Abstract
Network neuroscience is a thriving and rapidly expanding field. Empirical data on brain networks, from molecular to behavioral scales, are ever increasing in size and complexity. These developments lead to a strong demand for appropriate tools and methods that model and analyze brain network data, such as those provided by graph theory. This brief review surveys some of the most commonly used and neurobiologically insightful graph measures and techniques. Among these, the detection of network communities or modules, and the identification of central network elements that facilitate communication and signal transfer, are particularly salient. A number of emerging trends are the growing use of generative models, dynamic (time-varying) and multilayer networks, as well as the application of algebraic topology. Overall, graph theory methods are centrally important to understanding the architecture, development, and evolution of brain networks.
Keywords: connectome, functional MRI, graph theory, neuroanatomy, neuroimaging
Abstract
La neurociencia de la red es un campo próspero y de rápida expansión. Los datos empíricos sobre las redes cerebrales, desde niveles moleculares hasta niveles conductuales, son cada vez más grandes en tamaño y complejidad. Estos desarrollos llevan a una fuerte demanda de herramientas y métodos apropiados que modelen y analicen los datos de la red cerebral, como los proporcionados por la teoría de grafos. Esta breve revisión examina algunas de las medidas y técnicas gráficas más comúnmente empleadas y neurobiológicamente más discriminadoras. Entre estas, son particularmente importantes la detección de módulos o comunidades de redes, y la identificación de elementos de redes centrales que facilitan la comunicación y la transferencia de señales. Algunas tendencias emergentes son el empleo creciente de modelos generativos, de redes dinámicas (de tiempo variable) y de multicapa, así como la aplicación de topología algebraica. En general, los métodos de la teoría de grafos son especialmente importantes para comprender la arquitectura, el desarrollo y la evolución de las redes cerebrales.
Abstract
La neuroscience des réseaux est un domaine florissant qui s'étend rapidement. Les données empiriques sur les réseaux cérébraux, de l'échelle moléculaire à comportementale, ne cessent d'augmenter en volume et en complexité. Ces développements génèrent une demande forte d'outils et de méthodes appropriés pour modéliser et analyser les données des réseaux cérébraux, comme celles fournies par la théorie des graphes. Dans cette rapide analyse, nous examinons certaines des techniques et mesures de graphes les plus couramment utilisées et les plus signifiantes neurobiologiquement. Parmi elles, la détection des modules ou communautés de réseaux et l'identification des éléments de réseau central qui facilite la communication et le transfert du signal, sont particulièrement marquantes. Dans les tendances émergentes, on note l'utilisation croissante de modèles génératifs, dynamiques (variables avec le temps) et les réseaux multi-couches, ainsi que l'application de la topologie algébrique. Globalement, les méthodes de la théorie des graphes sont essentielles pour comprendre l'architecture, le développement et l'évolution des réseaux cérébraux.
Introduction
Some of the most daunting scientific challenges of the 21st century involve complex social, technological, and biological systems—from the stability of global financial and economic networks to the spreading of epidemics, the web of biotic interactions in an ecosystem, and metabolic and transcriptional processes inside cells and tissues. An important theoretical foundation for understanding complexity is network science,1-3 which focuses on the structure and function of systems that are composed of numerous elements and their interactions. Over the past couple of decades, the network perspective has gained considerable ground in neuroscience.4-11 Brain networks have become a fertile area of research, now called network neuroscience, ranging across different scales, from interacting biomolecules all the way to social behavior. A major driving force has been the application of mathematical and computational network science tools to neurobiological systems, especially models and measures of graph theory.1,5,9
Graph theory is a branch of mathematics that dates back to the 18th century. Today, applications of graph theory pervade all scientific disciplines as well as many modern information and computing technologies. The brain is a natural fit for graph theory approaches as it is readily represented as a network (a graph) of elements and their pairwise interconnections, also called nodes and edges. Comprehensive maps of brain connectivity have given rise to the emerging field of connectomics,12,13 a central focus of which is the systematic and quantitative study of brain networks and graphs.
Graph theory methods, when applied properly, can offer important new insights into the structure and function of networked brain systems, including their architecture, evolution, development, and clinical disorders. This brief review surveys some of the most relevant graph theory methods and illustrates their application in various neurobiological contexts. Comprehensive coverage of the topic is beyond the scope of this article (see a recent textbook9). Instead, the emphasis here is on highlighting some new methodological trends, discussing their application to brain data, and suggesting future avenues for graphical models and measures.
Basic concepts
Networks or graphs are collections of elements (nodes, vertices) and their pairwise links (edges, connections) which, in their simplest form, can be summarized in the form of a connection (or adjacency) matrix. The complete set of all pairwise connections defines the graph's topology, providing a complete map of all relations among nodes and edges. Brain nodes may be individual neurons or entire brain regions, depending on the measurement technique. Edges can take on binary or weighted values, and they can be directed or undirected, depending on how interactions are estimated from empirical data. The selection of appropriate graph theory methods for modeling and analyzing empirical data requires that the nature of the edge representation is taken into account.6,9 Put differently, not all graph theory methods fit all purposes.
The two most common species of brain graphs describe structural and functional connectivity among neural elements. Structural graphs are generally sparse (most possible structural connections in a given nervous systems do not exist) and temporally relatively stable (but subject to plasticity and development). In contrast, functional graphs record statistical dependencies among neuronal time series, and hence are often dense and highly variable across time. The dichotomy of structural and functional graphs is important to consider, as each domain draws on a specific subset of graph theory methods that are commensurate with the origin of the data.
The definition of nodes generally requires sophisticated data-processing pipelines and is the subject of much methodological discussion. In whole-brain networks acquired with neuroimaging, nodes are often derived from a parcellation of the original voxel-level data, designed to extract coherent brain “areas” that form the building blocks of structural or functional brain networks. Some approaches leverage anatomical templates, others pursue parcellation in a data-driven manner, eg, by boundary detection and clustering patterns of connectivity.14 A recent multimodal study employed machine learning to extract coherent brain regions on the basis of several different anatomical and functional criteria.15 Node definition is considerably more straightforward in studies of circuits and neuronal populations, where individual neurons are natural candidates for individual network nodes.
A major simplification inherent in most current applications of graph theory is the assumption that, within a given network representation, all nodes and edges are identical and homogeneous. Annotation of nodes and edges can address this limitation of simple graphs, by allowing additional layers of data to be linked to network elements, which can be useful for identifying biologically meaningful network communities.16 A further elaboration of simple graphs is the inclusion of multidimensional relationships which can be expressed in multilayer networks,17 composed of different layers that encode different types of interactions (eg, synaptic links, temporal correlations, transmitter systems, or gene coexpression).
Graphs can be investigated at different levels of scale, and specific measures capture graph attributes at local (nodal) and global (network-wide) scales.6 Nodal measures include simple statistics such as node degree or strength, while global measures express network-wide attributes such as the path length or the efficiency. Intermediate scales can be accessed via hierarchical neighborhoods around single graph elements,18 or by considering subgraphs or motifs.19,20 Motifs are defined as subsets of network nodes and their mutual edges whose patterns of connectivity can be classified into distinct motif categories. In empirical networks, these categories often occur in characteristic frequencies that can be compared with distributions from appropriate (random) null models. In the brain, motif analysis has been applied to structural20 and functional graphs.21
Most highly resolved structural brain networks are not fully, or even densely, connected. In such sparsely connected graphs, the minimal topological distance between two nodes, ie, the length of the shortest path, often involves multiple steps. Network paths are composed of unique edges that are traversed only once, while the usually much more abundant walks between two nodes can use edges any number of times. Paths and walks are considered important for the flow of signals and communication,22 and are the basis for popular graph metrics such as the so-called efficiency which defines the global capacity of a graph to pass information via short paths. Importantly, concepts of path lengths and efficiency are most naturally applied to brain graphs that represent structural connectivity. In contrast, they have rather different (and potentially problematic) interpretations when applied to functional connectivity. The distinction derives from the nature of functional connectivity which reports statistical associations among neural time series rather than a web of physical links that propagates neural signals.
Modularity
Among the most widely encountered and biologically meaningful aspects of brain networks is their organization into distinct network communities or modules6,10,23 (Figure 1). Modules are useful to partition larger networks into basic “building blocks,” ie, internally densely connected clusters that are more weakly interconnected amongst each other. Modular partitions have neurobiological significance as their boundaries separate functionally related neural elements, define critical bridges and hubs that join communities, channel and restrict the flow of neural signals and information, and limit the uncontrolled spread of perturbations.23
Figure 1. Modularity. (A) Schematic network plot showing a set of nodes and edges interconnected to form two relatively distinct modules (communities). Note that the two modules are linked via a single hub node (black) that maintains two bridges between the two modules. Panels (B) to (E) use a 77-node data set from reference 74, representing the 77 areas and directed weighted projections of the rat cerebral cortex. (B) The plot at the top illustrates the varying number of modules as the value of the resolution parameter is increased from 0.1 to 4.0. The number of detected modules increases from 1 to 22 within this range. (C) The matrix plot represents the variation of information between all detected partitions within the range of the resolution parameter plotted above. Dark blue corresponds to a variation of information (distance) of zero, ie, identity. The region around gamma=0.7 is the most homogeneous region within the range. (D) The rat cerebral cortex connection matrix (weights displayed on log-scale), reordered by module assignment for gamma=0.7. The three modules are indicated with white boundaries. (E) The multiscale co-assignment matrix, computed using the method described in ref. 37. Co-assignment varies between 1 (node pair in same module at all scales) to 0 (node pair never co-assigned at any scale). Tree plot at the bottom shows all hierarchically clustered solutions, with the top one corresponding to the same three modules shown in panel (C). Within each of the three modules, additional modular structure is detected.
There are numerous computational techniques for extracting communities and modules from complex networks.24,25 One of the most widely used approaches in network neuroscience is modularity maximization which aims to divide a given network into a set of non-overlapping communities by maximizing a global objective function, the modularity metric.26 Originally, this metric was formulated to detect communities whose internal density of connections is maximal, relative to a degree-preserving null model. More recently, variants of the metric that can be applied to directed27 and signed28 networks have been proposed. Of special note are variants of the scheme that are designed to deal with correlation matrices,29 a data type that is often encountered in studies of functional connectivity.
Modularity maximization faces important methodological limitations that need to be addressed. One limitation refers to the existence of several, often quite numerous, distinct partitions that are almost equally optimal (ie, degenerate) under the modularity metric. Given the inherent noisiness of empirical estimates of the network topology, it seems arbitrary to pick a single “optimal” partition as the sole representative of the network's community structure. Instead, multiple degenerate solutions should be aggregated into consensus partitions, for example by forming an agreement matrix that can then be reclustered until a single consensus partition emerges.30 The application of such a consensus approach can also reveal additional facets of community organization, including the degree to which individual nodes are affiliated with their host community.28,31
Another fundamental limitation is the inability of the original modularity metric to detect modules below a certain size. This resolution limit32 can be addressed in a number of ways. One common avenue is the inclusion of an additional resolution parameter into the modularity metric that rescales the intrinsic null model and allows the detection of smaller or larger communities.33 Varying the resolution parameter is important since many brain networks exhibit communities across different scales,34 which renders the selection of a single scale of analysis potentially problematic. The issue becomes of fundamental neurobiological importance when community detection methods are used to identify, for example, specific partitions of the brain into resting-state functional networks or “functional systems.” While several landmark studies have proposed canonical partitions of such networks;35,36 that are now widely applied in the field, it should be noted that other partitions at both finer and coarser scales may represent additional levels of organization. Up to date, most studies circumvent full multiscale analysis by selecting one or a few settings of the resolution parameter, usually based on some criteria of partition stability.
One way to preserve and represent the full multiscale structure of brain networks is to perform consensus clustering across multiple spatial resolutions,37 an approach that combines sampling the entire range of possible spatial resolutions with a hierarchical consensus clustering procedure. The approach returns a coassignment matrix that captures the probability that each node pair remains associated within the same community as the scale is varied, together with a hierarchy that illustrates their mutual relations. Multiresolution consensus clustering delivers a more complete picture of community structure than is provided by single partitions, and avoids complicated and often rather arbitrary models for selecting relevant scales in brain networks.
Finally, many extensions of the above framework for detecting communities in brain networks should be noted. While modularity maximization detects non-overlapping communities, it may also be useful to define modules as overlapping communities (eg, see ref 38), ie, sets of nodes where some, or all, nodes maintain multiple affiliations. Other methods, eg, multislice modularity,39 are designed to track modular partitions, and their nodal memberships, across time. Yet another promising avenue, with deep historical roots in the social sciences, is the use of block models.40 Block modeling attempts to fit a statistical model for generating networks to empirical data to identify those model parameters that provide the best match. For example, those parameters may correspond to a number of blocks and the corresponding within and between block connection probabilities. Since block models are not limited to strictly modular arrangements (maximizing within-module density while minimizing between-module density) they can detect more complex block structure in networks,41 including the existence of a dense core and a more weakly connected periphery. In addition to versatility, block models offer the advantage of fitting a generative model to data (see below), which may offer insight into the processes that underlie the observed network topology.
Centrality and communication
Empirical networks have architectures that differ significantly from those of classic random graph models—most importantly, their nodes and edges are not equal in the way they are connected with the rest of the network. Far from being “equipotential,” the ways in which nodes and edges are embedded within the overall topology play a major role in determining their specific contributions to network function. Thus, network theory reconciles functional specialization with distributed processing—a dichotomy that has in the past led to strongly polarized theories of brain function, to the detriment of scientific progress.
Indeed, a major rationale for mapping brain connectivity arises from the idea that connectivity drives the functional specialization of local network elements. This idea is inherent in the notion that different brain regions have unique connectivity fingerprints that are indicative of their network embedding and predictive of their functional roles.42 Similarities among connectivity fingerprints can be informative of functional groupings of areas and their mutual relations.
Numerous measures quantify the potential of individual nodes and edges to influence the global state of the network. Many of them allow the identification of network hubs,43,44 generally defined as highly central parts of the network. The number of connections maintained by a node (its degree) or the combined weight of these connections (its strength) often provides a strong indicator of influence or centrality. Other measures of centrality take advantage of the layout of the shortest paths within the network, and record the number of such paths that pass through a given node or edge, a measure called the betweenness. Yet another way to approach centrality is by referencing the relation of nodes and edges to a network's community structure. The participation coefficient quantifies the diversity of a given node's connections across multiple modules—high participation indicates that many of these connections are made across modules, thus linking structurally and functionally the distinct communities. This measure is particularly useful in brain networks, as it can be applied to both structural and functional network data.45
Most measures of node centrality are mutually related (and hence statistically correlated); for example, in most (but not all) networks nodes with many connections (high degree) also serve as intermediaries on many short paths (high betweenness). Since different measures of centrality index different aspects of network organization, it can be beneficial to rank nodes by aggregating multiple centrality measures.43
Centrality measures can be useful tools for charting the global architecture of a brain network. Of particular neurobiological interest is the mutual association among highly central (eg, high degree) nodes. If these nodes maintain interconnections that are found to be denser than expected by chance (ie, a suitable null model) the network is said to exhibit so-called rich club organization46 (Figure 2). Rich clubs have been found in virtually all structural connectivity data, from the connectomes of humans47 to those of invertebrates.48,49 Rich club organization tends to centralize network traffic, as the dense core formed by interconnected high-degree nodes attracts the bulk of short network paths that link lower degree nodes to each other.50 This important role in network communication is thought to boost network communication and efficiency, thus trading off against the high wiring cost involved in linking spatially distributed hub regions.
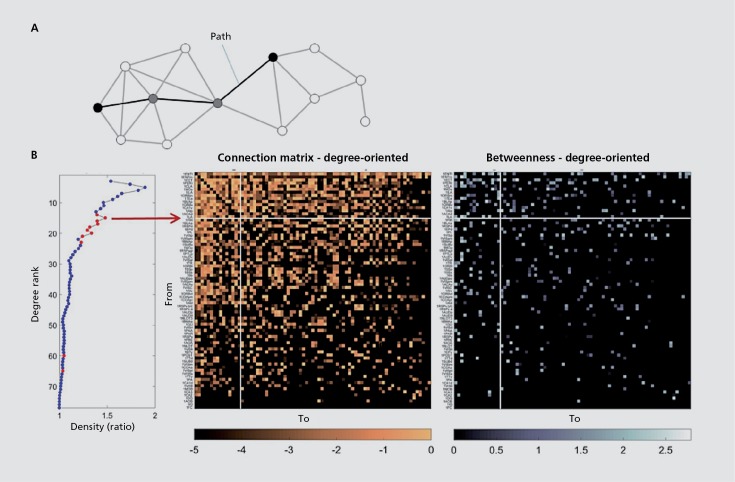
Figure 2. Paths and rich club organization. (A) Schematic network plot illustrating an optimally short path (length three steps) that links the two nodes shown in black; intermediate nodes are shown in gray. (B) Left: Using the rat cerebral cortex data set from ref 74, this plot shows the density of subgraphs, compared with a degree-sequence preserving null model, with subgraphs increasing in size from 1 to N (N=77) and with nodes arranged by total degree. Subgraph of size 1 comprises the single node with highest degree, subgraph size 2 the one comprising the two highest degree nodes, and so forth. Red data points indicate subgraphs for which the density is significantly above that of the null model (P<0.001, false discovery rate-corrected). Middle: Rat cerebral cortex connection matrix, with node arranged by total degree (highest degree node in the top row and left-most column). Note dense (nearly full) connectivity among the top 15 high-degree nodes (white lines). Right: Edge betweenness displayed in the same node ordering as middle panel. Note that there are numerous edges with high edge betweenness in upper left section of the matrix. These edges link high-degree nodes and they also participate in a large number of shortest paths across the network.
While much of the interest in centrality is based on the putative role of hubs in network communication, it should be noted that the mechanisms by which brain networks communicate remain obscure. Most models and their associated graph theory metrics assume that communication unfolds along the most efficient and shortest paths available. However, this idea ignores the fact that these paths cannot be discovered by neural elements or signals in the absence of global information about the network topology.22 Hence, alternative models based on spreading and diffusion51 and path ensembles52 are important to explore in future work.
Emerging trends
This final section briefly reviews several new directions that have great potential for future applications in brain networks.
Generative models
Most current graph theory methods applied to brain data deliver descriptive statistics that capture various aspects of network architecture. While such measures can be informative about the topological characteristics of a specific empirically measured network, their estimation should always be accompanied by some estimate of statistical significance or evidence. Every graph, even one generated by an entirely random process, will exhibit some graph attributes, including some chance level of clustering and modularity. Null models are important adjuncts of descriptive graph analysis as they allow discriminating which graph attributes are due to chance, and which exceed the expected values given by the null model. The choice of a proper null model is crucial for any descriptive analysis, as it will determine which graph features survive statistical comparison. Classic null models involve the rewiring of an empirical graph by swapping connections such that local degree is preserved while the global graph architecture is randomized. Other null models preserve subgraph frequencies to estimate the significance of larger subgraph distributions, or null models that preserve spatial locations of nodes.
Null models that fix a number of different factors such as local node measures, spatial locations, and wiring cost effectively become generative models of the empirical data. They generate graphs that may become indistinguishable from the empirical network, and in that sense can account for its topological properties. Hence, generative models can provide important insights into the factors that have shaped the emergence of specific architectural or performance characteristics. As such they are important reminders that not all graph attributes are the product of adaptation, and instead may have arisen as “spandrels” along the way,53 a mere by-product of other more basic generative factors such as degree distributions or spatial embedding. Another important insight that can be gleaned from the design of generative models is that many graph attributes are mutually dependent and arise jointly from a common set of driving factors. For example, high clustering is invariably linked to particular statistics of subgraphs that favor triangles, and strongly favored by spatial embedding and wiring conservation.
Spatial embedding as an important generative principle behind the organization of brain graphs deserves special mention, as it is a fundamental constraint on brain architecture.54,55 Much evidence points to distance-dependence as a major rule that governs the topology of anatomical brain connectivity within local circuits in single brain regions as well as for inter-areal projections. Generative models that combine spatial embedding with other, nonspatial, topological rules have suggested that their mutual trade-off may more fully account for characteristics of brain topology than any single (spatial or topological) factor alone.56
Generative models are also crucial for understanding the emergence of dynamic states and functional connectivity.57 Many classic models explored in computational neuroscience are generative models, in that they attempt to generate neuronal activity and dynamics from simple biophysical and structural ingredients. In human neuroimaging, the relationship between structural and functional connectivity has been illuminated by the use of computational models that can capture some of the patterns exhibited in brain dynamics. Some of these generative models are simple, as they can be computed analytically on top of structural graphs,58 or make minimal assumptions (eg, linearity) about the nature of neuronal dynamics.59 Other models utilize detailed biophysical mechanisms to generate neuronal time series and population activity.60 Implementation of large numbers of generative models with varying model parameters combined with model selection are central to estimate variations in network parameters, including causal effects, in the course of varying conditions of stimulus and task.
Dynamic networks
Brain networks are not immutable, static constructs—rather, their structural and functional connectivity patterns change on multiple time scales. Data on time-varying brain graphs generally takes on the form of time series (or stacks) of graphs that form an ordered series of snapshots, for example recorded in the course of learning or across developmental stages. Changes in network topology can be tracked by computing graph measures on each time point followed by the subsequent examination of the resultant time courses of graph metrics. Another analysis approach involves arranging this stack into a series of time slices that are mutually coupled and can be analyzed as a single graph construct.39 This allows the derivation of nodal measures of flexibility which can pinpoint parts of the network that are more variable across learning or development.61
Much effort has recently been expended to track fast changes in graph topology and organization of functional networks recorded with fMRI.62 Most commonly, a long time series of fMRI activations is partitioned into shorter “windows” that are then analyzed separately. Possible confounds are measurement artifacts such as physiological noise and increased uncertainty of estimating the magnitudes of functional connections on short samples.63 In resting state, fMRI networks appear to undergo fluctuations between states of higher integration and segregation,64 or modularity.65 Similar transitions between different network states occur during task switching66 and in the course of cognitively demanding spontaneous stimulation.67
Multilayer networks
The arrival of multiomic data has enabled the joint analysis of networks between elements of neurobiological systems at different levels of organization. Prime examples are recent studies that combine maps of anatomical and functional networks, as well as studies that combine large-scale brain connectivity data with spatially registered data on patterns of gene expression. The latter have yielded important insights into relations between the centrality of network elements, for example their membership in a dense core or rich club, and distinct genetic signatures in energy metabolism.68 In most studies so far, graph theoretic analysis proceeds through simple comparison or correlation of graph metrics across different levels (eg, anatomy and genetics). In the future, more explicit use of a multi-layer graphical framework is likely to occur. A few early examples are studies that place structural and functional connectivity into a multilayer model, eg, with data from human neuroimaging69 and magnetoencephalography.70
Algebraic topology
All graph theory approaches discussed so far build on networks that are composed of pairwise (dyadic) interactions. However, higher-order interactions can be highly informative for understanding non-random attributes of brain networks. Such higher-order relations can be represented with tools from applied algebraic topology, such as so-called simplicial complexes or simplices.71 Simplices reframe the problem of relational data in terms of collections of vertices: a 0-simplex is a node, a 1-simplex is an edge, and a 2-simplex is a filled (connected) triangle. Simplices can be used to locate cliques (all-to-all connected subgraphs) or cavities. Recent applications of simplices to human connectome data have shown the utility of the approach for identifying both densely connected groups of nodes as well as other patterns of connectivity (eg loop-like paths) that would facilitate parallel processing.72 Finally, the related area of topological data analysis attempts to detect, quantify and compare mesoscale structure present in complex network data. Essentially, the approach attempts to embed the data in a way that provides an optimal summary of its global structure. A recent example used topological data analysis to reveal dynamical organization in multitask fMRI time series, by creating graphical representations of relations among single image frames at the level of individual participants.73 These representations allow a comparison of how individuals transition among multiple cognitive tasks and states and could provide useful markers for clinical diagnosis and treatment. Overall, the arrival of these topological methods capitalizes on higher-order and high-dimensional features in brain data that have so far been inaccessible with simple graph methods, and are therefore promising avenues for future investigation.
Conclusion
The growth of network neuroscience over the past decade or two has been nothing short of astonishing. A major driving force for this rapid expansion is the availability of relational data recording couplings and interactions among elements of neural systems. For now, most studies remain descriptive and focus entirely on pairwise interactions resulting in graphs composed of dyadic links. But graph theory is much more powerful than current methods applied to brain networks suggest. Generative models, dynamic networks, multilayer models, and algebraic topology are just a few of the promising directions that are currently pursued. With time, these new approaches will likely find applications not only in basic, but also in clinical and translational research. For years to come graph theory methods will remain indispensable tools to further our understanding of the brain as a complex interconnected system.
Acknowledgments
The author's research was supported by the US National Institutes of Health (R01-AT009036). The author has no conflict of interest to declare.
REFERENCES
- 1.Newman M. Networks: An Introduction. Oxford, UK: Oxford University Press; 2010. [Google Scholar]
- 2.Barabási AL. Network Science. Cambridge, UK: Cambridge University Press; 2016. [Google Scholar]
- 3.Estrada E. The Structure of Complex Networks: Theory and Applications. New York, NY: Oxford University Press; 2012. [Google Scholar]
- 4.Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 5.Sporns O. Networks of the Brain. Cambridge, MA: The MIT Press; 2010. [Google Scholar]
- 6.Rubinov M., Sporns O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage. 2010;529(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Fornito A., Zalesky A., Breakspear M. Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage. 2013;80:426–444. doi: 10.1016/j.neuroimage.2013.04.087. [DOI] [PubMed] [Google Scholar]
- 8.Sporns O. Structure and function of complex brain networks. Dialogues Clin Neurosci. 2013;15(3):247–262. doi: 10.31887/DCNS.2013.15.3/osporns. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fornito A., Zalesky A., Bullmore E. Fundamentals of Brain Network Analysis. Boston, MA: Academic Press; 2016. [Google Scholar]
- 10.Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014;17(5):652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- 11.Bassett DS., Sporns O. Network neuroscience. Nat Neurosci. 2017;20(3):353–364. doi: 10.1038/nn.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sporns O., Tononi G., Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1(4):e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sporns O. The human connectome: a complex network. Ann N Y Acad Sci. 2011;1224(1):109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- 14.Gordon EM., Laumann TO., Adeyemo B., Petersen SE. Individual variability of the system-level organization of the human brain. Cereb Cortex. 2017;27(1):386–399. doi: 10.1093/cercor/bhv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glasser MF., Coalson TS., Robinson EC., et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536(7615):171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy AC., Gu S., Khambhati AN., et al. Explicitly linking regional activation and function connectivity: community structure of weighted networks with continuous annotation. 2016;arXiv:1611.07962. [Google Scholar]
- 17.Kivelä M., Arenas A., Barthelemy M., Gleeson JP., Moreno Y., Porter MA. Multilayer networks. J Compl Netw. 2014;2:203–271. [Google Scholar]
- 18.da Fontoura Costa L., Silva FN. Hierarchical characterization of complex networks. J Stat Phys. 2006;125(4):841–872. [Google Scholar]
- 19.Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298(5594):824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 20.Sporns O., Kötter R. Motifs in brain networks. PLoS Biol. 2004;2(11):e369. doi: 10.1371/journal.pbio.0020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan SE., Achard S., Termenon M., Bullmore ET., Vertes PE. Low dimensional morphospace of topological motifs in human fMRI brain networks. Netw Neurosci. 2018;https://doi.org/10.1162/NETN_a_00038. doi: 10.1162/netn_a_00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avena-Koenigsberger A., Misic B., Sporns O. Communication dynamics in complex brain networks. Nat Rev Neurosci. 2018;19:17–33. doi: 10.1038/nrn.2017.149. [DOI] [PubMed] [Google Scholar]
- 23.Sporns O., Betzel RF. Modular brain networks. Annu Rev Psychol. 2016;67:613–640. doi: 10.1146/annurev-psych-122414-033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortunato S. Community detection in graphs. Phys Rep. 2010;486(3):75–174. [Google Scholar]
- 25.Fortunato S., Hric D. Community detection in networks: A user guide. Phys Rep. 2016;659:1–44. [Google Scholar]
- 26.Newman MEJ., Girvan M. Finding and evaluating community structure in networks. Phys Rev. 2004;E69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 27.Leicht EA., Newman ME. Community structure in directed networks. Phys Rev Lett. 2008;100(11):118703. doi: 10.1103/PhysRevLett.100.118703. [DOI] [PubMed] [Google Scholar]
- 28.Rubinov M., Sporns O. Weight-conserving characterization of complex functional brain networks. Neuroimage. 2011;56(4):2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- 29.MacMahon M., Garlaschelli D. Community detection for correlation matrices. Phys Rev X. 2015;5(2):021006. [Google Scholar]
- 30.Lancichinetti A., Fortunato S. Consensus clustering in complex networks. Sci Rep. 2012;2:336. doi: 10.1038/srep00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinn M., Romero-Garcia R., Seidlitz J., Váša F., Vértes PE., Bullmore E. Versatility of nodal affiliation to communities. Sci Rep. 2017;7:4273. doi: 10.1038/s41598-017-03394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fortunato S., Barthélemy M. Resolution limit in community detection. Proc Natl Acad Sci U S A. 2007;104:36–41. doi: 10.1073/pnas.0605965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichardt J., Bornholdt S. Statistical mechanics of community detection. Phys Rev E. 2006;74(1):016110. doi: 10.1103/PhysRevE.74.016110. [DOI] [PubMed] [Google Scholar]
- 34.Betzel RF., Bassett DS. Multi-scale brain networks. Neuroimage. 2017;160:73–83. doi: 10.1016/j.neuroimage.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Power JD., Cohen AL., Nelson SM., et al. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo BTT., Krienen FM., Sepulchre J., et al. The organization of the human cerebral cortex estimated by functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeub LG., Sporns O., Fortunato S. Multiresolution consensus clustering in networks. Sci Rep. 2018;8:3259. doi: 10.1038/s41598-018-21352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn YY., Bagrow JP., Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466(7307):761–764. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 39.Mucha PJ., Richardson T., Macon K., Porter MA., Onnela JP. Community structure in time-dependent, multiscale, and multiplex networks. Science. 2010;328:876–878. doi: 10.1126/science.1184819. [DOI] [PubMed] [Google Scholar]
- 40.Karrer B., Newman ME. Stochastic blockmodels and community structure in networks. Phys Rev E. 2011;83(1):016107. doi: 10.1103/PhysRevE.83.016107. [DOI] [PubMed] [Google Scholar]
- 41.Betzel RF., Medaglia JD., Bassett DS. Diversity of meso-scale architecture in human and non-human connectomes. Nat Comm. 2018;9:346. doi: 10.1038/s41467-017-02681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passingham RE., Stephan KE., Kötter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3(8):606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- 43.Sporns O., Honey CJ., Kötter R. Identification and classification of hubs in brain networks. PLoS ONE. 2007;2:e1049. doi: 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Heuvel MP., Sporns O. Network hubs in the human brain. Trends Cogn Sci. 2013;17:683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Power JD., Schlaggar BL., Lessov-Schlaggar CN., Petersen SE. Evidence for hubs in human functional brain networks. Neuron. 2013;79(4):798–813. doi: 10.1016/j.neuron.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colizza V., Flammini A., Serrano MA., Vespignani A. Detecting rich-club ordering in complex networks. Nature Phys. 2006;2:110–115. [Google Scholar]
- 47.van den Heuvel MP., Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31(44):15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Towlson EK., Vértes PE., Ahnert SE., Schafer WR., Bullmore ET. The rich club of the C. elegans neuronal connectome. J Neurosci. 2013;33(15):6380–6387. doi: 10.1523/JNEUROSCI.3784-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shih CT., Sporns O., Yuan SL., et al. Connectomics-based analysis of information flow in the Drosophila brain. Current Biol. 2015;25(10):1249–1258. doi: 10.1016/j.cub.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 50.van den Heuvel MP., Kahn RS., Goñi J., Sporns O. High-cost, high-capacity backbone for global brain communication. Proc Natl Acad Sci U S A. 2012;109(28):11372–11377. doi: 10.1073/pnas.1203593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mišić B., Betzel RF., Nematzadeh A., et al. Cooperative and competitive spreading dynamics on the human connectome. Neuron. 2015;86(6):1518–1529. doi: 10.1016/j.neuron.2015.05.035. [DOI] [PubMed] [Google Scholar]
- 52.Avena-Koenigsberger A., Mišic B., Hawkins RX., et al. Path ensembles and a tradeoff between communication efficiency and resilience in the human connectome. Brain Struct Func. 2017;222(1):603–618. doi: 10.1007/s00429-016-1238-5. [DOI] [PubMed] [Google Scholar]
- 53.Rubinov M. Constraints and spandrels of interareal connectomes. Nature Comm. 2016;7:13812. doi: 10.1038/ncomms13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bullmore E., Sporns O. The economy of brain network organization. Nature Rev Neurosci. 2012;13(5):336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- 55.Horvát S., Gǎmǎnut R., Ercsey-Ravasz M., et al. Spatial embedding and wiring cost constrain the functional layout of the cortical network of rodents and primates. PLoS Biol. 2016;14:e1002512. doi: 10.1371/journal.pbio.1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Betzel RF., Avena-Koenigsberger A., Goni J., et al. Generative models of the human connectome. Neuroimage. 2016;124:1054–1064. doi: 10.1016/j.neuroimage.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Betzel RF., Bassett DS. Generative models for network neuroscience: prospects and promise. J Roy Soc Interface. 2017;14(136):20170623. doi: 10.1098/rsif.2017.0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goñi J., van den Heuvel MP., Avena-Koenigsberger A., et al. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc Natl Acad Sci U S A. 2014;111(2):833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdelnour F., Voss HU., Raj A. Network diffusion accurately models the relationship between structural and functional brain connectivity networks. Neuroimage. 2014;90:335–347. doi: 10.1016/j.neuroimage.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deco G., Jirsa VK., Robinson PA., Breakspear M., Friston K. The dynamic brain: from spiking neurons to neural masses and cortical fields. PLoS Comput Biol. 2008;4(8):e1000092. doi: 10.1371/journal.pcbi.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bassett DS., Wymbs NF., Porter MA., Mucha PJ., Carlson JM., Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci U S A. 2011;108(18):7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hutchison RM., Womelsdorf T., Allen EA., et al. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Betzel RF., Fukushima M., He Y., Zuo XN., Sporns O. Dynamic fluctuations coincide with periods of high and low modularity in resting-state functional brain networks. Neuroimage. 2016;127:287–297. doi: 10.1016/j.neuroimage.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shine JM., Koyejo O., Poldrack RA. Temporal metastates are associated with differential patterns of time-resolved connectivity, network topology, and attention. Proc Natl Acad Sci U S A. 2016;113(35):9888–9891. doi: 10.1073/pnas.1604898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukushima M., Betzel RF., He Y., et al. Fluctuations between high-and low-modularity topology in time-resolved functional connectivity. Neuroimage. 2017;https://doi.org/10.1016/j.neuroimage. 2017.08.044. doi: 10.1016/j.neuroimage.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez-Castillo J., Hoy CW., Handwerker DA., et al. Tracking ongoing cognition in individuals using brief, whole-brain functional connectivity patterns. Proc Natl Acad Sci U S A. 2015;112(28):8762–8767. doi: 10.1073/pnas.1501242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gilson M., Deco G., Friston K., et al. Effective connectivity inferred from fMRI transition dynamics during movie viewing points to a balanced reconfiguration of cortical interactions. Neuroimage. 2017;https://doi.org/10.1016/j.neuroimage. . 2017.09.061. doi: 10.1016/j.neuroimage.2017.09.061. [DOI] [PubMed] [Google Scholar]
- 68.Fulcher BD., Fornito A. A transcriptional signature of hub connectivity in the mouse connectome. Proc Natl Acad Sci U S A. 2016;113(5):1435–1440. doi: 10.1073/pnas.1513302113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Battiston F., Nicosia V., Chavez M., Latora V. Multilayer motif analysis of brain networks. Chaos. 2017;27(4):047404. doi: 10.1063/1.4979282. [DOI] [PubMed] [Google Scholar]
- 70.Tewarie P., Hillebrand A., van Dijk BW., et al. Integrating cross-frequency and within band functional networks in resting-state MEG: a multi-layer network approach. Neuroimage. 2016;142:324–336. doi: 10.1016/j.neuroimage.2016.07.057. [DOI] [PubMed] [Google Scholar]
- 71.Giusti C., Ghrist R., Bassett DS. Two's company, three (or more) is a simplex: Algebraic-topological tools for understanding higher-order structure in neural data. J Comput Neurosci. 2016;41(1):1–14. doi: 10.1007/s10827-016-0608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sizemore AE., Giusti C., Kahn A., Vettel JM., Betzel RF., Bassett DS. Cliques and cavities in the human connectome. J Comput Neurosci. 2018;44(1):115–145. doi: 10.1007/s10827-017-0672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saggar M., Sporns O., Gonzalez-Castillo J., et al. Towards a new approach to reveal dynamical organization of the brain using topological data analysis. Nature Comm. 2018;9:1399. doi: 10.1038/s41467-018-03664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swanson LW., Hahn JD., Sporns O. Organizing principles for the cerebral cortex network of commissural and association connections. Proc Natl Acad Sci U S A. 2017;114(45):E9692–E9701. doi: 10.1073/pnas.1712928114. [DOI] [PMC free article] [PubMed] [Google Scholar]