Abstract
Objective: Construction of a resource that provides semantic information about words and phrases to facilitate the computer processing of medical narrative.
Design: Lexemes (words and word phrases) in the Specialist Lexicon were matched against strings in the 1997 Metathesaurus of the Unified Medical Language System (UMLS) developed by the National Library of Medicine. This yielded a “semantic lexicon,” in which each lexeme is associated with one or more syntactic types, each of which can have one or more semantic types. The semantic lexicon was then used to assign semantic types to lexemes occurring in a corpus of discharge summaries (603,306 sentences). Lexical items with multiple semantic types were examined to determine whether some of the types could be eliminated, on the basis of usage in discharge summaries. A concordance program was used to find contrasting contexts for each lexeme that would reflect different semantic senses. Based on this evidence, semantic preference rules were developed to reduce the number of lexemes with multiple semantic types.
Results: Matching the Specialist Lexicon against the Metathesaurus produced a semantic lexicon with 75,711 lexical forms, 22,805 (30.1 percent) of which had two or more semantic types. Matching the Specialist Lexicon against one year's worth of discharge summaries identified 27,633 distinct lexical forms, 13,322 of which had at least one semantic type. This suggests that the Specialist Lexicon has about 79 percent coverage for syntactic information and 38 percent coverage for semantic information for discharge summaries. Of those lexemes in the corpus that had semantic types, 3,474 (12.6 percent) had two or more types. When semantic preference rules were applied to the semantic lexicon, the number of entries with multiple semantic types was reduced to 423 (1.5 percent). In the discharge summaries, occurrences of lexemes with multiple semantic types were reduced from 9.41 to 1.46 percent.
Conclusion: Automatic methods can be used to construct a semantic lexicon from existing UMLS sources. This semantic information can aid natural language processing programs that analyze medical narrative, provided that lexemes with multiple semantic types are kept to a minimum. Semantic preference rules can be used to select semantic types that are appropriate to clinical reports. Further work is needed to increase the coverage of the semantic lexicon and to exploit contextual information when selecting semantic senses.
Increasingly, medical institutions have access to patient records through computers. Much of the available data are in textual form as a result of transcription of dictated reports, use of speech recognition technology, and direct entry by health care providers. While textual data are convenient for tasks such as review by clinicians, they present significant obstacles for graphic presentation, searching, summarization, and statistical analysis. The techniques of natural language processing can be applied to transform medical narrative into a form more suitable for information processing and management.1,2,3,4,5,6,7,8,9,10,11 While there are many important issues in the processing of medical text (sentence parsing, discourse analysis, document structure),12,13,14 one of the most fundamental issues is how the computer represents the meanings of individual words and phrases.
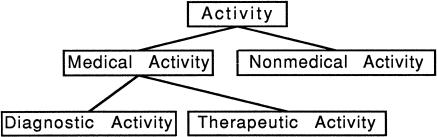
For humans, the meaning of a given word can be obtained by consulting a dictionary. The computer cannot make use of textual dictionary definitions, but instead requires a semantic representation that is simpler and more precise. Natural language processing systems represent the meaning of a given word or phrase using a symbol or code. For example, the verb treat (and its variant forms treats, treated, and treating) might be assigned the symbolic meaning THERAPEUTIC-ACTIVITY, while the noun form would not have a semantic representation in the medical domain. Symbolic meanings are made precise through some systematic organization, which may be a simple catalog of distinct meanings or a formal definition using a set of axioms. For example, various verbs (treat, assess, purchase, care for) can be classified according to the hierarchy depicted in ▶. In formal terms, a systematic arrangement of symbols is called an ontology, and the symbols that it organizes are called types or semantic types.
Figure 1.
Simple ontology of activities.
A resource that maps a lexical item (word or phrase) to one or more semantic types can be termed a semantic lexicon.15 This is in contrast to a lexicon that provides syntactic information about words, such as part of speech (noun, verb, adjective) or number (singular, plural). The semantic lexicon associates the different forms of words (e.g., singular and plural nouns, tenses of verbs) with one or more semantic types. A separate ontology defines the meaning of the semantic types. For example, a semantic lexicon using the ontology shown in ▶ might include the entries listed in ▶.
Table 1.
Simple Semantic Lexicon Showing Verb Entries
| Verb | Semantic Type |
|---|---|
| purchase | Non-medical-activity |
| purchases | Non-medical-activity |
| assess | Diagnostic-activity |
| assessed | Diagnostic-activity |
| treat | Therapeutic-activity |
| treating | Therapeutic-activity |
| care for | Medical-activity |
| cares for | Medical-activity |
At the present time, developers of a natural language processing system must construct the semantic lexicon by hand—an extremely labor-intensive task, requiring both linguistic and medical knowledge. While there are ontologies in use for general language processing,16,17,18,19,20 these lack sufficient medical content. Controlled medical vocabularies21,22,23,24 classify medical terms and therefore focus almost exclusively on nouns, omitting information about adjectives, verbs, and other parts of speech that are essential for sentence analysis.
This paper investigates how a semantic lexicon for medical language processing can be constructed by building on existing standards, specifically the Specialist Lexicon25,26 in the Unified Medical Language System (UMLS) of the National Library of Medicine (NLM).27 Standards for representing semantics in medical language processing systems have the following advantages:
Reduction of the tremendous intellectual labor required to build a medical language processing system
Reduction of differences in representations used by natural language processing systems, making them easier to understand and compare
Facilitation in mapping the output produced by natural language processing systems to various target information systems and applications
The following section of the paper describes some of the background work relating to medical semantics and language processing and discusses the issues that must be addressed in building a semantic lexicon. Then methods are described for building a semantic lexicon using the UMLS Specialist Lexicon and Metathesaurus. Results describe how well these resources can be combined and the extent to which the number of lexemes with multiple semantic types can be reduced. The discussion covers some problems in the methods, and focuses on design criteria for classifying medical lexemes (the ontology), to aid in choosing and assigning semantic types to lexical items.
Background
Several medical language processing systems in existence can analyze and structure information in medical reports.1,2,3,4,5,6,7,8,9,10,11 Each of these systems uses a semantic lexicon developed specially for that system. Some of the differences can be explained by different domains (e.g., radiology vs. surgery), target information system, intended uses of the data, and the parsing algorithm used to analyze sentences. However, construction of these lexicons still involves significant duplication of effort. For example, the list of anatomic terms could be expected to be largely the same across all systems.
Since a lexicon can contain thousands or even hundreds of thousands of entries, the effort involved is considerable. The NLM offers a number of resources that may help reduce this effort.28 To exploit these resources effectively, several issues must be considered:
Coverage of full range of semantic types (medical and nonmedical)
Formal vs. informal semantic representation
Granularity of semantic types
Combination of semantic and syntactic information
Minimization of multiply classified lexemes
Relative independence from applications
The following sections examine these issues and discuss related work.
Representing the Full Range of Medical Semantics
To analyze and extract information from medical text, an NLP system needs to know how sentences are put together and how to represent their information content. In general, deep knowledge about the world, e.g., CYC,20 is not necessary to carry out the extraction process. Ontologies developed for natural language processing—e.g., Penman17 and Mikrokosmos16—have a simpler organization and make distinctions more relevant to the linguistic domain. However, these ontologies do not possess sufficient medical content for processing medical language. In contrast, controlled medical vocabularies21,22,23,24 represent mostly medical terms (e.g., diseases, procedures, anatomic structures) but lack a detailed classification of qualifiers (e.g., size, degree, certainty) and relations (e.g., temporal, causal, spatial). Snomed23 does represent some modifiers (e.g., disease severity, topography, administration routes) and some spatial relations, both of which are included in the UMLS Metathesaurus.
Medical texts contain many words that need to be analyzed but that may not be considered medical terms. For example, the UMLS Metathesaurus contains many examples of specific difficulties (e.g., “Difficulty seeing at night)” but does not have the word “difficulty” as a term by itself. A semantic type for this word is needed to process the sentence (from a discharge summary) “Patient walked without difficulty.”
For general semantic types (what is referred to as a “upper level ontology”), the UMLS Semantic Network29 is very close to what is needed for medical language processing. It is medically oriented and makes distinctions that are useful for language processing. Interestingly, the Semantic Network is similar in some ways to the upper levels of the Mikrokosmos ontology, which is also used in language processing (language generation). While the UMLS Semantic Network makes fine distinctions in some areas (e.g., chemicals), qualifiers and relations are not well developed. The granularity of the Semantic Network is too broad for its use as a complete meaning representation for medical language processing (granularity is discussed further below). For detailed semantic information, a controlled medical vocabulary (such as SNOMED) must be used.
Formal Representation of Semantics
When a computer program analyzes a natural language sentence and produces some representation, one may ask in what sense this captures the meaning of the sentence.30 This paper adopts the approach that the meaning is the information content of the sentence that can be useful for a given computer application. For example, if the task of the program is to populate a database using information extracted from text reports, then only facts pertinent to the database are considered meaningful. Even this pragmatic definition defines a very broad area of research involving complex semantic relationships, time, modality, argumentation, and such. Therefore, the current study considers only the meanings of words (and technical phrases made up of multiple words). If a computer program is to analyze a sentence at all, it must at least understand the lexical items present.
Natural language processing systems represent the meanings of lexical items (words and phrases) using a special set of symbols or codes. In what sense are these codes meanings? Using the pragmatic definition above, the codes must correspond to data values that can be used by a computer application. But how do we know what these values mean? If there are a small number of values that are well understood by those using the computer application, this is not a significant issue. However, when there are a relatively large number of values, they can be made meaningful only through some kind of systematic organization—an “ontology.” The larger the number of values, and the more individuals using the application, the greater the need to formalize the ontology. Without formalization, there will be constant confusion about what a given code or symbol means. For example, the symbol SODIUM could represent the element, a dietary supplement being ordered for a patient, or the level of chemical present in a body substance (e.g., blood).
For natural language processing, the advantages of using a formal ontology to represent the lexicon are:
Using the lexical ontology in more than one natural language processing system
Mapping the results of a natural language processing system into the databases and knowledge bases used by other applications
Maintaining the lexicon ontology and adding new items in a consistent manner
The degree of formalization in ontologies (both medical and nonmedical) can vary. The ontology may consist of a list of concepts with text definitions used to distinguish one from another; an arrangement into a classification hierarchy (e.g., International Classification of Diseases,22 Medical Subject Headings thesaurus21); a “semantic net” with subsumption and other semantic relations (e.g., UMLS Semantic Network,29 Medical Entities Dictionary31); or a formal system with axioms (e.g., CYC,20 Mikrokosmos16). If an ontology is too informal, the possibility of inconsistency is greater, and if it is too formal (many complex axioms), the ontology will be difficult to understand and maintain. For natural language processing, a semantic network appears to be the right degree of formalization. Concepts should be organized into a subsumption hierarchy, with semantic relations used to define differentiae between a supertype and a given subtype, and between any pair of subtypes of a supertype.31,32 Since the main use of the ontology is to help the lexicon administrator to add new entries, the only semantic relations needed are those that help differentiate concepts in order to assign the proper semantic type. These relations will largely be a subset of those used in controlled medical terminologies.31,32,33 However, additional semantic relations will be required to adequately classify abstractions, relations, and qualifiers that are often found in natural language but not in controlled vocabulary.
Granularity of Semantic Types
The semantic types used by a natural language processing system may have different “granularities” depending on the application. For example, the ontology shown in ▶ does not distinguish drug administration from surgery; a hierarchy with finer granularity could have distinct types for these activities. Natural language processing systems make use of semantic information at two distinct levels of granularity:
Broad—to enforce semantic constraints during sentence processing
Narrow—to specify particular meanings of words and phrases in the final representation produced by the natural language processing system.
An ontology with very broad types is useful for parsing, to aid in determining how words and phrases combine to form a medically meaningful sentence. A purely syntactic approach will have significant problems.10 For example, when a computer program considers only syntactic information, it finds dozens of possible parses for the following sentence: “He negotiated five steps with both hand rails on stairs with close supervision upon discharge.” This is because the computer (unlike a human) sees many ways in which a prepositional phrase could modify a preceding noun or verb. For example the prepositional phrase “upon discharge” could modify “supervision,” “stairs,” “rails,” “steps,” or “negotiated.” Because of their semantic knowledge, humans tend not to notice such structural ambiguity.
However, when the computer knows the semantic types of lexical items and has rules that restrict how these semantic types can combine, the number of parses is reduced (often to a single possibility). For example, the computer might avoid the above ambiguities by applying “semantic patterns”1,5,9 such as:
Behavior with-instrument Manufactured-Object
Behavior on-location Manufactured-Object
Behavior occurs-with Activity
Behavior occurs-upon Activity
These patterns allow the word “discharge” (Activity) to combine with “negotiated” (Behavior) but not with “rails” and “stairs” or “steps” (Manufactured-Objects). Very broad semantic types such as these have been shown to be very effective in reducing parsing ambiguity.
If an ontology with very narrow types is used, far more relationships are possible and the number of possible combinations increases exponentially. Thus, a very specific ontology does not help much in the parsing phase.
On the other hand, broad types are not useful in producing the final representation, because they do not make sufficient distinctions. For example, the representation of the sentence above using broad types might appear in a slot-value notation (▶). The semantic types in the “value” column do not convey sufficient detail about the original sentence for the purposes of most clinical applications, e.g., storage in a patient database. Instead, the specific terms such as “hand rails,” “supervision,” and “discharge” must be mapped to fine-grained concepts.
Table 2.
Hypothetical Semantic Representation of a Sentence from a Discharge Summary
| Slot | Value | Words from Sentence |
|---|---|---|
| Event | Behavior | negotiated |
| Agent | Patient | he |
| Location | Manufactured-Object | stairs |
| Instrument | Manufactured-Object | hand rails |
| Means | Activity | supervision |
| At-Time | Health-Care-Activity | discharge |
The creation of a detailed semantic ontology for all words of medical texts is a task far beyond the ability of any single institution. The practical solution is for natural language processing applications to parse sentences using generic semantic types, then map these representations into a standard controlled vocabulary such as the UMLS Metathesaurus or SNOMED.
Reducing Multiply Classified Words
True homonyms (words having multiple meanings that are mutually exclusive in a given text) are rare in general language.34 In technical and scientific domains such as medicine, homonyms are even less common, because of the restricted use of language in narrow semantic domains.35,36,37,38 Homonyms require multiple semantic types in the lexicon to represent each distinct meaning. For example, the two distinct meanings of the medical homonym “growth” are represented by the UMLS semantic types Physiologic-Process and Acquired-Abnormality.
Many lexemes have a number of different “senses” that reflect distinct patterns of usage. These differ from homonyms because the senses of the lexeme are determined in a systematic way by context. For example, the word “surgery” has the sense Therapeutic-or-Preventive Procedure in the sentence “Patient underwent surgery October 5th,” and the sense Health-Care-Organization in “Surgery was asked for a consult.” The senses are closely related: Surgery is the department that conducts surgical procedures.
A semantic lexicon can represent multiple senses of lexical items using distinct semantic types. However, greater richness in representing senses can lead to a decrease in parsing efficiency. The more semantic types assigned to each lexical entry, the more potential combinations found by the parser when analyzing a sentence. Thus, the semantic lexicon should avoid assigning multiple semantic types as much as possible.
The concern with parsing efficiency distinguishes a semantic lexicon from a general-purpose controlled medical vocabulary. For example, the UMLS Metathesaurus assigns to the term “digoxin” the semantic types Steroid, Carbohydrate, Pharmacologic-Substance, Biologically-Active-Substance, and Laboratory-Procedure. However, to parse the sentence “Patient was put on digoxin” it is sufficient to know that “digoxin” is a pharmaceutical. Because the structural perspective of chemicals is seldom relevant to clinical narrative, the Steroid and Carbohydrate senses may be dropped. Since all pharmaceuticals are biologically active, the Biologically-Active-Substance sense is redundant. While not useful for parsing, this additional knowledge about digoxin is still available in the controlled vocabulary for exploitation by other applications.
The grammar of the parsing system can often account for the different senses of a lexeme by means of syntactic and semantic patterns. This relieves the lexicon of the burden of representing all possible senses of a lexical item. For example, in the phrase “slightly elevated digoxin of 2.6,” the qualifier “elevated” indicates a laboratory procedure, whereas in “digoxin p.o.” the route modifier “p.o.” signals medication administration. If two semantic patterns are employed to capture this distinction, “digoxin” and the names of many other medications can be represented in the lexicon with just a single sense, Pharmacologic-Substance.
Combining Semantic and Syntactic Information
When parsing a sentence, the syntactic type of a word (noun, verb, adjective, adverb) is often a useful determinant of semantic type. If we know that a word is being used as a verb in a sentence, the semantic type of the word must be a subtype of the semantic type “Event”; if the word is an adjective, it is likely to be an abstract semantic type (not having a concrete location in space and time), since most qualities are abstractions. Thus, syntactic information can be used to reduce semantic ambiguity. For example, the word “left” can occur as an adjective in “opacity seen in left lung” and as a verb in “patient left hospital.” Similarly, “cold” is an adjective in “Patient had cold hands” and a noun in “Patient had a cold.” A semantic lexicon that incorporates syntactic information for these sentences might appear as shown in ▶. If this lexicon is used in processing the above sentences, syntactic information can be used to select the appropriate semantic type, increasing parsing efficiency. Processing systems that do not make use of syntactic information can ignore these constraints but must then assign “cold” and “left” two semantic types each and disambiguate the sentences using semantic patterns.
Table 3.
Lexical Entries Combining Syntactic and Semantic Information
| Lexeme | Syntactic Type | Semantic Type |
|---|---|---|
| cold | noun | Disease-or-Syndrome |
| cold | adjective | Qualitative-Concept |
| left | verb | Activity |
| left | adjective | Spatial-Concept |
Application Independence
If the output of a natural language processing system is to be used by other applications, the ontology of that system must be well defined. The better the formalization of the semantic types, the easier it will be to map the natural language processing results into a database or knowledge base.39,40 As discussed above, the lexical ontology must make only those distinctions that aid in parsing; too much detail does not help. However, the natural language processing output must also make sufficient distinctions to be mappable to controlled vocabularies and databases. If the ontology of the lexicon is highly detailed, it will approach the size of a controlled vocabulary and will be extremely difficult to maintain. If an existing controlled vocabulary (e.g., SNOMED) is used as the ontology, the natural language processing system becomes dependent on a knowledge structure that could change.
An alternative approach is for the natural language processing system to generate an intermediate representation41 consisting of general semantic types, plus the original words that occurred in the natural language sentence. This hybrid requires a relatively small ontology to organize the semantic types. By retaining the original words of the sentence, rules can be constructed to map the lexical items into a given target vocabulary. In this way, the natural language parser can be made independent of particular coding structures and databases.1,6
Methods
A semantic lexicon suitable for computer processing of clinical narrative can be constructed using resources available in the UMLS. Steps for building a semantic lexicon appropriate for discharge summaries are described briefly below. The following sections explain each of these methods in greater detail.
Lexemes in the Specialist Lexicon were matched against terms in the Metathesaurus to create a lexicon in which entries are assigned both syntactic and semantic types.
Lexemes in the semantic lexicon were then matched against word sequences in a corpus of discharge summaries, to determine which entries might be relevant to clinical narrative.
Lexemes having two or more semantic types were examined, and pairs of co-occurring semantic types were collected and ranked according to frequency of occurrence in the corpus.
A “semantic preference rule” was proposed whenever one member of a semantic pair was found to be consistently preferred over the other in the corpus.
The resulting set of semantic preference rules was generalized using the type hierarchy of the UMLS Semantic Network.
Semantic preference rules were applied to the initial semantic lexicon to create a lexicon customized for analysis of discharge summaries.
Lexical Matching
The UMLS Specialist Lexicon25 was used as the primary source of lexical items. These entries were matched against terms in the UMLS Metathesaurus (1997 version) in order to assign a list of possible semantic types to each lexeme. The syntactic types (noun, verb, adjective, etc.) of each lexeme are maintained in addition to the semantic information (as suggested in ▶).
Each variant of each lexeme in the Specialist Lexicon was matched against the strings in the Metathesaurus. A lexeme can match exactly, or it can be matched by making the first letter of the first word uppercase, or by making the first letter of each word uppercase, or by matching the “head noun.” In head noun matching, all words in the Metathesaurus string are checked, starting with the first preposition (in, of, with, etc.). If this term does not match, the leftmost word is discarded until a match occurs or no words remain. In addition, Metathesaurus terms can have “NOS” (not otherwise specified) or numbers in brackets (“<1>”, “<2>”, etc.) appended at the end. Examples of matches are shown in ▶.
Table 4.
Matching Specialist Lexemes to Metathesaurus Strings
| Specialist Lexeme | Metathesaurus String |
|---|---|
| cough | cough |
| fever | Fever |
| root canal | Root canal |
| medication | Medication, NOS |
| blood | Blood <1> |
| smear | PAP Smear |
| smear | Bacterial smear of specimen from eye |
Once a match was obtained, the semantic types for the Metathesaurus concept were retrieved. All derivational and inflectional variants of the lexical item were then generated, and each was assigned this list of semantic types. For example, the lexical item “suffocating” is a verb, and matches a Metathesaurus concept with semantic types Finding, Injury-or-Poisoning, and Disease-or-Syndrome. Using information in the Specialist Lexicon, lexical variants for this verb are generated, as shown in ▶.
Table 5.
Lexical Variants of the Verb “Suffocate”
| Lexeme | Syntactic Type | Semantic Types |
|---|---|---|
| suffocate | verb | Finding, Injury-or-Poisoning, Disease-or-Syndrome |
| suffocates | verb | Finding, Injury-or-Poisoning, Disease-or-Syndrome |
| suffocated | verb | Finding, Injury-or-Poisoning, Disease-or-Syndrome |
| suffocating | verb | Finding, Injury-or-Poisoning, Disease-or-Syndrome |
Corpus Matching
A corpus of discharge summaries for one year of hospital visits was collected from a database of online reports by selecting narrative sections such as “course in hospital” and “history of present illness.” These sections were broken into 603,306 separate sentences. Contiguous word sequences (up to five words) in the discharge summary sentences were matched against the semantic lexicon to determine which lexemes might be useful for analysis of the corpus. The resulting list of lexical items was ordered by the number of times each item occurred in the corpus.
Lexemes with Multiple Semantic Types
Lexemes occurring in the discharge summaries that had two or more semantic types were collected. Pairs of co-occurring semantic types were listed and ranked according to the number of occurrences of the lexical item in the corpus. The ten most frequently occurring semantic pairs are shown in ▶.
Table 6.
Ten Most Frequent Pairs of Co-occurring Semantic Types in Discharge Summaries
| Occurrences | Type 1 | Type 2 | Lexeme Examples |
|---|---|---|---|
| 65781 | Functional-Concept | Qualitative-Concept | positive, secondary, general |
| 60505 | Organic-Chemical | Pharmacologic-Substance | lasix, coumadin, aspirin |
| 43872 | Qualitative-Concept | Spatial-Concept | right, round, open |
| 36305 | Finding | Pathologic-Process | weakness, complications, bleeding |
| 35630 | Qualitative-Concept | Temporal-Concept | history, long, persistent |
| 33013 | Qualitative-Concept | Quantitative-Concept | secondary, all, total |
| 25814 | Sign-or-Symptom | Therapeutic-or-Preventive-Procedure | pulse, analgesic, sedation |
| 25384 | Occupation-or-Discipline | Temporal-Concept | history, histories |
| 25384 | Occupation-or-Discipline | Qualitative-Concept | history, histories |
| 22407 | Health-Care-Organization | Manufactured-Object | hospital, operating room, recovery room |
Semantic Preference Rules
The lexical items assigned to each semantic pair were examined in context in the corpus of discharge summaries. The contextual information was used to determine whether both semantic types can be assigned to some of the lexemes in the list or whether one type is consistently preferred to the other. A concordance program was used to find lexical items and show the surrounding words aligned on the right and left. This technique enables quick determination of similarities and differences of usage.
For example, ▶ shows how concordance information can be used to verify multiple senses of the word “drainage.” The UMLS Metathesaurus lists three semantic types for this word: Therapeutic-or-Preventive-Procedure, Pathologic-Process, and Body-Substance. These different senses can be detected in the corpus on the basis of surrounding words. The therapeutic sense is indicated when a lexeme is the object of verbs such as “performed” or a modifier of nouns such as “catheter” or “bag.” The pathologic sense occurs with other disease words such as “anastomosis” and “fistula” and with verbs such as “persisted” or “tapered.” The substance sense occurs with verbs such as “cultured” and adjectives such as “brown” or “bloody.”
Table 7.
Concordance for Lexeme “Drainage,” Grouped by Distinct Senses
| Therapeutic-or-Preventive-Procedure: | ||
| A | drainage | catheter was left in place. |
| ... as consulted who opined that a | drainage | procedure was required. |
| ... with a right ureteral stent to a | drainage | bag as well as a Malecot drain... |
| ... the Operating Room for abscess | drainage | , abscess was not reachable via... |
| ... Room and had incision and | drainage | of the vulvar hematoma. |
| ... the patient had an incision and | drainage | performed in the Emergency Room... |
| Pathologic-Process: | ||
| ... primary anastomosis and abscess | drainage | . |
| ... mediastinal tube were removed after | drainage | tapered off to 2 cubic centimeters... |
| The fistula and | drainage | persisted throughout the hospital... |
| ... onset of fever with redness and | drainage | of the superior portion of his... |
| ... with continued minimal bilious | drainage | for a period of one or two days... |
| Body-Substance: | ||
| The abscess | drainage | was cultured. |
| It did not have any | drainage | from it. |
| ... was completely dry without any | drainage | . |
| ... therapy and gradually the bloody | drainage | from her knee decreased. |
| Abdomen: obese; dark brown | drainage | at PEG site; some erythema. |
When one member of a pair of semantic types is found to be preferred for all lexical items assigned to that pair, a semantic preference rule is proposed. For example, the semantic pair consisting of Occupation-or-Discipline and Temporal-Concept is assigned to the lexemes “history” and “histories” (see ▶). Examination of usage in discharge summaries shows that only the temporal sense occurs. This observation suggests that when a lexical item has both a temporal sense and an occupational sense, the temporal sense should be preferred. This rule can be written as:
Occupation-or-Discipline → Temporal-Concept
A similar rule can be proposed for the pair Occupation-or-Discipline and Qualitative-Concept. Notice that these rules affect tens of thousands of occurrences in the corpus.
Generalizing Preference Rules
As semantic preference rules are discovered for the corpus, generalizations become apparent, which can result in a simpler set of rules. For example, when examining the usage of lexemes pertaining to chemicals (▶), we see a consistent preference for the functional sense of a lexeme over the structural sense. This is not surprising, since chemicals are discussed from a clinical perspective rather than a basic science perspective.
Table 8.
Pairs of Semantic Types for Chemical Lexemes
| Organic-Chemical | Pharmacologic-Substance | Lasix, Coumadin, aspirin |
| Organic-Chemical | Biologically-Active-Substance | creatinine, aspirin, glucose |
| Amino-Acid-or-Peptide-or-Protein | Immunologic-Factor | immunoglobulin, interferon, vaccine |
| Carbohydrate | Pharmacologic-Substance | heparin, digoxin, glucose |
These specific rules for chemicals can be simplified into a single general rule using more general semantic types from the UMLS Semantic Net:
Chemical-Viewed-Structurally → Chemical-Viewed-Functionally
The meaning of this rule is that any subtype of Chemical-Viewed-Functionally should be preferred over any subtype of Chemical-Viewed-Structurally.
The type hierarchy of the Semantic Network can also be used to make a second type of generalization about preference rules. A fair number of lexemes are assigned a pair of semantic types in which one is the supertype of the other (▶). This is redundant, because the super type can be inferred from the subtype. A useful generalization of these pairs is to prefer the subtype, which simplifies the lexicon without loss of information.
Table 9.
Pairs of Co-occurring Semantic Types Exhibiting Supertype/Subtype Relationship
| Pathologic-Process | Disease-or-Syndrome | atrophy, atelectasis, heart failure |
| Finding | Sign-or-Symptom | nausea, weakness, diaphoresis |
| Diagnostic-Procedure | Laboratory-Procedure | biopsy, biopsies, esophageal manometry |
A similar generalization can be made about pairs of semantic types that are siblings in the Semantic Network hierarchy (▶). An alternative to assigning both types to the lexemes is to assign their parent semantic type. For example, rather than stating that the word “care” is both individual and social behavior, it is simpler to assign to the lexeme the more general semantic type Behavior.
Table 10.
Pairs of Co-occurring Semantic Types That Are Siblings in Type Hierarchy
| Individual-Behavior | Social-Behavior | care, speech, singing |
| Diagnostic-Procedure | Therapeutic-or-Preventive-Procedure | electrocardiography, cardiac catheterization, colonoscopy |
| Mental-Process | Tissue-Function | sensation, touch, hearing |
The complete set of generalized semantic preference rules obtained by these methods is shown in ▶. For brevity, UMLS semantic type names have been shortened to four letter codes, which are listed in ▶. The preference rules are ordered by the number of occurrences in the corpus that are affected. The number of ambiguous lexemes in the semantic lexicon that are affected is also shown, along with a few examples of lexemes to which the rule applies.
Table 11.
Semantic Preference Rules, Ordered by Frequency of Occurrence in Discharge Summaries
| No. of Occurrences Affected | Frequency of Occurrence | Preference Rule | No. of Lexemes Affected | Lexeme Examples |
|---|---|---|---|---|
| 141666 | 0.21469 | CHVS→CHVF | 1158 | Lasix, Coumadin, creatinine |
| 97372 | 0.14757 | FIND→THNG | 631 | vital signs, wound, weakness |
| 79034 | 0.11977 | SIBS→PRNT | 393 | history, blood, care |
| 69529 | 0.10537 | FUNC→THNG | 130 | x-ray, wound, prophylaxis |
| 46048 | 0.06978 | PRNT→CHLD | 298 | nausea, procedure, diaphoresis |
| 41733 | 0.06325 | QUAL→SPAT | 30 | right, round, open |
| 32711 | 0.04957 | MPRO→THNG | 52 | will, evaluation, tolerance |
| 28161 | 0.04268 | ASUB→PHAR | 204 | heparin, aspirin, digoxin |
| 25384 | 0.03847 | OCCU→TEMP | 2 | history, histories |
| 20960 | 0.03176 | QUAL→QUAN | 15 | all, total, complete |
| 18368 | 0.02784 | LABP→CHEM | 103 | digoxin, glucose, vancomycin |
| 17549 | 0.02660 | TFUN→DIAG | 25 | blood pressure, vital capacity, diastolic pressure |
| 14481 | 0.02195 | OATR→BIOF | 22 | pulse, birth weight, visual acuity |
| 13293 | 0.02015 | THER→SSYM | 16 | pulse, sedation, analgesia |
| 11556 | 0.01751 | POPG→OATR | 4 | female, females, males |
| 10943 | 0.01658 | NPHE→DIAG | 5 | x-ray, ultrasound |
| 9550 | 0.01447 | PATH→ABNO | 263 | abnormalities, scar, aneurysm |
| 9227 | 0.01398 | QUAN→TEMP | 3 | first, second |
| 8946 | 0.01356 | QUAN→PATH | 4 | secondary, death |
| 7390 | 0.01120 | PHEN→BEHA | 20 | speech, drinking, suicide |
| 6277 | 0.00951 | SPAT→BIOF | 20 | cesarean section, deviation, displacement |
| 6169 | 0.00935 | DIAG→BSUB | 2 | urine, urines |
| 4520 | 0.00685 | NPHE→INJR | 1 | wound |
| 3786 | 0.00574 | RACT→THER | 23 | resection, removal, isolation |
| 3296 | 0.00500 | PLNT→THNG | 59 | guaiac, coffee, tobacco |
| 3079 | 0.00467 | OCCU→NBIO | 4 | ultrasound, chemistry |
| 2719 | 0.00412 | PHAR→HAZR | 39 | cocaine, phencyclidine hydrochloride, heroin |
| 2161 | 0.00327 | BPAR→BLOC | 16 | left arm, right arm, lower leg |
| 1499 | 0.00227 | THER→MDEV | 25 | prosthesis, toilet, sutures |
| 1422 | 0.00216 | FULL→ABNO | 25 | scar, callus, hammer toes |
| 1246 | 0.00189 | SPAT→POBJ | 27 | airway, stoma, rectosigmoid |
| 1086 | 0.00165 | THER→TFUN | 5 | touch, ultrafiltration, imagery |
| 1004 | 0.00152 | IPRD→MANU | 22 | films, manual, book |
| 939 | 0.00142 | REGU→THNG | 3 | patient, Medicaid, law |
| 798 | 0.00121 | PHAR→BPAR | 3 | vessel |
| 597 | 0.00090 | ANIM→THNG | 26 | gag, spots, turkey |
| 462 | 0.00070 | HACT→INJR | 7 | cut, puncture, avulsion |
| 411 | 0.00062 | QUAL→INJR | 6 | side effects, adverse effects |
| 363 | 0.00055 | QUAN→FAMG | 9 | twin, triplets, quadruplets |
| 339 | 0.00051 | TFUN→DISS | 4 | cystic fibrosis, vasospasm |
| 279 | 0.00042 | DISS→OFUN | 12 | breech presentation, grip, amenorrhea |
Figure 2.
Simplified UMLS semantic-type hierarchy.
The first row of this table states that chemicals viewed functionally are preferred to those viewed structurally. This rule applies to 141,666 occurrences in the corpus of discharge summaries (21 percent), and 1,558 of those lexical items found to be ambiguous (33 percent). The second rule captures the observation that the semantic types for findings (FIND, SSYM, RSLT) do not contribute additional meaning to lexical items. This is represented using the notation (FIND→THNG) where THNG (“thing”) represents the most general type in the semantic hierarchy (the parent of ENTY and EVNT). In all cases where another semantic type was present, this more specific type was preferred. For example, the lexeme “vital signs” is typed as a diagnostic procedure, and since any diagnostic procedure can be a finding, a semantic type for finding adds no additional meaning. The third rule merges sibling types into a parent type (this is written as SIBS→PRNT). The fourth rule prefers any semantic type to the functional concept type (FUNC→THNG rule). Finally, the fifth rule prefers subtypes to supertypes (written as PRNT→CHLD).
Customized Semantic Lexicon
The 40 semantic preference rules from ▶ were applied to the original semantic lexicon (described under “Lexical Matching” above). This greatly reduces the number of entries with multiple semantic types. When this customized lexicon is used to assign semantic types to sentences from discharge summaries, the number of occurrences of lexemes with multiple types is accordingly reduced.
Results
The Specialist Lexicon contains 155,759 distinct lexical forms (e.g., “cough,” coughs,” and “coughing” are three distinct forms). However, since the syntactic type is considered relevant in the methods described here (“cough” is both a noun and a verb), these can be counted as 164,850 different lexical forms. These were matched against the 630,658 strings in the Metathesaurus, resulting in a semantic lexicon with 75,711 entries.
Narrative sections were pulled from one year's worth of discharge summaries (22,596 reports) and broken into 603,306 separate sentences. This corpus contained 48,575 unique single words. Assuming that the average length of a lexeme is 1.39 words (based on the Specialist Lexicon), the total number of lexemes in the corpus can be estimated at 34,946 distinct forms. When contiguous word sequences in the corpus were matched against lexemes in the Specialist Lexicon, 27,633 matches were found. This number suggests that the 1997 version of the Specialist Lexicon has syntactic coverage about 79.1 percent of the total number of distinct lexical forms in the corpus. Using the semantic lexicon, it was determined that 13,322 of the lexical forms in the corpus had at least one semantic type, implying a semantic coverage of 38.1 percent.
The semantic lexicon contained 22,805 lexical forms that had two or more semantic types (30.1 percent of the entries). Of the 27,633 lexical forms occurring in discharge summaries, 3,474 (12.6 percent) had multiple semantic types. The number of distinct pairs of semantic types that occur together in lexical entries was 638. Pairs in which one semantic type was consistently preferred to another resulted in a preliminary set of semantic preference rules, which were ultimately reduced to the 40 more general rules shown in ▶.
When these semantic preference rules were applied to the semantic lexicon, the number of entries with multiple semantic types was reduced to 423 (1.5 percent). In the discharge summaries, occurrences of lexemes with multiple semantic types were reduced from 9.41 to 1.46 percent.
The benefits of reducing multiple semantic types can be seen in the paragraph below, randomly selected from the corpus. In this example, lexical items with semantic types are marked in angle brackets. When multiple semantic types are possible for a lexeme, they are separated by commas. The semantic type selected by preference rules is underlined.
The <PATG patient> remained on <THER suction>, however he continued to have a <TEMP,QUAL persistent> <SUBS air> leak and a <TEMP,QUAL persistent> <DISS,THER pneumothorax>. A <BLOC chest> CT was obtained which showed about 20% <DISS,THER pneumothorax> on the <SPAT,QUAL right> side in the <SPAT,QUAL right> <SPAT apical> <BPAR lung> fields. There were <QUAN some> <FIND cystic> <SPAT areas> suggestive of <AQAB,FIND blebs>. The <PATG patient> was taken to the <MANU,HORG Operating Room> for a blebectomy on 12/15/97. The <PATG patient> tolerated the <HACT,OACT procedure> <QUAL well>, without <FIND,PATH complications>. He was admitted to the <BOCC Surgical> <HORG Intensive Care Unit> that night and was <PFUN,THER transferred> out on postoperative day #1 <QUAL after> an uneventful <BOCC Surgical> <HORG Intensive Care Unit> course. The <PATG patient> on the <MANU Floor> was comfortable. His <BLOC chest> <MDEV tube> showed no <SUBS air> leak. <DIAG,FUNC,NPHE x-rays> showed a markedly decreased <DISS,THER pneumothorax>.
Discussion
The semantic preference rules described above result in a substantial reduction in the semantic ambiguity of clinical narrative. Although the rules were developed using the 1997 version of UMLS Knowledge Sources, the same rule set can be applied to the 1998 version. In this manner, new versions of the semantic lexicon can be generated as UMLS resources are updated. Because new lexemes are added each year to the Specialist Lexicon and Metathesaurus, the matching process must also be repeated. If new semantic types are added to the Semantic Network and Metathesaurus that significantly increase the number of lexemes with multiple types, additional semantic preference rules will need to be considered.
The number of matches between the Specialist Lexicon and Metathesaurus is smaller than might be expected. More sophisticated techniques for matching lexemes to Metathesaurus terms (by extending the head word matching methods) will produce many more entries in the semantic lexicon. The current approach is based on words and word sequences and simply associates semantic information with a given word form. Additional techniques using morphology and combinatory approaches can help manage the size of the semantic lexicon.43 For example, it might be possible to automatically provide semantic types for verbs that are derived from nouns whose semantic types are known (e.g., the verb “intubate” and its noun form “intubation”).
It is encouraging that an estimated 79 percent of lexemes in discharge summaries can be assigned syntactic information using UMLS resources. However, it is somewhat disappointing that the estimated semantic coverage is only 38 percent. Semantic coverage is not expected to reach 100 percent, because a large number of lexemes are general English and do not have medical semantic content. However, the current study was limited by adopting the Specialist Lexicon as the gold standard for medical lexemes. It could be argued that a semantic lexicon should include complex medical terms that might not be considered lexical items in a traditional sense. One possibility for increasing semantic coverage is to add terms from the Metathesaurus that occur in clinical text to the semantic lexicon. The difficulty with this approach is that syntactic information about these terms would be unknown and would have to be inferred automatically. Since the vast majority of terms in the Metathesaurus are nouns, this method could be successful.
The current approach attempts to assign semantic types to lexical items in clinical text simply by looking up the lexical items in the semantic lexicon. More sophisticated methods, known as “semantic tagging,”44,45,46 try to exploit contextual information. In a sense, these methods try to automate the manual techniques based on a concordance, as described under “Semantic Preference Rules” above). The current work can serve as a starting point for such approaches by providing data for co-occurrence statistics and other techniques for discovering semantic patterns.
Conclusion
The purpose of the present work is to create a lexical resource to aid in computer analysis of clinical reports. Semantic information is vital to the process but is effective only if the majority of lexical items have a single semantic type. Our results demonstrate that it is possible to construct a semantic lexicon of more than 75,000 entries automatically from UMLS resources and that precise semantic preference rules can be written to reduce the number of multiply typed lexemes to less than two percent. Further work is needed to increase the coverage of the semantic lexicon and to exploit contextual information in the selection of semantic senses.
This work was supported in part by grant IR29-LM05783 from the National Library of Medicine.
References
- 1.Sager N, Friedman C, Lyman MS. Medical Language Processing: Computer Management of Narrative Data. Reading, Mass.: Addison Wesley, 1987.
- 2.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994;1:142-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabrieli E, Speth D. Automated analysis of the discharge summary. Clin Comput. 1985;15(1):1-28. [PubMed] [Google Scholar]
- 4.Pietrzyk PM. A medical text analysis system for German syntax analysis. Methods Inf Med. 1991;30(4):275-83. [PubMed] [Google Scholar]
- 5.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C, Johnson SB, Forman B, Starren S. Architectural requirements for a multipurpose natural language processor in the clinical environment. Proc 19th Annu Symp Comput Appl Med Care. 1995:347-51. [PMC free article] [PubMed]
- 7.Haug PJ, Koehler S, Lau LM, Wang P, Rocha R, Huff SM. Experience with a mixed semantic/syntactic parser. Proc 19th Annu Symp Comput Appl Med Care. 1995:284-8. [PMC free article] [PubMed]
- 8.Haug P, Koehler S, Lau LM, Wang P, Rocha R, Huff S. A natural language understanding system combining syntactic and semantic techniques. Proc 18th Annu Symp Comput Appl Med Care. 1994:247-51. [PMC free article] [PubMed]
- 9.Do Amaral Marcio B, Satomura Y. Associating semantic grammars with the SNOMED: processing medical language and representing clinical facts into a language-independent frame. Medinfo. 1995;8(1):18-22. [PubMed] [Google Scholar]
- 10.Baud RH, Rassinoux AM, Lovis C, Wagner J, Griesser V, Michel PA. Knowledge sources for natural language processing. Proc AMIA Annu Fall Symp. 1996:70-4. [PMC free article] [PubMed]
- 11.Baud R, Lovis C, Alpay L, et al. Modeling for natural language understanding. Proc 17th Annu Symp Comput Appl Med Care. 1993:289-93. [PMC free article] [PubMed]
- 12.Friedman C, Johnson SB. Medical text processing: past achievements, future directions. In: Ball MJ, Collen MF (eds). Aspects of the Computer-based record. New York: Springer-Verlag, 1992.
- 13.Johnson SB. Natural language processing in biomedicine. In: Bronzino JD (ed). The Handbook of Biomedical Engineering. Boca Raton, Fla.: CRC Press, 1995:2768-73.
- 14.Spyns P. Natural language processing in medicine: an overview. Methods Inf Med. 1996;35(4-5):285-301. [PubMed] [Google Scholar]
- 15.Baud RH, Rassinoux AM, Wagner JC, et al. Representing clinical narratives using conceptual graphs. Methods Inf Med. 1995;34(1-2):176-86. [PubMed] [Google Scholar]
- 16.Mahesh K, Nirenburg S. A Situated Ontology for Practical NLP. In: Proceedings of the Workshop on Basic Ontological Issues in Knowledge Sharing. International Joint Conference on Artificial Intelligence; 1995; Montreal, Canada.
- 17.Bateman JA, Kasper RT, Moore JD, Whitney RA. A General Organization of Knowledge for Natural Language Processing: The Penman Upper Model [unpublished research report]. Marina del Rey, Calif.: USC Information Sciences Institute, 1989.
- 18.Knight K, Luk S. Building a large knowledge base for machine translation. Proc Am Assoc Artif Intell Conf., 1994: xxx-xx.
- 19.Miller GA, Beckwith R, Fellbaum C, Gross D, Miller KJ. Introduction to WordNet: an on-line lexical database. Int J Lexicography. 1990;3(special issue):235-312. [Google Scholar]
- 20.Guha RV, Lenat DB, Pittman K, Pratt D, Shepherd M. CYC: a midterm report. Commun ACM. 1990;33(8):xxx-xx. [Google Scholar]
- 21.National Library of Medicine. Medical Subject Headings Thesaurus. Bethesda, Md., NLM, 1989.
- 22.United States National Center for Health Statistics. The International Classification of Diseases, 9th Revision, with Clinical Modifications. Washington, D.C.: NCHS, 1980.
- 23.Coté RA (ed). Systematized Nomenclature of Medicine, 2nd ed. Skokie, Ill.: College of American Pathologists, 1982.
- 24.Tuttle NS, Olson NE, Campbell KE, Sherertz DD, Nelson SJ, Cole WG. Formal properties of the Metathesaurus. Proc 18th Annu Symp Comput Appl Med Care. 1994:145-9. [PMC free article] [PubMed]
- 25.Natural Language Systems Group. The Specialist Lexicon. Bethesda, Md.: National Library of Medicine, 1993.
- 26.McCray AT, Sponsler J, Brylawski B, Browne A. The role of lexical knowledge in biomedical text understanding. Proc 11th Annu Symp Comput Appl Med Care. 1986:103-7.
- 27.Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. In: van Bemmel JH, McCray AT (eds). 1993 Yearbook of Medical Informatics, Amsterdam, The Netherlands: International Medical Informatics Association, 1993:41-51. [DOI] [PMC free article] [PubMed]
- 28.McCray AT, Aronson AR, Browne AC, Rindflesch TC, Razi A, Srinivasan S. UMLS knowledge for biomedical language processing. Bull Med Libr Assoc. 1993:81(2):184-94. [PMC free article] [PubMed] [Google Scholar]
- 29.McCray AT. The UMLS Semantic Network. Proc 13th Annu Symp Comput Appl Med Care. 1989:503-7.
- 30.Kiuchi T, Kaihara S. On the linguistic representation of medical information: natural language, controlled language, and formal language. Medinfo. 1995(1):23-7. [PubMed]
- 31.Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. J Am Med Inform Assoc. 1994; 1:35-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zweigenbuam P, Bachimont B, Bouaud J, Charlet J, Boisvieux J-F. Issues in the structuring of an ontology for medical language understanding. Methods Inf Med. 1995;34(1-2):15-24. [PubMed] [Google Scholar]
- 33.Rector AL, Rogers JE, Pole P. The GALEN high level ontology. Proc Med Inform Europe. 1996; Copenhagen, Denmark.
- 34.Buitelaar P. CoreLex: Systematic polysemy and underspecification [PhD thesis]. Waltham, Mass.: Brandeis University, 1998.
- 35.Grishman R, Kittredge R (eds). Analyzing Language in Restricted Domains: Sublanguage Description and Processing. Hillsdale, N.J.: Erlbaum Associates, 1986.
- 36.Kittredge R, Lehrberger J (eds). Sublanguage: Studies of Language in Restricted Semantic Domains. New York: De Gruyter, 1982.
- 37.Sager N. Sublanguage: linguistic phenomenon, computational tool. In: Grishman R, Kittredge R (eds). Analyzing Language in Restricted Domains: Sublanguage Description and Processing. Hillsdale, N.J.: Erlbaum Associates, 1986.
- 38.Harris Z. A Theory of Language and Information: A Mathematical Approach. Oxford, England: Clarendon Press, 1991.
- 39.Sowa JF. Conceptual analysis for knowledge-base design. Methods Inf Med. 1995;34(1-2):165-71. [PubMed] [Google Scholar]
- 40.Gruber TR. Toward principles for the design of ontologies used for knowledge sharing. Stanford, Calif.: Stanford University, 1993. Technical report KSL 93-04.
- 41.Wehrli E, Clark R. Natural language processing, lexicon and semantics. Methods Inf Med. 1995;34(1-2):68-74. [PubMed] [Google Scholar]
- 42.Lovis C, Michel PA, Baud R, Scherrer JR. Word segmentation processing: a way to exponentially extend medical dictionaries. Medinfo. 1995(1):28-32. [PubMed]
- 43.Wilks Y, Stevenson M. Sense tagging: semantic tagging with a lexicon. SIGLEX ANLP-97 Workshop on Tagging Text with Lexical Semantics, 1997.
- 44.Resnik P. Selectional preference and sense disambiguation. SIGLEX ANLP-97 Workshop on Tagging Text with Lexical Semantics, 1997.
- 45.Basili R, Della Rocca M, Pazienza MT. Towards a bootstrapping framework for corpus semantic tagging. SIGLEX ANLP-97 Workshop on Tagging Text with Lexical Semantics, 1997.