Abstract
Regular exercise is an exogenous factor of gene regulation with numerous health benefits. The study aimed to evaluate human genes linked to physical exercise in an ‘omic scale, addressing biological questions to the generated database. Three literature databases were searched with the terms ‘exercise’, ‘fitness’, ‘physical activity’, ‘genetics’ and ‘gene expression’. For additional references, papers were scrutinized and a text-mining tool was used. Papers linking genes to exercise in humans through microarray, RNA-Seq, RT-PCR and genotyping studies were included. Genes were extracted from the collected literature, together with information on exercise protocol, experimental design, gender, age, number of individuals, analytical method, fold change and statistical data. The ‘omic scale dataset was characterized and evaluated with bioinformatics tools searching for gene expression patterns, functional meaning and gene clusters. As a result, a physical exercise-related human gene compendium was created, with data from 58 scientific papers and 5.147 genes functionally correlated with 17 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. While 50.9% of the gene set was up-regulated, 41.9% was down-regulated. 743 up- and 530 down-regulated clusters were found, some connected by regulatory networks. To summarize, up- and down-regulation was encountered, with a wide genomic distribution of the gene set and up- and down-regulated clusters possibly assembled by functional gene evolution. Physical exercise elicits a widespread response in gene expression.
Keywords: Physical exercise transcriptome, Molecular exercise physiology, Genes linked to physical exercise, Exercise gene ontology, Exercise genomics
INTRODUCTION
A large number of molecular signalling mechanisms have been linked to the adaptations in skeletal muscle by regular exercise [1]. Earlier studies in the fields of human exercise genomics identified polymorphisms, mainly single nucleotide polymorphisms (SNPs), linked to elite athletes [2]. However, while the genome holds the information, the expressed gene set defines the metabolic responses in the biological systems. Endogenous and exogenous gene expression regulation can modulate the production of mRNA and proteins, eventually leading to physiological changes [3]. Regular exercise is an exogenous gene regulation factor with numerous health benefits [1], whose organismal multiple regulatory systems can be visible at the ‘omic level.
Timmons [4] stated that “genes work in complex, nonlinear, redundant networks, and thus we need to take a fresh approach to molecular physiology”. Bouchard [5] recommended that exercise genomics studies use the power of genomics, epigenomics and transcriptomics, using computational biology and bioinformatics expertise. A wider, ‘omic scale, view on molecular exercise physiology is needed. With lowering costs of large-scale RNA analyses, researchers are publishing large data sets of genes linked to different categories of exercise (endurance / mixed / power), evaluating distinct groups of individuals (males / females; young adults / middle aged / elderly). Analysing gene expression data on a large scale, ‘omic screening methods can point towards biological molecules responsible for physiological alterations.
Most of the recent genome-wide gene expression studies in the physical science area are based on microarrays [6]. This technique requires previous knowledge on the genetic targets but can be used for several purposes. Multi-factorial approaches could be used, such as sex-specific transcription [7], the ageing body’s response associated with endurance exercise [8], chronic strength training [9], and exercise aerobic capacity [10].
The ever decreasing cost of sequencing is bringing an explosion of genome-wide sequencing studies [6]. RNA-Seq is a methodology where all the mRNA in the cell is sequenced and quantified. However, in exercise molecular physiology, studies using second generation sequencing techniques are still incipient. Lindholm and colleagues used RNA-Seq to obtain exercise transcriptome associated with DNA methylation in skeletal muscle in males and females who underwent chronic endurance training. They attempted to understand the causal relationship between methylation and transcription in relation to physical exercise. Significant differential expression was observed in approximately 4000 genes [11, 12].
Enrichment of the pathway-associated gene network arising from massive RNA expression data could help to identify genetic variance for different physical training phenotypes from a biotechnological perspective. Based on this premise, we propose the Fitnome Catalogue (FitC), aiming to aggregate statistically relevant systematic information in order to evaluate genes linked to physical exercise in the context of multiple different studies of microarrays, RNA-Seq, RT-PCR, as well as genotyping studies. Gene expression data are usually shown as fold changes, a measure of differential expression, when two physiological situations are compared. A positive sign reveals that the treated individual has an expression level above the control sample; this up-regulation shows that the gene is activated by the studied situation (e.g. disease, drug treatment, physical stimulus). A negative fold change indicates a lower expression level when compared to control samples; this down-regulation shows that expression is being suppressed. A fold change of 3 linked to acute resistance exercise, for example, means that the RNA for that particular gene was found to be three times more concentrated after one session of resistance exercise. These and other genomic data were used as a basis to study aspects of: (a) spatial distribution within the human genome, (b) the presence and regulation of gene clusters, and (c) functional networks affected by the stimulus. In order to perform such analyses, bioinformatics tools were used in an effort to bring biotechnological advances to physical exercise evaluation.
MATERIALS AND METHODS
Genes linked to exercise were gathered from research papers identified by a systematic review using PubMed, PMC and Google Scholar as literature databases and the following search terms: ‘exercise’, ‘fitness’, ‘physical activity’, ‘genetics’, and ‘gene expression’. Papers were also manually investigated for additional references, and a text-mining approach (Genie [13]) was used to search for additional research papers linking human genes to physical exercise. As eligibility criteria, we selected papers linking genes to exercise in humans through microarray, RNA-Seq, RT-PCR and genotyping studies. Duplicates were removed using Microsoft Excel, using functions such as “==” and “IF”. The following data were collected from the studies: exercise protocol, experimental design (acute/chronic), gender, age range, number of individuals tested, analytical method, gene symbol, differential expression data, statistical significance, and reference. As an ‘omic approach, all reported genes linked to physical exercise, not restricting to statistically significant correlations, were incorporated in the study. Around 60 papers linking genes to exercise in humans were used in order to compose the Fitnome gene set (Supplementary Table S1).
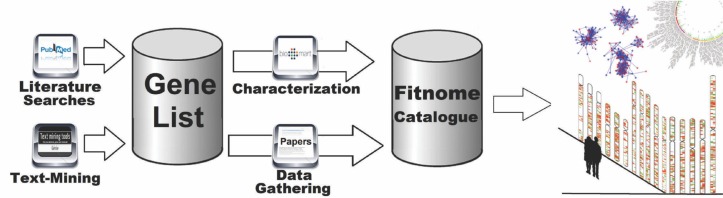
A wide variety of bioinformatic tools addressing researchers’ specific needs have been made available recently. Using freely available bioinformatic tools and techniques, the data-set was characterized and inspected in search of gene expression patterns, functional meaning and possible gene clusters (Figure 1).
FIG. 1.
Study pipeline.
Information on genetic architecture and gene product type was collected with Ensembl’s BioMart MartView tool (http://biomart.org/biomart/martview) [14], generating a spreadsheet with gene attributes. Meanwhile, study design and gene expression data were gathered from research papers manually, and another spreadsheet was produced, this one with information associating them with physical exercise. When one gene was reported as linked to exercise more than once, gene expression and statistical data were integrated by sequential insertion in the Excel cell. Data from the two produced spreadsheets were merged using Microsoft Excel’s decision tools and functions, such as == and IF. The gene set was evaluated with EnrichNet (www.enrichnet.org) [15], a web-based enrichment tool that uses an integrative analysis approach in the evaluation of pathways in which the gene set plays a role. We chose to classify the gene set using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [16]. For the analysis, the metabolic pathways were considered linked to physical exercise when there were relevant XD scores (association scores between the gene set and pathways >1.06) and significant q-values (<0.05).
After reorganizing the data to the appropriate format, a gene map was produced using Idiographica (www.ncrna.org/idiographica) [17]. Visualizing the idiogram, it was decided to evaluate the occurrence of clustering within the gene set. Co-localization analyses were performed using Kerfuffle, a web-based tool designed to obtain genomic organization data, discovering clusters within gene sets in the selected species (www.atwallab.org/kerfuffle) [18]. In this analysis, we evaluated two gene sets (up- and down-regulated) in humans, with the p-value defined as <0.01 and considering clusters of two or more genes. The resulting clusters were shown in circular idiograms, produced with Circos [19], through an interaction between the two platforms.
Co-localized genes were further analysed over aspects of molecular interactions that may affect transcription and translation using GeneMania (www.genemania.org) [20], with the following advanced options: Homo sapiens genes and transcriptional-factor-targets-2013 networks. For this bioinformatics tool, the physical interaction described is based on protein-protein interaction data collected from primary studies found in protein interaction databases, including BioGRID (www.thebiogrid.org) and Pathway Commons (www.pathwaycommons.org). Genetic interaction data were also analysed where two genes are functionally associated if the effects of one gene were found to alter expression of a second gene. These data are collected from primary studies and from BioGRID. All bioinformatic analyses were based on human genome assembly GRCh38, and the number of annotated genes was obtained by browsing Ensembl on 07/04/2015 [21]. The full name (and other information mentioned) of all Fitnome genes cited throughout the text can be found in Supplementary Table S1.
RESULTS
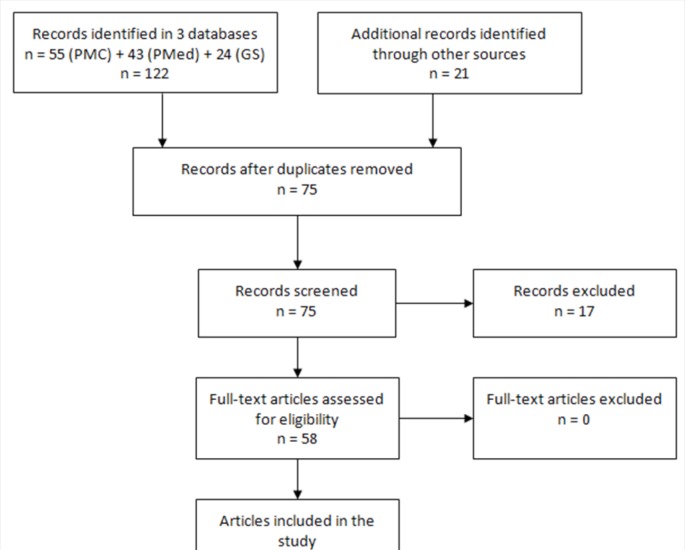
After filtering through 122 records in three scientific literature databases (PMC, Pub Med and Google Scholar), 75 papers were screened and 58 were included in the study (2003 to 2014 period) (Figure 2).
FIG. 2.
Summary of systematic review results
Extracting the information from these papers, physical exercise was linked to 5,147 gene accession numbers in data obtained from more than 4500 individuals in several countries. Supplementary material includes information on nomenclature (gene symbol and full official name), Ensembl ID, location (chromosome, band, strand, position) and transcript count as well as genetic expression data of these 5,147 genes, totalling 101,262 records.
The resulting spreadsheet showed the preferential use of aerobic and chronic exercise protocols in research projects (Table 1, Supplementary Table S1). This could occur due to easier management of experimental variables, such as exercise volume and intensity, or to the impact this type of exercise has on cardiovascular health [22], for example. The studies spanned all age ranges, from children to elderly subjects.
TABLE 1.
Physical exercise literature used for FitC characterization: exercise protocol, experimental design and sex of participants.
| Category | Characteristic | Number of genes |
|---|---|---|
| Exercise protocol | Endurance Resistance Resistance & endurance |
4497 158 401 |
| Exercise experimental design | Chronic Acute Chronic & acute |
4128 226 159 |
| Sex | Males Females Males & females |
583 06 3774 |
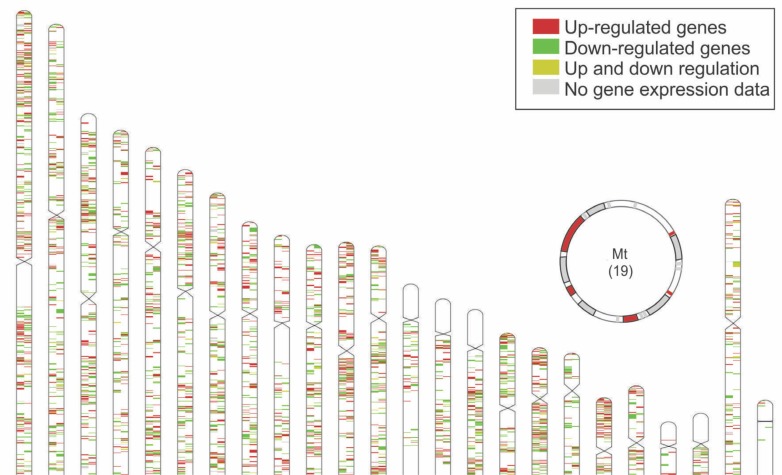
This gene set corresponded to 8.63% of all known human genes, and within the protein coding gene category the FitC was even more highly represented, corresponding to 23.66% of the total number of human genes in the current genomic databases (Human Genome Assembly GRCh38) [21]. Regarding gene biotype, most of the exercise-related genes in the catalogue were protein-coding (4803 genes), but there were also 190 long non-coding, 75 short non-coding, 77 pseudogenes and 2 not yet classified (FLJ42393 and GABARAPL3). FitC genes are distributed across the genome, with most genes located in the autosomes, 182 on the X, 11 on the Y chromosome and 19 mitochondrial genes (Figure 3).
FIG. 3.
Location of human genes related to exercise. In brackets are the numbers of Fitnome genes in each chromosome. The mitochondrial genome (Mt) is depicted in the upper portion of the figure. Different shades represent gene expression data.
Evaluating gene expression data after exercise, it could be observed that while 50.9% of the gene set was up-regulated, 41.9% of the genes were found to be down-regulated. The most extreme fold changes were recorded for myosin binding protein H (MYBPH, 50.5 times more expressed) [7], early growth response 1 (EGR1, up to 40.9 times over-expressed after exercise) [7], PDK4 (over-expressed 18 times) [23], CDK2 (up-regulated 14.29 times) [9] and MIR1270, down-regulated 11.5 times through exercise [24].
Chromosomal co-localization analyses of 2 subsets of the FitC, dividing the gene set into up-regulated and down-regulated genes, revealed 743 up-regulated clusters and 503 down-regulated clusters (Supplementary Figure 1). Most of the up-regulated clusters were composed of two genes, but there were also 178 clusters with three, 43 with four and 33 clusters with five or more genes (Supplementary Figure 1A). Within the down-regulated genes, 371 clusters with two genes were detected, 93 with three, 28 with four and 11 clusters with five or more genes (Supplementary Figure 1B). Some of the largest FitC clusters are further scrutinized in the Discussion section.
In a quest for functional meaning in all these data, enrichment analyses revealed that FitC genes were significantly correlated with 17 KEGG pathways (KPs) (Table 2). KEGG hierarchical levels are layers of pathways organized in stratified functional relationships [16]. The statistically significant KPs, classification and coverage can be seen in Table 2, the latter expressed as percentage of genes in the pathway linked to exercise, as well as the proportion of up- and down-regulation.
TABLE 2.
Functional classification of FitC pathways. Metabolic pathways placed in hierarchical levels with statistical coverage and percentage of FitC genes in the KEGG pathways (KPs). Expression level is expressed as percentage of up-regulated (↑), down-regulated (↓) and genes with up- and down-regulation (↕).
| 1st hierarchical KEGG level | 2nd hierarchical KEGG level | KEGG Pathway (pathway ID) | Fisher test q-value | % of pathway genes | % Up/Down expression |
|---|---|---|---|---|---|
| Metabolism | Amino acid metabolism | Valine, leucine and isoleucine degradation (hsa00280) | 1.8X10-6 | 70 | ↑81 ↓06 ↕13 |
| Arginine and proline metabolism (hsa00330) | 8.6 X10-3 | 52 | ↑56 ↓30 ↕11 |
||
| Carbohydrate metabolism | Citrate cycle (TCA cycle) (hsa00020) | 1.6 X10-4 | 70 | ↑100 | |
| Propanoate metabolism (hsa00640) | 2.4 X10-3 | 63 | ↑75 ↓15 ↕10 |
||
| Pyruvate metabolism (hsa00620) | 4.4 X10-2 | 50 | ↑80 ↓15 ↕05 |
||
| Energy metabolism | Oxidative phosphorylation (hsa00190) | 3.9 X10-16 | 68 | ↑92 ↓02 ↕00 |
|
| Lipid metabolism | Fatty acid metabolism (hsa00071) | 2.4 X10-3 | 59 | ↑63 ↓13 ↕25 |
|
| Organismal Systems | Circulatory system | Cardiac muscle contraction (hsa04260) | 1.7 X10-7 | 64 | ↑66 ↓21 ↕02 |
| Endocrine system | PPAR signaling pathway (hsa03320) | 1.3 X10-4 | 57 | ↑44 ↓15 ↕26 |
|
| Human Diseases | Cardiovascular diseases | Viral myocarditis (hsa05416) | 8.5 X10-5 | 57 | ↑65 ↓28 ↕07 |
| Hypertrophic cardiomyopathy (hsa05410) | 8.5 X10-5 | 55 | ↑49 ↓33 ↕11 |
||
| Immune diseases | Allograft rejection (has05330) | 1.8 X10-2 | 54 | ↑55 ↓15 ↕15 |
|
| Neurodegenerative diseases | Parkinson’s disease (hsa05012) | 1.6 X10-17 | 70 | ↑86 ↓07 ↕01 |
|
| Alzheimer’s disease (hsa05010) | 3.9 X10-16 | 64 | ↑79 ↓12 ↕04 |
||
| Huntington’s disease (hsa05016) | 1.9 X10-15 | 61 | ↑78 ↓10 ↕07 |
||
| Environmental Information Processing | Signal transduction | mTOR signaling pathway (hsa04150) | 1.4 X10-4 | 61 | ↑19 ↓55 ↕23 |
| Notch signaling pathway (hsa04330) | 1.9 X10-2 | 51 | ↑54 ↓38 ↕08 |
KPs of FitC genes in the respective pathways were mostly up-regulated by exercise rather than down. FitC genes linked to the TCA cycle (hsa00020), for example, were 100% up-regulated. Only one KP in Table 2 was mostly down-regulated: the mTOR signalling pathway (hsa04150).
DISCUSSION
FitC data suggest that ‘omic-scale physical exercise studies need to be more extensive for a complex and multi-factorial process where all biological animal systems contribute. For the animal genome evolution, the contractile function uses systems and tissues with an inherent ability of adaptation for the organism, providing the power of locomotion and life-sustaining processes. In animal locomotion capacity, the contractile function and physical exercise phenotype has evolved by activation of a subset and repression of another subset of genes. Global alterations and muscular adaptations produced in response to regular exercise are modulated by gene expression events [25].
Genes and transcripts are classified into several biotypes: protein coding, pseudogene, long non-coding and short non-coding [21]. Exercise-related genes were found to be protein-coding, long non-coding, short non-coding, pseudogenes and genes not yet classified. The vast majority were found to be protein-coding; however, this enrichment of protein-coding genes could also reflect a bias in detection levels of certain gene biotypes, such as short non-coding genes, by the assays. Since protein synthesis is intrinsically related to exercise, the studies tend to emphasize protein-coding genes, perhaps leaving others aside.
Quantitative expression and genetic location
Assessing gene expression data after exercise, it could be observed that 50.9% of the gene set was up-regulated and 41.9% was down-regulated. These data suggest that gene transcription activation is slightly more common than repression following exercise. Some genes were noted to be extremely affected, with 36 showing up- or down-regulation by 5 times or over. 4.8% of the genes showed conflicting evidence, with up- and down-regulation detected. The latter indicates that other factors are involved, which is not surprising given the multi-factorial and complex nature of gene expression. For 2.4% of the gene set (119 genes) there were no gene expression data linked to exercise through genotyping studies. In Supplementary Table S1 these genes are clearly indicated.
MYBPH and EGR1 are outliers, with extreme up-regulation in the present analysis, and have remarkable remodelling functions. Together with CDK2, also up-regulated, they showed a typical frame of adaptive modifications of physical exercise. These proteins have been targets for extensive physical exercise and sports literature [4, 7, 9, 10]. MYBPH protein is a myosin filament with function not yet well characterized, but associated with muscular contractility [26], elevated expression in cardiac hypertrophy and a muscular regeneration premature signal [27]. Muscular regeneration is also associated with EGR1 protein, which is a transcription regulatory factor, in response to stress signals [28], such as the increase of muscle contractile activity stimulating muscular regeneration and mitochondrial biogenesis [29]. PDK4 expression is related to pyruvate dehydrogenase, a key step to the glucose input in the TCA cycle and the lipid utilization in energetic dynamics [30], also a classical adaptation to aerobic exercise. Increased gene expression of PDK4 is predominant in the oxidative skeletal muscle [31]. CDK2 is a member of the serine/threonine protein kinase family that participates in cell cycle regulation, specially the G1-S phase involved in cell and tissue development, an important regulator of the eukaryotic cell cycle [32].
The mitochondrial genome has 37 genes, with 19 linked to physical exercise: 6 up-regulated and 13 genes linked by genotyping studies. Mitochondria encode tRNA and rRNA genes, as well as genes linked to oxidative phosphorylation, an important pathway for energy metabolism.
FitC genes are widely distributed across the human genome. When observing the gene map (Figure 3), it could be seen that some regions had a higher concentration of exercise-related genes than others. The genes seem to be more densely distributed in some areas of the genome, such as the short arm of chromosome 1, part of chromosome 11’s long arm, and chromosomes 17 and 19. The concept of synteny describes the physical co-localization of genetic loci on the same chromosome within an individual or species. The genetic synteny based on chromosome proximity could be related to biochemical and genetic regulation. Since some agglomerations of genes could be observed with a visual analysis, the presence of co-localized, possibly clustered genes was investigated. Co-regulation of genes, with clustered organization of functionally related genes, in units known as operons, is frequent in prokaryotes, though in eukaryotes much lower numbers of functionally related clusters are observed. Studying the connection between human gene function and localization, Thevenin et al. [33] stated that functionally related genes are aggregated, both within chromosomes and in space.
Evaluating co-localization within the chromosomes of two subsets of the FitC revealed 743 up-regulated clusters and 503 down-regulated clusters (Supplementary Figure 1). Most up-regulated clusters were composed of a few genes, but there were clusters with up to eight genes, such as the one in chromosome 11 (TRPT1, DNAJC4, NUDT22, VEGFB, FKBP2, PPP1R14B, PLCB3, and GPR137), here referred to as Fitnome cluster 1 (FCL1, Supplementary Figure 1A). This cluster was found to be co-expressed, corroborating data in the present study. The network within genes in the clusters probably occurs as result of indirect relations mediated by transcription factors, as shown by GeneMania evaluations [20], based on data from Molecular Signature Database and Gene Set Enrichment Analysis databases tools. The genes FFBP2, VEGFB, DNAJC4 and PLCB3 were physically clustered and possible targets of a few transcription factors: GTF3A, MAZ and VDR. Some of these transcription factors recognize genes with promoter regions near (-2kb, 2kb) transcription start sites containing the specific gene motif GSCCSCRGGCNRNRNN (GTF3A) or GGGKNARNRRGGWSA (VDR). Both of these transcription factors regulate the transcription of PLCB3. Four genes (DNAJC4, FKBP2, VEGFB, and PLCB3) possessed a motif for the same transcription factor, MAZ.
Another exercise-related cluster is FCL2 (Supplementary Figure 1A), found in chromosome 7, cytoband q36, with seven co-localized genes up-regulated by exercise: GIMAP1, GIMAP2, GIMAP4, GIMAP5, GIMAP6, GIMAP8, and TMEM176B. While TMEM176B codes for a transmembrane protein in the nuclear envelope, the proteins encoded by the GIMAP genes belong to the immuno-associated nucleotide subfamily of nucleotide-binding proteins, part of the GTP-binding superfamily. Key cellular functions are regulated by GTP binding proteins, as they are involved in signal transduction mechanisms that regulate gene expression [34]. The activation of these genes by exercise could indicate a molecular link between exercise and a rise in transcription levels. The GIMAP human gene cluster has been known for some time [35], but its association with exercise is novel. The six GIMAP genes were found to be correlated through physical interactions.
Other higher physical interactions percentages were obtained in five of six gene clusters of six genes each: 64.66% (MRPL39, JAM2, ATP5J, APP, CYYR1, and ADAMTS1); 63.14% (PRDM1, AIM1, RTN4IP1, QRSL1, C6orf203, and PDSS2); 60.26% (LXN, GFM1, RARRES1, MFSD1, IQCJ-SCHIP1, and SCHIP1); 55.84% (COLQ, BTD, HACL1, ANKRD28, RFTN1, and OXNAD1); 27.88% (CCL22, CX3CL1, COQ9, CIAPIN1, DOK4, and CCDC102A). Genetic interaction showed lower values in these sets of genes: 0.92%; 2.25%; 2.17%; 2.14%; 1.99% and 0.99% respectively. Containing genes in multiple pathway functions, these up-regulated clusters apparently depict post-translational control of regulation as well as chromosomal co-localization.
Considering now the down-regulated genes, this research revealed around 500 clusters ranging from two to six co-localized genes. One of these clusters (FCL3, Supplementary Figure 1B) was located in chromosome 10, cytoband q23, comprising: ANKRD1, PCGF5, HECTD2, TNKS2, BTAF1, and CPEB3. All except PCGF5 were found to be correlated through co-expression and were linked by a transcription factor network. Transcription of ANKRD1, CPEB3 and BTAF1 can be mediated by the same transcription factor, RAB11. All genes in this cluster, except ANKRD1, were also connected by a miRNA regulatory network, where Mir-524 regulates TNKS2 and CPEB3, Mir-202 regulates CPEBP3 and HECT2, Mir-23a/23b interact with BTAF1 and TNKS2 and Mir-103/107 target CREBP3 and PCGF5. This evidence points to an intricate relationship between these genes, since they are co-localized and seem to be related in terms of transcription and translation regulation.
Functional aspects of physical exercise metabolism
The capacity of exercise in gene expression activation is probably due to the multiple metabolic demands of the organism. Physical exercise increases protein turnover, owing to the renovation overcompensation of structural proteins, damaged by muscular contraction’s mechanical stimuli [4]. De novo production of enzymes to meet energetic demand and signalling pathways between tissues, organs and systems in response to the immediate and adaptive regulation to exercise occurs, in a global view, through structural interactions [1, 36]. However, physical exercise does not lead only to increases in protein production or up-regulation of the genome; the response to this environmental stimulus is an organic homeostasis-directed system for complex events of genetic activation and inhibition [9, 11, 12]. Structural alterations lead to compensatory skeletal muscular hypertrophy and a rise in protein levels through antagonistic but complementary signalling pathways [37]. Physical exercise is a stimulator for the adaptive response resulting from up- and down-regulation of genes and metabolic pathways [9, 11].
FitC genes were significantly correlated with 17 KPs (Table 2), with pathways mostly up-regulated by exercise, the TCA cycle being an example with 100% up-regulation. Amino acid, carbohydrate, energy and lipid metabolism are particularly well-defined pathways and exercise produces alterations in protein and amino acid metabolism [38]. All seven pathways linked to exercise and metabolism were mostly up-regulated by physical exercise. Global muscle has a large proportion (40%) of the total protein in the adult human body and contributes in a great proportion to the protein turnover, including oxidation and the flow of amino acids, particularly of the branched chain amino acids such as valine and leucine (KP: hsa00280). Most of these metabolic pathways show impressive up-regulation induced by physical exercise.
Numerous metabolic pathways of glycolysis (KP: hsa00190) cover thousands of chemical transformations including the oxidative capacity of the body. This oxidative capacity is also a mitochondrial function, increased by the peroxisome proliferator-activated receptor (PPAR) signalling pathway (KP: hsa03320) [39], and is modulated by arginine and proline metabolism [40].
Showing higher up-regulation after physical activity, as already described in the relevant literature, TCA metabolism (KP: hsa00020 / hsa00620) is the major final common aerobic pathway for oxidation of carbohydrates and fatty acids. In the TCA intermediary pool are functional molecules necessary for the exercising human skeletal muscle. The TCA cycle is encoded by 44 genes, 31 being up-regulated by exercise, with no down-regulation observed after this stimulus in this path [41].
Target of rapamycin signalling (KP: hsa04150) is a conserved Ser/Thr kinase, an enzyme that regulates cell growth and tissue plasticity in response to hormones, environmental stress, nutrients, energy and other stimuli such as exercise [42].
Some human disease pathways were found to be strongly linked to the FitC, mainly related to cardiac (myocarditis, KP: hsa05416; hypertrophic cardiomyopathy, KP: hsa05410) and neural functions (Parkinson’s disease, KP: hsa05012; Alzheimer’s disease, KP: hsa05010; Huntington’s disease, KP: hsa05016). Global analysis of differential gene expression showed a new overlap between physical exercise and some important neurodegenerative and cardiac diseases. Genes with a high degree of differential expression after exercise linked to cardiac function (e.g. PRKAG2, HLA-DQA1 and CCND1) and neurodegenerative diseases (e.g. PPARGC1A, NDUFB3 and ITPR1) are potential targets for studies focusing on the molecular interactions between physical exercise and these afflictions.
FitC analysis showed enrichment, high q-values and a large quantity of genes belonging to metabolic pathways with strong up-regulation. The catalogue can serve as a starting point for the development of new biodiagnostic methods based on gene expression patterns for the sports biotechnology field.
CONCLUSIONS
Physical exercise incites a widespread response in the human body, mainly in the expression of protein-coding genes. Within the 5,147 exercise-related genes, up- and down-regulation was observed, and some genes seemed to be extremely affected by this stimulus. Using the FitC as a basis to ask physiological questions, gene localization was evaluated, revealing a wide genomic distribution of the gene set as well as a number of up- and down-regulated clusters probably assembled by functional gene evolution. Functional pathways linked to exercise-related genes were mainly related to metabolism, followed by human diseases, organismal systems and environmental information processing. Genetic analyses in this ‘omic scale can open the doors for a new paradigm in physical exercise molecular physiology and biotechnology, with the creation of transcription pattern based training-level tests.
Supplementary material
Supplementary Table S1. The Fitnome catalogue – human genes linked to physical exercise Table_S1_Pacheco_et_al.xlsx (link)
Supplementary Figure 1. Exercise related human co-localized genes. A. Clusters of genes up-regulated by exercise; B. Clusters of genes down-regulated by exercise. (link)
Acknowledgements
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP). The funders had no role in the design of the study, decision to publish or preparation of the manuscript. The authors declare that there are no conflicts of interest.
Conflict of interests
The authors declared no conflict of interests regarding the publication of this manuscript.
REFERENCES
- 1.Coffey VG, Hawley JA. The molecular bases of training adaptation. Sports Med. 2007;37(9):737–63. doi: 10.2165/00007256-200737090-00001. [DOI] [PubMed] [Google Scholar]
- 2.Bray MS, Hagberg JM, Perusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The Human Gene Map for Performance and Health-Related Fitness Phenotypes: The 2006–2007 Update. Med Sci Sports Exerc. 2009;41(1):34–72. doi: 10.1249/mss.0b013e3181844179. [DOI] [PubMed] [Google Scholar]
- 3.Conesa A, Mortazavi A. The common ground of genomics and systems biology. BMC Syst Biol. 2014;8(2):1–10. doi: 10.1186/1752-0509-8-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timmons JA. Variability in training-induced skeletal muscle adaptation. J Appl Physiol. 2011;110:846–53. doi: 10.1152/japplphysiol.00934.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchard C. Exercise genomics – a paradigm shift is needed: a commentary. Br J Sports Med. 2015;49:1492–6. doi: 10.1136/bjsports-2015-095294. [DOI] [PubMed] [Google Scholar]
- 6.Palotie A, Widen E, Ripatti S. From genetic discovery to future personalized health research. N Biotechnol. 2013;30(3):291–5. doi: 10.1016/j.nbt.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Sartor MA, Nader GA, Gutmann L, Treutelaar MK, Pistilli EE, IglayReger HB, Burant CF, Hoffman EP, Gordon PM. Skeletal muscle gene expression in response to resistance exercise: sex specific regulation. BMC Genomics. 2010;11:659. doi: 10.1186/1471-2164-11-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee K, Edgett BA, Burrows HW, Castro C, Griffin JL, Schwertani AG, Gurd BJ, Funk CD. Whole blood transcriptomics and urinary metabolomics to define adaptive biochemical pathways of high-intensity exercise in 50-60 year old masters athletes. PloS One. 2014;9(3):e92031. doi: 10.1371/journal.pone.0092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stepto NK, Coffey VG, Carey AL, Ponnampalam AP, Canny BJ, Powell D, Hawley JA. Global gene expression in skeletal muscle from well-trained strength and endurance athletes. Med Sci Sports Exerc. 2009;41(3):546–65. doi: 10.1249/MSS.0b013e31818c6be9. [DOI] [PubMed] [Google Scholar]
- 10.Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, Keller P, Scheele C, Vollaard NBJ, Nielsen S, Åkerström T, MacDougald OA, Jansson E, Greenhaff PL, Tarnopolsky MA, van Loon LJC, Pedersen BK, Sundberg CJ, Wahlestedt C, Britton SL, Bouchard C. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol. 2010;108:1487–96. doi: 10.1152/japplphysiol.01295.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindholm ME, Marabita F, Gomez- Cabrero D, Rundqvist H, Ekström T, Tegnér J, Sundberg CJ. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics. 2014a;9(12):1557–69. doi: 10.4161/15592294.2014.982445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindholm ME, Huss M, Solnestam BW, Kjellqvist S, Lundeberg J, Sundberg CJ. The human skeletal muscle transcriptome: sex differences, alternative splicing, and tissue homogeneity assessed with RNA sequencing. FASEB J. 2014b;28(10):4571–81. doi: 10.1096/fj.14-255000. [DOI] [PubMed] [Google Scholar]
- 13.Fontaine JF, Priller F, Barbosa-Silva A, Andrade-Navarro MA. Genie: literature-based gene prioritization at multi genomic scale. Nucleic Acids Res. 2011;39:W455–61. doi: 10.1093/nar/gkr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, Kersey P, Flicek P. Ensembl BioMart: a hub for data retrieval across taxonomic space. Database. 2011 doi: 10.1093/database/bar030. bar230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaab E, Baudot A, Krasnogor N, Schneider R, Valencia A. EnrichNet: network-based gene set enrichment analysis. Bioinformatics. 2012;28(18):451–7. doi: 10.1093/bioinformatics/bts389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kin T, Ono Y. Idiographica: a general-purpose web application to build idiograms on-demand for human, mouse and rat. Bioinformatics. 2007;23(21):2945–6. doi: 10.1093/bioinformatics/btm455. [DOI] [PubMed] [Google Scholar]
- 18.Aboukhalil R, Fendler B, Atwal GS. Kerfuffle: a web tool for multi-species gene colocalization analysis. BMC Bioinformatics. 2013;14(1):22. doi: 10.1186/1471-2105-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an Information Aesthetic for Comparative Genomics. Genome Res. 2009;19(9):1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl.1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinxton: European Bioinformatics Institute; Ensembl Web site [Internet] [cited 2015 April 07]. Available from: www.ensembl.org/Homo_sapiens/Info/Annotation. [Google Scholar]
- 22.Baldwin KM. Research in the exercise sciences: where do we go from here? J Appl Physiol. 2000;88(1):332–6. doi: 10.1152/jappl.2000.88.1.332. [DOI] [PubMed] [Google Scholar]
- 23.Psilander N, Wang L, Westergreen J, Tonkonogi M, Sahlin K. Mitochondrial gene expression in elite cyclists: effects of high-intensity interval exercise. Eur J Appl Physiol. 2010;110(3):597–606. doi: 10.1007/s00421-010-1544-1. [DOI] [PubMed] [Google Scholar]
- 24.Chilton WL, Marques FZ, West J, Kannourakis G, Berzins ST, O’Brien BJ, Charchar FJ. Acute Exercise Leads to Regulation of Telomere-Associated Genes and Micro RNA Expression in Immune Cells. Plos One. 2014;9(4):e92088. doi: 10.1371/journal.pone.0092088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fluck M. Functional, structural and molecular plasticity of mammalian skeletal muscle in response to exercise stimuli. J Exp Biol. 2006;209(12):2239–48. doi: 10.1242/jeb.02149. [DOI] [PubMed] [Google Scholar]
- 26.Palstra AP, Rovira M, Rizo-Roca D, Torrella JR, Spaink HP, Planas JV. Swimming-induced exercise promotes hypertrophy and vascularization of fast skeletal muscle fibres and activation of myogenic and angiogenic transcriptional programs in adult zebrafish. BMC Genomics. 2014;15:1136–56. doi: 10.1186/1471-2164-15-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conti A, Riva N, Pesca M, Iannaccone S, Cannistraci CV, Corbo M, Previtali SC, Quattrini A, Alessio M. Increased expression of Myosin binding protein H in the skeletal muscle of amyotrophic lateral sclerosis patients. Biochim Biophys Acta. 2014;1842(1):99–106. doi: 10.1016/j.bbadis.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Fan YY, Ye GH, Lin KZ, Yu LS, Wu SZ, Dong MW, Han JG, Feng XP, Li XB. Time-dependent expression and distribution of Egr-1 during skeletal muscle wound healing in rats. J Mol Hist. 2013;44(1):75–81. doi: 10.1007/s10735-012-9445-8. [DOI] [PubMed] [Google Scholar]
- 29.Irrcher I, Hood DA. Regulation of Egr-1, SRF, and Sp1 mRNA expression in contracting skeletal muscle cells. J Appl Physiol. 2004;97:2207–13. doi: 10.1152/japplphysiol.00388.2004. [DOI] [PubMed] [Google Scholar]
- 30.Sugden MC, Holness MJ. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol Metab. 2003;284:E855–62. doi: 10.1152/ajpendo.00526.2002. [DOI] [PubMed] [Google Scholar]
- 31.Jeong JY, Jeoung NH, Park KG, Lee IK. Transcriptional Regulation of Pyruvate Dehydrogenase Kinase. Diabetes Metab. J. 2012;36:328–35. doi: 10.4093/dmj.2012.36.5.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Zhang J, Gao W, Zhang L, Pan Y, Zhang S, Wang Y. Insights on Structural Characteristics and Ligand Binding Mechanisms of CDK2. Int J Mol Sci. 2015;16(5):9314–40. doi: 10.3390/ijms16059314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thevenin A, Ein-Dor L, Ozery-Flato M, Shamir R. Functional gene groups are concentrated within chromosomes, among chromosomes and in the nuclear space of the human genome. Nucleic Acids Res. 2014;42(15):9854–61. doi: 10.1093/nar/gku667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filén S, Lahesmaa R. GIMAP Proteins in T-Lymphocytes. J Signal Transduct. 2010;268589 doi: 10.1155/2010/268589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krücken J, Schroetel RM, Müller IU, Saïdani N, Marinovski P, Benten WP, Stamm O, Wunderlich F. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene. 2004;341:291–304. doi: 10.1016/j.gene.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Mooren FC, Volker K. Molecular and cellular exercise physiology. New York, NY: Human Kinetics; 2005. Human Kinetics; p. 464p. [Google Scholar]
- 37.Fyfe JJ, Bishop DJ, Stepto NK. Interference between concurrent resistance and endurance exercise: molecular bases and the role of individual training variables. Sports Med. 2014;44(6):743–62. doi: 10.1007/s40279-014-0162-1. [DOI] [PubMed] [Google Scholar]
- 38.Wagenmakers AJ. Muscle amino acid metabolism at rest and during exercise: role in human physiology and metabolism. Exerc Sport Sci Rev. 1998;26:287–314. [PubMed] [Google Scholar]
- 39.Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, Holloszy JO. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1 alpha expression. J Biol Chem. 2007;282(1):194–9. doi: 10.1074/jbc.M606116200. [DOI] [PubMed] [Google Scholar]
- 40.Hatazawa Y, Tadaishi M, Nagaike Y, Morita A, Ogawa Y, Ezaki O, Takai-Igarashi T, Kitaura Y, Shimomura Y, Kamei Y, Miura S. PGC-1α-mediated branched-chain amino acid metabolism in the skeletal muscle. PLoS One. 2014;9(3):e91006. doi: 10.1371/journal.pone.0091006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowtell JL, Marwood S, Bruce M, Constantin-Teodosiu D, Greenhaff PL. Tricarboxylic acid cycle intermediate pool size: functional importance for oxidative metabolism in exercising human skeletal muscle. Sports Med. 2007;37(12):1071–88. doi: 10.2165/00007256-200737120-00005. [DOI] [PubMed] [Google Scholar]
- 42.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–9. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]