SUMMARY
Gliomas account for the majority of primary tumors of the CNS, of which glioblastoma (GBM) is the most common and malignant, and for which survival is very poor. Despite significant inter- and intra-tumor heterogeneity, all patients are treated with a standardized therapeutic approach. While some clinical features of GBM patients have already been established as classic prognostic factors (e.g., patient age at diagnosis and Karnofsky performance status), one of the most important research fields in neuro-oncology today is the identification of novel molecular determinants of patient survival and tumor response to therapy. Here, we aim to review and discuss some of the most relevant and novel prognostic biomarkers in adult and pediatric GBM patients that may aid in stratifying subgroups of GBMs and rationalizing treatment decisions.
Practice Points.
Glioblastoma multiforme (GBM) is the most frequent and lethal tumor of the CNS, for which curative therapies are not available. Classic prognostic factors such as patient's age and performance status, together with tumor characteristics, including grade and molecular features, predict survival in GBM patients.
The methylation status of the MGMT gene promoter and mutation in IDH1 and IDH2 genes are among the most promising prognostic biomarkers in GBM.
GBMs may be stratified into four molecular subtypes – classical, mesenchymal, proneural and neural – each displaying different underlying genetic alterations and gene expression signatures. The assessment of the subtype of each GBM might be important while designing therapeutic approaches.
A variety of putative prognostic biomarkers have been identified in adult GBM patients. Some examples include the presence of mutations or the expression levels of receptor tyrosine kinases, growth factors and intracellular targets (e.g., PI3K); miRNA gene signatures; and serum concentrations of the YKL-40 protein. More recently, the expression of HOX and cancer stem cell-associated genes, and loss of chromosome 10 were suggested as novel putative biomarkers predictive of survival in adult GBM, but further studies are required to validate their value.
Mutations in the H3F3A gene are specific to pediatric GBMs, highlighting that pediatric and adult GBMs present a distinct underlying biology. H3F3AK27M mutant tumors have a significantly shorter overall survival than H3F3AG34R/V or wild-type tumors.
The fast-accumulated knowledge on new putative biomarkers of GBM aggressiveness and prognosis holds reason for both optimism and caution. While many of these biomarkers have been validated in independent studies, their clinical applicability to highly heterogeneous GBMs is still limited.
Tumors of the CNS comprise of a broad variety of entities, which range from benign to highly malignant. Typically, their classification is based on their location and histopathological features [1]. Gliomas are the most frequent CNS primary tumors in adults, whose main histological subtypes include astrocytomas, oligodendrogliomas and ependymomas. Astrocytomas represent approximately 70% of all diagnosed gliomas, and are graded from I to IV according to the WHO [1]. Of these, glioblastoma multiforme (GBM; WHO grade IV) is the most frequent and lethal, accounting for more than 50% of all glial tumor types, with an estimated global incidence rate of approximately five per 100,000 people/year [2]. GBMs are characterized by rapid growth and diffuse invasiveness of the adjacent brain parenchyma, and their histopathological features include cellular polymorphism, mitotic activity, nuclear atypia, vascular thrombosis, microvascular proliferation and marked necrosis [1]. Despite several efforts, the treatment for GBM remains mostly palliative, with a median survival of only 15 months [3]. Standard treatment uses a combination of maximum surgical resection, radiation, and concurrent and adjuvant chemotherapy with the alkylating agent temozolomide [3]. Molecular stratification with biomarkers predictive of patient response and outcome may prove crucial in rationalizing treatment decisions. Currently, the most consistently reported and best-established prognostic factors include patient age and performance status, tumor grade and histology, and extent of surgical resection (Box 1) [4–6]. Age is among the most consistent variables associated with GBM patient survival, as older patients fare worse than young patients [4,5,7]. In addition, patients that present higher Karnofsky performance status scores have increased overall survival and better responses to chemotherapy [5–7]. Among the intrinsic tumor characteristics, gliomas of higher WHO grades of malignancy typically have shorter survivals than those of lower grades [8]. Clinically, the extent of tumor surgical resection has also been reported as crucial on influencing GBM patient prognosis, as more complete resections are associated with better outcomes [4,6,7]. In this sense, these classic factors must be clearly assessed when assigning patients for randomized clinical trials, but may also be crucial in aiding clinicians in the refinement of treatment decisions. Nonetheless, in the last decade, studies have identified molecular features that might be prognostically valuable [9–32]. The current most relevant prognostic biomarkers in GBM are summarized in Figure 1 and Box 2, and the most promising ones will be discussed.
Box 1. . Selected clinical prognostic markers for glioblastoma.
Classic prognostic factor
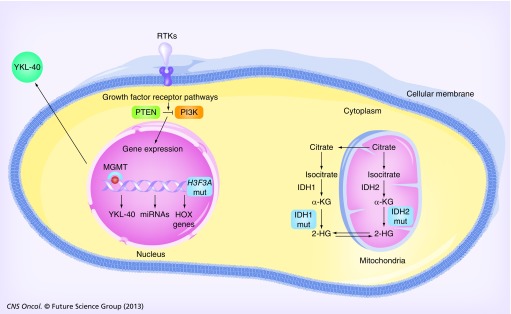
Figure 1. Current putative prognostic biomarkers in glioblastoma.
The deregulation of receptor tyrosine kinase pathways leads to aberrant intracellular signaling, including the PI3K pathway, that, among other effects, induces the transcription of genes responsible for sustaining several cancer hallmarks (e.g., HOX genes). Loss of PTEN function by mutation or loss of heterozygosity has been correlated with poor glioblastoma multiforme (GBM) patient survival. MGMT expression and promoter methylation levels are markers of GBM patient prognosis. Mut IDH1R132H and IDH2R172K enzymes are able to convert α-KG into 2-HG, and GBM patients with tumors presenting IDH mutations have longer survival. Deregulation of several miRNAs has been implicated in both the initiation and progression of GBM, and many have been reported to present prognostic value. YKL-40 is produced by GBM cells and released into the serum, where its levels are predictive of an aggressive phenotype and associated with poor overall patient survival. Mut H3F3AK27M is highly specific to pediatric GBM, and associates with worse prognosis in these patients. Green and orange boxes indicate loss or increased function, respectively. Blue boxes indicate mut IDHs and H3F3A.
2-HG: 2-Hydroxyglutarate; α-KG: α-ketoglutarate; Mut: Mutant; RTK: Receptor tyrosine kinase.
Box 2. . Selected molecular prognostic markers for glioblastoma.
Molecular prognostic marker
MGMT promoter methylation [21,38,40,43,44]
IDH1 and IDH2 mutations [28,45,49,59]
Loss of chromosome 10 [27,123]
Activation of the PI3K/AKT pathway [14,29,96]
Aberrant p53/RB pathway [12,32,129]
HOX gene signatures [19,26]
HOXA9 overexpression [17,19]
CHI3LI (YKL-40) expression [22,27]
Single miRNA/miRNA expression signatures [67–77]
EGFR expression/EGFR mutation (EGFRvIII) [9,20,25,26,84–87]
PTEN expression (wild-type) [25]
Molecular signatures [30,54]
MET overexpression [23]
High expression of angiogenic genes (e.g., VEGF and VEGFR) [30,94]
Stem cell-like gene expression signatures [10,11,24,26,119–122,137]
Activation of MAPK members [29]
Glioma-CpG island methylator phenotype [52,63]
NFKBIA deletion [13]
PTEN and DLL3 expression [30]
H3F3A mutation [31]
MGMT promoter methylation status
The methylation status of the MGMT gene promoter region has been shown by many studies as one of the most promising prognostic biomarkers in GBM, although it has not yet reached worldwide clinical applicability [21,33]. MGMT encodes a DNA repair enzyme that removes alkyl groups from the O6 position of guanine, an important site for DNA alkylation after treatment of tumor cells with alkylating agents. If left unrepaired, these lesions trigger cytotoxicity and apoptosis leading to cell death [34]. Two groups showed that epigenetic silencing of MGMT by promoter methylation induced loss of MGMT expression [35,36], and, therefore, reduced DNA repair activity. Thereafter, Hegi et al. showed that this silencing leads to increased sensitivity of the tumor cells to temozolomide treatment [21]. This sensitivity translated into differences in patient survival, with MGMT methylation being associated with greater overall survival (median: 21.7 months), as well as higher 2-year survival rates (46%), in comparison with patients with unmethylated MGMT (median survival: 12.7 months; 2-year survival: 13.8%). This landmark study suggests that MGMT promoter methylation is an independent and favorable prognostic biomarker in GBM patients, and predictive of response to temozolomide [21]. A follow-up study by Stupp et al. evaluated adult patients with newly diagnosed GBM, who were treated with standard radiotherapy or radiotherapy combined with concomitant and adjuvant temozolomide [37]. In this study, the methylation status of MGMT was evaluated in 206 patients from both cohorts and revealed to be a strong predictor of patient outcome and response to chemoradiation [37]; patients with a methylated MGMT promoter not only presented longer survivals than patients with unmethylated MGMT, but also seemed to benefit more from combined chemoradiotherapy [37]. The value of MGMT methylation status is also supported by a recent clinical trial comparing radiotherapy and temozolomide-based treatment in elderly patients [38]. This reported an association between good outcome and MGMT methylation in the temozolomide cohort, but not in the radiotherapy cohort [38]. These results were further supported by another study showing that elderly GBM patients presenting with methylated MGMT promoters and treated with temozolomide had a significantly longer survival than those who did not present with MGMT promoter methylation, or those on the radiotherapy branch irrespective of MGMT promoter methylation [39]. Thus, the authors of both reports state that treatment decisions for elderly GBM patients would be aided by assessing MGMT promoter methylation [38,39]. A meta-analysis performed by Olson et al. that included 20 different studies and a total of 2018 patients, showed that silencing of MGMT expression was highly associated with improved overall survival in patients receiving adjuvant chemotherapy, a mild association in patients that received adjuvant radiotherapy and no benefit in those submitted to surgery alone [40]. Nonetheless, other reports have not supported a statistically significant association between MGMT methylation and survival. For example, a study by Costa et al. that analyzed the methylation status of MGMT in a set of 90 GBM patients treated with postoperative temozolomide-based chemoradiation, observed a trend for longer progression-free and overall survival in GBM patients presenting with MGMT promoter methylation, but the differences did not reach statistical significance [15]. Similar results were observed in other studies and reviewed by Costa et al. [15]. Another study by van der Bent et al. also showed that the methylation status of MGMT did not present prognostic significance in GBM patients, and was not able to predict the responses to adjuvant procarbazine, lomustine and vincristine chemotherapy [41]. This is mainly due to the heterogeneity of the study participants, not only with respect to grade, histology and treatment, but also analysis of MGMT mRNA expression, methylation status and protein levels [15]. Moreover, sample classification as methylated or unmethylated for a certain gene is still debatable, as the relationship between the CpG methylation at individual sites; overall CpG island methylation and their effects on gene silencing is highly dependent on the location within the gene [42]. Bady et al. evaluated the relationship between MGMT expression, the specific location of CpG methylation and the outcome of patients treated with alkylating agents [43]. In this study, two regions of methylated CpGs negatively correlated with MGMT gene expression, and were strongly associated with patient survival [43]. This is consistent with MGMT expression silencing via CpG methylation, leading to sensitization to alkylating agents [43]. Similarly, Shah et al. also identified three regions of methylated CpGs on MGMT that correlate with favorable patient progression-free survival, within a population of 44 GBM patients treated with radiotherapy and concomitant and adjuvant temozolomide [44].
Although the accumulated knowledge on MGMT has increased in the last few years, its true clinical significance remains unclear. In fact, MGMT methylation status is not yet typically used by clinicians to aid in therapy decisions. Therefore, it is still important to conduct novel clinical trials in prospectively followed patients, and by investigating different drugs and dosages. Indeed, MGMT depletion using pseudosubstrates, such as O6-(4-bromophenyl)guanine or O6-benzylguanine, may be able to improve GBM patient response to temozolomide therapy. If so, overcoming temozolomide resistance due to MGMT promoter methylation will be a major advance in GBM therapy. In the next few years our understanding of MGMT prognostic value and its ability to predict tumor response to different therapies will be very much improved due to the ongoing clinical trials, which may definitively establish it as a major biomarker for GBM patient management.
IDH mutations
Recent genomic studies revealed the presence of mutations in IDH1 and IDH2 genes (IDH when referring to both) as important prognostic factors for GBM [28,45,46]. These NADP-dependent enzymes are able to catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate, with the simultaneous production of NADPH [47]. High-throughput sequencing studies of GBM revealed a new IDH1 mutation, consisting of changing a guanine to an adenine at position 395 of the gene (G395A), thus leading to the replacement of an arginine with a histidine at residue 132 of the protein (R132H) [28]. This heterozygous and somatic mutation was found in 12% of all GBM patients. Similarly, the evaluation of the IDH2 exon sequence revealed a point mutation changing a guanine to an adenine at position 515 of the gene (G515A), causing the substitution of an arginine to a lysine at residue 172 (R172K). This is analogous to the R132 residue in IDH1 [45]. IDH mutations are highly frequent in secondary GBM (up to 80%), but are rare in primary GBM (less than 10%) [45,48]. Importantly, IDH mutations occur more frequently in younger patients, and are associated with greater patient survival [45,49,50]. In addition to demonstrating the capacity of IDH1 mutations on distinguishing anaplastic astrocytomas and GBMs into clinically meaningful prognostic subgroups, a report by Hartmann et al. showed that the distribution of IDH1 mutations is associated with patient's age, thus impacting the prognosis of high-grade astrocytoma patients [51]. Interestingly, in grade III anaplastic astrocytoma patients over 60 years old, the absence of IDH1 mutations was associated with low overall survival, which was comparable with the overall survival of grade IV GBM patients with wild-type IDH1 [51].
IDH1 and IDH2 mutations are mutually exclusive and generally associate with specific genetic and clinical characteristics, when compared with gliomas that present wild-type IDH. Particularly, it was shown that IDH mutations and amplification of EGFR in GBMs are mutually exclusive events [50], and that IDH mutations are often associated with the methylation of the MGMT gene promoter [50,52]. However, these associations are yet to be clarified as they might represent a direct consequence of the mutant IDH activity, or alternative markers for epigenetic changes in tumors presenting IDH mutations [53]. Therefore, the understanding of the link between common genetic events and IDH mutations in GBMs might provide insights into their roles in gliomagenesis [45,54]. Dang et al. showed that cells presenting the IDH1R132H mutation have the capacity to reduce α-ketoglutarate into 2-hydroxyglutarate, while converting NADPH to NADP+, which contributes to tumorigenesis [55]. Some hypotheses have been raised on how the mutant IDH may contribute to gliomagenesis; for example, as dioxygenases require α-ketoglutarate as cofactors and its structure is similar to 2-hydroxyglutarate, the former may compete for the binding site of dioxygenases, thus inhibiting their activity [56]. An example of dioxygenases critical in the context of cancer are PHDs, which are dependent on α-ketoglutarate, and is responsible for the negative regulation of HIF-1α; a transcription factor that stimulates tumor growth under hypoxic conditions by modulating apoptosis, cell survival and angiogenesis [57]. In addition, the production of 2-hydroxyglutarate by mutant IDH inhibits Jmjc domain-containing histone demethylases [58] and TET 5-methylcytosine hydroxylases [56], leading to altered genome-wide histone and DNA methylation. Mutant IDH may also contribute to gliomagenesis by increasing DNA methylation, an effect termed glioma-CpG island methylator phenotype (G-CIMP; as discussed in the ‘Glioma-CpG island methylator phenotype’ section).
Although the understanding of IDH mutations is far from complete, its incorporation into prognostic models might possibly improve the clinical management of a subset of glioma patients. Furthermore, a recent study by Songtao et al. evaluated the response of 86 secondary GBMs to temozolomide treatment, and associated several markers of GBM (including IDH mutations and MGMT promoter methylation status) with overall and progression-free survival [59]. In this study IDH mutations were found in 73.4% of patients, and an association between these mutations and higher progression-free survival was implied [59]. These authors found that patients presenting IDH mutations and MGMT promoter methylation had a better response to temozolomide treatment, and that IDH mutations may increase chemosensitivity in secondary GBMs [59]. In addition, the possibility of therapeutically targeting mutant IDH proteins is conceptually feasible, and might be a strategy to specifically target tumor cells. In fact, very recent studies used small molecule inhibitors of the most common IDH mutants in acute myeloid leukemia (IDH2R140Q) [60] and in gliomas (IDH1R132H) [61]. The treatment of a patient-derived oligodendroglioma cell line harboring IDH1R132H with the small molecule inhibitor AGI-5198 reduced growth in soft agar, and inhibited the growth of xenograft tumors derived from that cell line [61]. At the genome-wide level, several genes associated with glial differentiation were found to be upregulated and to have lost repressive histone marks at their promoter, indicating the putative capacity of IDH1 mutant inhibitors to erase histone modifications [61]. In the acute myeloid leukemia study, the treatment with AGI-6780 of an erythroleukemia cell line expressing IDH2R140Q lowered the 2-hydroxyglutarate production nearly to physiological levels, and the treatment of patient-derived samples induced differentiation of leukemic blasts in samples harboring IDH2R140Q [60]. However, one must take into account the fact that, although promising, these targeted drugs will be a limited approach in primary GBM patients due to the low frequency of these mutations, as well as due to the high intratumor heterogeneity that characterizes these malignancies. In the future, when IDH-targeting drugs become available [62], it will be interesting to determine if the use of these IDH-targeting drugs, individually or combined with other therapies, presents a significant anti-tumor effect in established gliomas. Despite the well-established relevance of IDH mutations in the prognosis of secondary GBM and lower-grade gliomas, their use in the prognostication of primary GBM patients is limited by their low frequency.
Molecular subtypes of GBM
Verhaak et al. stratified 200 GBMs from The Cancer Genome Atlas (TCGA) into four molecular subtypes – classical, mesenchymal, proneural and neural – each displaying different underlying genetic alterations and expression signatures [54]. The classical subtype was defined by displaying the most common genomic aberrations of GBM, with 93% of samples displaying chromosome 7 amplifications and chromosome 10 deletions, 95% showing EGFR amplification, and 95% with homozygous deletion on the Ink4a/ARF locus [54]. The mesenchymal subtype was mainly characterized by high expression levels of CHI3L1 (or YKL-40) and the MET proto-oncogene [30]; NF1 mutation or deletions were also found to be characteristic of this mesenchymal GBM [54]. Hallmarks of the proneural subtype include PDGFRα amplification, IDH1 mutations, loss of heterozygosity and mutations at TP53. Importantly, the proneural subtype was associated with younger age and longer survival [54]. The neural subtype was defined by the differential expression of neuronal markers, such as GABRA1, SLC12A5, NEFL and SYT1 [54]. This molecular classification is relevant because each GBM subtype responds differently to treatment [54]. For example, while aggressive treatment protocols significantly delayed mortality in GBM patients with classical and mesenchymal subtypes, and a tendency for a longer outcome was observed in the neural subtype, patients with proneural GBMs do not seem to benefit from this highly aggressive therapeutic approach [54]. In this sense, some of the genetic events underlying the different GBM subtypes could be used to stratify patients and rationalize treatment decisions, ultimately contributing to more personalized therapies. Moreover, publicly available resources, such as TCGA or Oncomine®, which systematically integrate genomic and clinical data from large cancer patient datasets, have been critical in exploring a wide spectrum of genomic alterations characteristic of each tumor type and evaluating their clinical value.
Glioma-CpG island methylator phenotype
A very recent report by Turcan et al. suggested that the accumulation of 2-hydroxyglutarate might inhibit the α-ketoglutarate-dependent dioxygenase family of enzymes, which in turn will cause histone and DNA hypermethylation – termed the G-CIMP – that results in epigenetic deregulation [63]. Noushmehr et al. reported that 24 out of 272 GBMs from the TCGA dataset were G-CIMP-positive tumors [52]. Of these, 21 were classified within the proneural expression group, which accounts for 30% of all the proneural GBMs in that dataset [52]. Moreover, these authors reported a significantly increased survival for proneural G-CIMP-positive GBM patients in comparison with proneural G-CIMP-negative patients, and indeed to all other nonproneural GBM patients [52]. In Cox multivariate analysis, G-CIMP remained a significant predictor of patient outcome after adjusting for patient age, tumor recurrence status and primary versus secondary GBM status [52]. Of note, G-CIMP-positive tumors were associated with recurrent or secondary tumors, and strongly associated with the IDH1 mutation [52]. In fact, this last association is in agreement with the report of Turcan et al., showing that the mutation of a single gene – IDH1 – is sufficient to establish the G-CIMP by remodeling the methylome, which results in the reorganization of the transcriptome [63]. The single introduction of mutant IDH1 into primary human astrocytes induced the alteration of specific histone marks and extensive DNA hypermethylation, as well as the reshaping of the methylome in a way that resembles the alterations observed in G-CIMP-positive lower-grade gliomas [63]. Moreover, the epigenomic alterations induced by mutant IDH1 activate important gene expression programs that characterize the G-CIMP-positive proneural GBMs, but not other proneural GBMs, and predict increased survival [63]. In this sense, the authors argue that IDH1 mutation is the molecular basis of the G-CIMP [63]. Considering the frequent co-occurrence of G-CIMP and IDH mutations, which is also a putative prognostic biomarker, future studies are necessary to clarify if the putative prognostic value of G-CIMP is independent of the IDH.
miRNAs
ncRNAs have recently emerged as important players in the deregulation of signaling pathways and gene expression in several tumor types, including GBMs. Indeed, the transcriptome is vastly more complex than initially anticipated at the time of the first genome-wide studies; for example, the number of noncoding transcripts is four-times higher than coding sequences. Of all ncRNAs, miRNAs are the most extensively studied, and are key regulators of several biological processes through negative control of gene expression at the post-transcriptional level [64]. Alterations in miRNA genes have been implicated in the initiation and progression of several cancers, either as tumor suppressors or oncogenes depending on their target genes [65]. Specifically, deregulation of these miRNAs has been detected in GBM, with a wide variety of functional roles in cell proliferation, apoptosis, cell cycle regulation, invasion, angiogenesis and glioma stem cell behavior [66]. Many reports have been published describing the ability of miRNAs to predict GBM patient survival [67–77]. For example, Ben-Hamo and Efroni studied five independent datasets and identified a gene–miRNA network comprising of p38 and its associated miRNA miR-9, which can stratify GBM patients into prognostic subgroups [67]. Other studies focused on the miR-10b expression in human glioma tumors and cell lines, and showed that increased expression correlates with increased glioma grade [78,79], and also with increased expression of the G-protein RhoC and the urokinase receptor uPAR, which have been implicated in migration and invasion [78]. Survival of GBM patients with high miR-10b expression was significantly shorter than those patients with low miR-10b expression [68]. In addition, miR-10b downregulates the expression of several tumor suppressor genes, and is associated with poorer GBM patient survival [69]. Silber et al. report that miR-124a is significantly downregulated in grade III and IV astrocytomas relative to non-neoplastic brain tissue [80]. More recently, in a retrospective review of 119 GBM samples, the downregulation of miR-124a was associated with poor patient prognosis [70]. Additionally, another study showed reduced expression of miR-451 in GBMs compared with normal brains [81], suggesting that miR-451 might be a tumor suppressor in the brain. Godlewski et al. later reported that high levels of miR-451 are associated with a poorer survival of GBM patients in the TCGA dataset [73].
In addition to studies addressing the prognostic value of individual miRNAs, others have tried to define miRNA expression signatures that may have higher discriminatory power concerning the prediction of GBM patient survival. Zhi et al. evaluated the expression profile of 200 miRNAs in a set of 84 astrocytoma samples of different WHO grades, and 20 normal adjacent tissue samples and reported a seven-miRNA (miR-24, miR-21, miR-30c, miR-124, miR-181b, miR-137 and miR-106a) differential expression signature in astrocytoma samples in comparison with normal adjacent tissue [74]. Importantly, this finding was validated in an independent set of 40 astrocytomas and 40 matched tissue samples [74]. Additionally, the authors observed an association between the downregulation of miR-137 and advanced state of the disease, and the low expression of miR-181b and miR-106a, and the high expression of miR-21 were significantly associated with shorter patient survival, independent of other clinicopathological factors [74]. Therefore, these authors suggest that miRNA profiling may be a powerful prognostic and diagnostic marker in astrocytomas [74]. Another study evaluated the levels of 365 miRNAs in eight GBM and four anaplastic astrocytomas, revealing 16 candidate markers associated with glioma progression, of which miR-196a and miR-196b presented the most significant differences [71]. High expression of miR-196 was shown to be an independent prognostic factor in a set of 39 GBM patients [71]. This result was reinforced by a recent study that evaluated the expression of miR-196b in 198 glioma tissues [72]. Functional analysis of miR-196b showed that it is a promoter of cellular proliferation, and, as such, its levels are inversely correlated with GBM patient survival [72]. A study performed by Srinivasan et al. assessed the miRNA expression data of 222 GBM patients from the TCGA dataset, and found that a ten-miRNA expression signature was an independent predictor of patient survival [75]. This expression signature was also able to segregate GBM patients into low- and high-risk cohorts [75]. Of the ten-miRNA signature, seven were found to be in the high-risk group (miR-31, miR-222, miR-148a, miR-221, miR-146b, miR-200b and miR-193a) and three were in the low-risk group (miR-20a, miR-106a and miR-17-5p). These are thought to either inhibit or promote several traits of cancer cells [75]. Another study by Zhang et al. performed whole-genome miRNA expression profiling in 82 Chinese GBM patients [76]. The authors identified a five-miRNA signature, comprising of miR-181d, miR-518b, miR-524-5p, miR-566 and miR-1227, that was able to predict patient survival [76]. Patients scoring high with the five-miRNA signature presented poorer overall and progression-free survival when compared with patients presenting low-risk scores [76]. Moreover, this signature was found to be independent of other prognostic factors [76]. Lakomy et al. evaluated the expression of eight miRNAs (miR-21, miR-128a, miR-181c, miR-195, miR-196a, miR-196b, miR-221 and miR-222), as well as the methylation status of MGMT promoters in a group of 38 patients with primary GBMs [77]. In addition to the significant associations between the methylation status of the MGMT promoter and longer overall and progression-free survivals, the authors also found that the expression of miR-195 and -196b was negatively correlated with overall survival. Moreover, miR-181c in combination with miR-21 was highly sensitive and specific in the prediction of tumor progression within 6 months of diagnosis. However, the remaining miRNAs (miR-128a, miR-196a, miR-221 or miR-222) presented no prognostic or predictive value in GBM patients [77]. Importantly, of all the miRNAs reviewed, miR-196b and the miR-181 family have been more consistently reported to be of relevance in glioma. However, when evaluating miRNA expression profiles, special attention should be paid to the non-neoplastic reference due to variations on basal miRNA expression levels inherent to each individual, or to the fact that commercial references are usually RNAs pooled from several non-neoplastic tissues [82]. Nevertheless, the studies presented here demonstrate not only the potential for using single miRNA genes or miRNAs signatures in the prediction of patient outcome, but also how miRNAs may be crucial to the understanding of GBM biology and to the development of new therapeutics.
Growth factor signaling pathways
Overexpression or mutations in receptor tyrosine kinases (RTKs), growth factors and intracellular RTK targets greatly contribute to the tumorigenic process, and may represent important prognostic factors in GBM. EGFR is frequently amplified in GBM, and approximately 50% of these express the truncated form EGFRvIII, which is constitutively active and induces cell proliferation, survival and motility [9,83]. EGFR amplification and EGFRvIII mutants were associated with increased aggressiveness, and pointed to by some authors as prognostically valuable in GBM, as they are associated with shorter patient survival [9,20,25,26,84–87]. However, it is important to highlight that other authors state that these EGFR alterations did not associate with survival [88,89]. Similar to EGFR, the expression of PDGFR was reported to be frequently altered in GBM. Specifically, the phosphorylation of the PDGFRα subunit was associated with shorter survival in recurrent GBM patients [90], while other studies stated that the amplification of PDGFRα did not predict GBM patient survival [91,92]. Another RTK frequently altered in GBM is MET, which was rarely found amplified in GBM (only ∼5% of GBMs), but presented a high frequency of overexpression (∼29%) [23]. Moreover, these authors found that MET overexpression was associated with GBM shorter patient survival time [23]. In addition to RTKs, soluble growth factors are also important during tumorigenesis; an important example in GBM is VEGF, which is a prominent angiogenic factor [93]. Specifically the VEGF-A isoform, the best characterized isoform, was reported to be more frequently expressed in higher glioma grades, and was associated with poor GBM patient prognosis [93,94].
Moreover, the abnormal expression of intracellular targets of RTK signaling may associate with GBM patient prognosis. In particular, NF-κB is a transcription factor that is activated by the EGFR pathway [95]. The NFKBIA is a repressor of NF-κB, which was shown to be deleted in up to 24% of GBMs [13]. The deletion of NFKBIA was associated with poor GBM patient prognosis [13]. Interestingly, the authors found a pattern of mutual exclusiveness between NFKBIA deletion and EGFR amplification, and that the restoration of NFKBIA expression lessened the malignant phenotype and increased susceptibility to chemotherapeutic treatment in GBM cell lines [13].
The activation of the PI3K pathway is frequently deregulated in cancer, including GBMs. The pathway activation is associated with increased tumor grade, decreased apoptosis and poor patient outcomes [14,29]. In addition, several of the pathway intermediates per se presented prognostic significance. For example, phosphorylated AKT was reported as a biomarker of poor outcome in GBM patients [29,96]. The decreased expression of PTEN (at RNA or protein levels), or loss of heterozygosity of chromosome 10q, which encompasses the PTEN gene, were reported as indicators of shorter GBM patient survival [97]; however, this is still controversial [98]. Another recent example is the prognostic value of RAF kinase inhibitor, the expression of which was reported to associate with longer overall survival of GBM patients [99]. In the future, novel studies aiming to understand how the integrated analysis of several molecular components of growth signaling pathways may help clarify the true prognostic value of these biomarkers and lead to their integration into the clinical management of GBM patients.
Serum biomarkers of prognosis
Access to primary tumor samples is essential in evaluating tumor-specific genetic and epigenetic features. Biopsy, debulking or serial sampling may be difficult in the scenario of GBM, and moreover, a variety of imaging modalities are used to monitor tumor progression and response to treatment. MRIs can show an increase in tumor volume up to four weeks following completion of radiotherapy. In 50% of cases this is due to an increase in vascular permeability (treatment related) – an effect called pseudoprogession [100] – and might not necessarily translate into poor treatment response. This confounder [101], in addition to the unfeasibility of multiple tumor sampling during the course of the malignancy [102,103], clearly highlights the need for establishing less invasive predictive and prognostic markers. Serological markers mirroring tumor properties might be very good candidates. Serological biomarkers that correlate with patient survival in GBM include cathepsin D [104], AHSG [105], MMP-9[22] and YKL-40, which is the most widely studied [22,27,106–109]. A study conducted by Tanwar et al. evaluated gene expression microarray data of glioma tumor tissue and showed that the most highly expressed gene was YKL-40 [106]. The role of YKL-40 is not well-established; evidence suggests it may be implicated in cell differentiation, angiogenesis and proliferation, decreasing apoptosis and extracellular matrix remodeling [110]. Serum concentrations of YKL-40 seem to be a strong predictor of an aggressive phenotype in GBM [22,106]. Moreover, increased expression has been associated with glioma grade, shorter time to progression, resistance to radiotherapy and poor patient overall survival [22,107–109]. However, for the establishment of YKL-40 serum levels as a prognostic marker, further prospective studies with repeated measurements of YKL-40 levels before and after surgery are required. The high reproducibility of YKL-40 measurements in serum, as well as the fact that this biomarker is already well established for routine use, indicates that its inclusion in clinical practice should be relatively straightforward, and might provide crucial information on tumor progression and patient survival. In general, the use of serum biochemical markers that correlate with the biological traits of the tumor may be important during the design of treatment strategies and evaluating response to treatment. Equally important, these biomarkers may be able to detect disease progression or relapse early.
Histone mutations
GBM occurs comparatively less frequently in children than in adults but still remains a devastating disease with an incidence of 0.5 per 100,000 in Europe. Presenting symptoms, neurological sequelae, radiological and histological appearances are identical in both adults and children. What is particularly unique to the pediatric setting, however, is the occurrence of diffuse intrinsic pontine glioma; a form of malignant glioma specific to the pons and which, due to its location, is a challenge to treat. Pediatric tissue samples are scarce relative to adult counterparts and it has, therefore, been difficult to draw definitive conclusions about the underlying biology. As a result, they have been viewed as virtually indistinct from adults, contributing to a universal treatment strategy of surgery, radiotherapy and chemotherapy with temozolomide.
An increasing number of molecular profiling studies had hinted at the distinct underlying biology of pediatric cases [111–115], which was definitively proven with the identification of specific mutations in the H3F3A gene [116,117]. These were the first mutations described in histone genes in cancer and are highly specific to pediatric GBM. The H3F3A gene encodes the histone variant H3.3, and the mutations produce the amino acid substitutions glycine to arginine or valine at position 34 (G34R/V) or substitution of lysine to methionine at position 27 (K27M). Diffuse intrinsic pontine gliomas may also harbor K27M mutations in the gene encoding histone H3.1, HIST1H3B [117]. G34R/V mutant tumors peak at approximately 13–14 years, and are restricted to the cerebral hemispheres, while K27M mutant tumors arise at 6–7 years and are located in the pons and midline structures, especially the thalamus [31,118]. Although difficult to separate from the effects of anatomical location, K27M mutant tumors have a significantly worse overall survival than G34R/V or wild-type tumors [31,118]. This is clinically important, as thalamic tumors are currently treated on supratentorial protocols, although they may instead need to be considered along with diffuse intrinsic pontine glioma in terms of novel molecularly targeted therapies.
Other putative prognostic factors
In addition to the abovementioned prognostic biomarkers, other molecular characteristics of GBM have been suggested in some studies to associate with patient survival. Some examples include abnormal p53 and RB functions, expression of cancer stem cell markers [119–122], loss of chromosome 10 [27,123], codeletion of 1p/19q [124,125] and activation of HOX genes [17,19,26].
The p53 and RB tumor suppressor pathways are frequently altered in GBM (∼80%). Nonetheless, their prognostic value is still controversial, with some studies reporting an impact of p53 pathway in patient survival [12,126], while others did not replicate this [127,128]. Similarly, decreased expression of the RB gene in GBM has not been clearly established as a prognostic factor [32,129,130]. In addition, wild-type p16 was associated with improved survival of GBM patients treated with chemoradiotherapy [131], while the homozygous deletion of p16 was associated with poor survival in male GBM patients [132].
Studies have shown the presence of a subpopulation of cells within GBM, termed glioma stem cells, that present abnormal characteristics regarding proliferation and differentiation [133]. This population of cells was associated with tumor recurrence [134] and therapy resistance [11], and characterized by several stem cell markers, including CD133, CD44, ID1, Nestin and SOX-2 [135]. Several studies have tried to correlate the expression of these markers with GBM patient prognosis, but no consistent associations have currently been established [136]. For example, some studies report an association between CD133, Nestin, cJun, CD44 and ID1 expression and GBM patient poor prognosis [10,119–121,135,137], while others suggest an association of CD133 and SOX-11 expression with longer prognosis [24,122]; other studies report no effects on GBM patient survival due to the expression of Nestin, CD133 and CD15 [138,139]. The contradictory findings regarding the clinical relevance of these putative stem cell markers in GBM warrants further investigation.
The clinical relevance of some chromosomal copy number aberrations has also been investigated in GBM. Loss of chromosome 10 is highly frequent in GBM [27,123], and emerged as an important influence on global changes in the tumor gene expression, being reported as the most important copy number alteration for GBM classification and associated with a negative prognosis in GBM [27]. In addition, although codeletions of 1p/19q in oligodendrogliomas have been established as clinically relevant prognostic markers associated with increased patient survival time [140], these codeletions are uncommon in GBM, and the studies concerning their prognostic value in GBM have currently reported controversial findings [124,125].
Homeobox genes have also been recently studied in the context of glioma, particularly GBM. These genes encode transcription factors that play critical roles during normal development and differentiation [141], and have been found to be deregulated in cancer [141]. Recently, the expression of several HOX genes was found altered in gliomas [142]. A subsequent report identified the expression of a HOX-dominated gene cluster in GBM, enriched for stem cell-like properties, as an independent predictor factor for shorter survival time in patients treated with radiotherapy and concomitant chemotherapy [26]. Costa et al. showed that HOXA genes are differentially activated in GBM when compared with lower-grade gliomas and normal brain tissue, and identified GBMs with an abnormal chromosomal domain of transcriptional activation that includes the HOXA cluster [17]. This gene cluster is reversibly regulated by the PI3K pathway via an epigenetic mechanism regulating the levels of histone H3 lysine 27 trimethylation [17]. Of all HOXA genes, HOXA9 expression was predictive of GBM poor patient survival in two independent datasets, and was shown to have proproliferative and antiapoptotic functions in GBM cells [17]. More recently, Gaspar et al. showed that pediatric GBM cell lines resistant to temozolomide present a coordinated expression of several HOX genes, of which HOXA9 and HOXA10 are crucial effectors, and also suggested that the HOX-enriched signature is regulated by the PI3K pathway [19]. Importantly, pediatric patients with high-grade gliomas that express HOXA9 and HOXA10 had significantly shorter survival [19]. Overall, these studies suggest some HOXA genes may be prognostically valuable in both pediatric and adult GBM patients.
Conclusion
In conclusion, the work on prognostic factors in GBMs provides reasons for both optimism and caution in dealing with this highly malignant cancer. To date, the most relevant and still promising biomarker of prognosis in adult GBM patients is the status of MGMT promoter methylation, which has frequently been associated with patient survival and therapeutic responses. The multiplicity of techniques available to evaluate MGMT methylation status (including methylation-specific PCR) allows its routine establishment in the clinics, but, of equal importance, these methods may be applied in formalin-fixed paraffin-embedded tissues that is the standard format of samples deposited in tumor banks. New putative biomarkers, such as the expression levels of HOXA genes and the presence of IDH mutations, may be performed in this sample format using routine techniques such as immunohistochemistry or PCR followed by sequencing, respectively. However, the evaluation of IDH mutations in patient prognostication is limited to secondary GBM or lower-grade gliomas due to its low frequency in primary GBM (<10%). Concerning the pediatric setting, mutations of the H3F3A gene are as highly important, and may be established in the prognostication of these patients. Nonetheless, true clinical benefit will most likely only be seen with careful patient selection based on the presence of such biomarkers within clinical trials (the so-called precision medicine model). The increasing integration of molecular and clinical data through contemporary bioinformatics tools, will hasten the introduction of such biomarkers into the clinic leading to tailored treatment according to molecular subgroups. This also allows timely identification of patients unlikely to respond to standard therapies, permitting rapid entry into clinical trials, while avoiding the adverse unnecessary side effects of ineffective therapies. The challenge ahead is to identify and truly translate their relevance into effective, targeted drug therapies as well as to continue the discovery of further molecular markers of GBM.
Future perspective
The true clinical benefit of prognostic markers in GBM will probably only be perceived upon careful selection of patients based on the evaluation of tumor biomarkers and their integration in clinical trials. The cumulative integration of molecular and clinical data due to developments in bioinformatics tools will most certainly lead to the faster introduction of molecular biomarkers into the clinical routine, and thus to patient-tailored treatments according to the molecular alterations of GBM subgroups. Critically, this will conceptually allow the timely identification of patients that may not positively respond to conventional therapeutics, allowing their informed choice of entering clinical trials, while being spared the side effects and significant costs of ineffective therapies. The challenge in the neuro-oncology field for the next decade is to discover novel biomarkers of prognosis and therapy response, and to translate and integrate this knowledge into the development of targeted and effective therapies.
Acknowledgements
The authors would like to extend their appreciation to S Abreu and M Pojo for helpful assistance regarding the figure design.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: ▪ of interest ▪▪ of considerable interest
- 1.Kleihues P, Sobin LH. World Health Organization classification of tumors. Cancer. 2000;88(12):2887. doi: 10.1002/1097-0142(20000615)88:12<2887::aid-cncr32>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 2.Lino MM, Merlo A. PI3Kinase signaling in glioblastoma. J. Neurooncol. 2010;103(3):417–427. doi: 10.1007/s11060-010-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, Van Den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Curran WJ, Jr, Scott CB, Horton J, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J. Natl Cancer Inst. 1993;85(9):704–710. doi: 10.1093/jnci/85.9.704. [DOI] [PubMed] [Google Scholar]
- 5.Lamborn KR, Chang SM, Prados MD. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro Oncol. 2004;6(3):227–235. doi: 10.1215/S1152851703000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filippini G, Falcone C, Boiardi A, et al. Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. Neuro Oncol. 2008;10(1):79–87. doi: 10.1215/15228517-2007-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmadloo N, Kani AA, Mohammadianpanah M, et al. Treatment outcome and prognostic factors of adult glioblastoma multiforme. J. Egypt Natl Cancer Inst. 2013;25(1):21–30. doi: 10.1016/j.jnci.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Buckner JC, Schomberg PJ, McGinnis WL, et al. A Phase III study of radiation therapy plus carmustine with or without recombinant interferon-α in the treatment of patients with newly diagnosed high-grade glioma. Cancer. 2001;92(2):420–433. doi: 10.1002/1097-0142(20010715)92:2<420::aid-cncr1338>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Aldape KD, Ballman K, Furth A, et al. Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. J. Neuropathol. Exp. Neurol. 2004;63(7):700–707. doi: 10.1093/jnen/63.7.700. [DOI] [PubMed] [Google Scholar]
- 10.Ardebili SY, Zajc I, Gole B, et al. CD133/prominin1 is prognostic for GBM patient's survival, but inversely correlated with cysteine cathepsins’ expression in glioblastoma derived spheroids. Radiol. Oncol. 2011;45(2):102–115. doi: 10.2478/v10019-011-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao S, Wu Q, Mclendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 12.Birner P, Piribauer M, Fischer I, et al. Prognostic relevance of p53 protein expression in glioblastoma. Oncol. Rep. 2002;9(4):703–707. [PubMed] [Google Scholar]
- 13.Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in glioblastomas. N. Engl. J. Med. 2011;364(7):627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarti A, Zhai G, Suzuki Y, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J. Clin. Oncol. 2004;22(10):1926–1933. doi: 10.1200/JCO.2004.07.193. [DOI] [PubMed] [Google Scholar]
- 15.Costa BM, Caeiro C, Guimaraes I, et al. Prognostic value of MGMT promoter methylation in glioblastoma patients treated with temozolomide-based chemoradiation: a Portuguese multicentre study. Oncol. Rep. 2010;23(6):1655–1662. doi: 10.3892/or_00000808. [DOI] [PubMed] [Google Scholar]
- 16.Costa BM, Ferreira P, Costa S, et al. Association between functional EGF+61 polymorphism and glioma risk. Clin. Cancer Res. 2007;13(9):2621–2626. doi: 10.1158/1078-0432.CCR-06-2606. [DOI] [PubMed] [Google Scholar]
- 17.Costa BM, Smith JS, Chen Y, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70(2):453–462. doi: 10.1158/0008-5472.CAN-09-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]; Unraveling of the mechanism behind HOXA gene cluster activation in glioblastoma multiforme (GBM), and association of HOXA9 with poor prognosis in GBM patients.
- 18.Costa BM, Viana-Pereira M, Fernandes R, et al. Impact of EGFR genetic variants on glioma risk and patient outcome. Cancer Epidemiol. Biomarkers Prev. 2011;20(12):2610–2617. doi: 10.1158/1055-9965.EPI-11-0340. [DOI] [PubMed] [Google Scholar]
- 19.Gaspar N, Marshall L, Perryman L, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010;70(22):9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas-Kogan DA, Prados MD, Lamborn KR, Tihan T, Berger MS, Stokoe D. Biomarkers to predict response to epidermal growth factor receptor inhibitors. Cell Cycle. 2005;4(10):1369–1372. doi: 10.4161/cc.4.10.2105. [DOI] [PubMed] [Google Scholar]
- 21.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]; ▪Associates the methylation status of MGMT promoter with GBM patient survival and response to temozolomide.
- 22.Hormigo A, Gu B, Karimi S, et al. YKL-40 and matrix metalloproteinase-9 as potential serum biomarkers for patients with high-grade gliomas. Clin. Cancer Res. 2006;12(19):5698–5704. doi: 10.1158/1078-0432.CCR-06-0181. [DOI] [PubMed] [Google Scholar]
- 23.Kong DS, Song SY, Kim DH, et al. Prognostic significance of c-Met expression in glioblastomas. Cancer. 2009;115(1):140–148. doi: 10.1002/cncr.23972. [DOI] [PubMed] [Google Scholar]
- 24.Korkolopoulou P, Levidou G, El-Habr EA, et al. Sox11 expression in astrocytic gliomas: correlation with nestin/c-Met/IDH1-R132H expression phenotypes, p-Stat-3 and survival. Br. J. Cancer. 2013;108(10):2142–2152. doi: 10.1038/bjc.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 26.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related ‘self-renewal’ signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]; Report of a HOX-dominated gene cluster in GBM with stem cell properties as a marker of poor prognosis in patients treated with chemoradiotherapy.
- 27.Nigro JM, Misra A, Zhang L, et al. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65(5):1678–1686. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 28.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪First report on IDH1 mutations in GBM and its association with patient survival.
- 29.Pelloski CE, Lin E, Zhang L, et al. Prognostic associations of activated mitogen-activated protein kinase and Akt pathways in glioblastoma. Clin. Cancer Res. 2006;12(13):3935–3941. doi: 10.1158/1078-0432.CCR-05-2202. [DOI] [PubMed] [Google Scholar]
- 30.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 32.Backlund LM, Nilsson BR, Goike HM, et al. Short postoperative survival for glioblastoma patients with a dysfunctional Rb1 pathway in combination with no wild-type PTEN. Clin. Cancer Res. 2003;9(11):4151–4158. [PubMed] [Google Scholar]
- 33.Colman H, Aldape K. Molecular predictors in glioblastoma: toward personalized therapy. Arch. Neurol. 2008;65(7):877–883. doi: 10.1001/archneur.65.7.877. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Markowitz S, Gerson SL. Mismatch repair mutations override alkyltransferase in conferring resistance to temozolomide but not to 1,3-bis(2-chloroethyl)nitrosourea. Cancer Res. 1996;56(23):5375–5379. [PubMed] [Google Scholar]
- 35.Qian XC, Brent TP. Methylation hot spots in the 5´ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997;57(17):3672–3677. [PubMed] [Google Scholar]
- 36.Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol. Cell Biol. 1997;17(9):5612–5619. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised Phase III study: 5-year analysis of the EORTC–NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 38.Wick W, Platten M, Meisner C, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, Phase 3 trial. Lancet Oncol. 2012;13(7):707–715. doi: 10.1016/S1470-2045(12)70164-X. [DOI] [PubMed] [Google Scholar]
- 39.Malmstrom A, Gronberg BH, Marosi C, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, Phase 3 trial. Lancet Oncol. 2012;13(9):916–926. doi: 10.1016/S1470-2045(12)70265-6. [DOI] [PubMed] [Google Scholar]
- 40.Olson RA, Brastianos PK, Palma DA. Prognostic and predictive value of epigenetic silencing of MGMT in patients with high grade gliomas: a systematic review and meta-analysis. J. Neurooncol. 2011;105(2):325–335. doi: 10.1007/s11060-011-0594-5. [DOI] [PubMed] [Google Scholar]
- 41.van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J. Clin. Oncol. 2009;27(35):5881–5886. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Vlodrop IJ, Niessen HE, Derks S, et al. Analysis of promoter CpG island hypermethylation in cancer: location, location, location! Clin. Cancer Res. 2011;17(13):4225–4231. doi: 10.1158/1078-0432.CCR-10-3394. [DOI] [PubMed] [Google Scholar]
- 43.Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah N, Lin B, Sibenaller Z, et al. Comprehensive analysis of MGMT promoter methylation: correlation with MGMT expression and clinical response in GBM. PLoS One. 2011;6(1):e16146. doi: 10.1371/journal.pone.0016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathway. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koshland DE, Jr, Walsh K, Laporte DC. Sensitivity of metabolic fluxes to covalent control. Curr. Top Cell Regul. 1985;27:13–22. doi: 10.1016/b978-0-12-152827-0.50009-8. [DOI] [PubMed] [Google Scholar]
- 48.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 49.Ichimura K, Pearson DM, Kocialkowski S, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11(4):341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 51.Hartmann C, Hentschel B, Wick W, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120(6):707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- 52.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of a subgroup of GBM that present with the glioma-CpG island methylator phenotype and have a significantly improved outcome.
- 53.Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29(49):6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- 54.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪Robust classification of GBM into subtypes based on gene expression profiles with different responses to therapy.
- 55.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science. 2009;324(5924):261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 59.Songtao Q, Lei Y, Si G, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103(2):269–273. doi: 10.1111/j.1349-7006.2011.02134.x. [DOI] [PubMed] [Google Scholar]
- 60.Wang F, Travins J, Delabarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 61.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]; Selective small molecule inhibitor of mutant IDH1R132H in glioma blocked the capacity of the mutant enzyme to produce 2-hydroxyglutarate.
- 62.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 63.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32(Database issue):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 66.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 67.Ben-Hamo R, Efroni S. Gene expression and network-based analysis reveals a novel role for hsa-miR-9 and drug control over the p38 network in glioblastoma multiforme progression. Genome Med. 2011;3(11):77. doi: 10.1186/gm293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gabriely G, Yi M, Narayan RS, et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71(10):3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin J, Teo S, Lam DH, Jeyaseelan K, Wang S. MicroRNA-10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death Dis. 2012;3:e398. doi: 10.1038/cddis.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fowler A, Thomson D, Giles K, et al. miR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur. J. Cancer. 2011;47(6):953–963. doi: 10.1016/j.ejca.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 71.Guan Y, Mizoguchi M, Yoshimoto K, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin. Cancer Res. 2010;16(16):4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 72.Ma R, Yan W, Zhang G, et al. Upregulation of miR-196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS One. 2012;7(6):e38096. doi: 10.1371/journal.pone.0038096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Godlewski J, Nowicki MO, Bronisz A, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell. 2010;37(5):620–632. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhi F, Chen X, Wang S, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur. J. Cancer. 2010;46(9):1640–1649. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 75.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6(3):e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang W, Zhang J, Yan W, et al. Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer. 2012;119(4):814–824. doi: 10.1002/cncr.27826. [DOI] [PubMed] [Google Scholar]
- 77.Lakomy R, Sana J, Hankeova S, et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 2011;102(12):2186–2190. doi: 10.1111/j.1349-7006.2011.02092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int. J. Cancer. 2009;125(6):1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 79.Sun L, Yan W, Wang Y, et al. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 80.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nan Y, Han L, Zhang A, et al. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 82.Visani M, De Biase D, Marucci G, Taccioli C, Baruzzi A, Pession A. Definition of miRNAs expression profile in glioblastoma samples: the relevance of non-neoplastic brain reference. PLoS One. 2013;8(1):e55314. doi: 10.1371/journal.pone.0055314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang HS, Nagane M, Klingbeil CK, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 1997;272(5):2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 84.Heimberger AB, Hlatky R, Suki D, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin. Cancer Res. 2005;11(4):1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 85.Liu L, Backlund LM, Nilsson BR, et al. Clinical significance of EGFR amplification and the aberrant EGFRvIII transcript in conventionally treated astrocytic gliomas. J. Mol. Med. (Berl.) 2005;83(11):917–926. doi: 10.1007/s00109-005-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shinojima N, Tada K, Shiraishi S, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63(20):6962–6970. [PubMed] [Google Scholar]
- 87.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc. Natl Acad. Sci. USA. 1990;87(21):8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heimberger AB, Suki D, Yang D, Shi W, Aldape K. The natural history of EGFR and EGFRvIII in glioblastoma patients. J. Transl. Med. 2005;3:38. doi: 10.1186/1479-5876-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huncharek M, Kupelnick B. Epidermal growth factor receptor gene amplification as a prognostic marker in glioblastoma multiforme: results of a meta-analysis. Oncol. Res. 2000;12(2):107–112. doi: 10.3727/096504001108747576. [DOI] [PubMed] [Google Scholar]
- 90.Paulsson J, Lindh MB, Jarvius M, et al. Prognostic but not predictive role of platelet-derived growth factor receptors in patients with recurrent glioblastoma. Int. J. Cancer. 2011;128(8):1981–1988. doi: 10.1002/ijc.25528. [DOI] [PubMed] [Google Scholar]
- 91.Nobusawa S, Stawski R, Kim YH, Nakazato Y, Ohgaki H. Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: a population-based study. Neuropathology. 2011;31(6):583–588. doi: 10.1111/j.1440-1789.2011.01204.x. [DOI] [PubMed] [Google Scholar]
- 92.Alentorn A, Marie Y, Carpentier C, et al. Prevalence, clinico-pathological value, and co-occurrence of PDGFRA abnormalities in diffuse gliomas. Neuro Oncol. 2012;14(11):1393–1403. doi: 10.1093/neuonc/nos217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou YH, Tan F, Hess KR, Yung WK. The expression of PAX6, PTEN, vascular endothelial growth factor, and epidermal growth factor receptor in gliomas: relationship to tumor grade and survival. Clin. Cancer Res. 2003;9(9):3369–3375. [PubMed] [Google Scholar]
- 94.Flynn JR, Wang L, Gillespie DL, et al. Hypoxia-regulated protein expression, patient characteristics, and preoperative imaging as predictors of survival in adults with glioblastoma multiforme. Cancer. 2008;113(5):1032–1042. doi: 10.1002/cncr.23678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sethi G, Ahn KS, Chaturvedi MM, Aggarwal BB. Epidermal growth factor (EGF) activates nuclear factor-κB through IκBα kinase-independent but EGF receptor-kinase dependent tyrosine 42 phosphorylation of IκBα. Oncogene. 2007;26(52):7324–7332. doi: 10.1038/sj.onc.1210544. [DOI] [PubMed] [Google Scholar]
- 96.Suzuki Y, Shirai K, Oka K, et al. Higher pAkt expression predicts a significant worse prognosis in glioblastomas. J. Radiat. Res. 2010;51(3):343–348. doi: 10.1269/jrr.09109. [DOI] [PubMed] [Google Scholar]
- 97.Srividya MR, Thota B, Shailaja BC, et al. Homozygous 10q23/PTEN deletion and its impact on outcome in glioblastoma: a prospective translational study on a uniformly treated cohort of adult patients. Neuropathology. 2011;31(4):376–383. doi: 10.1111/j.1440-1789.2010.01178.x. [DOI] [PubMed] [Google Scholar]
- 98.Carico C, Nuno M, Mukherjee D, et al. Loss of PTEN is not associated with poor survival in newly diagnosed glioblastoma patients of the temozolomide era. PLoS One. 2012;7(3):e33684. doi: 10.1371/journal.pone.0033684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maresch J, Birner P, Zakharinov M, Toumangelova-Uzeir K, Natchev S, Guentchev M. Additive effect on survival of Raf kinase inhibitor protein and signal transducer and activator of transcription 3 in high-grade glioma. Cancer. 2010;117(11):2499–2504. doi: 10.1002/cncr.25799. [DOI] [PubMed] [Google Scholar]
- 100.Brandsma D, Stalpers L, Taal W, Sminia P, Van Den Bent MJ. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol. 2008;9(5):453–461. doi: 10.1016/S1470-2045(08)70125-6. [DOI] [PubMed] [Google Scholar]
- 101.Wen PY, Kesari S. Malignant gliomas in adults. N. Engl. J. Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 102.Sorensen AG, Batchelor TT, Wen PY, Zhang WT, Jain RK. Response criteria for glioma. Nat. Clin. Pract. Oncol. 2008;5(11):634–644. doi: 10.1038/ncponc1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Van Den Bent MJ, Vogelbaum MA, Wen PY, Macdonald DR, Chang SM. End point assessment in gliomas: novel treatments limit usefulness of classical Macdonald's Criteria. J. Clin. Oncol. 2009;27(18):2905–2908. doi: 10.1200/JCO.2009.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fukuda ME, Iwadate Y, Machida T, et al. Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res. 2005;65(12):5190–5194. doi: 10.1158/0008-5472.CAN-04-4134. [DOI] [PubMed] [Google Scholar]
- 105.Petrik V, Saadoun S, Loosemore A, et al. Serum α2-HS glycoprotein predicts survival in patients with glioblastoma. Clin. Chem. 2008;54(4):713–722. doi: 10.1373/clinchem.2007.096792. [DOI] [PubMed] [Google Scholar]
- 106.Tanwar MK, Gilbert MR, Holland EC. Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer Res. 2002;62(15):4364–4368. [PubMed] [Google Scholar]
- 107.Bernardi D, Padoan A, Ballin A, et al. Serum YKL-40 following resection for cerebral glioblastoma. J. Neurooncol. 2012;107(2):299–305. doi: 10.1007/s11060-011-0762-7. [DOI] [PubMed] [Google Scholar]
- 108.Iwamoto FM, Hottinger AF, Karimi S, et al. Serum YKL-40 is a marker of prognosis and disease status in high-grade gliomas. Neuro Oncol. 2011;13(11):1244–1251. doi: 10.1093/neuonc/nor117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pelloski CE, Mahajan A, Maor M, et al. YKL-40 expression is associated with poorer response to radiation and shorter overall survival in glioblastoma. Clin. Cancer Res. 2005;11(9):3326–3334. doi: 10.1158/1078-0432.CCR-04-1765. [DOI] [PubMed] [Google Scholar]
- 110.Johansen JS, Schultz NA, Jensen BV. Plasma YKL-40: a potential new cancer biomarker? Future Oncol. 2009;5(7):1065–1082. doi: 10.2217/fon.09.66. [DOI] [PubMed] [Google Scholar]
- 111.Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J. Clin. Oncol. 2010;28(18):3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bax DA, Mackay A, Little SE, et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin. Cancer Res. 2010;16(13):3368–3377. doi: 10.1158/1078-0432.CCR-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zarghooni M, Bartels U, Lee E, et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor α and poly (ADP-ribose) polymerase as potential therapeutic targets. J. Clin. Oncol. 2010;28(8):1337–1344. doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 114.Paugh BS, Broniscer A, Qu C, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J. Clin. Oncol. 2011;29(30):3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jones C, Perryman L, Hargrave D. Paediatric and adult malignant glioma: close relatives or distant cousins? Nat. Rev. Clin. Oncol. 2012;9(7):400–413. doi: 10.1038/nrclinonc.2012.87. [DOI] [PubMed] [Google Scholar]
- 116.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]; ▪First report highlighting mutations in a regulatory histone in humans that may outline pediatric and young adult GBM pathogenesis.
- 117.Wu G, Broniscer A, Mceachron TA, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44(3):251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bjerke L, Mackay A, Nandhabalan M, et al. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-12-0426. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Metellus P, Nanni-Metellus I, Delfino C, et al. Prognostic impact of CD133 mRNA expression in 48 glioblastoma patients treated with concomitant radiochemotherapy: a prospective patient cohort at a single institution. Ann. Surg. Oncol. 2011;18(10):2937–2945. doi: 10.1245/s10434-011-1703-6. [DOI] [PubMed] [Google Scholar]
- 120.Nduom EK, Hadjipanayis CG, Van Meir EG. Glioblastoma cancer stem-like cells: implications for pathogenesis and treatment. Cancer J. 2012;18(1):100–106. doi: 10.1097/PPO.0b013e3182452e0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pallini R, Ricci-Vitiani L, Banna GL, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin. Cancer Res. 2008;14(24):8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 122.Pallini R, Ricci-Vitiani L, Montano N, et al. Expression of the stem cell marker CD133 in recurrent glioblastoma and its value for prognosis. Cancer. 2011;117(1):162–174. doi: 10.1002/cncr.25581. [DOI] [PubMed] [Google Scholar]
- 123.Mohapatra G, Bollen AW, Kim DH, et al. Genetic analysis of glioblastoma multiforme provides evidence for subgroups within the grade. Genes Chromosomes Cancer. 1998;21(3):195–206. [PubMed] [Google Scholar]
- 124.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J. Clin. Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 125.Smith JS, Perry A, Borell TJ, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J. Clin. Oncol. 2000;18(3):636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 126.Schiebe M, Ohneseit P, Hoffmann W, Meyermann R, Rodemann HP, Bamberg M. Analysis of mdm2 and p53 gene alterations in glioblastomas and its correlation with clinical factors. J. Neurooncol. 2000;49(3):197–203. doi: 10.1023/a:1006410702284. [DOI] [PubMed] [Google Scholar]
- 127.Kraus JA, Wenghoefer M, Glesmann N, et al. TP53 gene mutations, nuclear p53 accumulation, expression of Waf/p21, Bcl-2, and CD95 (APO-1/Fas) proteins are not prognostic factors in de novo glioblastoma multiforme. J. Neurooncol. 2001;52(3):263–272. doi: 10.1023/a:1010684203704. [DOI] [PubMed] [Google Scholar]
- 128.Rich JN, Hans C, Jones B, et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65(10):4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- 129.Goldhoff P, Clarke J, Smirnov I, et al. Clinical stratification of glioblastoma based on alterations in retinoblastoma tumor suppressor protein (RB1) and association with the proneural subtype. J. Neuropathol. Exp. Neurol. 2012;71(1):83–89. doi: 10.1097/NEN.0b013e31823fe8f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmidt MC, Antweiler S, Urban N, et al. Impact of genotype and morphology on the prognosis of glioblastoma. J. Neuropathol. Exp. Neurol. 2002;61(4):321–328. doi: 10.1093/jnen/61.4.321. [DOI] [PubMed] [Google Scholar]
- 131.Ang C, Guiot MC, Ramanakumar AV, Roberge D, Kavan P. Clinical significance of molecular biomarkers in glioblastoma. Can. J. Neurol. Sci. 2010;37(5):625–630. doi: 10.1017/s0317167100010805. [DOI] [PubMed] [Google Scholar]
- 132.Kamiryo T, Tada K, Shiraishi S, et al. Analysis of homozygous deletion of the p16 gene and correlation with survival in patients with glioblastoma multiforme. J. Neurosurg. 2002;96(5):815–822. doi: 10.3171/jns.2002.96.5.0815. [DOI] [PubMed] [Google Scholar]
- 133.Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23(43):7267–7273. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 134.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 135.Brescia P, Richichi C, Pelicci G. Current strategies for identification of glioma stem cells: adequate or unsatisfactory? J. Oncol. 2012:376894. doi: 10.1155/2012/376894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dahlrot RH, Hermansen SK, Hansen S, Kristensen BW. What is the clinical value of cancer stem cell markers in gliomas? Int. J. Clin. Exp. Pathol. 2013;6(3):334–348. [PMC free article] [PubMed] [Google Scholar]
- 137.Mangiola A, Lama G, Giannitelli C, et al. Stem cell marker nestin and c-Jun NH2-terminal kinases in tumor and peritumor areas of glioblastoma multiforme: possible prognostic implications. Clin. Cancer Res. 2007;13(23):6970–6977. doi: 10.1158/1078-0432.CCR-07-1229. [DOI] [PubMed] [Google Scholar]
- 138.Chinnaiyan P, Wang M, Rojiani AM, et al. The prognostic value of nestin expression in newly diagnosed glioblastoma: report from the Radiation Therapy Oncology Group. Radiat. Oncol. 2008;3:32. doi: 10.1186/1748-717X-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim KJ, Lee KH, Kim HS, et al. The presence of stem cell marker-expressing cells is not prognostically significant in glioblastomas. Neuropathology. 2011;31(5):494–502. doi: 10.1111/j.1440-1789.2010.01194.x. [DOI] [PubMed] [Google Scholar]
- 140.Brandes AA, Tosoni A, Cavallo G, et al. Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: a prospective GICNO study. J. Clin. Oncol. 2006;24(29):4746–4753. doi: 10.1200/JCO.2006.06.3891. [DOI] [PubMed] [Google Scholar]
- 141.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 142.Abdel-Fattah R, Xiao A, Bomgardner D, Pease CS, Lopes MB, Hussaini IM. Differential expression of HOX genes in neoplastic and non-neoplastic human astrocytes. J. Pathol. 2006;209(1):15–24. doi: 10.1002/path.1939. [DOI] [PubMed] [Google Scholar]
- 143.Green SB, Byar DP, Walker MD, et al. Comparisons of carmustine, procarbazine, and high-dose methylprednisolone as additions to surgery and radiotherapy for the treatment of malignant glioma. Cancer Treat. Rep. 1983;67(2):121–132. [PubMed] [Google Scholar]