Short abstract
Objective
To develop a combinatorial panel of salivary cytokines that manifests the presence of non-small cell lung cancer (NSCLC) that will eventually improve prognosis by facilitating the early diagnosis and management of this common cancer.
Methods
We performed a case-control study comparing salivary cytokine profiles of 35 adult subjects with NSCLC with those of 35 matched, healthy nonsmokers. Multiplex bead array assays were used to quantify 27 cytokines in saliva, serum, and oral mucosal transudate samples. Logistic regression analysis was used to develop an informative cytokine panel. Receiver operating characteristic (ROC) curves were generated to evaluate the discriminant ability of the panel.
Results
A combinatorial 12-cytokine panel (interleukin receptor antagonist [IL1RN], IL1B, IL6, IL7, IL8, IL10, C-C motif chemokine ligand 11 [CCL11], tumor necrosis factor, C-X-C motif chemokine ligand 10 [CXCL10], C-C motif chemokine ligand 3, C-C motif chemokine ligand 4, and platelet-derived growth factor-BB) distinguished patients with NSCLC from healthy controls. Further, ROC analysis revealed that a cytokine panel comprising IL10 (odds ratio, 1.156) and CXCL10 (odds ratio, 1.000) discriminated NSCLC with a sensitivity of 60.6% and specificity of 80.8% (area under the ROC curve, 0.701).
Conclusion
A combinatorial panel of select salivary cytokines indicates the presence of NSCLC.
Keywords: Cytokine, saliva, non-small cell lung cancer, logistic regression analysis, oral mucosal transudate, combinatorial panel, multiplex analysis
Introduction
Lung cancer is one of the most common malignancies worldwide, with considerable associated morbidity and mortality.1 In many countries, including Japan, lung cancer is the leading cause of cancer-related mortality.2 The majority (85%) of these malignancies are classified as non-small cell lung cancer (NSCLC). Surgery, with other adjunctive treatment, is the preferred therapeutic option for patients with early stages of NSCLC (5-year survival rate approximately 70% to 80%).3 Yet, more than half of patients with NSCLC initially present with more advanced stages of the disease. Although newer therapeutic strategies such as molecular targeted agents have marginally improved survival rates, the prognosis of advanced-stage NSCLC is stubbornly poor, with an overall 5-year survival rate of 17%.4 Given that early detection of NSCLC greatly increases 5-year survival rates, screening and identification of NSCLC during its earliest stages has been a major interest of the oncology community.
Among the pathways to tumor formation and progression, the inflammatory pathway has stimulated particular interest.5 Considerable evidence indicates that inflammation may be an ancillary, or even inseparable, aspect of tumor development.6–10 Chronic inflammatory states associated with infection and irritation may lead to environments that promote genomic lesions and the development of dysplasia.11 In particular, cytokines are secreted by tumor and stromal cells in the tumor microenvironment, and these factors can function in an autocrine or paracrine manner (or both) to perpetuate the tumor and facilitate local invasion and metastasis.12
The putative association of cytokines with tumor development and progression has led to interest in exploiting cytokines as biological indicators of the presence or progression of malignancies. Although most attention focuses on developing distinct serum cytokine profiles of malignancies, interest has increasingly pivoted to the use of saliva as a diagnostic medium. Saliva has several compelling attributes as a biofluid for investigating systemic diseases.13 Although the oral mucosal transudate (OMT) is not regularly used in the field of oral health, its use will increase when commercial collecting devices are introduced. Saliva can be easily, repeatedly, and noninvasively obtained and stored without the use of specialized equipment. Saliva poses a much smaller risk of exposing clinicians and laboratory technicians to pathogens such as human immunodeficiency virus or hepatitis viruses.14
Numerous salivary analytes of interest in biobehavioral research are constituents of serum that are synthesized, stored, and released from the granules within the secretory cells of the saliva glands. Others contribute to humoral immunity, such as antibodies or cytokines secreted by neutrophils, macrophages, or lymphocytes in the oral mucosa.15 Multiple molecules isolated from saliva have been used as screening and diagnostic tools or as prognostic biomarkers of diseases, including cancer.16–18 However, the role of cytokines in saliva as a distant manifestation of lung malignancies has been largely unexplored.
The primary aim of the present study was to evaluate the association between salivary cytokine levels and the presence of NSCLC. We hypothesized that the levels of salivary cytokines in patients with NSCLC vary with the severity of disease. Moreover, combinatorial assessment of cytokine levels may prove useful for screening for NSCLC. For this purpose, we characterized cytokine profiles of the saliva of patients with NSCLC and compared them with those of normal, healthy matched subjects. We further explored the relationships between cytokine levels in saliva, serum, and OMT in patients with NSCLC with those of their matched controls.
Materials and methods
Subjects
The Ethics Committee of Shinshu University and Shinshu University Hospital approved this study. Written informed consent was obtained from all patients. Between March 2016 and July 2017, 35 adult patients (13 men and 22 women, aged 42 to 80 years; mean, 67.5 years, median 69.0 years) with Stages I–IV NSCLC were recruited from the Shinshu Cancer Center and the Department of Thoracic Surgery of Shinshu University Hospital. Diagnosis and staging of NSCLC was confirmed by cytology and histopathology according to the WHO criteria. Stages I, II, III, and IV tumors were defined according to the Union for International Cancer Control’s TNM staging criteria for lung cancer (Ver. 7). Twenty-one (60%) patients were stage I (IA and IB), two (6%) were stage IIA, two (6%) were stage IIIB, and 10 (28%) were stage IV. Biological samples of patients with NSCLC were obtained before pretreatment such as chemotherapy or surgery. We recruited 35 normal, healthy nonsmokers who were not taking medication (17 men and 18 women, aged 42 to 69 years; mean, 53.6 years; median, 52.0 years) to serve as controls. An independent investigator, unaware of the subjects’ laboratory data, selected controls from a pool of hospital staff and patients who were matched for age, sex, and smoking habits to the NSCLC patients.
Sample collection
Immunological data were acquired using saliva, blood, and OMT. To assess diurnal variations, saliva samples were collected from patients three-times daily as follows: morning (6:00–9:00), noon (11:00–13:00), and evening (15:00–16:00). Saliva, blood, and OMT samples were collected from all subjects once daily at noon. Biological samples were collected before treatment. It is possible that chronic periodontitis influences the concentrations of inflammatory and anti-inflammatory cytokines.19 Therefore, before procuring a biospecimen, any active periodontal disease was excluded using a test paper-strip method that detects occult blood in saliva (Perioscreen; Sunstar Inc., Osaka, Japan). Subjects did not drink or chew gum in the 30 minutes before saliva collection. Commonly used cotton-based absorbent materials can alter quantitative estimates of the salivary analytes through nonspecific binding, cross-linking, or filtering constituent biomolecules.20,21 The saliva samples were provided by the participants using the passive-drool method, in which participants allow saliva to pool in the mouth and then drool it into a tube (MS-50, Japan Medical Ltd., Tokyo, Japan). Approximately 150 µL of whole saliva was collected during 1 minute. Next, 4.5 mL of whole blood was collected by a nurse using a vacuum blood collection tube (anticoagulant, 3.2% sodium citrate; Terumo Co., Tokyo, Japan) from each participant.22,23 OMT samples were collected by the participants for 5 minutes using an OraSure collection device (OraSure Technologies, Inc., Bethlehem, PA, USA).24,25 The saliva, blood, and OMT samples were stored at −80°C.
Clinical information and the results from testing the samples were collected and entered into a database by research staff not directly involved in diagnosis, treatment, or sample analysis. Patients’ diagnostic and pathological data were masked during sample collection and biomarker detection.
Multiplex analysis of cytokines
We used a multiplex bead array assay (Cat. #M500KCAF0Y, Bi-Plex Pro Human Cytokine Grp I Panel 27-Plex; Bio-Rad Laboratories, Inc., Tokyo, Japan) that has been used to measure salivary cytokines.26,27 The cytokines were as follows: interleukin (IL) receptor antagonist (RN); IL1β; IL2; IL4; IL5; IL6; IL7; IL8; IL9; IL10; IL12 (p70); IL13; IL15; IL17A; C-C motif chemokine ligand 11 (CCL11); fibroblast growth factor 2 (FGF2); colony stimulating factor 3 (CSF3); colony stimulating factor 2 (CSF2); interferon gamma (IFN-γ); tumor necrosis factor (TNF); C-X-C motif chemokine ligand 10 (CXCL10); C-C motif chemokine ligand 2 (CCL2); C-C motif chemokine ligand (CCL3); C-C motif chemokine ligand 4 (CCL4); platelet-derived growth factor-BB (PDGF-BB); regulated on activation, normal T cell expressed and secreted (RANTES); and vascular endothelial growth factor (VEGF). All were analyzed according to the manufacturer’s instructions. The Bio-Plex Multiplex Immunoassay employed human cytokine panels, and the plates were read using a Bio-Plex Array Reader (Bio-Plex 200 System and Bio-Plex Manager Version 6.1, Bio-Rad Laboratories, Inc.).
Saliva, blood, and OMT were prepared using the same pretreatment. The biofluids were frozen at −80°C, thawed at 4°C in a refrigerator, equilibrated to room temperature (24°C), and then added to the assay plates. The samples were then centrifuged at 1,500 ×g for 15 minutes, and a micropipette was used to sample a 50-µL aliquot of each sample for subsequent analysis.
Statistical analysis
Statistical analyses were performed using IBM SPSS Statistics, version 25 (IBM Corp., Armonk, NY, USA). Within-group comparisons of diurnal variations in cytokine levels were performed using one-way analysis of variance (ANOVA). Comparisons between two groups of cytokine samples acquired at noon were evaluated using the Mann–Whitney test. Unless otherwise stated, data are expressed as the mean ± standard deviation (SD), and P < 0.05 indicates statistical significance. Statistical analysis was not performed if the number of data points missing from one group was >18.
We used logistic regression analysis to evaluate the association between the levels of salivary cytokines and the presence of NSCLC.28 The outcome variables for the development of NSCLC were 0 for “no” and 1 for “yes”. To generate a multivariable logistic regression model, the results of the Mann–Whitney test (P < 0.05) and the backward elimination method were used. The influence of a variable on the likelihood of detecting NSCLC is expressed as the odds ratio (OR), 95% confidence interval (CI).
The performance of the model was assessed using a discrimination test combined with receiver operating characteristic (ROC) analysis.29 ROC curves were generated to verify the discriminatory power of the salivary cytokine levels. The areas under the curves (AUCs) were calculated to provide an overall summary of the diagnostic accuracy of the comparisons of salivary cytokine levels. Diagnostic performance was classified as follows: poor, 0.50 ≤ AUC < 0.69; good, 0.70 ≤ AUC < 0.89; and excellent, 0.90 ≤ AUC < 1.
Results
Multiplex analysis of cytokines in biofluids
Table 1 summarizes the backgrounds of the subjects and the number of samples. Of the 27 target cytokines, only 21 salivary cytokines were detected in patients with NSCLC because of insufficient sensitivity of the Bio-Plex system for detecting IL2, IL4, IL15, IL17A, FGF2, and CSF2. When the diurnal variations in the salivary cytokines of the NSCLC patients were evaluated using one-way ANOVA, significant differences were observed for eight salivary cytokines as follows: IL1RN, IL7, IL8, IL10, IL13, CCL11, IFN-γ, and TNF (P < 0.05). With the exception of IL6, CSF3, CXCL10, and CCL3, the levels of salivary cytokines in the morning generally were the highest.
Table 1.
Summary of subjects’ backgrounds and biofluid samples
| Group | Male/Female | Smoking status, |
Sample number, n |
||||||
|---|---|---|---|---|---|---|---|---|---|
| (n = 35) | n (%) | Histology, n (%) | Morning | Noon | Evening | ||||
| NSCLC | 13/22 | Never | 21 (60) | Adenocarcinoma | 32 (91) | serum | – | 33 | – |
| Former | 14 (40) | Squamous cell carcinoma | 3 (9) | saliva | 35 | 35 | 35 | ||
| OMT | – | 35 | – | ||||||
| Control | 17/18 | Never | 27 (77) | serum | – | 35 | – | ||
| Former | 8 (23) | saliva | – | 35 | – | ||||
| OMT | – | 35 | – | ||||||
Abbreviation: non-small cell lung cancer (NSCLC).
The correlation coefficients of biofluid levels were calculated using data from the patients with NSCLC and the controls. The correlation coefficients of the serum vs saliva levels ranged between −0.20 (PDGF-BB) and 0.55 (CCL11). For the positive values, the maximum, minimum, and mean were 0.55 (CCL11), 0.02 (IL8), and 0.18, respectively. For the negative values, the maximum, minimum, and mean were −0.01 (VEGF), −0.20 (PDGF-BB), and −0.09, respectively. The correlation coefficients of the serum vs OMT levels ranged between −0.18 (CCL2) and 0.26 (IL6). For the positive values, the maximum, minimum, and mean were 0.26 (IL6), 0.01 (VEGF), and 0.07, respectively. For the negative values, the maximum, minimum, and mean were −0.01 (IL4), −0.18 (CCL2), and −0.09, respectively. The correlation coefficients of saliva vs OMT levels ranged between −0.13 (IL5) and 0.91 (CCL3). For the positive values, the maximum, minimum, and mean were 0.91 (CCL3), 0.19 (IL1RN), and 0.46, respectively. For the negative values, the maximum, minimum, and mean were −0.01 (IL17A), −0.13 (IL5), and −0.07, respectively.
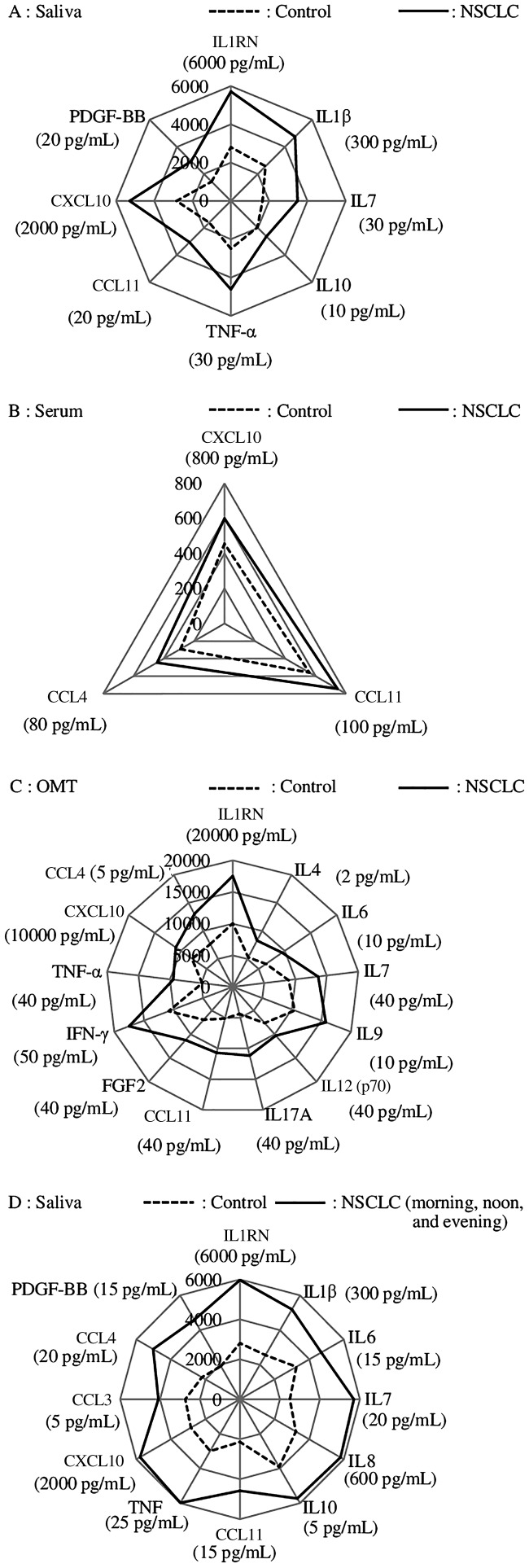
Table 2 shows the results of multiplex analysis of cytokine levels in biofluids at noon and the two-group comparisons using the Mann–Whitney test (n = 35). For statistical analysis, cytokine levels less than the limit of detection are presented as half the limit of detection.30,31 The levels of salivary cytokines of the patients with NSCLC ranged between 0.01 (IL13) and 29,519.09 (IL1RN) pg/mL. In contrast, the cytokine levels of the control subjects ranged between 0.01 (IL4, IL6, IL13, and PDGF-BB) and 9,257.21 (IL1RN) pg/mL. For the noon samples, significant differences were observed between the salivary cytokine levels of the patients with NSCLC and controls as follows: IL1RN, IL1B, IL7, IL10, CCL11, TNF, CXCL10, and PDGF-BB (P < 0.05) (Figure 1a). Significant differences in serum cytokine levels at noon between patients with NSCLC and controls were as follows: CCL11, CXCL10, and CCL4 (P < 0.05) (Figure 1b). IL2 and IL15 were undetectable. There were significant differences in OMT cytokine levels at noon between patients with NSCLC and the controls as follows: IL1RN, IL4, IL6, IL7, IL9, IL12 (p70), IL17A, CCL11, FGF2, IFN-γ, TNF, CXCL10, and CCL4 (P < 0.05) (Figure 1c). IL2, IL15, and CSF2 were undetectable.
Table 2.
Results of multiplex analysis of cytokines in biofluids at noon and comparisons between NSCLC patients and healthy controls (Mann–Whitney test)
| Cytokine | Serum (pg/mL) |
P | Saliva (pg/mL) |
P | OMT (pg/mL) |
P | LOD (pg/mL) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | NSCLC | Control | NSCLC | Control | NSCLC | |||||
| IL1β | 1.45 ± 1.01 | 1.71 ± 2.03 | 0.736 | 128 ± 321 | 237 ± 352 | 0.044* | 42.8 ± 59.8 | 59.9 ± 105 | 0.470* | |
| IL1RN | 119 ± 100 | 119 ± 86.6 | 0.463 | 2810 ± 2400 | 5717 ± 4189 | 0.001* | 10043± 7782 | 17537±17725 | 0.001* | |
| IL2 | – | – | – | – | – | – | – | – | – | |
| IL4 | 1.19 ± 0.72 | 1.44 ± 1.12 | 0.606 | 0.45 ± 0.46 | – | – | 0.53 ± 0.30 | 0.83 ± 0.43 | 0.001* | 0.01 |
| IL5 | 7.64 ± 5.09 | 10.5 ± 9.43 | 0.379 | 1.86 ± 1.93 | 1.47 ± 1.81 | 0.145 | 1.23 ± 0.89 | 2.07 ± 2.21 | 0.679 | |
| IL6 | 2.80 ± 3.34 | 4.46 ± 7.12 | 0.544 | 8.15 ± 12.9 | 11.9 ± 14.4 | 0.090 | 3.21 ± 3.08 | 4.76 ± 2.88 | 0.010* | 0.02 |
| IL7 | 6.50 ± 6.82 | 5.19 ± 5.81 | 0.281 | 8.29 ± 6.05 | 17.4 ± 15.3 | 0.001* | 17.8 ± 14.3 | 27.2 ± 15.9 | 0.002* | 0.13 |
| IL8 | 11.9 ± 10.7 | 14.3 ± 9.29 | 0.053 | 323 ± 399 | 467 ± 688 | 0.179 | 144 ± 152 | 219 ± 381 | 0.810 | |
| IL9 | 21.4 ± 48.0 | 18.4 ± 27.4 | 0.559 | 5.02 ± 5.46 | 6.19 ± 7.41 | 0.432 | 5.23 ± 3.54 | 7.88 ± 4.94 | 0.017* | 0.04 |
| IL10 | 5.47 ± 9.60 | 10.0 ± 21.8 | 0.178 | 3.26 ± 3.99 | 4.37 ± 3.12 | 0.024* | 4.15 ± 2.93 | 6.37 ± 5.76 | 0.149 | |
| IL12 (p70) | 10.4 ± 22.3 | 12.2 ± 19.9 | 0.106 | 19.2 ± 14.4 | 22.2 ± 11.5 | 0.171 | 15.2 ± 7.66 | 20.5 ± 9.35 | 0.010* | |
| IL13 | 17.2 ± 15.4 | 25.2 ± 23.5 | 0.171 | 0.70 ± 0.60 | 0.78 ± 0.66 | 0.638 | 3.31 ± 3.10 | 4.47 ± 4.25 | 0.347 | |
| IL15 | – | – | – | 0.86 ± 0.82 | – | – | 0.58 ± 0.06 | – | – | 0.57 |
| IL17A | 15.1 ± 41.7 | 15.5 ± 51.0 | 0.387 | 1.19 ± 2.53 | – | – | 8.72 ± 9.03 | 22.3 ± 22.6 | 0.004* | 0.07 |
| CCL11 | 69.9 ± 36.1 | 93.1 ± 44.6 | 0.048* | 5.33 ± 3.92 | 10.2 ± 10.8 | 0.010* | 10.3 ± 6.83 | 21.5 ± 14.3 | 0.000* | 1.02 |
| FGF2 | 49.8 ± 49.1 | 37.3 ± 19.1 | 0.205 | 5.84 ± 7.43 | 8.35 ± 8.77 | 0.459 | 13.8 ± 16.3 | 22.4 ± 17.9 | 0.011* | 0.36 |
| CSF3 | 16.0 ± 11.8 | 22.8 ± 15.8 | 0.124 | 23.3 ± 43.5 | 33.9 ± 67.8 | 0.142 | 12.1 ± 8.15 | 23.5 ± 47.5 | 0.068 | 0.01 |
| CSF2 | 48.0 ± 37.4 | 35.7 ± 25.5 | 0.183 | – | – | – | – | – | – | |
| IFN-γ | 42.0 ± 23.7 | 51.1 ± 40.6 | 0.564 | 28.8 ± 21.9 | 28.2 ± 19.4 | 0.952 | 27.1 ± 16.9 | 43.8 ± 27.1 | 0.000* | |
| CXCL10 | 456 ± 458 | 600 ± 408 | 0.013* | 949 ± 1488 | 1765 ± 1883 | 0.020* | 3897 ± 4812 | 5477 ± 5101 | 0.037* | |
| CCL2 | 6.09 ± 5.77 | 15.6 ± 25.4 | 0.294 | 125 ± 124 | 101 ± 99.6 | 0.456 | 61.9 ± 53.6 | 54.6 ± 50.2 | 0.582 | 0.42 |
| CCL3 | 5.14 ± 7.94 | 3.90 ± 2.61 | 0.946 | 2.28 ± 1.59 | 3.90 ± 6.99 | 0.118 | 1.91 ± 1.05 | 3.26 ± 4.40 | 0.028* | 0.12 |
| PDGF-BB | 144 ± 270 | 36.4 ± 31.5 | 0.053 | 4.79 ± 6.05 | 9.77 ± 7.97 | 0.001* | 27.4 ± 20.4 | 35.7 ± 26.6 | 0.240 | 0.52 |
| CCL4 | 29.1 ± 12.9 | 44.5 ± 45.0 | 0.018* | 7.34 ± 9.22 | 16.9 ± 28.4 | 0.118 | 10.7 ± 11.9 | 13.5 ± 17.3 | 0.217 | 0.17 |
| RANTES | 1582± 1000 | 1361± 1036 | 0.262 | – | 1.88 ± 2.07 | – | 3.67 ± 2.77 | 5.78 ± 4.80 | 0.165 | 0.10 |
| TNF | 137 ± 167 | 107 ± 117 | 0.544 | 12.5 ± 11.0 | 23.1 ± 32.4 | 0.013* | 9.74 ± 8.58 | 18.9 ± 14.0 | 0.000* | 0.45 |
| VEGF | 14.8 ± 27.2 | 10.2 ± 7.27 | 0.740 | 1259 ± 1748 | 1337 ± 1432 | 0.183 | 965 ± 643 | 915 ± 411 | 0.703 | 0.62 |
*P < 0.05.
LODs were calculated from the measured results of concentrations of negative control and standard solutions provided by Bio-Rad Laboratories, Inc.
Abbreviations: oral mucosal transudate (OMT); limit of detection (LOD); non-small cell lung cancer (NSCLC): interleukin (IL); C-C motif chemokine ligand (CCL); fibroblast growth factor 2 (FGF2); colony stimulating factor (CSF); interferon (IFN); C-X-C motif chemokine ligand (CXCL); regulated on activation, normal T cell expressed and secreted (RANTES); tumor necrosis factor (TNF); vascular endothelial growth factor (VEGF).
Figure 1.
Comparison of the salivary cytokine levels between patients with non-small cell lung cancer and controls (Mann–Whitney test)
To verify the possibility of screening for NSCLC using the levels of salivary cytokines, we used the data for morning, noon, and evening of patients with NSCLC (35 subjects × 3 data points = 105). Significant differences between the salivary cytokine levels of patients and those of the controls were as follows: IL1RN, IL1B, IL6, IL7, IL8, IL10, CCL11, TNF, CXCL10, CCL3, CCL4, and PDGF-BB (P < 0.05) (Figure 1d).
Logistic regression analysis
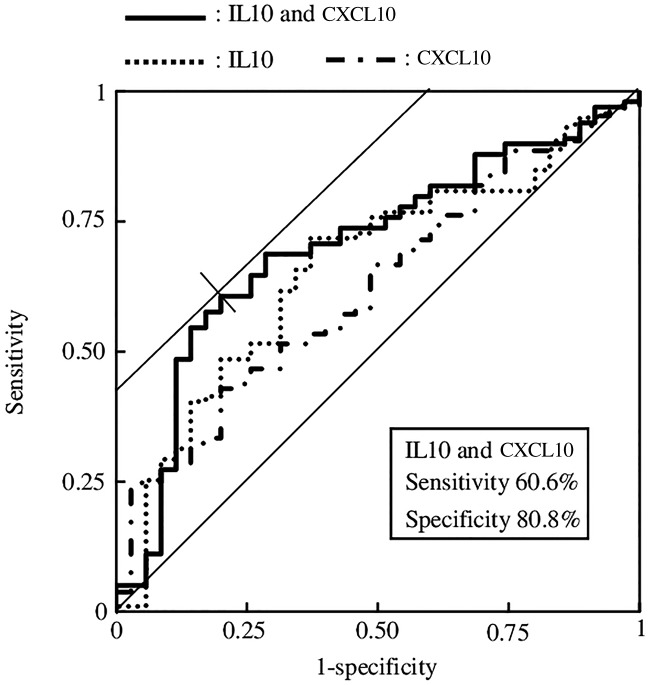
The association between the salivary cytokine levels of 12 cytokines and the risk of NSCLC was evaluated using logistic regression analysis. The backward elimination method identified the combination of salivary cytokines IL10 and CXCL10 as the most highly associated with NSCLC (P < 0.05). In comparison, the P values and ORs for all possible combinations of analytes were calculated using a direct (simultaneous) method. Cytokines significantly associated (P < 0.05) with NSCLC were present in three combinations when two types of cytokines were used (Table 3). A significant association with NSCLC was not observed when more than three cytokines were used. Ultimately, the combination of IL10 and CXCL10 had the maximum ORs as (1.156 for IL10 [95% CI 1.048–1.275] and 1.000 for CXCL10 [95% CI 1.000–1.001]).
Table 3.
Outcomes of the logistic regression analysis of patients with NSCLC
| Cytokines | Regression coefficient, β | Standard error | P value | Odds ratio | 95% confidence interval |
|---|---|---|---|---|---|
| IL10 | 0.145 | 0.050 | 0.004 | 1.156 | 1.048–1.275 |
| CXCL10 | 0.000 | 0.000 | 0.021 | 1.000 | 1.000–1.001 |
| IL1B | 0.002 | 0.001 | 0.018 | 1.002 | 1.000–1.004 |
| CXCL10 | 0.000 | 0.000 | 0.011 | 1.000 | 1.000–1.001 |
| IL6 | 0.041 | 0.020 | 0.039 | 1.042 | 1.002–1.083 |
| CXCL10 | 0.000 | 0.000 | 0.006 | 1.000 | 1.000–1.001 |
Abbreviations: non-small cell lung cancer (NSCLC); interleukin (IL); C-X-C motif chemokine ligand 10 (CXCL10).
A multivariable prediction model (logistic model) for the risk of NSCLC was built using IL10 and CXCL10 as follows:
| (1) |
| (2) |
where z is an index that combines the criteria variables, and f(z) is a logistic function.
ROC analysis
To confirm the preceding findings, we performed ROC analysis. Figure 2 shows the ROC of the presence of NSCLC when IL10 and CXCL10 were used. The AUC was 0.701 (P = 0.000, 95% CI 0.602–0.801), which demonstrates “good” capability for discriminating between patients with NSCLC and controls. The optimal cutoff values of sensitivity and specificity were 60.6% and 80.8%, respectively. The cutoff values of the logistic function f(z) between the true positives and false negatives ranged from 0.698 to 0.708. The cutoff values for the logistic function between the true negatives and false positives ranged from 0.702 to 0.719. Thus, the cutoff values of the logistic function ranged from 0.702 to 0.708.
Figure 2.
Receiver operating characteristic analysis of interleukin 10 and C-X-C motif chemokine ligand 10 levels to determine the risk of non-small cell lung cancer
For comparison, the ROCs of IL10 and CXCL10 are shown in Figure 2. The values of the AUC, sensitivity, and specificity of IL10 were 0.660, 71.7, and 62.9, respectively. The values of the AUC, sensitivity, and specificity of CXCL10 were 0.622, 42.9, and 80.0, respectively. The maximum AUC was achieved when the maximum values of IL10 and CXCL10 were combined.
Discussion
The results of the present study support our working hypothesis that differential levels of certain salivary cytokines in patients with NSCLC and combinatorial assessment of cytokine levels are useful for screening for NSCLC. Our findings are limited by the inability to study age-matched, healthy nonsmokers and subjects with a mean age of 67.5 years to serve as our control group. Even after controlling for diurnal variation, the levels of IL1RN, IL7, IL8, IL10, IL13, CCL11, IFN-γ, and TNF were significantly different. The cytokines were grouped as inflammatory (IL7, IL8, CCL11, IFN-γ, and TNF) and anti-inflammatory (IL1RN, IL10, and IL13) cytokines. The levels of salivary cytokines that varied with a circadian rhythm in patients with NSCLC did not depend on their functional category or concentrations in saliva. Typically, the levels of anti-inflammatory cytokines that vary with a circadian rhythm are higher shortly after waking and lower before sleeping.32,33 In the present study, the levels of all salivary anti-inflammatory cytokines were higher in the morning, consistent with previous findings.
Multivariable analysis of cytokine levels in saliva, serum, and OMT revealed that saliva and OMT had incremental beneficial utility compared with serum. Salivary cytokines are being assessed for their associations with oral diseases such as periodontitis, gingivitis, cancer, and other systemic diseases. For example, Khan detected numerous cytokines and chemokines in saliva samples of normal subjects.34 Compared with matched controls, the samples from patients with NSCLC studied here showed significant differences in cytokine levels in saliva (8 types), serum (3 types), and OMT (13 types). There were fewer significant differences in cytokine levels in serum compared with saliva or the OMT. In contrast, the cytokine levels in saliva and the OMT were highly concordant, judged by the correlation coefficients of cytokine levels in serum and saliva compared with those of serum and the OMT. Saliva is a major component of the OMT, and one can therefore surmise that the ease of collection may serve as the major determinant when choosing between the two biofluids. Generally, the passive-drool method for collecting saliva samples is quick and simple compared with techniques for collecting the OMT.
The combinatorial assessment of salivary cytokine levels shows promise for screening for the presence of NSCLC. The levels of cytokines at all sampling times differed significantly between patients with NSCLC and controls as follows: IL1RN, IL1B, IL6, IL7, IL8, IL10, TNF, CCL11, CXCL10, CCL3, CCL4, and PDGF-BB (P < 0.05) (Figure 1d). The levels of eight of these salivary cytokines differed significantly in the morning (Figure 1a). However, the morning levels were not representative of the differences between patients with NSCLC and healthy controls. ROC analysis supports the assertion that NSCLC can be detected by measuring the levels of salivary cytokines when using a selective model employing IL10 and CXCL10 (Figure 2).
Our present data provide preliminary support for utilizing differential levels of salivary IL10 and CXCL10 as indicators of existing NSCLC. Emerging research supports our strategy that employed a panel of inflammatory cytokines to detect NSCLC. For example, Daly et al.35 measured the expression levels of candidate biomarkers of individuals at high-risk for lung cancer and identified a panel of seven analytes, mainly inflammatory cytokines such as IL6, IL10, IL1RN, and TNF. DeCotiis et al.36 measured inflammatory cytokines in serum and concluded that cytokine biomarkers may serve as a promising tool for the detection of lung cancer. However, these two studies35,36 did not identify CXCL10.
IL10 is an anti-inflammatory pleotropic cytokine that is present in the exhaled breath of patients with lung cancer.37 IL10 is produced by normal and neoplastic cells and likely contributes to autoimmunity, transplantation tolerance, and tumorigenesis.38 The levels of serum IL10 present in patients with NSCLC are higher compared with those of healthy control subjects.39 An emerging body of research supports the plausibility of our findings.35,36 Thus, CXCL10 is a potent angiostatic cysteine-X amino acid-cysteine (CXC) chemokine40 that regulates NSCLS-induced angiogenesis, tumor growth, and spontaneous metastasis.41 The levels of CXCL10 detected in surgical specimens of NSCLC tumors are significantly higher compared with specimens of adjacent normal lung tissue.42 These findings may explain why CXCL10 was identified using the Mann–Whitney test when we included patients with stages II–IV NSCLC (Figure 1d).
Despite its small sample size, our study shows potential utility for measuring the levels of salivary biomolecules that significantly differ among patients with NSCLC and controls. NSCLC comprises the adenocarcinoma, squamous cell carcinoma (SCC), and large cell carcinoma subtypes. The mechanisms of tumorigenesis and molecular characteristics of SCC differ from those of adenocarcinoma. The relationships of histological types and the immune environment with the levels of associated cytokines seem worthy of future study. The discovery of a saliva-based biomarker panel may represent a significant source of discriminatory biomarkers for detecting NSCLC. Equally important, or study shows the translational promise of the emerging field of salivary diagnostics for systemic diseases. The ease of saliva collection and continuing advances in high-throughput technologies that allow quick and incisive insights into saliva’s constituents are advancing the use of oral fluids as a credible diagnostic alternate to other biofluids.
Our study shows the promise of utilizing differential levels of salivary cytokines for point-of-care testing, thus allowing NSCLC to be detected to facilitate therapeutic intervention and improve survival rates. The results of our study will likely serve as a foundation for validation studies that evaluate the temporal changes in salivary cytokine levels and investigate their associations with response to treatment, relapse, complications, and survival. It is conceivable that the levels of IL10 and CXCL10 will return to normal after surgical treatment of NSCLC.
Conclusions
This study revealed significant differences in the levels of certain salivary cytokines between patients with NSCLC and control groups. In particular, a 12-cytokine panel (IL1RN, IL1B, IL6, IL7, IL8, IL10, TNF, CCL11, CXCL10, CCL3, CCL4, and PDGF-BB) distinguished patients with NSCLC from healthy controls. ROC analysis revealed that a two-cytokine panel (IL10 and CXCL10) indicated the presence of NSCLC with high sensitivity and specificity.
Acknowledgments
We thank the patients and healthy subjects for their support and participation in this study. We also thank the staff in the Department of Thoracic Surgery, Shinshu University Hospital, Mariko Miyagawa, and Nanae Mitsui for their helpful support.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This work was supported in part by grants for the Development of Medical and Welfare Devices, Development Of Low-Cost, Rapid, and Noninvasive Diagnostic Technology for Cancer using Salivary Biomarkers, Fukushima, Japan; and from the Japan Society for the Promotion of Science, Visualization of Cancer by Cytokine Coding Method and Biosensor-Array with Microcapsules [grant number 16H03166].
References
- 1.Dubey S, Powell CA. Update in lung cancer 2008. Am J Respir Crit Care Med 2009; 179: 860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuda T, Marugame T, Kamo KI, et al. Cancer incidence and incidence rates in Japan in 2005: based on data from 12 Population-based Cancer Registries in the Monitoring of Cancer Incidence in Japan (MCIJ) Project. Jpn J Clin Oncol 2001; 41: 139–147. [DOI] [PubMed] [Google Scholar]
- 3.Rami-Porta R, Ball D, Crowley J, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the T descriptors in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol 2007; 2: 593–602. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014; 64: 9–29. [DOI] [PubMed] [Google Scholar]
- 5.Brenner DR, Scherer D, Muir K, et al. A review of the application of inflammatory biomarkers in epidemiologic cancer research. Cancer Epidemiol Biomarkers Prev 2014; 23: 1729–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballaz S, Mulshine JL. The potential contributions of chronic inflammation to lung carcinogenesis. Clin Lung Cancer 2003; 5: 46–62. [DOI] [PubMed] [Google Scholar]
- 7.Allavena P, Garlanda C, Borrello MG, et al. Pathways connecting inflammation and cancer. Curr Opin Genet Dev 2008; 18: 3–10. [DOI] [PubMed] [Google Scholar]
- 8.Engels EA. Inflammation in the development of lung cancer: epidemiological evidence. Expert Rev Anticancer Ther 2008; 8: 605–615. [DOI] [PubMed] [Google Scholar]
- 9.Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature 2008; 454: 436–444. [DOI] [PubMed] [Google Scholar]
- 10.Azad N, Rojanasakul Y, Vallyathan V. Inflammation and lung cancer: roles of reactive oxygen/nitrogen species. J Toxicol Environ Health B Crit Rev 2008; 11: 1–15. [DOI] [PubMed] [Google Scholar]
- 11.Tahara E. Growth factors and oncogenes in human gastrointestinal carcinomas. J Cancer Res Clin Oncol 1990; 116: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agnola CD, Biragyn A. Clinical utilization of chemokines to combat cancer: the double-edged sword. Expert Rev Vaccines 2007; 6: 267–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong DT. Salivary Diagnostics: Amazing as it might seem, doctors can detect and monitor diseases using molecules found in a sample of spit . Am Sci 2008; 96: 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaguchi M, Shetty V. Salivary Sensors for Quantification of Stress Response Biomarker. Electrochemistry 2011; 79: 442–446. [Google Scholar]
- 15.Granger DA, Johnson SB, Szanton SL, et al. Incorporating salivary biomarkers into nursing research: an overview and review of best practices. Biol Res Nurs 2012; 14: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrne ML, O'Brien-Simpson NM, Reynolds EC, et al. Acute phase protein and cytokine levels in serum and saliva: a comparison of detectable levels and correlations in a depressed and healthy adolescent sample. Brain Behav Immun 2013; 34: 164–175. [DOI] [PubMed] [Google Scholar]
- 17.Kamatani T, Shiogama S, Yoshihama Y, et al. Interleukin-1 beta in unstimulated whole saliva is a potential biomarker for oral squamous cell carcinoma. Cytokine 2013; 64: 497–502. [DOI] [PubMed] [Google Scholar]
- 18.Rathnayake N, Akerman S, Klinge B, et al. Salivary biomarkers for detection of systemic diseases. PLoS One 2013; 8: e61356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomas I, Arias-Bujanda N, Alonso-Sampedro M, et al. Cytokine-based predictive models to estimate the probability of chronic periodontitis: Development of diagnostic nomograms. Sci Rep 2017; 7: 11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granger DA, Kivlighan KT, Fortunato C, et al. Integration of salivary biomarkers into developmental and behaviorally-oriented research: problems and solutions for collecting specimens. Physiol Behav 2007; 92: 583–590. [DOI] [PubMed] [Google Scholar]
- 21.Shirtcliff EA, Granger DA, Schwartz E, et al. Use of salivary biomarkers in biobehavioral research: cotton-based sample collection methods can interfere with salivary immunoassay results. Psychoneuroendocrinol 2001; 26: 165–173. [DOI] [PubMed] [Google Scholar]
- 22.Biancotto A, Feng X, Langweiler M, et al. Effect of anticoagulants on multiplexed measurement of cytokine/chemokines in healthy subjects. Cytokine 2012; 60: 438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan VV, Ravindran R, Wun T, et al. Multiplexed measurements of immunomodulator levels in peripheral blood of healthy subjects: Effects of analytical variables based on anticoagulants, age, and gender. Cytometry B Clin Cytom 2014; 86: 426–435. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Botran R, Miller JJ, Burns VE, et al. Correlations among inflammatory markers in plasma, saliva and oral mucosal transudate in post-menopausal women with past intimate partner violence. Brain Behav Immun 2011; 25: 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang JJ, Eisenberger NI, Seeman TE, et al. Negative and competitive social interactions are related to heightened proinflammatory cytokine activity. Proc Natl Acad Sci USA 2012; 109: 1878–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arellano-Garcia ME, Hu S, Wang J, et al. Multiplexed immunobead-based assay for detection of oral cancer protein biomarkers in saliva. Oral Dis 2008; 14: 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorman R, Lundahl J, Yucel-Lindberg T, et al. Inflammatory cytokines in saliva: early signs of metabolic disorders in chronic kidney disease. A controlled cross-sectional study. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2010; 110: 597–604. [DOI] [PubMed] [Google Scholar]
- 28.Kleinbaum DG, Klein M. Logistic regression. 3rd ed New York: Springer Science+Business Media LCC, 2010, pp. 1–32. [Google Scholar]
- 29.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: 837–845. [PubMed] [Google Scholar]
- 30.Kezic S, O’Regan GM, Lutter R, et al. Filaggrin loss-of-function mutations are associated with enhanced expression of IL-1 cytokines in the stratum corneum of patients with atopic dermatitis and in a murine model of filaggrin deficiency. J Allergy Clin Immunol 2012; 129: 1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Attur M, Wang HY, Kraus VB, et al. Radiographic severity of knee osteoarthritis is conditional on interleukin 1 receptor antagonist gene variations. Ann Rheum Dis 2010; 69: 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agorastos A, Hauger RL, Barkauskas DA, et al. Circadian rhythmicity, variability and correlation of interleukin-6 levels in plasma and cerebrospinal fluid of healthy men. Psychoneuroendocrinol 2014; 44: 71–82. [DOI] [PubMed] [Google Scholar]
- 33.Geiger SS, Fagundes CT, Siegel RM. Chrono-immunology: progress and challenges in understanding links between the circadian and immune systems. Immunology 2015; 146: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan A. Detection and quantitation of forty eight cytokines, chemokines, growth factors and nine acute phase proteins in healthy human plasma, saliva and urine. J Proteomics 2012, 75: 4802–4819. [DOI] [PubMed] [Google Scholar]
- 35.Daly S, Rinewalt D, Fhied C, et al. Development and validation of a plasma biomarker panel for discerning clinical significance of indeterminate pulmonary nodules. J Thorac Oncol: IASLC 2013, 8: 31–36. [DOI] [PubMed] [Google Scholar]
- 36.DeCotiis C, Hu Y, Greenberg AK, et al. Inflammatory cytokines and non-small cell lung cancer in a CT-scan screening cohort: Background review of the literature. Cancer Biomark 2016; 16: 219–233. [DOI] [PubMed] [Google Scholar]
- 37.Kullmann T, Barta I, Csiszér E, et al. Differential cytokine pattern in the exhaled breath of patients with lung cancer. Pathol Oncol Res 2008; 14: 481–483. [DOI] [PubMed] [Google Scholar]
- 38.Shih CM, Lee YL, Chiou HL, et al. The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung Cancer 2005; 50: 291–297. [DOI] [PubMed] [Google Scholar]
- 39.Arenberg DA, White ES, Burdick MD, et al. Improved survival in tumor-bearing SCID mice treated with interferon-γ-inducible protein 10 (IP-10/CXCL10). Cancer Immunol Immunother 2001; 50: 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Vita F, Orditura M, Galizia G, et al. Serum interleukin-10 levels as a prognostic factor in advanced non-small cell lung cancer patients. Chest 2000; 117: 365–373. [DOI] [PubMed] [Google Scholar]
- 41.Arenberg DA, Kunkel SL, Polverini PJ, et al. Interferon-y-inducible Protein 10 (IP-10) Is an Angiostatic Factor That Inhibits Human Non-small Cell Lung Cancer (NSCLC) Tumorigenesis and Spontaneous Metastases. J Exp Med 1996; 184: 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belperio JA, Keane MP, Arenberg DA, et al. CXC chemokines in angiogenesis. J Leukocyte Biol 2000; 68: 1–8. [PubMed] [Google Scholar]