Abstract
The HER2 (official name ERBB2) gene encodes a membrane receptor in the epidermal growth factor receptor family amplified and overexpressed in adenocarcinoma. Activating mutations also occur in several cancers. We report mutation analyses of the HER2 kinase domain in 7497 histologically diverse cancers. Forty-five genes, including the kinase domain of HER2 with HER2 IHC and dual in situ hybridization, were analyzed in tumors from 7497 patients with cancer, including 850 breast, 770 colorectal, 910 non–small cell lung, 823 uterine or cervical, 1372 ovarian, and 297 pancreatic cancers, as well as 323 melanomas and 2152 other solid tumors. Sixty-nine HER2 kinase domain mutations were identified in tumors from 68 patients (approximately 1% of all cases, ranging from absent in sarcomas to 4% in urothelial cancers), which included previously published activating mutations and 13 novel mutations. Fourteen cases with coexisting HER2 mutation and amplification and/or overexpression were identified. Fifty-two of 68 patients had additional mutations in other analyzed genes, whereas 16 patients (23%) had HER2 mutations identified as the sole driver mutation. HER2 mutations coexisted with HER2 gene amplification and overexpression and with mutations in other functionally important genes. HER2 mutations were identified as the only driver mutation in a significant proportion of solid cancers. Evaluation of anti-HER2 therapies in nonamplified, HER2-mutated cancers is warranted.
HER2 [official name ERBB2; human epidermal growth factor receptor (EGFR) 2/Homo sapiens v-erb-b2 erythroblastic leukemia viral oncogene homolog 2] gene amplification and overexpression are found in a variety of human cancers. This alteration is associated with a worse overall survival in patients with breast, ovarian, endometrial, salivary gland, and gastric carcinomas.1, 2, 3, 4, 5 With the advent of HER2-targeted therapeutics, such as trastuzumab, lapatinib, pertuzumab, and trastuzumab emtansine, patients with HER2-amplified or -overexpressed breast6, 7, 8, 9, 10 and gastric11 cancers have had significant improvements in outcome when treated with these drugs, especially in combination with chemotherapy. Recently, activating mutations have been identified in the extracellular and tyrosine kinase domains of HER2 in breast,12, 13, 14, 15, 16, 17, 18, 19, 20 lung,15, 21, 22, 23, 24, 25, 26 gastric,16, 25 colorectal,16, 27 liver,28 ovarian,25, 29 cervical,30 urothelial,31, 32 and brain25, 33 cancers, predominantly among those lacking HER2 gene amplification.13 Most of these mutations have been found in cultured cells and xenografts to be associated with activation of the HER2 pathway. Currently available HER2 tyrosine kinase inhibitors inhibit HER2 in model systems with specific activating mutations.13 Because of the potential importance of HER2 mutations in a broad spectrum of human cancers, we report our experience with these mutations.
Materials and Methods
Tissue Specimens
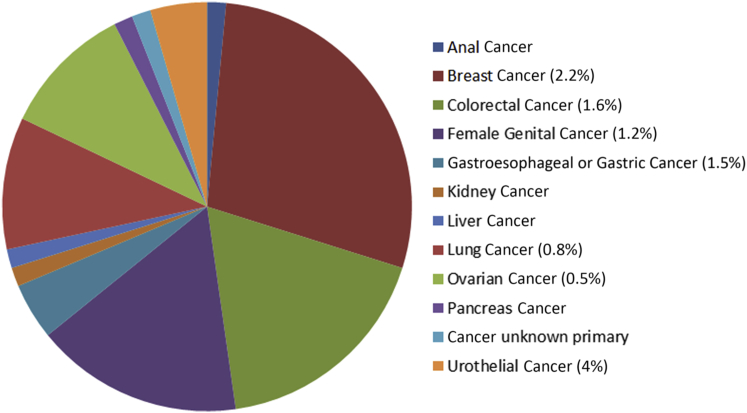
Formalin-fixed, paraffin-embedded (FFPE) tissue blocks from 7497 cancer patients were used for this investigation. The study included 850 breast carcinomas, 770 colorectal carcinomas, 966 lung carcinomas (913 non–small cell lung cancers and 53 small cell lung cancers), 823 uterine cancers, 1372 ovarian epithelial carcinomas, 297 pancreatic cancers, 323 melanomas, and 2152 other solid malignant tumors (Figure 1).
Figure 1.
Patient distribution and relative frequency of the HER2 mutations by cancer type and lineage (frequency is only indicated in lineages with more than three mutations detected).
Next-Generation Sequence Analysis
Direct sequence analysis of HER2 was performed on genomic DNA isolated from a FFPE tumor sample using the MiSeq platform (Illumina, San Diego, CA). DNA was isolated from FFPE tissue using the QIAamp DNA FFPE tissue kit (Qiagen, Valencia, CA). Before extraction samples were evaluated for tumor density by a board-certified pathologist (W.W. or Z.G.) and manually microdissected. Library preparation was performed using the Illumina TruSeq Amplicon Cancer panel and sequenced using the Illumina MiSeq platform. Specific regions of the genome were amplified using the Illumina TruSeq Amplicon Cancer Panel, which includes analysis of 212 amplicons from 45 different cancer-related genes (ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2/HER2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, VHL, EZH2, and IDH2). The cancer panel covers HER2 from amino acids 746 to 827 and 832 to 883. Sequencing reads from multiplex sequencing reactions were assigned to the corresponding biological samples based on a sequence barcode using Illumina software. Initial variant candidates were identified using Illumina miSeq Reporter software version 2.1.40, including Illumina somatic mutation detection software. The mutations that were flagged as low quality by Illumina software (MiSeq Reporter version 2.1.40, Somatic Variant Caller version 2.1.7.0, and PINDEL version 024t) were subsequently removed. For final mutation calls, only mutations with variant allele frequency that was statistically significantly above an internally determined threshold were called mutations (>99% confidence); those that do not reach the level of significance were deemed indeterminate. For a gene to be called wild type, all target amplicons for that gene must have at least 100-fold coverage. All variants reported by this analysis are detected with >99% confidence based on the frequency of the mutation present and the amplicon coverage. Small insertions or deletions of up to 27 bp are detected by this assay.
HER2 Protein Status by IHC
HER2 protein status was determined using the Ventana PATHWAY anti-HER2 (4B5) rabbit monoclonal primary antibody (Ventana Medical Systems, Inc., Tucson, AZ) with an immunohistochemistry (IHC) staining protocol (CC2 mild; iView DAB Detection Kit) on the Ventana Benchmark ULTRA automated platform. The immunostaining was interpreted as no (0), weak (1+), moderate (2+), or strong (3+) immunostaining, with 0 and 1+ considered HER2 negative, 3+ considered HER2 positive, and 2+ considered indeterminant according to requirements described elsewhere.34, 35
HER2 Gene Amplification Status by Dual in Situ Hybridization
HER2 gene amplification was determined with bright field microscopy using tissue sections processed with the INFORM HER2 Dual ISH DNA Probe Cocktail Assay (Ventana Medical Systems, Inc.) according to the manufacturer's instructions as described elsewhere.5 Automated staining was performed using the Ventana Benchmark ULTRA platform (Ventana Medical Systems, Inc.) with reagents supplied by the manufacturer. HER2-to-chromosome 17 centromere ratios and amplification status were calculated in an identical fashion to fluorescence in situ hybridization with ratios ≥2.0 considered as HER2 amplification.34, 35 Tissue sections were systematically evaluated for the presence of intratumoral HER2 genomic heterogeneity, defined as the presence of both amplified and not amplified HER2 subpopulations of carcinoma cells in a single tumor arranged in geographically distinct areas. No genomic heterogeneity of HER2 amplification or overexpression was observed in 68 patients with HER2 mutations.
Results
Breast Cancer
Nineteen of 850 breast carcinomas (2.2%) had mutations in HER2 in exons 19, 20, or 21 that encode the tyrosine kinase intracellular domain (Figures 1 and 2). The histopathologic findings of these 19 breast carcinomas were as follows: eight ductal, three lobular, two pleomorphic lobular, one micropapillary, and five poorly differentiated carcinomas. The most frequently altered codon was L755 (10 mutations, L755S) followed by D769 (four mutations, D769Y/H/E), and one of each of the following mutations: S779F, P780_Y781insGSP, R814C, L869R, and T875I (Table 1). Three of the breast carcinomas were estrogen receptor (ER) positive and progesterone receptor (PR) positive, five were ER positive and PR negative, and eight were triple negative. The ER and PR status of the other three cases is unknown. Five of the 19 breast cancer cases (26%) with HER2 mutations also had HER2 amplification and two additional cases had HER2 overexpression by IHC but not amplification (Table 1). Interestingly, both these latter cases, as well as one with HER2 amplification, had mutation on codon D769 (D769Y/H), although another breast cancer with a D769Y did not have HER2 amplification or overexpression (Table 1). All seven of the breast cancers with HER2 mutations and either HER2 amplification or overexpression had mutations in one or more of the other 44 genes analyzed in the cancer panel (six of the seven had TP53 mutations), whereas only seven of the 12 cases with HER2 mutations but without HER2 amplification or overexpression had mutations in the other genes analyzed (P = 0.068).
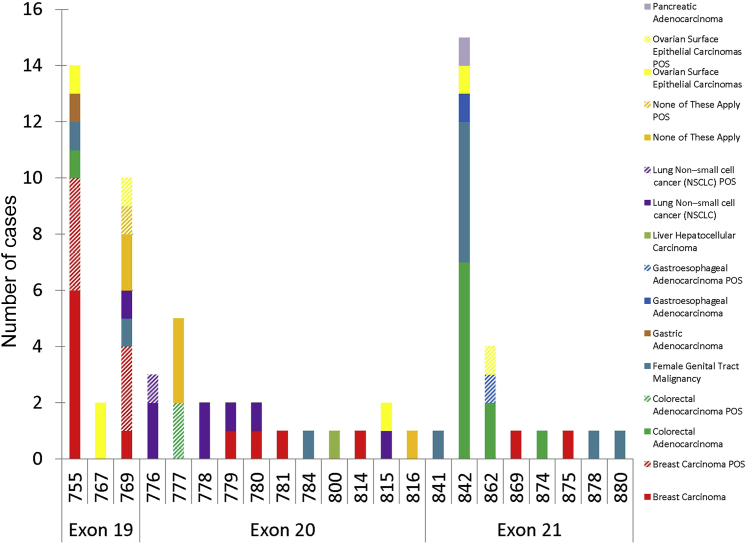
Figure 2.
HER2 mutation spectrum in solid tumors. POS, HER2-positive by HER2 fluorescent in situ hybridization or HER2 overexpression by immunohistochemistry (3+).
Table 1.
Diversity of Human Cancers with HER2 Mutations
| Case no. | Cancer type | HER2 mutation∗ | Exon | Activation status | HER2 IHC protein status† | HER2 status by DISH† | Other mutant genes | Encoded amino acid change of other mutant genes |
|---|---|---|---|---|---|---|---|---|
| 1 | Breast carcinoma | L755S | 19 | Activating | Equivocal | Amplified | TP53 | E204X |
| 2 | Breast carcinoma | L755S | 19 | Activating | Equivocal | Not amplified | ||
| 3 | Breast carcinoma | L755S | 19 | Activating | Equivocal | Amplified | PIK3CA; TP53; GNAS | G1049R; K292X; R201G |
| 4 | Breast carcinoma | L755S | 19 | Activating | Negative (1+) | Not amplified | ||
| 5 | Breast carcinoma | L755S | 19 | Activating | Positive | Amplified | PIK3CA; TP53; GNAS | G1049R; K292X; R201G |
| 6 | Breast carcinoma | L755S | 19 | Activating | Equivocal | Amplified | PIK3CA; TP53 | H1047R; H179L |
| 7 | Breast carcinoma | L755S | 19 | Activating | Negative (1+) | Not amplified | PIK3CA; APC; TP53 | E545K; R1146C; E271X |
| 8 | Breast carcinoma | L755S | 19 | Activating | Negative (0) | Not amplified | TP53 | R306X |
| 9 | Breast carcinoma | L755S | 19 | Activating | Negative (1+) | Not amplified | ||
| 10 | Breast carcinoma | L755S | 19 | Activating | Negative (1+) | Not amplified | ||
| 11 | Colorectal cancer | L755S | 19 | Activating | Negative (1+) | Not amplified | ||
| 12‡ | Endometrial cancer | L755S | 19 | Activating | Negative (0) | Not amplified | TP53; HNF1A; FGFR2 | R273C; P291fs; K292N; |
| 13 | Gastric cancer | L755S | 19 | Activating | Negative (0) | Not amplified | PIK3CA | H1047R |
| 14 | Ovarian epithelial carcinomas | L755S | 19 | Activating | Equivocal | Not amplified | ||
| 15 | Ovarian epithelial carcinomas | I767F | 19 | VUS | Negative (0) | Not amplified | FBXW7; CTNNB1 | R479Q; S37C |
| 16 | Ovarian epithelial carcinomas | I767M | 19 | No effect on MCF10A cell line13 | Negative (1+) | Not amplified | TP53 | D281E |
| 17 | Breast carcinoma | D769Y | 19 | Activating13 | Positive | Amplified | TP53 | Y220S |
| 18 | Breast carcinoma | D769Y | 19 | Activating13 | Positive | Not amplified | PIK3CA | H1047R |
| 19 | Breast carcinoma | D769H | 19 | Activating13 | Positive | Not amplified | TP53; HNF1A | E285K; P289fs |
| 20 | Breast carcinoma | D769Y | 19 | Activating13 | Negative (1+) | Not amplified | TP53 | C238Y |
| 21 | Cervical squamous cell carcinoma | D769Y | 19 | Activating13 | Negative (0) | Not amplified | ||
| 22 | Non–small cell lung cancer | D769E | 19 | VUS | Negative (0) | Not amplified | ||
| 23 | Bladder or urothelial carcinoma | D769H | 19 | Activating13 | Positive | Amplified | ||
| 24 | Bladder or urothelial carcinoma | D769H | 19 | Activating13 | Negative (0) | Not amplified | TP53 | E180K |
| 25 | Bladder or urothelial carcinoma | D769H | 19 | Activating13 | Equivocal | Not amplified | APC | P1458S |
| 26 | Ovarian epithelial carcinomas | D769Y | 19 | Activating13 | Equivocal | Amplified | TP53 | Y234C |
| 27 | Non–small cell lung cancer | G776delinsVC | 20 | Activating36 | Equivocal | Not amplified | TP53 | D281N |
| 28 | Non–small cell lung cancer | G776delinsVC | 20 | Activating36 | Equivocal | Amplified | TP53 | F270L |
| 29 | Non–small cell lung cancer | G776delinsVC | 20 | Activating36 | Negative (1+) | Not amplified | ||
| 30 | Colorectal cancer | V777L | 20 | Activating13 | Positive | Amplified | APC; KRAS; SMAD4 | R876X&R1450X; G13D; G365A |
| 31 | Colorectal cancer | V777L | 20 | Activating13 | Positive | Amplified | TP53 | R196X |
| 32 | Anal squamous cell carcinoma | V777L | 20 | Activating13 | Negative (0) | Not amplified | TP53 | Y234C |
| 33 | Neurofibroma | V777L | 20 | Activating13 | Negative (0) | Not amplified | ||
| 34 | Cancer of unknown primary | V777L | 20 | Activating13 | Negative (1+) | Not amplified | BRAF | G464V |
| 35 | Non–small cell lung cancer | G778C | 20 | VUS | Negative (1+) | Not amplified | PTEN | Y16fs&K267fs |
| 36 | Non–small cell lung cancer | G778_P780dup§ | 20 | Activating13 | Equivocal | Not amplified | TP53 | N239D |
| 37 | Breast carcinoma | S779F | 20 | VUS | Negative (0) | Not amplified | PIK3CA | E545K |
| 38 | Breast carcinoma | P780_Y781insGSP | 20 | Activating13 | Equivocal | Not amplified | TP53 | C275F |
| 39 | Endometrial malignant mixed mullerian tumor | R784C | 20 | VUS | Negative (0) | Not amplified | TP53; PTEN | R282W; C250del |
| 40 | Liver hepatocellular carcinoma | L800F | 20 | VUS | Negative (0) | Not amplified | ||
| 41 | Breast carcinoma | R814C | 20 | VUS | Negative (0) | Not amplified | ||
| 42 | Non–small cell lung cancer | G815R | 20 | VUS | Negative (0) | Not amplified | TP53; EGFR | C238S; E746;_A750delinsAP |
| 43 | Ovarian epithelial carcinomas (Fallopian tube) | G815R | 20 | VUS | Negative (1+) | Not amplified | TP53 | L111fs |
| 44 | Kidney (renal cell carcinoma) | R816H | 20 | VUS | Negative (0) | Not amplified | ATM | N870D (VUS) |
| 45 | Uterine rhabdomyosarcoma | L841V | 21 | VUS | Negative (0) | Not amplified | TP53 | I195T |
| 46 | Colorectal cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | TP53; KRAS; APC | S94X; A146T; R1450X |
| 47 | Colorectal cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | KRAS; APC; PIK3CA | G12A; L1302fs; Q546K |
| 48 | Colorectal cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | HNF1A; VHL | G292fs; R167W |
| 49 | Colorectal cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | TP53 | R342X |
| 50 | Colorectal cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | FBXW7; APC | R278X; I1516fs |
| 51 | Colorectal cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | KRAS | G12V |
| 52 | Colorectal cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | PIK3CA; APC | H1047R; R1450X |
| 53 | Endometrial cancer | V842I | 21 | Activating13 | Negative (1+) | Not amplified | PTEN | D268fs |
| 54 | Endometrial cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | PIK3CA | C420R |
| 55 | Endometrial cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | PTEN; | N323fs |
| 56 | Endometrial cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | TP53; FBXW7 | R273H; R465H |
| 57† | Endometrial cancer (same patient as 12) | V842I | 21 | Activating13 | Negative (0) | Not amplified | TP53; HNF1A; FGFR2 | R273C; P291fs; K292N |
| 58 | Gastroesophageal junction adenocarcinoma | V842I | 21 | Activating13 | Negative (0) | Not amplified | ||
| 59 | Ovarian epithelial carcinomas | V842I | 21 | Activating13 | Negative (0) | Not amplified | FGFR2; CTNNB1; PTEN | N549D; S37C; R14fs; |
| 60 | Pancreatic cancer | V842I | 21 | Activating13 | Negative (0) | Not amplified | PTEN; APC; | K267fs; S1465fs |
| 61 | Colorectal cancer | T862A | 21 | VUS | Negative (0) | Not amplified | APC; APC; PIK3CA; NRAS | Q879X; S1465fs; E542K; G12D |
| 62 | Colorectal cancer | T862A | 21 | VUS | Negative (1+) | Not amplified | KRAS | G13D |
| 63 | Gastroesophagel junction adenocarcinoma | T862A | 21 | VUS | Positive | Amplified | TP53 | E221X |
| 64 | Ovarian epithelial carcinomas | T862A | 21 | VUS | Positive | Amplified | TP53 | H193R |
| 65 | Breast carcinoma | L869R | 21 | VUS | Equivocal | Not amplified | PIK3CA; CTNNB1 | N345I; S23T |
| 66 | Colorectal cancer | E874K | 21 | VUS | Negative (1+) | Not amplified | APC; KRAS | Q1429X; G12C |
| 67 | Breast carcinoma | T875I | 21 | VUS | Negative (1+) | Not amplified | PIK3CA; TP53 | H1047R; Y220S |
| 68 | Endometrial cancer | H878Y | 21 | VUS | Negative (0) | Not amplified | ||
| 69 | Cervical squamous cell carcinoma | D880H | 21 | VUS | Negative (0) | Not amplified |
DISH, dual in situ hybridization; IHC, immunohistochemistry; VUS, variant of unknown significance.
Novel mutations not previously reported in the literature are in bold.
The overall concordance between IHC and DISH is 96% (55/57, excluding IHC equivocal cases) (IHC 0/DISH not amplified: 33; IHC 1+/DISH not amplified: 15; IHC equivocal (2+)/DISH not amplified, 7; IHC equivocal (2+)/DISH amplified, 5; IHC positive (3+)/DISH not amplified, 2; IHC positive (3+)/DISH amplified, 7). No association was observed between IHC immunostaining and HER2 mutation status.
Same patient with two HER2 mutations.
G778_P780dup is also known as P780_Y781insGSP mutation.
Ovarian Cancer
Seven of the 1372 ovarian carcinomas (0.5%) analyzed had mutations in the HER2 gene (Figure 1). These included three invasive serous, two endometrioid, and two mucinous carcinomas. The affected codons and mutations were L755S, I767F/M, D769Y, G815R, V842I, and T862A (Table 1). Two of these seven ovarian carcinomas (28%) had coexisting HER2 gene amplification, including the ovarian cancer with a D769Y mutation, as described above for breast cancer, and a not previously reported T862A mutation (Table 1). Both of them were mucinous carcinomas and also had TP53 mutations.
Female Genital Tract Cancer
There were 10 women whose female genital tract cancers had mutations in HER2 among 823 women (1.2%) with uterine malignant tumors (Figure 1). One of these women had an endometrial endometrioid carcinoma with two mutations in HER2, one in codon L755, and another in codon V842 (Table 1). The other nine women with HER2 mutations in their uterine cancers had two squamous cell carcinomas of the cervix (D769Y and D880H), a malignant mixed mullerian tumor (R784C), three clear cell carcinomas (all V842I), one rhabdomyosarcoma (L841V), and two primary endometrial serous carcinomas (V842I and H878Y) (Table 1). Altogether five mutations were V842I (three clear cell carcinomas, one serous, and one endometrioid carcinoma) with the other six mutations as L755S, D769Y, R784C, L841V, H878Y, and D880H. Interestingly, five of the seven endometrial carcinomas had the same mutation in the V842 codon, and three of these five cancers (60%) contained mostly a clear cell component, whereas one of these five (20%) had a serous component. None of these endometrial cancers had HER2 gene amplification; four had mutations in TP53 (Table 1).
Gastroesophageal Adenocarcinoma
Two esophageal adenocarcinomas of 104 esophageal or gastroesophageal adenocarcinomas (1.9%) had one mutation each (V842I and T862A). The one with T862A mutation also had HER2 gene amplification and a mutation in TP53 (Table 1).
Gastric Adenocarcinoma
One of 86 gastric adenocarcinomas (1.2%) had an L755S mutation and a coexisting PIK3CA mutation but no HER2 amplification or overexpression. The other 85 (98.8%) were wild-type HER2.
Colorectal Cancer
Thirteen of 771 colorectal adenocarcinomas (1.6%) had mutations in HER2, particularly V842I detected in seven cancers. Other mutations identified were L755S, V777L, T862A, and E874K (Table 1). Two of 13 colorectal carcinomas (15%) had HER2 amplification and overexpression as well as V777L mutations in the gene.
Hepatocellular Carcinoma
One hepatocellular carcinoma of 46 (2.2%) had an L800F mutation in HER2 (Figure 1).
Pancreatic Adenocarcinoma
One of 297 pancreatic adenocarcinomas (0.3%) had a mutation in HER2 (V842I) (Table 1).
Lung Cancer
Seven HER2 mutations (three G776delinsVC and one each D769E, G778C, G778_P780dup, and G815R) were identified among 913 non–small cell lung cancers (NSCLCs) (0.8%) (Figure 1 and Table 1), whereas no HER2 mutations were identified among 53 small cell lung cancers. One lung cancer with a HER2 G815R mutation also harbored an in-frame deletion/insertion (E746_A750) in exon 19 of EGFR. Two of seven NSCLCs (28%) had an HER2 mutation detected as the sole driver mutation (D769 and EG776delinsVC). All the NSCLCs with HER2 mutations were adenocarcinomas.
Urothelial Cancers
Three of the 69 urothelial carcinomas (4%) of the bladder had a D769H HER2 mutation. One of these three urothelial carcinomas with HER2 mutations also had amplification of the gene and protein overexpression (Table 1).
Kidney
A renal cell carcinoma of collecting duct variant histopathologic type had a R816H mutation, whereas the other 71 renal cell carcinomas did not. None of these had HER2 gene amplification.
Glioblastomas and Gliomas
None of the 195 glioblastomas or the 19 low-grade gliomas had HER2 mutations.
Sarcomas
None of 203 soft-tissue tumors had a mutation detected in HER2.
Other Cancers
Three other neoplasms (an anal squamous cell carcinoma, a neurofibroma, and an adenocarcinoma of unknown primary) had identical HER2 V777L mutations (Table 1).
Overall, 16 of these 68 cancers (24%) had an HER2 kinase domain mutation identified as the sole driver mutation among the 45 genes analyzed for mutations, whereas the other 52 cancers (76%) had mutation(s) detected in other driver genes (Table 1). These driver genes are listed as follows in descending frequency: TP53 (n = 29), PIK3CA (n = 13), APC (n = 10), KRAS (n = 6), PTEN (n = 6), FBXW7 (n = 3), HNF1A (n = 3), CTNNB1 (n = 3), FGFR2 (n = 2), GNAS (n = 2), SMAD4 (n = 1), NRAS (n = 1), BRAF (n = 1), EGFR (n = 1), ATM (n = 1), and VHL (n = 1) (Table 1).
Discussion
HER2 gene amplification is a well-known therapeutic target in breast6, 7, 8, 9 and gastric cancer11 and has been identified in several other cancer primary sites.1, 2, 3, 4, 37, 38 Mutations in the HER2 gene have been reported previously in breast,12, 13, 14, 15, 16, 17, 18, 19, 20 lung,15, 21, 22, 23, 24, 25, 26 gastric,16, 25 colorectal,16, 27 ovarian,25, 29 cervical,30 bladder,31, 32 and brain cancers25, 33 at an overall prevalence rate of approximately 2.2%. We report DNA sequence analyses of the ERBB2 kinase domain from 7497 cancers indicating the mutation spectrum of 69 HER2 mutations from various cancer lineages in 68 patients using the Illumina MiSeq platform (Figures 1 and 2 and Table 1).
Spectrum of Cancers with HER2 Mutations
Mutations were observed in carcinoma of patients with breast (n = 19/850), ovarian (n = 7/1372), uterine (n = 10/822), lung (n = 7/966), gastric and gastroesophageal junction (n = 3/190), colorectal (n = 13/771), liver (n = 1/46), pancreatic (n = 1/297), urothelial (n = 3), kidney (n = 1), and anal canal (n = 1) cancers, as well as an adenocarcinoma of unknown primary (n = 1) and a neurofibroma (n = 1). We did not identify HER2 mutations in prostate carcinomas (n = 62), cholangiocarcinomas (n = 81), extrahepatic biliary adenocarcinomas (n = 13), head and neck squamous carcinomas (n = 113), cutaneous melanomas (n = 323), ocular uveal melanomas (n = 13), soft-tissue sarcomas (n = 203), or neuroendocrine neoplasms (n = 192). We identified HER2 mutations in several primary cancers not previously reported to harbor HER2 mutations, including endometrial carcinomas (n = 7), gastric and gastroesophageal junction adenocarcinomas (n = 3), renal cell carcinoma (n = 1), and pancreatic adenocarcinoma (n = 1), which may be potential targets for HER2-targeted therapy.
HER2 Mutations Not Previously Reported
We identified 13 cancers with novel mutations not previously reported, including I767F, D769E, G778C, S779F, R784C, L800F, R814C, G815R, R816H, L841V, E874K, T875I, and D880H (Table 1). Mutations, previously characterized as HER2-activating mutations (D769Y, D769H, G776VinsC, V777L, P780_Y781insGSP, and V842I),13, 36 were also identified in this series in breast, lung (NSCLC), colorectal, cervical, endometrial, gastroesophageal, bladder, ovarian, pancreatic, and urothelial carcinomas (Table 1). A uterine rhabdomyosarcoma contained an HER2 mutation L841V and a TP53 mutation. Other identified mutations (L755S, I767M, T862A, L869R, and H878Y), previously reported by others,16, 18, 21, 28, 31, 39 have not yet been found to activate increased phosphorylation of HER2, downstream signaling, or altered biological behavior. Nevertheless, the L755S mutation, although not activating in the model systems used for characterization, confers resistance to lapatinib therapy.13
Protein tyrosine kinases in quiescent cells are maintained in an inactive state by a variety of inhibitory mechanisms. On stimulation, the inhibition is relieved, leading to kinase activation. A common activation mechanism involves phosphorylation of tyrosine residues in a segment of the kinase domain referred to as the activation loop. The HER2 kinase is stabilized in an inactive state by a loop (designated αC-β4 loop) between the αC helix and β4 sheet in its kinase domain (amino acids 776 to 783).36 Select mutations in this loop, especially in codons G776 and G778, can release the autoinhibitory interaction and lead to HER2 kinase activation. These mutations in the αC-β4 loop significantly increase the binding affinity for ATP and the catalytic rate and are associated with constitutive signaling.36
Variety of Additional Cancer Genes Mutated
Fifty-two of the 68 patients (76%) had mutations in additional genes (eg, TP53, APC, PIK3CA, PTEN, KRAS). Sixteen patients (24%) had HER2 mutations identified as the sole driver mutation in their tumors, including L755S in breast (n = 4), colorectal (n = 1) and ovarian (n = 1) cancer, D769H in bladder cancer, D769Y and D880H in two cervical squamous cell carcinomas, D769E in NSCLC, G776VinsC in NSCLC, L800F in hepatocellular carcinoma, R814C in breast cancer, and H878Y in endometrial carcinoma (Table 1). Although it has been suggested that mutations of the kinase domain of HER2 are mutually exclusive with alterations in EGFR, KRAS, and ALK,21 we observed a modest number of co-existing mutations in EGFR (n = 3) and KRAS (n = 6), as well as one mutation each in HRAS and NRAS. ALK gene translocations were not detected in NSCLC patients in this study.
Co-Occurrence of HER2 Gene Amplification and Mutation
HER2 mutations were observed across a broad spectrum of adenocarcinomas arising from diverse primary sites. Some of these primary sites (breast, endometrium, ovary, and stomach) are associated with adenocarcinomas having HER2 gene amplification at a prevalence rate of approximately ≥10%. Nevertheless, oncogenic activation of HER2 in these cancers can be by either gene amplification or mutation, and these changes, based on our observations, do not appear to be exclusive. There were 12 cases with coexisting HER2 mutations and amplification, which had the following sequence alterations: L755S (breast, n = 4), V777L (colorectal, n = 2), D769Y (breast and ovary), T862A (esophagus and ovary), D769H (bladder), and G776VinsC (NSCLC). Five of the 19 breast cancer patients (26%) with HER2 mutations reported here also had amplification of the gene, a rate similar to that which we have observed overall in breast cancer cohort studies1, 3, 40 and among screened populations for entry to large clinical trials.41 Surprisingly, colorectal carcinomas, a primary cancer site that overall rarely has HER2 gene amplification, had two of the 13 cases with mutations also showing amplification.
Most breast cancers are ductal in origin and have a relatively high prevalence of HER2 gene amplification (20% to 25%), whereas the less frequent invasive lobular carcinomas (ILCs) have a low prevalence of HER2 gene amplification (<5%). In contrast to the low rate of HER2 mutations (2%) observed in invasive ductal carcinomas, ILCs have a relatively high rate of HER2 mutations (23%) among those patients with relapsed disease, validated as ILC with identification of a CDH1 mutation.17 Although this HER2 mutation rate in relapsed ILC is high,17 HER2 mutations in primary invasive lobular carcinomas, although significant, are lower (approximately 9%).14 Our study is consistent with these latter observations, finding that five (three classic and two pleomorphic lobular carcinomas, approximately 26% of the HER2 mutations) harbored kinase domain mutations. We also identified eight HER2 mutations in triple-negative breast cancers among the 19 breast cancers with mutations in our series, an observation consistent with the perspective that these patients may benefit from treatment with HER2 small molecule inhibitors, such as neratinib.13
Study Limitations
One of the limitations of our study is related to our focus on the HER2 hotspots in the kinase domain (exons 19, 20, and 21). Therefore, some activating mutations (eg, G309E, S310Y, S310F) in exon 8 encoding the extracellular domain cluster in subdomain II, a region characterized by 11 disulfide bonds, were not included in the study. Mutations in this region have been found to alter intramolecular disulfide bonds formed by cysteine residues, resulting in the formation of intermolecular disulfide bonds and HER2 dimerization.42 These mutations may also be targeted by small molecule inhibitors of HER2. Our approach would not have identified fusion between HER2 and other genes as described recently by others.17, 31 Germline DNA was not available from any of these patients; therefore, we could not confirm that the novel mutations we identified are acquired somatic mutations and not rare inherited single-nucleotide polymorphisms.
In a study of 3800 NSCLC patients, 65 (1.7%) harbored an HER2 mutation, and among those who received HER2-based treatment, a disease control rate of 93% for trastuzumab-based therapies (n = 15) and a disease control rate of 100% for afatinib (n = 3) were observed but with no response to lapatinib (n = 3).23 Afatinib is clinically active in patients with lung adenocarcinomas that contain HER2 mutations in the kinase domain.43 On the basis of these results, the National Comprehensive Cancer Network guidelines now recommend trastuzumab for the treatment of NSCLC patients with HER2 mutations.
Potential Functional Significance of HER2 Mutations
Mutations in cancer have been categorized as either functional alterations affecting critical genes underlying the neoplastic process or as nonfunctional passenger changes.44 Although the incidence of HER2 mutations is relatively modest, the repetitive occurrence of a limited number of mutations encoding functionally important and highly conserved domains across a broad spectrum of cancers and the similarity of some HER2 tyrosine kinase domain mutations with the homologous EGFR mutations indicate that HER2 mutations are functionally important alterations, not passenger alterations. The functional and oncogenic importance of many of these mutations has already been revealed in experimental models.13, 36, 42, 45, 46 Furthermore, work with experimental models and anecdotal reports of responsiveness to targeted therapy in individual patients47 strongly indicate that HER2-activating mutations are suitable for treatment with HER2-targeted therapeutics. Neratinib in particular has been effective in growth inhibition on soft agar and inhibition of HER2 autophosphorylation and phosphorylation of downstream signaling proteins.13, 48 Trastuzumab has been effective at inhibiting growth of tumors bearing extracellular HER2 mutations.42 The potential for these activating mutations to be used for selection of patients to targeted therapies is currently being evaluated in clinical trials.
Footnotes
Supported by Caris Life Sciences and grants from the Breast Cancer Research Foundation (M.F.P.) and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (M.F.P.).
Disclosures: W.W., W.S.C., N.X., R.B., A.G., Z.T., J.S., S.Z.M., and Z.G. are employees of Caris Life Sciences and have stock options. G.B. is an employee of Caris Life Sciences. M.F.P. receives remuneration for consulting for F. Hoffmann La Roche Ltd., GlaxoSmithKline, Halozyme Therapeutics, NanoString Technologies, Inc., Oncomed Pharmaceuticals, Inc., Sanofi-Aventis, and Ventana Medical Systems, Inc. M.F.P. also receives travel sponsorship/reimbursements from Cepheid, F. Hoffmann La Roche Ltd., GlaxoSmithKline, NanoString Technologies, Inc., Sanofi-Aventis, and Ventana Medical Systems, Inc.
References
- 1.Press M.F., Bernstein L., Thomas P.A., Meisner L.F., Zhou J.Y., Ma Y., Hung G., Robinson R.A., Harris C., El-Naggar A., Slamon D.J., Phillips R.N., Ross J.S., Wolman S.R., Flom K.J. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol. 1997;15:2894–2904. doi: 10.1200/JCO.1997.15.8.2894. [DOI] [PubMed] [Google Scholar]
- 2.Press M.F., Pike M.C., Hung G., Zhou J.Y., Ma Y., George J., Dietz-Band J., James W., Slamon D.J., Batsakis J.G., El-Naggar A. Amplification and overexpression of HER-2/neu in carcinomas of the salivary gland: correlation with poor prognosis. Cancer Res. 1994;54:5675–5682. [PubMed] [Google Scholar]
- 3.Slamon D.J., Godolphin W., Jones L.A., Holt J.A., Wong S.G., Keith D.E., Levin W.J., Stuart S.G., Udove J., Ullrich A., Press M.F. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 4.Saffari B., Jones L.A., el-Naggar A., Felix J.C., George J., Press M.F. Amplification and overexpression of HER-2/neu (c-erbB2) in endometrial cancers: correlation with overall survival. Cancer Res. 1995;55:5693–5698. [PubMed] [Google Scholar]
- 5.Gordon M.A., Gundacker H.M., Benedetti J., Macdonald J.S., Baranda J.C., Levin W.J., Blanke C.D., Elatre W., Weng P., Zhou J.Y., Lenz H.J., Press M.F. Assessment of HER2 gene amplification in adenocarcinomas of the stomach or gastroesophageal junction in the INT-0116/SWOG9008 clinical trial. Ann Oncol. 2013;24:1754–1761. doi: 10.1093/annonc/mdt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slamon D.J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J., Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 7.Slamon D., Eiermann W., Robert N., Pienkowski T., Martin M., Press M., Mackey J., Glaspy J., Chan A., Pawlicki M., Pinter T., Valero V., Liu M.C., Sauter G., von Minckwitz G., Visco F., Bee V., Buyse M., Bendahmane B., Tabah-Fisch I., Lindsay M.A., Riva A., Crown J., Breast Cancer International Research G Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365:1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romond E.H., Perez E.A., Bryant J., Suman V.J., Geyer C.E., Jr., Davidson N.E., Tan-Chiu E., Martino S., Paik S., Kaufman P.A., Swain S.M., Pisansky T.M., Fehrenbacher L., Kutteh L.A., Vogel V.G., Visscher D.W., Yothers G., Jenkins R.B., Brown A.M., Dakhil S.R., Mamounas E.P., Lingle W.L., Klein P.M., Ingle J.N., Wolmark N. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 9.Baselga J., Cortes J., Kim S.B., Im S.A., Hegg R., Im Y.H., Roman L., Pedrini J.L., Pienkowski T., Knott A., Clark E., Benyunes M.C., Ross G., Swain S.M., Group C.S. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366:109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma S., Miles D., Gianni L., Krop I.E., Welslau M., Baselga J., Pegram M., Oh D.Y., Dieras V., Guardino E., Fang L., Lu M.W., Olsen S., Blackwell K., Group E.S. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bang Y.J., Van Cutsem E., Feyereislova A., Chung H.C., Shen L., Sawaki A., Lordick F., Ohtsu A., Omuro Y., Satoh T., Aprile G., Kulikov E., Hill J., Lehle M., Ruschoff J., Kang Y.K., ToGA Trial Investigators Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 12.Banerji S., Cibulskis K., Rangel-Escareno C., Brown K.K., Carter S.L., Frederick A.M. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bose R., Kavuri S.M., Searleman A.C., Shen W., Shen D., Koboldt D.C., Monsey J., Goel N., Aronson A.B., Li S., Ma C.X., Ding L., Mardis E.R., Ellis M.J. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan Z., Jaiswal B.S., Stinson J., Janakiraman V., Bhatt D., Stern H.M., Yue P., Haverty P.M., Bourgon R., Zheng J., Moorhead M., Chaudhuri S., Tomsho L.P., Peters B.A., Pujara K., Cordes S., Davis D.P., Carlton V.E., Yuan W., Li L., Wang W., Eigenbrot C., Kaminker J.S., Eberhard D.A., Waring P., Schuster S.C., Modrusan Z., Zhang Z., Stokoe D., de Sauvage F.J., Faham M., Seshagiri S. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.W., Soung Y.H., Seo S.H., Kim S.Y., Park C.H., Wang Y.P., Park K., Nam S.W., Park W.S., Kim S.H., Lee J.Y., Yoo N.J., Lee S.H. Somatic mutations of ERBB2 kinase domain in gastric, colorectal, and breast carcinomas. Clin Cancer Res. 2006;12:57–61. doi: 10.1158/1078-0432.CCR-05-0976. [DOI] [PubMed] [Google Scholar]
- 17.Ross J.S., Wang K., Sheehan C.E., Boguniewicz A.B., Otto G., Downing S.R., Sun J., He J., Curran J.A., Ali S., Yelensky R., Lipson D., Palmer G., Miller V.A., Stephens P.J. Relapsed classic E-cadherin (CDH1)-mutated invasive lobular breast cancer shows a high frequency of HER2 (ERBB2) gene mutations. Clin Cancer Res. 2013;19:2668–2676. doi: 10.1158/1078-0432.CCR-13-0295. [DOI] [PubMed] [Google Scholar]
- 18.Shah S.P., Morin R.D., Khattra J., Prentice L., Pugh T., Burleigh A., Delaney A., Gelmon K., Guliany R., Senz J., Steidl C., Holt R.A., Jones S., Sun M., Leung G., Moore R., Severson T., Taylor G.A., Teschendorff A.E., Tse K., Turashvili G., Varhol R., Warren R.L., Watson P., Zhao Y., Caldas C., Huntsman D., Hirst M., Marra M.A., Aparicio S. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 19.Shah S.P., Roth A., Goya R., Oloumi A., Ha G., Zhao Y. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens P.J., Tarpey P.S., Davies H., Van Loo P., Greenman C., Wedge D.C. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arcila M.E., Chaft J.E., Nafa K., Roy-Chowdhuri S., Lau C., Zaidinski M., Paik P.K., Zakowski M.F., Kris M.G., Ladanyi M. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18:4910–4918. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding L., Getz G., Wheeler D.A., Mardis E.R., McLellan M.D., Cibulskis K. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazieres J., Peters S., Lepage B., Cortot A.B., Barlesi F., Beau-Faller M., Besse B., Blons H., Mansuet-Lupo A., Urban T., Moro-Sibilot D., Dansin E., Chouaid C., Wislez M., Diebold J., Felip E., Rouquette I., Milia J.D., Gautschi O. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31:1997–2003. doi: 10.1200/JCO.2012.45.6095. [DOI] [PubMed] [Google Scholar]
- 24.Shigematsu H., Takahashi T., Nomura M., Majmudar K., Suzuki M., Lee H., Wistuba I.I., Fong K.M., Toyooka S., Shimizu N., Fujisawa T., Minna J.D., Gazdar A.F. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 25.Stephens P., Hunter C., Bignell G., Edkins S., Davies H., Teague J. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto H., Higasa K., Sakaguchi M., Shien K., Soh J., Ichimura K., Furukawa M., Hashida S., Tsukuda K., Takigawa N., Matsuo K., Kiura K., Miyoshi S., Matsuda F., Toyooka S. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J Natl Cancer Inst. 2014;106:djt338. doi: 10.1093/jnci/djt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bekaii-Saab T., Williams N., Plass C., Calero M.V., Eng C. A novel mutation in the tyrosine kinase domain of ERBB2 in hepatocellular carcinoma. BMC Cancer. 2006;6:278. doi: 10.1186/1471-2407-6-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojesina A.I., Lichtenstein L., Freeman S.S., Pedamallu C.S., Imaz-Rosshandler I., Pugh T.J. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross J.S., Wang K., Gay L.M., Al-Rohil R.N., Nazeer T., Sheehan C.E., Jennings T.A., Otto G.A., Donahue A., He J., Palmer G., Ali S., Nahas M., Young G., Labrecque E., Frampton G., Erlich R., Curran J.A., Brennan K., Downing S.R., Yelensky R., Lipson D., Hawryluk M., Miller V.A., Stephens P.J. A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma. Clin Cancer Res. 2014;20:68–75. doi: 10.1158/1078-0432.CCR-13-1992. [DOI] [PubMed] [Google Scholar]
- 33.The Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Press M.F., Slamon D.J., Flom K.J., Park J., Zhou J.Y., Bernstein L. Evaluation of HER-2/neu gene amplification and overexpression: comparison of frequently used assay methods in a molecularly characterized cohort of breast cancer specimens. J Clin Oncol. 2002;20:3095–3105. doi: 10.1200/JCO.2002.09.094. [DOI] [PubMed] [Google Scholar]
- 35.Wolff A.C., Hammond M.E., Hicks D.G., Dowsett M., McShane L.M., Allison K.H., Allred D.C., Bartlett J.M., Bilous M., Fitzgibbons P., Hanna W., Jenkins R.B., Mangu P.B., Paik S., Perez E.A., Press M.F., Spears P.A., Vance G.H., Viale G., Hayes D.F., American Society of Clinical Oncology. College of American Pathologists Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 36.Fan Y.X., Wong L., Ding J., Spiridonov N.A., Johnson R.C., Johnson G.R. Mutational activation of ErbB2 reveals a new protein kinase autoinhibition mechanism. J Biol Chem. 2008;283:1588–1596. doi: 10.1074/jbc.M708116200. [DOI] [PubMed] [Google Scholar]
- 37.Sauter G., Moch H., Moore D., Carroll P., Kerschmann R., Chew K., Mihatsch M.J., Gudat F., Waldman F. Heterogeneity of erbB-2 gene amplification in bladder cancer. Cancer Res. 1993;53:2199–2203. [PubMed] [Google Scholar]
- 38.Simon R., Atefy R., Wagner U., Forster T., Fijan A., Bruderer J., Wilber K., Mihatsch M.J., Gasser T., Sauter G. HER-2 and TOP2A coamplification in urinary bladder cancer. Int J Cancer. 2003;107:764–772. doi: 10.1002/ijc.11477. [DOI] [PubMed] [Google Scholar]
- 39.Kelly R.J., Carter C.A., Giaccone G. HER2 mutations in non-small-cell lung cancer can be continually targeted. J Clin Oncol. 2012;30:3318–3319. doi: 10.1200/JCO.2012.43.4902. [DOI] [PubMed] [Google Scholar]
- 40.Press M.F., Pike M.C., Chazin V.R., Hung G., Udove J.A., Markowicz M., Danyluk J., Godolphin W., Sliwkowski M., Akita R. Her-2/neu expression in node-negative breast cancer: direct tissue quantitation by computerized image analysis and association of overexpression with increased risk of recurrent disease. Cancer Res. 1993;53:4960–4970. [PubMed] [Google Scholar]
- 41.Press M.F., Sauter G., Bernstein L., Villalobos I.E., Mirlacher M., Zhou J.Y., Wardeh R., Li Y.T., Guzman R., Ma Y., Sullivan-Halley J., Santiago A., Park J.M., Riva A., Slamon D.J. Diagnostic evaluation of HER-2 as a molecular target: an assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clin Cancer Res. 2005;11:6598–6607. doi: 10.1158/1078-0432.CCR-05-0636. [DOI] [PubMed] [Google Scholar]
- 42.Greulich H., Kaplan B., Mertins P., Chen T.H., Tanaka K.E., Yun C.H., Zhang X., Lee S.H., Cho J., Ambrogio L., Liao R., Imielinski M., Banerji S., Berger A.H., Lawrence M.S., Zhang J., Pho N.H., Walker S.R., Winckler W., Getz G., Frank D., Hahn W.C., Eck M.J., Mani D.R., Jaffe J.D., Carr S.A., Wong K.K., Meyerson M. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109:14476–14481. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Greve J., Teugels E., Geers C., Decoster L., Galdermans D., De Mey J., Everaert H., Umelo I., In't Veld P., Schallier D. Clinical activity of afatinib (BIBW 2992) in patients with lung adenocarcinoma with mutations in the kinase domain of HER2/neu. Lung Cancer. 2012;76:123–127. doi: 10.1016/j.lungcan.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herter-Sprie G.S., Greulich H., Wong K.K. Activating mutations in ERBB2 and their impact on diagnostics and treatment. Front Oncol. 2013;3:86. doi: 10.3389/fonc.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S.E., Narasanna A., Perez-Torres M., Xiang B., Wu F.Y., Yang S., Carpenter G., Gazdar A.F., Muthuswamy S.K., Arteaga C.L. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 47.Vornicova O., Hershkovitz D., Yablonski-Peretz T., Ben-Itzhak O., Keidar Z., Bar-Sela G. Treatment of metastatic extramammary Paget's disease associated with adnexal adenocarcinoma, with anti-HER2 drugs based on genomic alteration ERBB2 S310F. Oncologist. 2014;19:1006–1007. doi: 10.1634/theoncologist.2014-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minami Y., Shimamura T., Shah K., LaFramboise T., Glatt K.A., Liniker E., Borgman C.L., Haringsma H.J., Feng W., Weir B.A., Lowell A.M., Lee J.C., Wolf J., Shapiro G.I., Wong K.K., Meyerson M., Thomas R.K. The major lung cancer-derived mutants of ERBB2 are oncogenic and are associated with sensitivity to the irreversible EGFR/ERBB2 inhibitor HKI-272. Oncogene. 2007;26:5023–5027. doi: 10.1038/sj.onc.1210292. [DOI] [PubMed] [Google Scholar]