Abstract
Over the past decades, network neuroscience has played a fundamental role in the understanding of large-scale brain connectivity architecture. Brains, and more generally nervous systems, can be modeled as sets of elements (neurons, assemblies, or cortical chunks) that dynamically interact through a highly structured and adaptive neurocircuitry. An interesting property of neural networks is that elements rich in connections are central to the network organization and tend to interconnect strongly with each other, forming so-called rich clubs. The ubiquity of rich-club organization across different species and scales of investigation suggests that this topology could be a distinctive feature of biological systems with information processing capabilities. This review surveys recent neuroimaging, computational, and cross-species comparative literature to offer an insight into the function and origin of rich-club architecture in nervous systems, discussing its relevance to human cognition and behavior, and vulnerability to brain disorders.
Keywords: anatomical connectivity, clinical neuroscience, comparative connectomics, complexity, connectome, evolution, functional integration, functional dynamics, graph analysis, neural network, neuroimaging, rich club
Abstract
En las últimas décadas, la neurociencia de las redes ha desempeñado un papel fundamental en la comprensión, a gran escala, de la arquitectura de las conexiones cerebrales. Los cerebros, y más en general los sistemas nerviosos, pueden ser modelados como conjuntos de elementos (neuronas, ensamblajes o fragmentos corticales) que interactúan dinámicamente mediante neu-rocircuitos altamente estructurados y adaptativos. Una propiedad interesante de las redes neurales es que los elementos ricos en conexiones son fundamentales para la organización de la red y tienden a interconectarse fuertemente entre sí, formando los llamados “clubes de ricos”. La ubicuidad de la organización de los clubes de ricos en diferentes especies y escalas de investigación sugiere que esta tipología tiene un papel clave en el desarrollo de procesos de red que están a la base de funciones neurales y que podría ser una característica distintiva de los sistemas que procesan información. Esta revisión analiza literatura comparada reciente entre especies, estudios de neuroimágenes y computacionales para ofrecer una idea acerca de la función y el origen de los clubes de ricos y la arquitectura central del sistema nervioso, al tiempo que discute la importancia de los clubes de ricos en la cognición y el comportamiento humano, como en la vulnerabilidad a los trastornos cerebrales.
Abstract
Ces dernières décennies, les neurosciences des réseaux ont joué un rôle fondamental dans la compréhension de l'architecture des connexions cérébrales à grande échelle. Le cerveau et plus généralement le système nerveux, peuvent être modelés comme des séries d'éléments (neurones, assemblages ou fragments corticaux) qui interagissent dynamiquement par l'intermédiaire d'un circuit neuronal hautement structuré et adapté. Les réseaux neuronaux ont une propriété intéressante : des éléments riches en connexions sont au centre de l'organisation en réseau et se connectent fortement entre eux, formant ce qu'on appelle un « club riche » (en connexions). L'omniprésence de l'organisation des « clubs riches » parmi les différentes espèces et échelles d'analyse suggère que cette topologie a un rôle clé dans le déploiement des processus en réseau sous-tendant les fonctions neuronales et qu'ils pourraient être une caractéristique distinctive des systèmes de traitement de l'information. Cet article étudie la neuro-imagerie récente, la littérature numérique et comparative à travers les espèces pour offrir un aperçu de la fonction et de l'origine des « clubs riches » et de l'architecture centrale du système nerveux, tout en analysant la pertinence des « clubs riches » pour le comportement et la cognition, ainsi que la vulnérabilité aux troubles cérébraux.
Introduction
The brain is a complex system composed of neural regions that dynamically interact at multiple spatial and temporal scales through a highly structured and adaptive neurocircuitry. This structural and dynamic organization forms the basis for all nervous-system capabilities, from input encoding to information processing and integration. In neural networks, segregated structural modules reflect brain functional specialization, while a minority of nodes with a high number of connections act as rich connectors between modules.1 This hierarchical architecture confers the advantage of evolvability and adaptability to nervous systems2,3 and has unique value in supporting complex brain dynamics underlying behavior and cognition.4,5
An intriguing characteristic of nervous systems architecture is the rich-club phenomenon, the tendency of central nodes in the network to densely connect to each other (Technical box). 6,7
Technical box. Rich-club architecture.
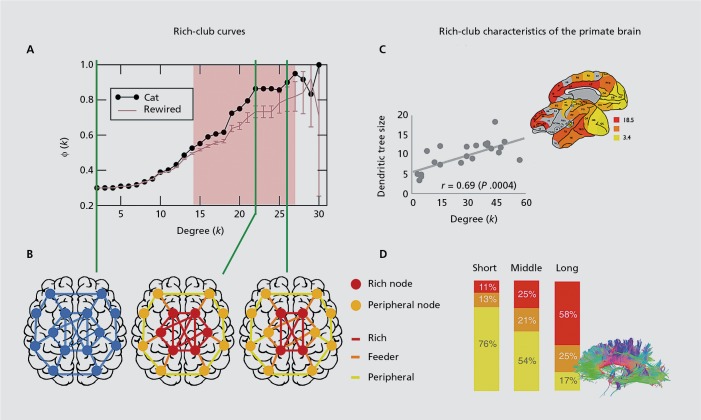
The term “rich club” refers to a topological property of brain and other real-world networks.6,7,15 A set of nodes is defined to form a “rich club” if their level of connectivity (ie, richness) exceeds what would be expected by chance alone, ie, what it would be expected to be in a randomized network with preserved richness sequence (Figure 1A and 1B).16,15 The nodal degree (ie, the number of connections of a node) is a common measure of nodal richness. Other measures can be considered to reflect richness, such as the weighted strength17 or nodal centrality measures.18 Within a network, the definition of rich clubs naturally selects subnetworks formed by topologically central nodes and their inter-connections. The “rich club” can also be intended as a topological feature of network architecture, without being associated to a univocally defined core subnetwork.
This organizational property is of particular interest because it dominates network topology8 and influences dynamic interactions among network elements.9 In the brain, rich nodes and their axonal projections enable short communication pathways and mediate connectivity among segregated functional systems.10,11 Rich connections span long distances in the white matter and have distinctive myelination and microstructural properties.12 Rich regions exhibit high oxidative metabolism and microstructural complexity in pyramidal neuron architecture, indicative of sustained activity and large computational capacity (Figure 1).13,14 These attributes suggest that rich clubs play a distinctive role in sustaining efficient communication across the whole-brain network, contributing to higher-order functions. However, the high connectivity density and metabolic demand make rich clubs costly features of brain networks and likely to be vulnerable to pathogenic agents.2 As a result, rich clubs express a trade-off between functional value, biophysical cost and vulnerability.
Figure 1. Rich-club curves. To assess whether a network demonstrates a rich-club architecture, a “richness” curve is drawn as a function of a nodal “richness” parameter (here, the nodal degree k) and compared with an equivalent curve drawn from a set of randomized networks. Panel A shows the rich-club curve of the cat structural connectome.6 Each point of the black curve represents the connection density Φ(k) between nodes of the cat connectome that have degree equal or larger than k. The “richness” red curve is derived from a series of randomized versions of the cat connectome, where the randomization procedure preserved the degree-sequence of the network. The pink area indicates the rich-club regime, where the connection density between high-degree nodes of the cat connectome is larger than expected in the randomized networks. The definition of network “rich-clubness” naturally selects nested rich subnetworks. Panel B sketches a whole-brain network (in blue) and rich-club subnetworks corresponding to two different values of the richness parameter k. Rich and peripheral nodes are represented in red and orange; rich, feeder, and peripheral connections are represented in red, orange, and yellow. Rich-club characteristics of the primate brain. Panel C: In the macaque connectome, the nodal richness parameter (k) significantly correlates with the dendritic tree size in layer III pyramidal neurons (right corner: color-coded macaque cortical map of dendritic tree size for 22 cortical regions).14 In humans, large proportions of rich and feeder brain connections span long distances in the white-matter. Panel D pictures the proportions of short- (<30 mm), middle- (30 to 90 mm) and long- range (>90 mm) connections belonging to the peripheral (yellow), feeder (orange), or rich (red) connection-classes10 (inset: example of white-matter tractography of the human brain, used for in vivo estimation of macroscale structural connections). Figures A, C, and D have been adapted with permission from ref 6: Zamora-Lopez G, Zhou C, Kurths J. Cortical hubs form a module for multisensory integration on top of the hierarchy of cortical networks. Front Neuroinform . 2010; 4:1 - ref 14: Scholtens LH, Schmidt R, de Reus, MA, van den Heuvel MP. Linking macroscale graph analytical organization to microscale neuroarchitectonics in the macaque connectome. J Neurosci. 2014; 34:12192-12205 - ref 10: van den Heuvel MP, Kahn RS, Goñi J, Sporns O. High-cost, high-capacity backbone for global brain communication. Proc Natl Acad Sci U S A. 2012; 109:11372-11377. Copyright © United States National Acadamy of Science, 2012.
How do rich clubs convey dynamic integration of brain processes? Why, and in response to which selection pressures, has rich-club architecture emerged in neural networks? What are the functional and evolutionary benefits and vulnerability drawbacks of the rich-club extra-connectivity? The field of network neuroscience offers an optimal framework to answer these questions. First, the conceptualization of nervous systems as networks of nodes (neurons, cortical columns, or gray-matter chunks) and connections (synapses, axons, or white-matter bundles) grants a sufficient level of abstraction for cross-scales and cross-species comparisons, allowing to integrate multimodal and multiscale data. Second, the mathematical graph-theoretical framework is well-suited to the study of collective dynamics of interlinked elements, both through mechanistic models and computational analyses. Third, network generative and developmental models enable uncovering the roots and the evolutionary aspects of nervous systems' organization.
The aim of this review is to provide an insight into the function and origin of the rich-club architecture observed in the human brain, and more generally in nervous systems, from a network science perspective. In the first part of the review, we mainly reason on the concept of functional integration and survey a selection of recent neuroimaging and computational studies that directly or indirectly investigate the contribution of rich-club architecture to whole-brain integrative dynamics. Next, we examine neural-network organizational principles across a series of animal species and scales of investigation, questioning the principles that contribute to the origin and evolution of rich clubs. Considering the relevance of rich-club architecture to clinical neuroscience, we conclude the review with a discussion on pathophysiological factors contributing to the vulnerability of rich clubs to neurological and psychiatric disorders, including schizophrenia and Alzheimer disease.
The functional role of the rich club
The topological characterization of neural networks positions the rich club at the center of brain communication pathways. From a mechanistic perspective, the short distance separating rich nodes from any other (peripheral) node in the network, and the gathering of shortest paths through rich edges make the rich club a putative core of neural integration. This view fosters a deeper conceptualization of functional integration in the brain, by means of both functional neuroimaging and computational studies, and in relation to the underlying anatomical substrate.
Functional integration
In functional neuroimaging experiments, the concept of functional integration has been associated with measures of statistical dependency (functional connectivity) between neural-related signals recorded across distinct locations in the brain. Interestingly, functional connectivity is partially predicted by structural connectivity,19 so that topologically close regions in the structural connectome also have the tendency to functionally connect. Paradoxically, the highly structurally connected rich club demonstrates low functional connectivity values at rest,12 questioning its role as an integrative core of the brain. While the shared structural connectivity,6,7 cytoarchitectural characteristics,20 and transcriptional profiles21,22 suggest a common function in the brain, it is not clear whether rich regions mainly act in concert or rather alternate over time to flexibly coordinate peripheral functional modules.23
To approach this question, it is necessary to consider a growing body of literature that investigates dynamical aspects of functional connectivity and time-dependent reconfiguration of brain states.24,25 During both resting and task conditions, the brain experiences time-varying functional connectivity and alternates periods of relatively localized activity/parallel processing (corresponding to low functional efficiency and high segregation) with periods of widespread synchronization (characterized by high functional efficiency).25-28 How does the rich club position itself within this dynamic landscape? As a first consideration, high-efficiency states consistently involve brain regions that partially overlap with the structural rich club27 and enable accurate performance of cognitive tasks requiring integration among multiple functional domains.28 As a second consideration, rich-club nodes and heteromodal cortices are particularly flexible over time, both at rest and in task settings.25,27,29 Flexibility refers to the property of some regions to frequently change their brain-wide functional connectivity profile, dynamically switching their allegiance to other functional communities.29 Structurally central regions, such as the frontoparietal system and default-mode-network areas, flexibly interact with other functional systems at rest26 and display variable patterns of functional connectivity across tasks, demonstrating high adaptability to changing demands.30 Interestingly, most flexible connections during rest show near-zero time-average functional correlations,27 which might explain the low levels of functional connectivity observed within the rich club at a time-average scale.12 Another possible explanation might be that, during rest, the brain tends to be less integrated than in cognitively demanding tasks and to “fall apart” into separate functional modules,28 so that the rich club could partially lose its integrative role and synchronization during rest.
Taken together, these observations indicate that functional integration cannot be ascribed to a single brain region or subnetwork, but should rather be regarded as a more complex and dynamic process, where flexibility and cross-system allegiance emerge as fundamental features. Structurally rich regions and connections might mediate those time-dependent integrative processes, with rich clubs forming an anatomical workspace for integration.
Computational underpinnings of rich-club function
Computational studies simulate the unfolding of functional dynamics on top of structural connectivity architectures, allowing for the assessment of the impact that structural topologies, presence of cores, or connectivity alterations have on global network dynamics.5 In the framework of this review, we are particularly interested in how a rich-club topology relates to the dynamic integrative processes characterizing nervous systems, and how rich clubs might support optimal information integration/segregation balance and functional complexity of the brain.
An early computational study on the cat connectome investigated how whole-brain activity, and in particular rich-club synchronization, is perturbed by an external input targeted toward primary sensory areas.6 This study showed that only a subset of the rich nodes tends to synchronize as a consequence of sensory stimulation, while the activities of the other rich nodes remain relatively independent. This result is compatible with above-reviewed experimental findings on rich-club flexibility and suggests that the rich club can functionally separate in response to changing inputs. This functional diversification may also relate to partially heterogeneous topological features of rich nodes, such as diverse participation coefficients to structural communities.18
Computational studies also show that a rich-club topology favors synchronization among peripheral areas (ie, integration of different and specialized brain units) and promotes complexity (richness) of brain dynamics.31-33 Structurally, single rich nodes tend to be (bidirectionally) connected with pairs of peripheral nodes not directly interconnected themselves, forming triadic resonant motifs. This particular topological configuration promotes synchronization between peripheral nodes through the central rich drivers.31 Moreover, rich nodes' signals possess particular spectral properties when compared with peripheral nodes. Computational models that allow heterogeneous local dynamics (ie, heterogeneous spectral characteristics) across brain regions34 show that, during task execution, the rich club tends to shift from a noisy to an oscillatory behavior that brings peripheral areas into coherence.33
Besides this integrative role, rich-club topology seems to promote dynamical complexity of networked systems. Complexity emerges when a dynamical system can access a large landscape of functional states that lie inbetween the two extremes of statistical independence (complete segregation) and global synchronization (complete integration) of its constituent elements.5 When considering the brain system, a condition of high functional complexity would correspond to a large landscape of accessible brain states and “optimal” metastable balance between segregation and integration, allowing both specialized processing and integration into conscious perception and higher-order functions.35 Simulations demonstrate that a rich-club topology achieves dynamic complexity levels larger than scale-free networks (ie, larger than networks with hubs but no rich-club phenomenon)36 and matching observations are reported in human functional data.32 Moreover, selective lesioning of the rich club entails a reduction in the system complexity,32 suggesting that rich-club organization subserves complexity and integration/segregation balance. This evidence has possible implications for the understanding of pathological mechanisms linking structural connectivity alterations, brain dynamics, and symptomatological/cognitive consequences.5,37
Overall, rich clubs are characterized by flexible and adaptive functional profiles. Rich nodes tend to mediate communication among distinct functional systems in virtue of their topological characteristics and spectral properties; they promote dynamic complexity and enable the coexistence of diverse integrative and localized functional patterns. The high neural cost of the formation of rich clubs may thus be justified by bringing advantageous integrative and dynamic properties to nervous systems.
An evolutionary and cross-species perspective
The wiring architecture of the human brain, and more generally of nervous systems observed in nature, are expressions of evolution, a process where different and often competing biological constraints and selection pressures shape biological organization. Understanding the mechanisms involved in the evolutionary path of nervous systems could provide valuable insights into the origin and function of complex features characterizing neural networks. A first question we might ask is whether a rich-club organization is an early feature of nervous systems, or a relatively recent “entry” in the evolutionary line. Subsequently, one might consider which selection pressures are compatible with the formation of rich clubs and/or lead to cross-species evolution of rich nodes and connections.
Rich clubs across species and neural scales
Connectivity studies highlight the presence of rich-club topology in a range of different species, ranging from small invertebrates to primates.3 The mesoscopic connectome of the nematode C. elegans has been mapped with electron microscopy and demonstrates a hierarchical structure, with modules interconnected through a dense rich club of neurons that are important for worm coordination and behavior.38 Similarly, the neurocircuitry of the Drosophila melanogaster, a fruit fly, includes local processing units (LPUs) organized in modules interconnected through a rich club.39 Rich LPUs lie deep in the center of the Drosophila brain, are heavily innervated by giant neurons, and constitute the sensorimotor integrative center of the organism, favoring nervous signals' spreading and coordination.39,40
At a macroscopic scale, vertebrate connectivity estimated from histological, neural tracing and in vivo MRI methods demonstrates defining features of hierarchical modular networks with rich-club organization.3 In the rodent, high-degree regions forming rich-clubs mainly overlap with associative cortices,41 shape brain functional patterns42 and have high co-expression of genes associated with oxidative energy metabolism, learning and organism behavior.22,43 The cat connectome displays spatially dislocated but topologically central hubs, forming a rich club that is responsible for inter-modular communication.10,11 Equivalent global organizational features are evident across mammals in general, as in the ferret,44 the macaque, and the chimpanzee.45
A comparative analysis of these findings demonstrates that functional specialization and integration are two fundamental aspects of neurocircuitry, preserved across species and scales of investigation.2,3,5 Interestingly enough, integration is consistently achieved through hierarchical rich-club architecture at different levels of network size and complexity, which makes “rich-clubness” a possible expression of convergent evolution. Furthermore, rich nodes correspond to functionally pivotal elements in the neurocircuitry of the different species, accomplishing tasks important for the survival and adaptability of the individual organisms (from sensory-motor coordination in the simplest invertebrates, to coherent perception and cognitive capabilities in mammals).
Finally, it is worthwhile to mention that rich clubs have been discovered in additional organizational scales of nervous systems. For example, in electrical recordings of spontaneous activity from in vitro mouse somatosensory cortex46 and in silico simulations of rat neocortical microcircuits,47 a minority of neurons forms rich clubs with high information transfer and a central role in shaping microcircuit synchronization patterns. “Rich-clubness” might be a scale-invariant organizational principle of natural systems with computational and information processing tasks.
Evolutionary and developmental principles
Rich-club formation thus emerges as a common organizational feature of nervous systems. This observation suggests that its formation might be driven by some fundamental evolutionary principles shared across different species. Generative models help elucidate this hypothesis by simulating network development under multiple constraints.48-50 In a generative experiment, network elements (nodes and/or connections) are progressively added to the system according to some predefined rule or to optimize some energy function. Alternatively, a real network can be progressively rewired according to some objective function. The resulting synthetic networks are then compared with real brain networks to identify possible evolutionary or developmental drivers.
A first evolutionary requirement shared by neural systems is the minimization of biological costs, which translates to limiting the overall number and length of connections. Cost-minimization is dictated by the finite spatial embedding, limited metabolic resources, and the necessity of bounding signal transmission delays in nervous systems. Generative experiments show that minimum-cost selection under geometrical constraints shapes highly clustered networks, but cannot reproduce long-distance connections, short path-length, and rich-club architecture.43,50 However, supplementing the model with additional rules, such as homophilic attraction (ie, the preferential formation of links between nodes that share a common neighborhood) results in cost-efficiency trade-off networks compatible with rich-club architecture.50,51
These considerations indicate that few simple generative rules can fairly reproduce a range of brain network properties, but the developmental/evolutionary interpretation of these results deserves further attention. It has been proposed that, at a microscopic scale, the homophily rule is compatible with Hebb's law for synaptic plasticity, so that neurons with common inputs (such as rich nodes) are more likely to be activated together, consolidating their interconnectivity and favoring rich-club formation in the long term.49 Other factors, such as the timing of the formation of nodes and connections, could also be a determinant for the resulting network architecture. In nonlinear growth models, new nodes are progressively added to the network, in numbers that increase exponentially with time. Nodes that develop early in the model tightly link together (as there are no other nodes for establishing connections), forming a dense core that contributes to shape the final network topology across later developmental stages.52 This exponential growth-model reproduces the same rich-club topologies observed in C. elegans and monkey, with no need for additional rules, such as preferential attachment or homophily.52 In the model, rich nodes appear early in the development of the network, similarly to what is observed in C elegans38 and in the human brain.53
In general, complex organizational features of nervous systems, such as a rich-club topology, might emerge as byproducts of simpler and (spatially or temporally) local developmental rules, which globally fit competing constraints and natural selection pressure. On the one hand, the fact that different species are subject to comparable developmental rules, dictated by comparable biological constraints, might partially explain the cross-species ubiquity of rich clubs. On the other hand, the subsistence of such constraints might limit the landscape of possible evolvable neural networks,3,47 questioning the mechanisms underlying cross-species differentiation and divergent evolution.
Cross-species differentiation
Rich nodes of nervous systems relate to crucial functions in the life of the organisms. In humans, rich regions have been associated with behavioral variability among individuals, including cognitive and intellectual performance.54 These functions are particularly developed in, or specific to, humans compared with their primate relatives. A fundamental question is therefore whether rich regions preserve their function among related species or whether they develop new or improved functions. Reasoning that brain functions are supported by the underlying cortical characteristics and white-matter wiring, one will also be interested in understanding whether rich regions and connections are spatially and morphologically preserved among related species, or whether they undergo substantial structural modifications.
Comparative studies of primate neuroimaging data show a significant overlap of rich regions among primate species, including the macaque, chimpanzee, and the human.45,55 Hubs have consistently been found in the insular, medial-parietal/precuneus, and ventro-lateral prefrontal cortices in all three species.45,56 However, hubs in the polar and medial prefrontal cortices are present in macaque and chimpanzee, but absent in human.45 Prefrontal regions undergo important morphological and microstructural changes between these species. Human brains have an expanded and more convoluted cortex and a larger white-matter volume in prefrontal areas,56,57 with lower neural density and higher number of dendrites.58 These structural variations suggest a functional specialization of the prefrontal cortex in human compared with other primates. This process might entail a partial reorganization of the brain-network topology, with a potential displacement of some network hubs (eg, the prefrontal hubs) and a possible reinforcement of other network hubs to achieve an adequate level of integration in a progressively more complex network. For example, the precuneus hub demonstrates an important expansion from chimpanzee to human59 and an altered connectivity pattern from macaque to human,55 which might suggest a topological reinforcement of this region. Further studies targeted to cross-species rich-club characterization might elucidate these aspects.
Rich clubs demonstrate cross-species structural variations. Many studies investigating primate-brain evolution have focused on cross-species morphological changes of the cortical mantle, highlighting a spatially nonuniform expansion centered on a few hot spots in frontal, parietal, and temporal areas.56,60 It remains to be understood whether rich regions are particularly involved in such morphological changes, and how these changes might relate to gray-matter microstructural reorganization (eg, increased/decreased neural density or arborization) and white-matter connectivity alterations (eg, reinforcement or diversification) in these regions. Moreover, it will be crucial to link cross-species structural differences with specific functional traits and increasing behavioral complexity across primates. Indeed, regions with the highest rates of cortical expansion from macaque and chimpanzee to human are involved in complex cognitive functions, such as relational thinking,58,60 and form resting-state networks that are present in humans, but absent in macaque,61 suggesting a relation between cross-species morphological evolution and functional development.
Finally, it should be noted that an expanding brain with increasing complexity and growing intelligence is also expected to be progressively more biologically expensive and susceptible to genetic and environmental insults.2 In particular, an evolving rich club, composed of highly active and functionally “stressed” cortices with long axonal projections, might reach large, and at worst unsustainable, biophysical costs. This aspect could have important evolutionary consequences, on the one hand impeding unlimited development of human intelligence,62 and on the other hand favoring disease susceptibility or the onset of human-specific brain disorders. For example, schizophrenia, a human-specific disorder, has been suggested to result from a “costly trade-off” between an increased connectivity complexity in humans compared with their ancestors and the development of valuable functions, such as social cognition.63
Rich-club vulnerability in pathological conditions
Over the last decades, the connectionist approach has caught on in the investigation of neurological and psychiatric disorders. Accordingly, symptoms and cognitive deficits can be read as faulty connectivity among brain areas.64,65 As already discussed, the rich club plays a distinctive role in sustaining overall functional coordination and is associated with higher-order cognitive abilities, which are impaired in the majority of brain disorders. One can therefore expect the rich club to be involved in a large number of pathologies. A recent meta-analysis, including 392 studies on 26 different disorders (Alzheimer disease, schizophrenia, and epilepsy, among others) indicates that gray-matter lesions are more likely to occur in brain hubs than in peripheral regions.66 MRI studies on large cohorts suggest that white-matter impairments also converge on rich and feeder connections in schizophrenia67 and other psychiatric and neurological disorders.68
Although different disorders may target different subsets of hubs,66 rich-club impairment and a parallel loss of network efficiency seem to be a general feature of brain pathologies.68 On one hand, a cross-disorder, rich-club impairment might express overlapping psychiatric and cognitive comorbidities.69 For example, depression, a mood disorder associated with hub impairment,70 is a comorbid factor of diverse pathologies, including multiple sclerosis, dementia, and epilepsy.71 On the other hand, a rich-club impairment might produce more pronounced symptoms by virtue of the rich-club functional importance.
The reasons underlying the vulnerability of rich regions and connections can be multiple and depend on the specific pathology under investigation. Rich regions demonstrate a continuously high baseline activity and glucose metabolism compared with peripheral regions.12 This aspect exposes the rich club to harmful mechanisms, such as oxidative stress and neuroinflammation, in a possibly preferential or selective way. Oxidative stress arises from a failure to maintain a correct balance between oxidative species and can lead to synaptic malfunction, deficits in myelination, alterations in cellular processes, and neuronal death. Different pathophysiological mechanisms, such as antioxidant system failure, metabolic alterations, and redox-species accumulation can jointly cause oxidative stress and ultimately converge on a rich-club vulnerability. For example, schizophrenia, a neurodevelopmental disorder characterized by a disruption of rich connections,72 has been associated with a deficit of glutathione synthesis, a major cellular antioxidant, and related to oxidative stress.68 In general, patients in the early stages of neurodegenerative disorders show increased compensatory brain activity, potentially concentrated in hub regions, which can lead to excitotoxicity and oxidative species accumulation.65 Among neurodegenerative disorders, Alzheimer's disease is characterized by deposition of β-amyloid (Aβ) plaques. The processing of amyloid precursor protein (APP), whose proteolysis generates Aβ, is activity-dependent and may therefore result in a preferential accumulation of Aβ; in high-metabolism rich regions.73 Furthermore, the topologically central rich club may mediate transneuronal spread of toxic substrate through axonal projections, accelerating disease progression or causing hub disruption as a secondary effect of unrelated pathological mechanisms. Indeed, different forms of brain dementia have been associated with “prion-like” transynaptic propagation of pathogenic agents, such as Aβ and other misfolded proteins.74 In those pathologies, the longitudinal spreading of brain lesions is highly predictable based on structural connectivity patterns,75 centralized through the rich-club circuitry.
The vulnerability of rich clubs may also relate to genetic factors. Patterns of similar gene expression in brain subnetworks can contribute to subnetworks' susceptibility to brain disorders in the context of transcriptional alterations or genetic risk factors. Notably, brain hubs demonstrate highly similar transcriptional profiles, enriched with genes relating to oxidative metabolism,22,76 synaptic signaling, and axonal structure.21 Hubs' transcriptional profiles are also enriched for schizophrenia-related genes,21 and the cortical expression patterns of those genes correlate with brain connectivity disruption in schizophrenia.77
The rich club also demonstrates characteristic developmental features. Rich regions and connections form early in the prenatal life: long-range corticocortical connections are established during the second and third semester of gestation, while feeder and peripheral connectivity develops over the third semester of gestation.53 After birth, long-range projections connecting associative hubs continue their maturation (myelination) longer than other peripheral connections, and until adolescence and adulthood.78 In parallel, cortical shrinkage and intracortical myelination rates are particularly high in hub regions during adolescence and early adulthood. Considering that hierarchical patterns of maturation in brain functional circuits relate to the development of specific cognitive functions, and that the timing of brain circuit maturation may determine windows of selective vulnerability,79 the developmental signature of the rich club, in combination with genetic and environmental adverse factors, could partially account for the involvement of the rich club in neurodevelopmental disorders.72
In summary, these observations demonstrate how different vulnerability factors (including biophysical and metabolic cost, topological centrality, genetic signature, and long maturational trajectories), combined with different pathological mechanisms (eg, primary vs secondary pathological pathways), can converge on a common end result, namely changes in rich-club characteristics. Large-scale, cross-disorder studies are required to elucidate cross-disorder differentiation from a network perspective.
Summary and perspectives
The research reviewed in this article identifies “rich clubness” as a scale invariant feature of nervous systems, an expression of convergent evolution, and a characterizing feature of biological systems with information processing capabilities. The rich club forms a flexible substrate promoting functional complexity and coordination among brain regions, ultimately supporting multisensory integration, coordination, and cognition. Further insights into the functional value of a rich club topology will require a better comprehension of brain dynamics and their cognitive and behavioral counterpart. Computational and neuroimaging studies able to explicitly model (directional) information flow through the structural network substrate might help clarify these aspects. This may require methodological development at multiple levels, including (i) multimodal and multi-scale integration methods (eg, functional and structural neuroimaging modalities); (ii) advanced network formalism (eg, multilayer and temporal networks for cross-frequency dynamics tracking); and (iii) high-complexity computational models (eg, including fine-grain intracortical characteristics, such as chemo-architecture and dynamics heterogeneity). Furthermore, it should be acknowledged that the “rich clubness” remains, per se, an abstract mathematical property of networks, which needs further validation and biological interpretation in the context of nervous systems' organization.7,80 On the one hand, this could be accomplished through animal studies where rich-club connectivity can be structurally and functionally manipulated or physically perturbed, for example with genetic or optogenetic techniques. On the other hand, future research could merge multiple fields of expertise, integrating genetic, neurobiological, and microstructural data with functional recordings and multi-scale analyses of in vivo and ex vivo brain connectivity information. Finally, the ascertained functional value of rich clubs comes at the price of a high biophysical cost that contributes to the vulnerability of rich-club resources to pathogenic agents. An intriguing hypothesis is that an increasing complexity of brain networks and (possible) expansion of rich clubs across primate species, might relate not only to the development of more sophisticated intellectual abilities, but also to the inception of human-specific brain disorders. Cross-species and cross-disorder network analyses on large datasets will help elucidate these aspects.
Acknowledgments
We thank Lianne Scholtens and Siemon de Lange for their critical reading of the manuscript and valuable comments. A. Griffa was funded by the Swiss National Science Foundation (SNSF grant P2ELP3_172087). M. P. van den Heuvel was funded by an ALW open (ALWOP.179) and VIDI (452-16-015) grant from the Netherlands Organization for Scientific Research (NWO) and a Fellowship of MQ. The authors have no conflicts of interest to declare.
Contributor Information
Alessandra Griffa, Dutch Connectome Lab, Department of Complex Trait Genetics, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, VU University Amsterdam, Amsterdam, The Netherlands.
Martijn P. Van den Heuvel, Dutch Connectome Lab, Department of Complex Trait Genetics, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, VU University Amsterdam, Amsterdam, The Netherlands; Department of Clinical Genetics, Amsterdam Neuroscience, VU University Medical Center, Amsterdam, the Netherlands.
REFERENCES
- 1.Sporns O. Network attributes for segregation and integration in the human brain. Curr Opin Neurobiol. 2013;23(2):162–171. doi: 10.1016/j.conb.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Bullmore E., Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13(5):336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- 3.van den Heuvel MP., Bullmore ET., Sporns O. Comparative connectomics. Trends Cogn Sci. 2016;20(5):345–361. doi: 10.1016/j.tics.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014;17(5):652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- 5.Deco G., Tononi G., Boly M., Kringelbach ML. Rethinking segregation and integration: contributions of whole-brain modelling. Nat Rev Neurosci. 2015;16(7):430–439. doi: 10.1038/nrn3963. [DOI] [PubMed] [Google Scholar]
- 6.Zamora-López G., Zhou C., Kurths J. Cortical hubs form a module for multisensory integration on top of the hierarchy of cortical networks. Front Neuroinform. 2010;4:1. doi: 10.3389/neuro.11.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Heuvel MP., Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31(44):15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu XK., Zhang J., Small M. Rich-club connectivity dominates assortativity and transitivity of complex networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82(4 Pt 2):046117. doi: 10.1103/PhysRevE.82.046117. [DOI] [PubMed] [Google Scholar]
- 9.Csermely P., London A., Wu L-Y., Uzzi B. Structure and dynamics of core/ periphery networks. J Complex Netw. 2013;1:93–123. [Google Scholar]
- 10.van den Heuvel MP., Kahn RS., Goni J., Sporns O. High-cost, high-capacity backbone for global brain communication. Proc Natl Acad Sci U S A. 2012;109(28):11372–11377. doi: 10.1073/pnas.1203593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.deReus MA., van den Heuvel MP. Rich club organization and intermodule communication in the cat connectome. J Neurosci. 2012;33(32):12929–12939. doi: 10.1523/JNEUROSCI.1448-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collin G., Sporns O., Mandl RC., van den Heuvel MP. Structural and functional aspects relating to cost and benefit of rich club organization in the human cerebral cortex. Cereb Cortex. 2014;24(9):2258–2267. doi: 10.1093/cercor/bht064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elston GN. Cortex, cognition and the cell: new insights into the pyramidal neuron and prefrontal function. Cereb Cortex. 2003;13(11):1124–1138. doi: 10.1093/cercor/bhg093. [DOI] [PubMed] [Google Scholar]
- 14.Scholtens LH., Schmidt R., de Reus MA., van den Heuvel MP. Linking macroscale graph analytical organization to microscale neuroarchitectonics in the macaque connectome. J Neurosci. 2014;34(36):12192–12205. doi: 10.1523/JNEUROSCI.0752-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alstott J., Panzarasa P., Rubinov M., Bullmore ET., Vértes PE. A unifying framework for measuring weighted rich clubs. Sci Rep. 2014;4:72–58. doi: 10.1038/srep07258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colizza V., Flammini A., Serrano MA., Vespignani A. Detecting rich-club ordering in complex networks. Nat Phys. 2006;2(2):110. [Google Scholar]
- 17.Opsahl T., Colizza V., Panzarasa P., Ramasco JJ. Prominence and control: the weighted rich-club effect. Phys Rev Lett. 2008;101(16):168702. doi: 10.1103/PhysRevLett.101.168702. [DOI] [PubMed] [Google Scholar]
- 18.Bertolero MA., Yeo BTT., D'Esposito M. The diverse club. Nat Commun. 2017;8(1):1277. doi: 10.1038/s41467-017-01189-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goñi J., van den Heuvel MP., Avena-Koenigsberger A., et al. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc Natl Acad Sci U S A. 2014;111(2):833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Heuvel MP., Scholtens LH., Feldman Barrett L., Hilgetag CC., de Reus MA. Bridging cytoarchitectonics and connectomics in human cerebral cortex. J Neurosci. 2015;35(41):13943–13948. doi: 10.1523/JNEUROSCI.2630-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitaker KJ., Vértes PE., Romero-Garcia R., et al. Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc Natl Acad Sci U S A. 2016;113(32):9105–9110. doi: 10.1073/pnas.1601745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulcher BD., Fornito A. A transcriptional signature of hub connectivity in the mouse connectome. Proc Natl Acad Sci U S A. 2016;113(5):1435–1440. doi: 10.1073/pnas.1513302113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Heuvel MP., Sporns O. An anatomical substrate for integration among functional networks in human cortex. J Neurosci. 2013;33(36):14489–14500. doi: 10.1523/JNEUROSCI.2128-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preti MG., Bolton TA., Van De Ville D. The dynamic functional connectome: state-of-the-art and perspectives. Neuroimage. 2017;160:41–54. doi: 10.1016/j.neuroimage.2016.12.061. [DOI] [PubMed] [Google Scholar]
- 25.de Pasquale F., Corbetta M., Betti V., Delia Penna S. Cortical cores in network dynamics. Neuroimage. 2017 Sep 30. Epub ahead of print. doi: 10.1016/j.neuroimage.2017.09.063. [DOI] [PubMed] [Google Scholar]
- 26.Fukushima M., Betzel RF., He Y., et al. Fluctuations between high- and low-modularity topology in time-resolved functional connectivity. Neuroimage. 2017 Aug 17. Epub ahead of print. doi: 10.1016/j.neuroimage.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zalesky A., Fornito A., Cocchi L., Gollo LL., Breakspear M. Time-resolved resting-state brain networks. Proc Natl Acad Sci U S A. 2014;111(28):10341–10346. doi: 10.1073/pnas.1400181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shine JM., Bissett PG., Bell PT., et al. The dynamics of functional brain networks: Integrated network states during cognitive task performance. Neuron. 2016;92(2):544–554. doi: 10.1016/j.neuron.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassett DS., Wymbs NF., Rombach MP., et al. Task-based core-periphery organization of human brain dynamics. PLoS Comput Biol. 2013;9(9):e1003171. doi: 10.1371/journal.pcbi.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole MW., Reynolds JR., Power JD., Repovs G., Anticevic A., Braver TS. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat Neurosci. 2013;16(9):1348–1355. doi: 10.1038/nn.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gollo LL., Zalesky A., Hutchison RM., van den Heuvel M., Breakspear M. Dwelling quietly in the rich club: brain network determinants of slow cortical fluctuations. Philos Trans R Soc Lond B Biol Sci. 2015;370(1668):20140165. doi: 10.1098/rstb.2014.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamora-López G., Chen Y., Deco G., Kringelbach ML., Zhou C. Functional complexity emerging from anatomical constraints in the brain: the significance of network modularity and rich-clubs. Sci Rep. 2016;6:38424. doi: 10.1038/srep38424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senden M., Reuter N., van den Heuvel MP., Goebel R., Deco G. Cortical rich club regions can organize state-dependent functional network formation by engaging in oscillatory behavior. Neuroimage. 2017;146:561–574. doi: 10.1016/j.neuroimage.2016.10.044. [DOI] [PubMed] [Google Scholar]
- 34.Deco G., Kringelbach ML., Jirsa VK., Ritter P. The dynamics of resting fluctuations in the brain: metastability and its dynamical cortical core. Sci Rep. 2017;7(1):3095. doi: 10.1038/s41598-017-03073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baars BJ. Global workspace theory of consciousness: toward a cognitive neuroscience of human experience. Prog Brain Res. 2005;150:45–53. doi: 10.1016/S0079-6123(05)50004-9. [DOI] [PubMed] [Google Scholar]
- 36.Senden M., Deco G., de Reus MA., Goebel R., van den Heuvel MP. Rich club organization supports a diverse set of functional network configurations. Neuroimage. 2014;96:174–182. doi: 10.1016/j.neuroimage.2014.03.066. [DOI] [PubMed] [Google Scholar]
- 37.Cabral J., Fernandes HM., van Hartevelt TJ., James AC., Kringelbach ML., Deco G. Structural connectivity in schizophrenia and its impact on the dynamics of spontaneous functional networks. Chaos. 2013;23(4):046111. doi: 10.1063/1.4851117. [DOI] [PubMed] [Google Scholar]
- 38.Towlson EK., Vértes PE., Ahnert SE., Schafer WR., Bullmore ET. The rich club of the C. elegans neuronal connectome. J Neurosci. 2013;33(15):6380–6387. doi: 10.1523/JNEUROSCI.3784-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih CT., Sporns O., Yuan SL., et al. Connectomics-based analysis of information flow in the Drosophila brain. Curr Biol. 2015;25(10):1249–1258. doi: 10.1016/j.cub.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 40.Worrell JC., Rumschlag J., Betzel RF., Sporns Olaf., Misic B. Optimized connectome architecture for sensorymotor integration. Netw Neurosci. 2017;1(4):415–430. doi: 10.1162/NETN_a_00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bota M., Sporns O., Swanson LW. Architecture of the cerebral cortical association connectome underlying cognition. Proc Natl Acad Sci U S A. 2015;112(16):E2093–E2101. doi: 10.1073/pnas.1504394112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethi SS., Zerbi V., Wenderoth N., Fornito A., Fulcher BD. Structural connectome topology relates to regional BOLD signal dynamics in the mouse brain. Chaos. 2017;27(4):047405. doi: 10.1063/1.4979281. [DOI] [PubMed] [Google Scholar]
- 43.Rubinov M., Ypma RJ., Watson C., Bullmore ET. Wiring cost and topological participation of the mouse brain connectome. Proc Natl Acad Sci U S A. 2015;112(32):10032–10037. doi: 10.1073/pnas.1420315112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sukhinin Dl., Engel AK., Manger P., Hilgetag CC. Building the ferretome. Front Neuroinform. 2016;10:16. doi: 10.3389/fninf.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L., Hu X., Preuss TM., et al. Mapping putative hubs in human, chimpanzee and rhesus macaque connectomes via diffusion tractography. Neuroimage. 2013;80:462–474. doi: 10.1016/j.neuroimage.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nigam S., Shimono M., Ito S., et al. Rich-club organization in effective connectivity among cortical neurons. J Neurosci. 2016;36(3):670–684. doi: 10.1523/JNEUROSCI.2177-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gal E., London M., Globerson A., et al. Rich cell-type-specific network topology in neocortical microcircuitry. Nat Neurosci. 2017;20(7):1004–1013. doi: 10.1038/nn.4576. [DOI] [PubMed] [Google Scholar]
- 48.Avena-Koenigsberger A., Goni J., Sole R., Sporns O. Network morphospace. J R Soc Interface. 2015;12(103):20140881. doi: 10.1098/rsif.2014.0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vértes PE., Alexander-Bloch A., Bullmore ET. Generative models of rich clubs in Hebbian neuronal networks and large-scale human brain networks. Philos Trans R Soc Lond B Biol Sci. 2014;369(1653):20130531. doi: 10.1098/rstb.2013.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Betzel RF., Avena-Koenigsberger A., Goni J., et al. Generative models of the human connectome. Neuroimage. 2016;124(Pt A):1054–1064. doi: 10.1016/j.neuroimage.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Csigi M., Kőrösi A., Biró J., Heszberger Z., Malkov Y., Gulyás A. Geometric explanation of the rich-club phenomenon in complex networks. Sci Rep. 2017;7(1):1730. doi: 10.1038/s41598-017-01824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bauer R., Kaiser M. Nonlinear growth: an origin of hub organization in complex networks. R Soc Open Sci. 2017;4(3):160691. doi: 10.1098/rsos.160691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ball G., Aljabar P., Zebari S., et al. Rich-club organization of the newborn human brain. Proc Natl Acad Sci U S A. 2014;111(20):7456–7461. doi: 10.1073/pnas.1324118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baggio HC., Segura B., Junque C., de Reus MA., Sala-Llonch R., Van den Heuvel MP. Rich club organization and cognitive performance in healthy older participants. J Cogn Neurosci. 2015;27(9):1801–1810. doi: 10.1162/jocn_a_00821. [DOI] [PubMed] [Google Scholar]
- 55.Goulas A., Bastiani M., Bezgin G., Uylings HB., Roebroeck A., Stiers P. Comparative analysis of the macroscale structural connectivity in the macaque and human brain. PLoS Comput Biol. 2014;10(3):e1003S29. doi: 10.1371/journal.pcbi.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rilling JK. Comparative primate neuroimaging: insights into human brain evolution. Trends Cogn Sci. 2014;18(1):46–55. doi: 10.1016/j.tics.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Schoenemann PT. Evolution of the size and functional areas of the human brain. Annu Rev Anthropol. 2006;35:379–406. [Google Scholar]
- 58.Vendetti MS., Bunge SA. Evolutionary and developmental changes in the lateral frontoparietal network: a little goes a long way for higher-level cognition. Neuron. 2014;84(5):906–917. doi: 10.1016/j.neuron.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruner E., Preuss TM., Chen X., Rilling JK. Evidence for expansion of the precuneus in human evolution. Brain Struct Funct. 2017;222(2):1053–1060. doi: 10.1007/s00429-015-1172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buckner RL., Krienen FM. The evolution of distributed association networks in the human brain. Trends Cogn Sci. 2013;17(12):648–665. doi: 10.1016/j.tics.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 61.Mantini D., Corbetta M., Romani GL., Orban GA., Vanduffel W. Evolutionary novel functional networks in the human brain? J Neurosci. 2013;33(8):3259–3275. doi: 10.1523/JNEUROSCI.4392-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofman MA. Evolution of the human brain: when bigger is better. Front Neuroanat. 2014;8:15. doi: 10.3389/fnana.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burns JK. An evolutionary theory of schizophrenia: cortical connectivity, metarepresentation, and the social brain. Behav Brain Sci. 2004;27(6):831–855. doi: 10.1017/s0140525x04000196. [DOI] [PubMed] [Google Scholar]
- 64.Uhlhaas PJ., Singer W. Neural synchrony in brain disorders: relevance for cognitive dysfunctions and pathophysiology. Neuron. 2006;52(1):155–168. doi: 10.1016/j.neuron.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 65.Fornito A., Zalesky A., Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16(3):159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- 66.Crossley NA., Mechelli A., Scott J., et al. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137(Pt 8):2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klauser P., Baker ST., Cropley VL., et al. White matter disruptions in schizophrenia are spatially widespread and topologically converge on brain network hubs. Schizophr Bull. 2017;43(2):425–435. doi: 10.1093/schbul/sbw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Griffa A., Baumann PS., Thiran JP., Hagmann P. Structural connectomics in brain diseases. Neuroimage. 2013;80:515–526. doi: 10.1016/j.neuroimage.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 69.Goodkind M., Eickhoff SB., Oathes DJ., et al. Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry. 2015;72(4):305–315. doi: 10.1001/jamapsychiatry.2014.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gong Q., He Y. Depression, neuroimaging and connectomics: a selective overview. Biol Psychiatry. 2015;77(3):223–235. doi: 10.1016/j.biopsych.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 71.Nuyen J., Schellevis FG., Satariano WA., et al. Comorbidity was associated with neurologic and psychiatric diseases: a general practice-based controlled study. J Clin Epidemiol. 2006;59(12):1274–1284. doi: 10.1016/j.jclinepi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 72.Collin G., Scholtens LH., Kahn RS., Hillegers MHJ., van den Heuvel MP. Affected anatomical rich club and structural-functional coupling in young offspring of schizophrenia and bipolar disorder patients. Biol Psychiatry. 2017;82(10):746–755. doi: 10.1016/j.biopsych.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 73.Buckner RL., Sepulcre J., Talukdar T., et al. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci. 2009;29(6):1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaufman SK., Diamond MI. Prion-like propagation of protein aggregation and related therapeutic strategies. Neurotherapeutics. 2013;10(3):371–382. doi: 10.1007/s13311-013-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mezias C., LoCastro E., Xia C., Raj A. Connectivity, not region-intrinsic properties, predicts regional vulnerability to progressive tau pathology in mouse models of disease. Acta Neuropathol Commun. 2017;5(1):61. doi: 10.1186/s40478-017-0459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vértes PE., Rittman T., Whitaker KJ., et al. Gene transcription profiles associated with inter-modular hubs and connection distance in human functional magnetic resonance imaging networks. Philos Trans R Soc Lond B Biol Sci. 2016;371(1705):20150362. doi: 10.1098/rstb.2015.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romme IA., de Reus MA., Ophoff RA., Kahn RS., van den Heuvel MP. Connectome disconnectivity and cortical gene expression in patients with schizophrenia. Biol Psychiatry. 2017;81(6):495–502. doi: 10.1016/j.biopsych.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 78.Hagmann P., Sporns O., Madan N., et al. White matter maturation reshapes structural connectivity in the late developing human brain. Proc Natl Acad Sci U S A. 2010;107(44):19067–19072. doi: 10.1073/pnas.1009073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meredith RM. Sensitive and critical periods during neurotypical and aberrant neurodevelopment: A framework for neurodevelopmental disorders. Neurosci Biobehav Rev. 2015;50:180–188. doi: 10.1016/j.neubiorev.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 80.van den Heuvel MP., Yeo BTT. A spotlight on bridging microscale and macroscale human brain architecture. Neuron. 2017;93(6):1248–1251. doi: 10.1016/j.neuron.2017.02.048. [DOI] [PubMed] [Google Scholar]