease led to the discovery of NPHS1. The NPHS1 gene has 29 exons and the encoded NEPHRIN protein (∼180 kDa) comprises eight extracellular immunoglobulin (Ig)-like domains characterized by cysteine bonds, followed by a fibronectin domain, transmembrane region, and cytoplasmic tail. The region between the sixth and seventh Ig-like domains is called the spacer region. Many mutations in the NPHS1 gene have been reported, including some that lead to protein truncation and others that result in amino acid substitutions (Beltcheva et al., 2001). The truncating mutations (Fin-major and Fin-minor types) result in absence of NEPHRIN expression, narrowing of filtration slits, and loss of the SD, although foot processes are formed (Patrakka et al., 2000, Ruotsalainen et al., 2000). These phenotypes are also observed in mice completely lacking NEPHRIN (Donoviel et al., 2001, Putaala et al., 2001).
There is little available information on the kidney histology induced by amino acid substitutions in NEPHRIN, and the mutation-dependent pathogenesis of the human disease has mainly been analyzed by overexpression of various types of NEPHRIN in heterologous cell lines (Liu et al., 2001). Because some mutant NEPHRIN proteins with amino acid substitutions fail to localize on the cell surface (Liu et al., 2001), it is hypothesized that the point mutations affect protein folding, resulting in retention of misfolded proteins in the endoplasmic reticulum (ER), incomplete glycosylation in the ER and Golgi apparatus, and eventually ER-associated degradation (Drozdova et al., 2013). However, other NEPHRIN point mutants are successfully localized on the cell surface following overexpression in cell lines, but still cause nephrotic disease in patients (Liu et al., 2001). In these settings, it is difficult to determine whether a particular amino acid substitution is a genuine disease-causing mutation or a SNP in human individuals. It is also difficult to predict which types of point mutants will be retained on the cell surface following expression in cell lines, and there is no clear relationship between mutation type and disease severity. Because heterologous cell lines do not express other SD-associated proteins or form the SD, they are not suitable for examining the effects of mutations on SD formation. Immortalized podocyte cell lines are also unable to form the SD, possibly due to the low expression levels of SD-associated proteins and two-dimensional culture settings (Chittiprol et al., 2011, Mundel et al., 1997, Saleem et al., 2002).
By redefining the origin of nephron progenitors that give rise to glomeruli and renal tubules, we previously succeeded in generating three-dimensional kidney tissues from human induced pluripotent stem cells (iPSCs) (Taguchi et al., 2014). The glomerular podocytes induced in vitro expressed NEPHRIN, and possessed nascent SD-like structures (Sharmin et al., 2016). Moreover, when the iPSC-derived nephron progenitors were transplanted into immunodeficient mice, human glomeruli were vascularized with mouse endothelial cells, and SD formation was observed between the foot processes of the podocytes (Sharmin et al., 2016). Therefore, we reasoned that our podocyte induction protocol in vitro and in vivo could reflect the diseased state resulting from NEPHRIN mutations more directly. Taking advantage of our expertise, we have clarified the initial phase of podocyte abnormalities using iPSCs established from a patient with a point mutation of NEPHRIN in the present study.
Results
Point Mutation Impairs Protein Processing toward the Cell Surface
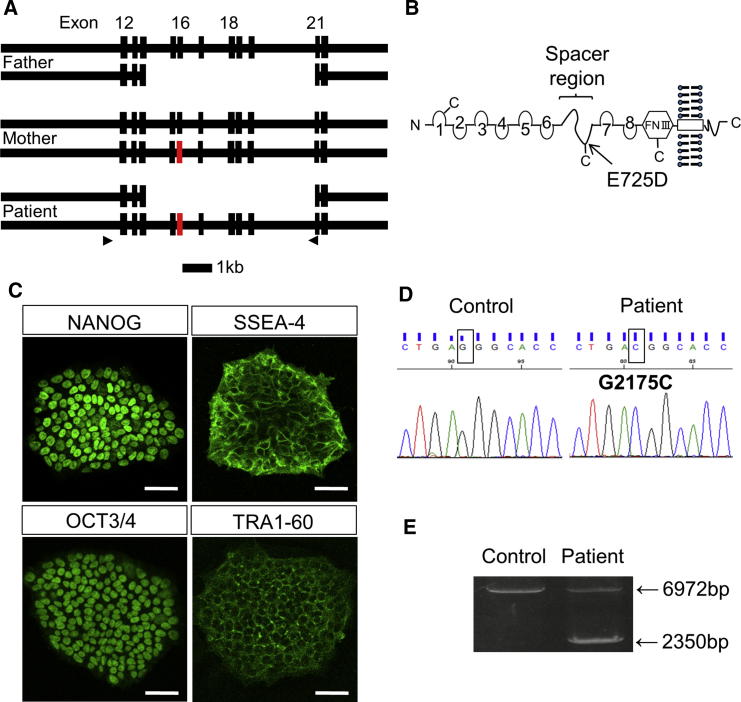
At 1 month after birth, a Japanese girl exhibited severe proteinuria (4+) and reduced serum albumin level (1.4 g/dL), and was diagnosed with congenital nephrotic syndrome. We identified that her paternal allele had a large deletion of multiple NPHS1 exons (exons 15–20), while the maternal allele contained a point mutation in exon 16 (G2175C), resulting in an amino acid substitution (E725D) in the extracellular spacer region of NEPHRIN (Figures 1A, 1B, and S1A–S1D). Thus, the maternal point mutation, combined with the non-functional paternal allele, was the likely cause of her nephrotic disease. Although this missense mutation (E725D) was previously reported in another patient (Beltcheva et al., 2001), no functional studies were available. Meanwhile, NEPHRIN with a neighboring mutation (S724C) was successfully localized on the cell surface when overexpressed in HEK293 cells (Liu et al., 2001).
Figure 1.
Establishment of iPSCs from a Patient with NPHS1 Mutations
(A) Exon/intron structures of the paternal, maternal, and patient alleles of NPHS1. A point mutation (G2175C) is located at exon 16 of the maternal allele (red). The arrowheads show the primers used for the experiment shown in (E). Scale bar, 1 kb.
(B) Putative NEPHRIN protein produced from the maternal allele. The arrow shows the amino acid substitution location in the spacer region. FNIII, fibronectin-like domain III.
(C) Stem cell markers expressed in the patient-derived iPSCs. Scale bars, 50 μm.
(D) NPHS1 sequences in the control and patient-derived iPSCs. The rectangular boxes show the nucleotide mutation at position 2175.
(E) Deletion of the paternal allele, as confirmed by genomic PCR. The primers shown in (A) were used.
See also Figure S1.
Thus, we initially overexpressed the wild-type and mutant (E725D) NEPHRIN proteins in HEK293 cells in a tetracycline-dependent manner. We employed site-directed stable integration to minimize clonal variations in expression levels (Figure S2A). Flow cytometry analysis using a monoclonal antibody (48E11) against the extracellular domain of NEPHRIN (Ruotsalainen et al., 2004) showed reduced levels of mutant NEPHRIN localization on the cell surface compared with the wild-type protein (Figures 2A and S2B). Immunostaining with another monoclonal antibody (50A9) against the extracellular domain of NEPHRIN (Ruotsalainen et al., 2004) in the absence of detergent also showed reduced signals for the mutant NEPHRIN protein (Figure 2B), while staining in the permeable condition (with detergent) showed comparable levels of NEPHRIN signals (Figure S2C). Western blotting using the antibody against the cytoplasmic domain of NEPHRIN identified two bands for wild-type NEPHRIN, but only the lower band for the mutant protein (Figure 2C). Biotin-mediated labeling of cell surface proteins showed that the upper band represented the majority of wild-type NEPHRIN on the cell surface, while the lower band of the mutant protein was only weakly detected on the cell surface (Figure 2C), suggesting that the mutant protein reached the cell surface less efficiently. The lower band was more sensitive to endoglycosidase H treatment than the upper band, suggesting that the mutant band did not have complex N-linked carbohydrate chains similar to the lower wild-type band (Figure 2D), consistent with overexpression studies on other mutant NEPHRIN proteins (Drozdova et al., 2013). We further monitored biotin-labeled surface proteins during the initial 48 hr after tetracycline addition (Figure 2E). While the lower band of wild-type NEPHRIN was detected weakly at 6 hr, the upper band was detected on the cell surface at 24 hr and its expression was increased at 48 hr. Similarly, the lower band of the mutant NEPHRIN was detected at 6 hr. Although it showed a mild increase thereafter, its relative detection level on the surface to the total input protein amount was low compared with the wild-type upper band. These results suggest that the missense mutation resulted in impaired glycosylation, which may have affected the proper cell surface translocation of the newly synthesized mutant protein.
Figure 2.
Point Mutation Impairs Protein Processing toward the Cell Surface
(A) Flow cytometry analysis of HEK293 cells overexpressing wild-type (WT) or mutant (MT) NEPHRIN using an antibody (48E11) against the extracellular domain of NEPHRIN. The y axis shows the cell counts normalized to 100% of the total cells. Blue, unstained HEK293 cells; red, tetracycline-induced HEK293 cells.
(B) Immunostaining of HEK293 cells overexpressing WT or MT NEPHRIN using an antibody (50A9) against the extracellular domain of NEPHRIN in the absence of detergent. Note the reduced signals for MT NEPHRIN. Wheat germ agglutinin (WGA) binds to the surface glycoproteins. Scale bars, 20 μm.
(C) Biotin-mediated cell surface NEPHRIN labeling in HEK293 cells. TfR, transferrin receptor (negative control). Biotinylated cell surface components were isolated with streptavidin-Sepharose and isolated proteins were detected by immunoblotting with the anti-NEPHRIN antibody that recognizes the cytoplasmic region of NEPHRIN (Bio/Strep-IP). Input represents 0.2% of the total protein used for analysis.
(D) Endoglycosidase H (endo H)-mediated cleavage of the lower NEPHRIN bands. Lysates of HEK293 cells overexpressing human WT or MT NEPHRIN were incubated with or without endo H. Endo H induced loss of the lower NEPHRIN band and appearance of a faster migrating band. The upper band of WT NEPHRIN was unaffected.
(E) Kinetics of NEPHRIN on the cell surface of HEK293 cells after tetracycline treatment. Cells were harvested at 0, 6, 24, and 48 hr, and cell surface components were detected by biotin-mediated labeling as described in (C).
See also Figure S2.
Establishment of iPSCs from a Patient with NPHS1 Mutations
It remained unclear whether the above findings held true in podocytes or how the mutation affected SD formation. Thus, we introduced reprogramming transcription factors into skin fibroblasts from the patient using a Sendai virus vector, and established iPSC lines expressing characteristic stem cell markers (Figure 1C). These cell lines exhibited normal karyotypes except for pericentric inversion within chromosome 9 (46, XX, inv(9) (p12q13)), which is found in a small percentage of normal individuals (Colls et al., 1997) and is irrelevant to chromosome 19 where NPHS1 is located (Figure S1E). The mutations on both the paternal and maternal alleles were preserved in the iPSCs (Figures 1D and 1E). When transplanted into immunodeficient mice, the iPSCs formed teratomas containing tissues derived from three germ layers: endoderm, ectoderm, and mesoderm (Figure S1F).
Mutant Podocytes Exhibit Reduced Cell Surface Localization of NEPHRIN
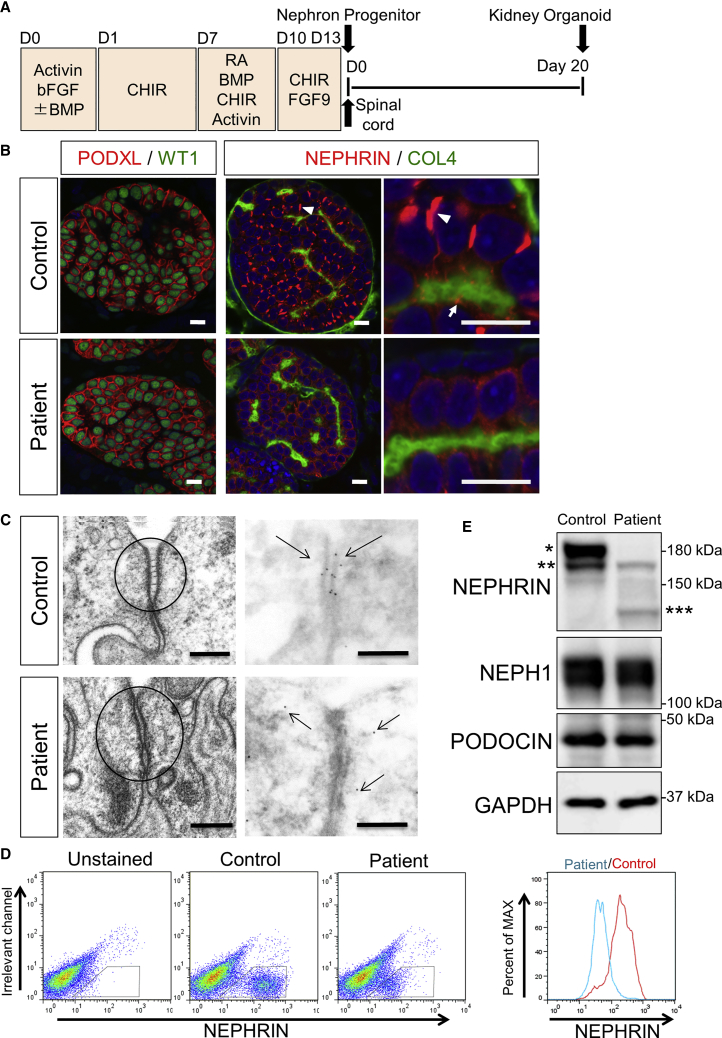
We induced the patient-derived iPSCs toward nephron progenitors, based on our published protocol (Taguchi and Nishinakamura, 2017, Taguchi et al., 2014), and combined these cells with mouse embryonic spinal cord, a potent inducer of nephrogenesis (Figure 3A). When cultured for 20 days, both the control (wild-type 201B7) and mutant clones formed kidney tissues containing glomeruli with podocytes that expressed WT1 and PODOCALYXIN (PODXL) (Figure 3B). However, the NEPHRIN localizations in the glomerular podocytes were markedly different. In the control clone, we observed prominent NEPHRIN+ rod-like structures on the lateral side of most podocytes (Figure 3B). NEPHRIN was also detected as smaller dots on the basal side adjacent to the type IV collagen+ (COL4) basement membrane. The lateral rods represented thickened parts of the filamentous structures encompassing from the basal to lateral side of the podocytes (Figure S1G). Electron microscopy of the control podocytes showed a widened intercellular space on the lateral side that was connected with ladder-like structures, with clustering of NEPHRIN to the cell membrane of these regions, as revealed by staining with an anti-NEPHRIN antibody (Figure 3C). These observations suggested that the rod-like structures may reflect a transit state of NEPHRIN protein shifting from the lateral to basal domain of the podocytes, and we refer to this structure as the “pre-SD domain” hereafter. Indeed, these structures were quite similar to the NEPHRIN+ ladder-like structures reported in embryonic human podocytes in vivo (Ruotsalainen et al., 2000). In contrast, NEPHRIN was weakly detected in the cytoplasm and pre-SD domains were rarely observed in the mutant podocytes (Figures 3B and S1H). On electron microscopy, the intercellular junctions showed tight adherence, and NEPHRIN was detected not at cellular junctions but in the cytoplasm in a scattered manner (Figure 3C).
Figure 3.
Mutant Podocytes Exhibit Reduced Cell Surface Localization of NEPHRIN
(A) Outline of the protocol to induce podocytes from iPSCs through nephron progenitors. CHIR, CHIR99021; RA, retinoic acid. Spinal cord was attached to nephron progenitors at day 13 to induce nephrogenesis, and the kidney organoids were cultured for a further 20 days.
(B) Section immunostaining of kidney organoids at day 20 after nephron induction, showing altered NEPHRIN localization in the patient-derived podocytes. The arrowheads point to the lateral pre-SD domains and the arrows point to the basal pre-SD domains. PODXL, PODOCALYXIN; COL4, type IV collagen. Scale bars, 10 μm.
(C) Electron microscopic images of podocytes in the kidney organoids. Left panels: ladder-like structures are formed in the control podocytes only. Right panels: immunoelectron microscopic images showing NEPHRIN localization at the intercellular junctions only in the control podocytes. The arrows show the NEPHRIN signals. Scale bars, 200 nm.
(D) Flow cytometry analysis of NEPHRIN in kidney organoids using an antibody (48E11) against the extracellular domain of NEPHRIN. The histograms show cell counts normalized to 100% of the total cells in the NEPHRIN+ gates.
(E) Western blots of NEPHRIN and SD-associated proteins from kidney organoids. ∗The upper band detected only in the control; ∗∗lower bands detected both in the control and the mutant; and ∗∗∗the additional band presumably representing the paternal truncated protein.
See also Figures S1 and S5.
Flow cytometry analysis of the induced podocytes using a monoclonal antibody (48E11) against the extracellular domain of NEPHRIN (Ruotsalainen et al., 2004) demonstrated that the mutant NEPHRIN was localized to the cell surface to a lesser extent than the control protein (Figure 3D). Western blotting using the antibody against the cytoplasmic domain of NEPHRIN also showed that expression of NEPHRIN, but not other SD-associated proteins, was significantly reduced in the mutant kidney organoids (Figure 3E). The upper band of the two dominated in the control organoids, while it was undetectable in the mutant organoids, which is consistent with the observations in HEK293 cells. The samples at earlier differentiation time points (day 6 and 10 instead of day 20 upon nephron progenitor induction) also showed gradual increases of the upper bands only in the controls (Figure S1I). These results suggest that the mutant NEPHRIN protein was not properly transferred to the cell surface, leading to impaired formation of pre-SD domains in the mutant podocytes. We also observed a further lower band in the mutant organoids that likely represented the paternal protein with in-frame deletion of the spacer region and the seventh and eighth Ig-like domains (Figures 3E and S1D), because the weak additional band did not disappear upon genetic correction of the maternal missense mutation as shown in Figure 5B.
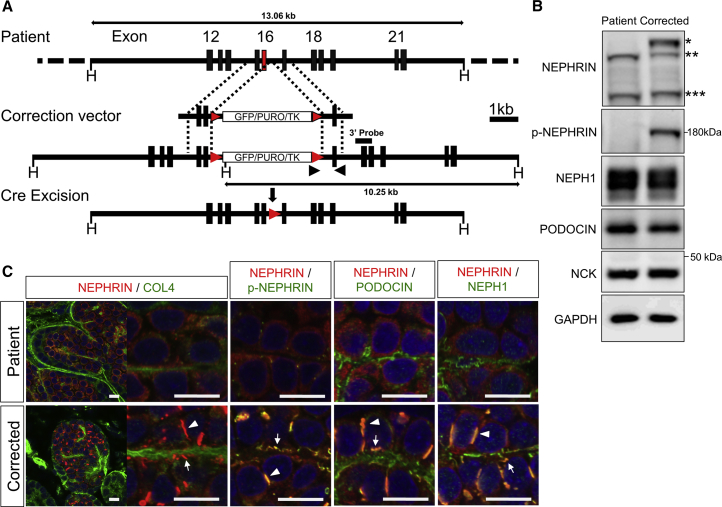
Figure 5.
Genetic Correction of the Point Mutation Restores NEPHRIN Localization
(A) Correction of the point mutation by homologous recombination, followed by Cre-mediated removal of the selection cassette. The puromycin-resistance cassette (GFP/PURO/TK) is flanked by loxP sites (red arrowheads). Black arrowheads show the primers used for screening. Scale bar, 1 kb.
(B) Western blots of NEPHRIN and SD-associated proteins from the kidney organoids in vitro (day 20). ∗The upper band detected only in the corrected podocytes; ∗∗lower bands detected in both samples; and ∗∗∗the additional bands presumably representing the paternal truncated protein.
(C) Restored localization and phosphorylation of NEPHRIN in the kidney organoids in vitro (day 20). NEPHRIN colocalizes with p-NEPHRIN, PODOCIN, and NEPH1 in the genetically corrected organoids. The arrowheads point to the lateral pre-SD domains and the arrows point to the basal pre-SD domains. Scale bars, 10 μm.
See also Figures S1 and S4.
Mutant Podocytes Form Foot Processes but Exhibit Impaired SD Formation
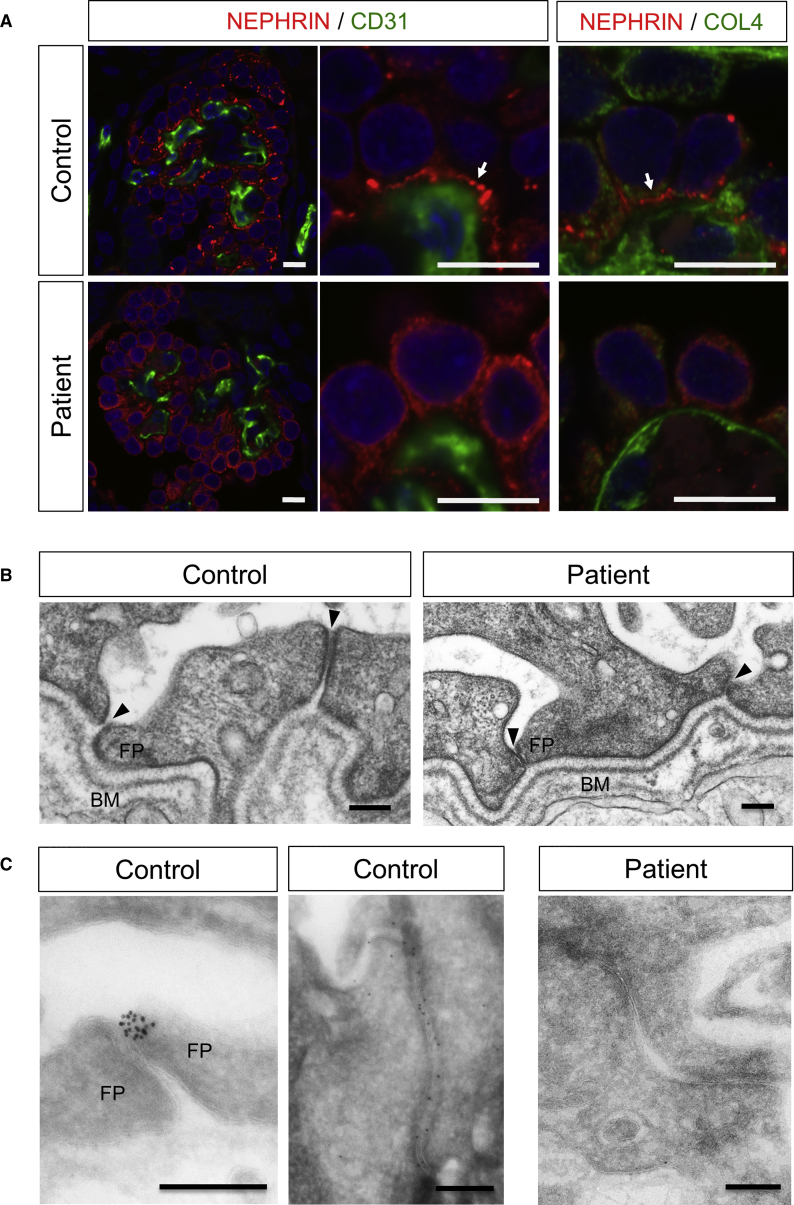
We previously reported that transplantation of iPSC-derived nephron progenitors resulted in SD formation in the induced podocytes (Sharmin et al., 2016). We utilized this system that combines in vitro and in vivo methods to examine the SD formation defects in the mutant podocytes. The iPSC-derived nephron progenitors were sorted as an ITGA8+/PDGFRA− fraction, induced by spinal cord incubation to initiate differentiation, and then transplanted beneath the kidney capsule of immunodeficient mice, as described (Kaku et al., 2017, Sharmin et al., 2016). At 20 days after transplantation, human glomeruli were vascularized, and the wild-type podocytes expressed NEPHRIN, which was linearly aligned with the CD31+ mouse vascular endothelial cells and COL4+ basement membrane (Figure 4A). In this setting, the lateral pre-SD domains observed in vitro were rarely detected, suggesting that the in vivo environment, possibly including the interaction with endothelial cells, may have enhanced the podocyte maturation, as we reported previously (Sharmin et al., 2016). However, NEPHRIN still remained in the cytoplasm in the mutant podocytes (Figure 4A). Expression of PODXL and PODOCIN was not markedly different (Figure S3). Electron microcopy of the control podocytes showed well-developed foot processes above the basement membrane (Figure 4B). The gaps between the processes (filtration slits) were enlarged (Figure 4B), and NEPHRIN was detected in these slits (Figure 4C), probably reflecting the initial phase of SD formation. In contrast, the foot processes of the mutant podocytes were closely attached to the neighboring cells, and NEPHRIN was not detected in the junctions (Figures 4B and 4C). These results demonstrate that the mutant podocytes formed foot processes, but exhibited impaired SD formation. Thus, the defects observed in vitro (Figure 3) and in vivo (Figure 4) are likely to represent the initial phase of this podocyte disease.
Figure 4.
Mutant Podocytes Form Foot Processes, but Exhibit Impaired SD Formation
(A) Immunostaining of iPSC-derived glomeruli and podocytes after transplantation. Glomeruli are vascularized with CD31+ endothelial cells in both groups, but the NEPHRIN distributions are different. The arrows point to the basal linear expression of NEPHRIN. COL4, type IV collagen. Scale bars, 10 μm.
(B) Widened filtration slits in the control podocytes, but not in mutant podocytes, as shown by electron microscopy. The right junction in the control is still at the pre-SD stage. The arrowheads show filtration slits. BM, basement membrane; FP, foot process. Scale bars, 200 nm.
(C) Failure of NEPHRIN localization between the foot processes (FP), as shown by immunoelectron microscopy. The control panels show two independent regions of filtration slits (left, slit between foot processes; right, pre-SD stage). Scale bars, 200 nm.
See also Figure S3.
Genetic Correction of the Point Mutation Normalizes the Phenotypes
We tried to correct the point mutation by homologous recombination (Figure 5A). For this, we constructed a pair of plasmids expressing transcription activator-like effector nucleases (TALENs) targeted in close proximity to the mutation. We introduced these TALEN plasmids, together with a targeting vector containing the homology arms with the wild-type sequences of exon 16, into the patient-derived iPSCs. The designed correction vectors successfully targeted the maternal allele because the paternal allele lacked exons 15–20. After verification of the normalized sequence (G at 2175) and Cre recombinase-mediated excision of the selection cassette (Figures S4A and S4B), the corrected clones were induced toward the kidney tissues. Western blotting confirmed that NEPHRIN protein expression was restored in the corrected kidney organoids in vitro: the upper band reappeared while the two lower bands remained unchanged (Figure 5B). It is known that cytoplasmic tyrosine residues of NEPHRIN on the cell surface are phosphorylated by intracellular kinases such as FYN, leading to the recruitment of various molecules including NCK and PODOCIN, and eventually to proper maintenance of podocyte morphology (Jones et al., 2006, New et al., 2016). Thus, we examined the phosphorylation status of NEPHRIN, and found that it was restored in the corrected clones (Figure 5B). The restoration kinetics at earlier time points were slightly slower than those in the control kidney organoids (Figure S1I), possibly because of the residual paternal allele or genetic background differences in the iPSC donors. Expression of NCK, as well as SD-associated molecules NEPH1 and PODOCIN, was relatively unaffected in the mutant and corrected clones. Histologically, lateral and basal NEPHRIN+ pre-SD domains were restored by correction of the mutation (Figures 5C and S4C). Some of the NEPHRIN+ accumulations formed linear structures on the basal side along the COL4+ basement membrane. These pre-SD domains almost completely overlapped with those of phosphorylated NEPHRIN, which was absent in the mutant podocytes. NEPH1 and PODOCIN were expressed on the basal side even in the mutant podocytes, while correction of the mutation led to colocalization of these proteins with NEPHRIN in both the lateral and basal regions (Figures 5C, S4D, and S4E). PODXL was excluded from the basal domains in both the mutant and corrected podocytes, but only excluded from the lateral pre-SD domains in the corrected podocytes (Figure S4F). Thus, the overall apicobasal distribution of PODXL, NEPH1, and PODOCIN was not affected by the NEPHRIN abnormalities, but NEPHRIN was required for recruitment of NEPH1 and PODOCIN to the pre-SD domains and exclusion of PODXL from the pre-SD domains. Electron microscopy confirmed restored accumulation of NEPHRIN at intercellular junctions of the kidney organoids in vitro (Figure 6A).
Figure 6.
Genetic Correction Restores the SD Formation Process
(A) Immunoelectron microscopic images showing the restored NEPHRIN localization (arrows) in the intercellular regions of the podocytes in the corrected kidney organoids in vitro (day 20). Scale bars, 200 nm.
(B) Transmission electron microscopic images showing filtration slits in the podocytes after transplantation (day 20). While the patient foot processes are tightly adhered, the corrected foot processes exhibit widened gaps. The arrowheads show filtration slits. BM, basement membrane; FP, foot process; EC, endothelial cells. Scale bars, 200 nm.
We further transplanted the patient and corrected nephron progenitors, and analyzed them by electron microscopy after 20 days. We selected the glomeruli with foot processes and assessed the junctions between the processes. While all 60 examined sites for the patient podocytes were tightly adhered, widened gaps were observed in 22 of 61 sites (36%) for the corrected podocytes (Figure 6B). The width of the gaps measured on high-magnification images was 35.0 ± 6.0 nm (n = 7), indicating recovery of filtration slit formation after mutation correction.
These data confirmed that the single amino acid substitution (E725D) was a disease-causing mutation, and that its correction restored not only the cell surface localization, but also the phosphorylation status of NEPHRIN, which presumably led to recruitment of NEPH1 and PODOCIN and proper formation of the SD structure.
Discussion
In the present study, we successfully identified podocyte abnormalities using iPSCs derived from a patient with an NPHS1 missense mutation. While CRISPR/Cas9-mediated null-type deletion of PODXL has been reported in iPSC-derived kidney organoids (Freedman et al., 2015, Kim et al., 2017), the present study is the demonstration of patient-derived iPSCs applied to glomerular diseases. In particular, our study shows that it is possible to analyze SD defects in nephrotic syndrome using iPSC-derived kidney organoids, but not in conventional cell lines. Our in vitro induction protocol, combined with transplantation in vivo, enables SD reconstitution and thus identification of the defects in the mutant podocytes. The induced glomerular podocytes in vitro (day 20 of differentiation from nephron progenitors) exhibited reduced NEPHRIN expression on the cell surface and impaired formation of pre-SD domains, which were observed on the lateral side of control podocytes. Upon transplantation, NEPHRIN accumulation was observed on the basal side adjacent to the endothelial cells, but such progression was absent in the patient-derived podocytes. Furthermore, genetic correction of the point mutation restored these abnormalities, clearly identifying the single amino acid substitution (E725D) as a disease-causing mutation.
Our data showed that this amino acid substitution (E725D) in the spacer region of NEPHRIN affected the cell surface localization of the protein, which likely resulted in the impaired SD formation. Two glycosylation sites in the spacer region are located close to the position of this mutation, which may have impaired the glycosylation of the protein required for its translocation to the cell surface. However, the precise mechanisms remain to be clarified. Our findings are consistent with the conventional model established using kidney sections from patients with truncation-type NEPHRIN mutations and null-type knockout mice, as well as overexpression in heterologous cell lines (Donoviel et al., 2001, Liu et al., 2001, Putaala et al., 2001, Ruotsalainen et al., 2000). Although a biopsy was not taken from our patient, our study using patient-derived iPSCs has likely identified abnormalities at the initial stage of the disease caused by the point mutation. Through progress in sequencing techniques, diagnosis of NEPHRIN mutations has become possible without a kidney biopsy, which can be complicated by tissue deterioration when performed at late stages of disease progression. Even in such cases, our protocol can reproduce the initial stage of the disease state. Because there are various mutant alleles of NPHS1, our approach is applicable for examining the disease mechanism in individual mutant alleles. Some mutant NEPHRIN proteins were reported to show normal localization on the cell surface when overexpressed in heterologous cell lines (Liu et al., 2001). Generation of podocytes from patients with such mutations will reveal distinct pathways that are dependent or independent of cell surface localization of NEPHRIN in this cell lineage. Furthermore, genetic correction of the mutations will discriminate between SNPs and disease-causing mutations.
Our data can not only reproduce the initial stage of congenital nephrotic syndrome, but also correlate with the developmental process of human podocytes in vivo. Podocyte development is classified into three stages: S-shaped body, capillary loop, and maturation stages. NEPHRIN+ ladder-like structures were reported to form at the lateral intercellular junctions in the former two stages (Ruotsalainen et al., 2000). These structures appear to migrate in the apical to basal direction as development progresses. In the maturation-stage podocytes, the gaps between the foot processes (filtration slits) are widened, and the SD is recognized as a NEPHRIN+ linear structure in the space containing filtration slits. The NEPHRIN+ ladder-like structures in our wild-type kidney organoids in vitro likely represent those in the S-shaped body and capillary loop stages, making the term pre-SD domain feasible. Podocytes in the organoids at 20 days after transplantation may correspond to the maturation stage. We observed that NEPHRIN was already phosphorylated and colocalized with NEPH1 and PODOCIN in the pre-SD domains. The affected distribution of these SD-associated molecules in the NEPHRIN mutant podocytes also indicates the importance of NEPHRIN in the recruitment of these proteins to the pre-SD domains in human podocytes. Thus, the development of iPSC-derived podocytes can faithfully reproduce that of podocytes in vivo, rendering our system useful for revealing the detailed mechanisms of SD formation in humans.
It is noteworthy that we detected abnormalities within 3 weeks after nephron progenitor differentiation. This time course is consistent with human kidney development in vivo, in which the initial sets of glomeruli are formed within 3–4 weeks after nephron progenitors become detectable (Lindström et al., 2018). Considering that nephron progenitors first appear at 4–5 weeks of gestation (Lindström et al., 2018), we may be observing nephrogenesis equivalent to 7–9 weeks of gestation. If this assumption is correct, manifestation of Finnish-type congenital nephrotic syndrome could occur at an unexpectedly early stage during embryogenesis. While this is intriguing, it would be preferable to monitor further progress of the disease state, including proteinuria and kidney failure. Our transplanted kidney tissues lack the urinary exit tract, because nephron progenitors only give rise to the glomeruli and renal tubules, and not the collecting duct or ureters. Without urinary flow, most glomeruli will eventually undergo degeneration. A recent report on subcutaneous transplantation of kidney organoids also showed limited formation of the urinary exit tract (Bantounas et al., 2018). We recently reported the generation of branching ureteric buds, as precursors of the collecting duct and ureters, from mouse embryonic stem cells (ESCs) and human iPSCs (Taguchi and Nishinakamura, 2017). We further showed that the combination of mouse ESC-derived nephron progenitors and ureteric buds, together with stromal progenitors from mouse embryonic kidneys, gave rise to higher-order kidney structures. The generation of such structures from human iPSCs and further maturation in vivo will enable analysis of not only Finnish-type congenital nephrotic syndrome but also other genetic kidney diseases with later onset.
It will also be possible to use iPSC-derived mutant podocytes to test the effects of candidate chemicals for restoration of proper SD formation. While large numbers of podocytes, as well as high-throughput screening methods, are required for this purpose, the impact of the present study is not limited to a specific congenital disease. Many devastating kidney diseases in both children and adults start with proteinuria, and NEPHRIN localization and SD formation are affected in these glomerular diseases. If mechanisms to restore NEPHRIN localization can be identified, they could be applied to the development of novel treatments that can cure proteinuria and eventually renal failure.
Taken together, we have reproduced the initial phase of congenital kidney disease leading to impaired SD formation, and identified reduced cell surface localization of NEPHRIN and subsequent histological and molecular events in human podocytes. These findings, as well as our established iPSCs and induction protocol, will serve as a basis for dissecting the mechanisms underlying glomerular diseases in humans, and eventually for drug development for therapies.
Experimental Procedures
Identification of NPHS1 Mutations in the Patient
Genomic DNA was extracted from peripheral leukocytes in whole-blood samples using a DNA isolation kit (Takara). Individual exons of NPHS1 were amplified by PCR. The primers for NPHS1 were designed on the basis of previously published information regarding intron-exon boundaries (Lenkkeri et al., 1999). The PCR products were purified and sequenced. A large heterozygous deletion mutation (LHDM) was detected by semi-qPCR amplification using capillary electrophoresis (Agilent, 2001 Bioanalyzer with DNA 1000 Lab Chips; Agilent Technologies). The breakpoint of the LHDM was detected by long PCR and direct sequencing using genomic DNA.
Ethics Status
All experiments using human samples were performed in accordance with institutional guidelines and approved by the Licensing Committee of Kumamoto University. The names of the ethics committees were Ethics Committee for Epidemiological and General Research at the Faculty of Life Science, Kumamoto University, Ethics Committee for Human Genome and Gene Analysis Research at the Faculty of Life Science, Kumamoto University, and Ethics Committee for Clinical Research and Advanced Medical Technology, Kumamoto University (approval numbers 335, 153, and 1,018, respectively). After explaining our study, the parents of the patient agreed to participate in the study and signed written informed consent forms. Human skin biopsies were collected from the patient after receiving informed consent.
Generation of iPSCs from the Patient
iPSCs were established from skin fibroblasts of the patient as described (Soga et al., 2015). In brief, four reprogramming transcription factors (OCT3/4, SOX2, KLF4, and MYC) were introduced using a Sendai virus vector. After establishment of the iPSC clones, the temperature-sensitive Sendai virus was eliminated by culture at 38°C for 24 hr. Among 14 clones, 3 were subjected to karyotyping and evaluated for teratoma formation (Soga et al., 2015). All three clones had normal karyotypes, but two clones (no. 1-5 and no. 3-3) exhibited teratoma formation with three germ layers. These two clones were adapted to feeder-free conditions (Nakagawa et al., 2014) and used for further studies. Clone no. 1-5 was mainly analyzed, but clone no. 3-3 showed consistent results. Wild-type 201B7 cells were used as control cells (Takahashi et al., 2007).
Kidney Induction from the Patient-Derived iPSCs
The patient-derived iPSC clones were induced toward nephron progenitors by a 13-day protocol as described (Taguchi and Nishinakamura, 2017) with a minor modification. At days 0–1, no BMP4 was added to the control 201B7 cells, while 3 ng/mL BMP4 was added to the patient-derived iPSC clones to optimize the percentage of the ITGA8+/PDGFRA− nephron progenitor fraction and the extent of nephrogenesis (Kaku et al., 2017). We confirmed that BMP4 concentrations at days 0–1 do not affect NEPHRIN localization, by comparing control, patient, and corrected clones with or without BMP4 treatment (Figure S5). The induced nephron progenitor aggregates were cultured at the air-fluid interface on a polycarbonate filter (0.8 μm; Whatman) supplied with DMEM containing 10% fetal calf serum, and with mouse embryonic spinal cord taken from embryonic day 12.5 (E12.5) embryos as described (Yoshimura et al., 2017).
Electron Microscopy
Transmission electron microscopy was performed as described (Kurihara et al., 2010). For immunoelectron microscopy, ultrathin cryosections were cut with an Ultracut UCT microtome equipped with the FCS cryoattachment (Leica) at −110°C as described (Tokuyasu, 1989). Sections were transferred to nickel grids (150 mesh) that had been coated with Formvar. After quenching of free aldehyde groups with PBS containing 0.01 M glycine, sections were incubated with primary antibodies overnight and then incubated with secondary antibodies coupled to 12-nm gold particles (diluted 1:100 with PBS containing 10% fetal calf serum) for 1 hr. After immunostaining, sections were fixed with 2.5% glutaraldehyde buffered with 0.1 M phosphate buffer (pH 7.4), contrasted with 2% neutral uranyl acetate solution for 30 min, and absorption-stained with 3% polyvinyl alcohol containing 0.2% acidic uranyl acetate for 30 min. Specimens were observed with a JEM1230 transmission electron microscope (JEOL). For in vitro studies, 16 control, 16 patient, and 4 corrected organoids were analyzed. For in vivo studies, 4 control, 6 patient, and 2 corrected organoids were analyzed after transplantation. The following primary antibodies were used: anti-C-terminal domain of NEPHRIN (Progen; GP-N2) for Figure 3C; and anti-N-terminal domain of NEPHRIN (Immuno-Biological Laboratories; 29050) for Figures 4C and 6A.
Generation of HEK293 Cell Lines Expressing the Mutant NEPHRIN Protein
For Flp recombinase-mediated site-direct integration of exogenous NEPHRIN, wild-type and mutant NPHS1 genes were cloned into the pcDNA5/FRT/TO vector, and transfected into Flp-In T-REx 293 cells with the pOG44 vector, according to the manufacturer's instructions (Thermo Fisher Scientific). The resultant colonies were picked up and expanded, followed by western blot analysis. At least three clones per construct were confirmed to express similar amounts of NEPHRIN protein at 48 hr after administration of tetracycline (1 μg/mL). The full-length NPHS1 cDNA was a kind gift from Dr. G. Walz (Gerke et al., 2003). Two DNA fragments containing the mutation (E725D) and the following part of the NPHS1 cDNA were amplified using two sets of primers: NEPHRIN mut-1&2 and NEPHRIN mut-3&4, respectively. The primer sequences are shown in Table S1. NEPHRIN mut-2 contained the E725D point mutation, marked in bold font in Table S1. The above-mentioned DNA fragments were ligated and cloned into the HindIII-BamHI site of the pBluescript II KS (−) plasmid using an In-Fusion Advanced PCR Cloning Kit (Takara-Clontech). A 1.6-kb Eam1105I-EcoRV fragment was used to replace the corresponding region of the wild-type NPHS1 cDNA in pcDNA5/FRT/TO.
Transplantation of iPSC-Derived Kidney Tissues
ITGA8+/PDGFRA− nephron progenitors were sorted and cultured as aggregates based on our published method (Kaku et al., 2017) with some modifications. The progenitor aggregates were cocultured with E12.5 mouse spinal cord in vitro for 3 days to initiate differentiation, and transplanted beneath the kidney capsules of immunodeficient NOD/SCID/JAK3-null mice (two spheres per animal) as described (Okada et al., 2008). Three independent experiments were performed and showed consistent results. All animal experiments were performed in accordance with institutional guidelines and approved by the Licensing Committee of Kumamoto University (no. A29-040).
Correction of the NPHS1 Point Mutation in Patient-Derived iPSCs
TALENs were designed to bind sequences and cleave DNA close to the point mutation of NPHS1: 5′-TCCGCCCACCCAGGGAT-3′ for the left TALEN and 5′-TTCCAGAACGGGAGGGTT-3′ for the right TALEN. A Platinum Gate TALEN Kit (Addgene; 1000000043) was used to construct a TALEN expression vector as described (Sakuma et al., 2013). An EF1α promoter-driven vector was used as the destination vector. To evaluate mutagenic efficiency, TALENs were transfected into HEK293 cells using Lipofectamine 2000 (Thermo Fisher Scientific). The target region was amplified, denatured, and annealed to examine the mutagenic effects.
To generate the targeting vector, the 5′ and 3′ homology arms (0.73 and 0.76 kb, respectively) of NPHS1 were amplified from the genomic DNA of the iPSCs. After sequence verification, the arms were cloned into the SpeI-BsrGI and NotI-SalI sites, respectively, of the HR210PA-1 vector (SBI System Biosciences).
Human iPSC line (clone no. 1-5) cells maintained on feeders were pretreated with Y27632 (10 μM) at 1 hr prior to electroporation, and dissociated into single cells with CTK solution (Reprocell) followed by Accutase (Merck Millipore). The targeting vector (5 μg) and the pair of TALEN plasmids (5 μg each) were electroporated into the dissociated human iPSCs using a Super Electroporator NEPA21 (Nepagene) under the following conditions: two poring pulses (125 V; 2.5 ms) followed by five transfer pulses (20 V; 50 ms). The electroporated cells were plated on puromycin-resistant DR4 feeders and puromycin (0.25 μg/mL) was added after 2 days as described (Tucker et al., 1997). Of 28 puromycin-resistant clones, 22 were examined for 3′ recombination by PCR (product size: 1,030 bp) and Southern blotting analyses. The 5′ recombination was confirmed by sequencing of the junctions between the host and donor DNA.
Among the 16 clones sequenced, mutations in 9 clones were corrected. Clone no. 1-5-9 was adapted to the feeder-free condition and further electroporated with a plasmid expressing Cre recombinase to delete the puromycin-resistant cassette. The resultant colonies were picked up in duplicate and puromycin-sensitive clones were expanded. The absence of the cassette was verified by PCR, and clones no. 1-5-9-1 and no. 1-5-9-10 were used for detailed analyses. At days 0–1, BMP4 (3 ng/mL) was added to optimize the percentage of the ITGA8+/PDGFRA− nephron progenitor fraction and the extent of nephrogenesis. Both clones showed comparable recovery of NEPHRIN localization.
Author Contributions
Conceptualization, R.N. and A.T.; Induction and Analysis of iPSC-Derived Podocytes, S.T.; Analysis of HEK293 Cells, M.I.; iPSC Establishment and Genetic Correction, S.S., F.H., and T.E.; Transplantation, H.N.; Protocol Optimization for iPSC Differentiation: Y.Y.; Patient Mutation Identification, H.N. and K.N.; TALEN Construction, T.S. and T.Y.; Electron Microscopy, H.K.; Manuscript Writing, S.T., M.I., A.T., and R.N.; Funding Acquisition, R.N.; Project Administration, R.N.
Acknowledgments
We thank K. Tryggvason for providing the anti-NEPHRIN antibodies (48E11 and 50A9), S. Okada for providing the immunodeficient mice, and T. Ohmori, S. Fujimura, K. Miike, and Y. Soejima for technical assistance. We also thank Alison Sherwin, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. The study was supported in part by a KAKENHI grant (JP17H06177) from the Japan Society for the Promotion of Science, a grant from the Research Center Network for Realization of Regenerative Medicine, Japan Agency for Medical Research and Development (AMED), and a grant from the Takeda Science Foundation.
Published: August 30, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.08.003.
Supplemental Information
References
- Bantounas I., Ranjzad P., Tengku F., Silajdžić E., Forster D., Asselin M.C., Lewis P., Lennon R., Plagge A., Wang Q. Generation of functioning nephrons by implanting human pluripotent stem cell-derived kidney progenitors. Stem Cell Reports. 2018;10:766–779. doi: 10.1016/j.stemcr.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltcheva O., Martin P., Lenkkeri U., Tryggvason K. Mutation spectrum in the nephrin gene (NPHS1) in congenital nephrotic syndrome. Hum. Mutat. 2001;17:368–373. doi: 10.1002/humu.1111. [DOI] [PubMed] [Google Scholar]
- Chittiprol S., Chen P., Petrovic-Djergovic D., Eichler T., Ransom R.F. Marker expression, behaviors, and responses vary in different lines of conditionally immortalized cultured podocytes. Am. J. Physiol. Renal Physiol. 2011;301:F660–F671. doi: 10.1152/ajprenal.00234.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colls P., Blanco J., Martínez-Pasarell O., Vidal F., Egozcue J., Márquez C., Guitart M., Templado C. Chromosome segregation in a man heterozygous for a pericentric inversion, inv(9)(p11q13), analyzed by using sperm karyotyping and two-color fluorescence in situ hybridization on sperm nuclei. Hum. Genet. 1997;99:761–765. doi: 10.1007/s004390050444. [DOI] [PubMed] [Google Scholar]
- Donoviel D.B., Freed D.D., Vogel H., Potter D.G., Hawkins E., Barrish J.P., Mathur B.N., Turner C.A., Geske R., Montgomery C.A. Proteinuria and perinatal lethality in mice lacking NEPH1, a novel protein with homology to NEPHRIN. Mol. Cell. Biol. 2001;21:4829–4836. doi: 10.1128/MCB.21.14.4829-4836.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdova T., Papillon J., Cybulsky A.V. Nephrin missense mutations: induction of endoplasmic reticulum stress and cell surface rescue by reduction in chaperone interactions. Physiol. Rep. 2013;1:e00086. doi: 10.1002/phy2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman B.S., Brooks C.R., Lam A.Q., Fu H., Morizane R., Agrawal V., Saad A.F., Li M.K., Hughes M.R., Werff R.V. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke P., Huber T.B., Sellin L., Benzing T., Walz G. Homodimerization and heterodimerization of the glomerular podocyte proteins NEPHRIN and NEPH1. J. Am. Soc. Nephrol. 2003;14:918–926. doi: 10.1097/01.asn.0000057853.05686.89. [DOI] [PubMed] [Google Scholar]
- Jones N., Blasutig I.M., Eremina V., Ruston J.M., Bladt F., Li H., Huang H., Larose L., Li S.S., Takano T. Nck adaptor proteins link NEPHRIN to the actin cytoskeleton of kidney podocytes. Nature. 2006;440:818–823. doi: 10.1038/nature04662. [DOI] [PubMed] [Google Scholar]
- Kaku Y., Taguchi A., Tanigawa S., Haque F., Sakuma T., Yamamoto T., Nishinakamura R. PAX2 is dispensable for in vitro nephron formation from human induced pluripotent stem cells. Sci. Rep. 2017;7:4554. doi: 10.1038/s41598-017-04813-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestilä M., Lenkkeri U., Männikkö M., Lamerdin J., McCready P., Putaala H., Ruotsalainen V., Morita T., Nissinen M., Herva R. Positionally cloned gene for a novel glomerular protein – nephrin – is mutated in congenital nephrotic syndrome. Mol. Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Refaeli I., Brooks C.R., Jing P., Gulieva R.E., Hughes M.R., Cruz N.M., Liu Y., Churchill A.J., Wang Y. Gene-edited human kidney organoids reveal mechanisms of disease in podocyte development. Stem Cells. 2017;35:2366–2378. doi: 10.1002/stem.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara H., Harita Y., Ichimura K., Hattori S., Sakai T. SIRP-α-CD47 system functions as an intercellular signal in the renal glomerulus. Am. J. Physiol. Renal Physiol. 2010;299:F517–F527. doi: 10.1152/ajprenal.00571.2009. [DOI] [PubMed] [Google Scholar]
- Lenkkeri U., Männikkö M., McCready P., Lamerdin J., Gribouval O., Niaudet P., Antignac C., Kashtan C.E., Holmberg C., Olsen A. Structure of the gene for congenital nephrotic syndrome of the Finnish type (NPHS1) and characterization of mutations. Am. J. Hum. Genet. 1999;64:51–61. doi: 10.1086/302182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström N.O., McMahon J.A., Guo J., Tran T., Guo Q., Rutledge E., Parvez R.K., Saribekyan G., Schuler R.E., Liao C. Conserved and divergent features of human and mouse kidney organogenesis. J. Am. Soc. Nephrol. 2018;29:785–805. doi: 10.1681/ASN.2017080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Doné S.C., Khoshnoodi J., Bertorello A., Wartiovaara J., Berggren P.O., Tryggvason K. Defective NEPHRIN trafficking caused by missense mutations in the NPHS1 gene: insight into the mechanisms of congenital nephrotic syndrome. Hum. Mol. Genet. 2001;10:2637–2644. doi: 10.1093/hmg/10.23.2637. [DOI] [PubMed] [Google Scholar]
- Mundel P., Reiser J., Borja A.Z.M., Pavenstädt H., Davidson G.R., Kriz W., Zeller R. Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp. Cell Res. 1997;236:248–258. doi: 10.1006/excr.1997.3739. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Taniguchi Y., Senda S., Takizawa N., Ichisaka T., Asano K., Morizane A., Doi D., Takahashi J., Nishizawa M. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 2014;4:3594. doi: 10.1038/srep03594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New L.A., Martin C.E., Scott R.P., Platt M.J., Keyvani Chahi A., Stringer C.D., Lu P., Samborska B., Eremina V., Takano T. NEPHRIN tyrosine phosphorylation is required to stabilize and restore podocyte foot process architecture. J. Am. Soc. Nephrol. 2016;27:2422–2435. doi: 10.1681/ASN.2015091048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Harada H., Ito T., Saito T., Suzu S. Early development of human hematopoietic and acquired immune systems in new born NOD/Scid/Jak3 null mice intrahepatic engrafted with cord blood-derived CD34+ cells. Int. J. Hematol. 2008;88:476–482. doi: 10.1007/s12185-008-0215-z. [DOI] [PubMed] [Google Scholar]
- Patrakka J., Tryggvason K. NEPHRIN – a unique structural and signaling protein of the kidney filter. Trends Mol. Med. 2007;13:396–403. doi: 10.1016/j.molmed.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Patrakka J., Kestilä M., Wartiovaara J., Ruotsalainen V., Tissari P., Lenkkeri U., Männikkö M., Visapää I., Holmberg C., Rapola J. Congenital nephrotic syndrome (NPHS1): features resulting from different mutations in Finnish patients. Kidney Int. 2000;58:972–980. doi: 10.1046/j.1523-1755.2000.00254.x. [DOI] [PubMed] [Google Scholar]
- Putaala H., Soininen R., Kilpeläinen P., Wartiovaara J., Tryggvason K. The murine NEPHRIN gene is specifically expressed in kidney, brain and pancreas: inactivation of the gene leads to massive proteinuria and neonatal death. Hum. Mol. Genet. 2001;10:1–8. doi: 10.1093/hmg/10.1.1. [DOI] [PubMed] [Google Scholar]
- Quaggin S.E., Kreidberg J.A. Development of the renal glomerulus: good neighbors and good fences. Development. 2008;135:609–620. doi: 10.1242/dev.001081. [DOI] [PubMed] [Google Scholar]
- Ruotsalainen V., Ljungberg P., Wartiovaara J., Lenkkeri U., Kestilä M., Jalanko H., Holmberg C., Tryggvason K. NEPHRIN is specifically located at the slit diaphragm of glomerular podocytes. Proc. Natl. Acad. Sci. USA. 1999;96:7962–7967. doi: 10.1073/pnas.96.14.7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruotsalainen V., Patrakka J., Tissari P., Reponen P., Hess M., Kestilä M., Holmberg C., Salonen R., Heikinheimo M., Wartiovaara J. Role of NEPHRIN in cell junction formation in human nephrogenesis. Am. J. Pathol. 2000;157:1905–1916. doi: 10.1016/S0002-9440(10)64829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruotsalainen V., Reponen P., Khoshnoodi J., Kilpeläinen P., Tryggvason K. Monoclonal antibodies to human NEPHRIN. Hybrid. Hybridomics. 2004;23:55–63. doi: 10.1089/153685904322772033. [DOI] [PubMed] [Google Scholar]
- Sakuma T., Ochiai H., Kaneko T., Mashimo T., Tokumasu D., Sakane Y., Suzuki K., Miyamoto T., Sakamoto N., Matsuura S. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci. Rep. 2013;3:3379. doi: 10.1038/srep03379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M.A., O’Hare M.J., Reiser J., Coward R.J., Inward C.D., Farren T., Xing C.Y., Ni L., Mathieson P.W., Mundel P. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 2002;13:630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- Schell C., Wanner N., Huber T.B. Glomerular development – shaping the multi-cellular filtration unit. Semin. Cell Dev. Biol. 2014;36:39–49. doi: 10.1016/j.semcdb.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Sharmin S., Taguchi A., Kaku Y., Yoshimura Y., Ohmori T., Sakuma T., Mukoyama M., Yamamoto T., Kurihara H., Nishinakamura R. Human induced pluripotent stem cell-derived podocytes mature into vascularized glomeruli upon experimental transplantation. J. Am. Soc. Nephrol. 2016;27:1778–1791. doi: 10.1681/ASN.2015010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga M., Ishitsuka Y., Hamasaki M., Yoneda K., Furuya H., Matsuo M., Ihn H., Fusaki N., Nakamura K., Nakagata N. HPGCD outperforms HPBCD as a potential treatment for Niemann-Pick disease type C during disease modeling with iPS cells. Stem Cells. 2015;33:1075–1088. doi: 10.1002/stem.1917. [DOI] [PubMed] [Google Scholar]
- Taguchi A., Nishinakamura R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell. 2017;21:730–746. doi: 10.1016/j.stem.2017.10.011. [DOI] [PubMed] [Google Scholar]
- Taguchi A., Kaku Y., Ohmori T., Sharmin S., Ogawa M., Sasaki H., Nishinakamura R. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tokuyasu K.T. Use of poly(vinylpyrrolidone) and poly(vinyl alcohol) for cryoultramicrotomy. Histochem. J. 1989;21:163–171. doi: 10.1007/BF01007491. [DOI] [PubMed] [Google Scholar]
- Tucker K.L., Wang Y., Dausman J., Jaenisch R. A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 1997;25:3745–3746. doi: 10.1093/nar/25.18.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y., Taguchi A., Nishinakamura R. Generation of a three-dimensional kidney structure from pluripotent stem cells. Methods Mol. Biol. 2017;1597:177–193. doi: 10.1007/978-1-4939-6949-4_13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.